Abstract
Therapeutic strategies are urgently needed for patients with acute myeloid leukemia (AML). Leukocyte immunoglobulin-like receptor B4 (LILRB4), which suppresses T cell activation and supports tissue infiltration of AML cells, represents an attractive drug target for anti-AML therapeutics. Here, we report the identification and development of an LILRB4-specific humanized mAb that blocks LILRB4 activation. This mAb, h128–3, showed potent activity in blocking development of monocytic AML in various models including patient-derived xenograft (PDX) mice and syngeneic immunocompetent AML mice. MAb h128–3 enhanced the anti-AML efficacy of chemotherapy treatment by stimulating mobilization of leukemia cells. Mechanistic studies revealed four concordant modes of action for the anti-AML activity of h128–3: 1) reversal of T cell suppression; 2) inhibition of monocytic AML cell tissue infiltration; 3) antibody-dependent cellular cytotoxicity (ADCC); and 4) antibody dependent cellular phagocytosis (ADCP). Therefore, targeting LILRB4 with antibody represents an effective therapeutic strategy for treating monocytic AML.
Introduction
Acute myeloid leukemia (AML), the most common adult acute leukemia, is characterized by clonal proliferation of immature myeloid hematopoietic cells in the bone marrow, blood, and other tissues (1). Each year in the United States, 19,000 new AML cases appear and there are about 10,000 AML-associated deaths (2). Despite increased understanding of the underlying biology of AML, the standard intervention of cytotoxic chemotherapy has not changed in the past 40 years. As many as 70% of patients 65 years or older die of their disease within a year of diagnosis (3). Moreover, immunotherapies, such as CTLA4 and PD-1/PD-L1 targeting strategies, have not yielded clinical benefits in AML patients (4). The FDA has approved several new therapeutics in 2017 and 2018 for AML, including inhibitors for IDH1, IDH2, and Flt3, liposome-encapsulated chemotherapeutics, and anti-CD33–drug conjugates that may benefit certain subsets of AML patients (5–7). Nevertheless, there remains an urgent need to develop new therapies with high therapeutic efficacy and low toxicity for various subtypes of AML.
The leukocyte Ig-like receptor subfamily B (LILRB) is a group of type I transmembrane glycoproteins, characterized by extracellular Ig-like domains for ligand binding and intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that can recruit tyrosine phosphatases SHP-1, SHP-2, or the inositol-phosphatase SHIP (8, 9). Because of their immune inhibitory functions, LILRBs are considered to be immune checkpoint proteins (8). In fact, LILRBs act on a broader array of immune cell types than the classical immune checkpoint proteins CTLA4 and PD-1 (10). We identified LILRB2 as a receptor for the hormone Angptl2 (11). Then, we demonstrated that a deficiency of the mouse ortholog of LILRB2, PirB, in AML models resulted in increased differentiation and decreased self-renewal of leukemia stem cells (11). In addition, we and others demonstrated that several LILRBs and a related ITIM receptor LAIR1 support AML development (12, 13). Using proteomics, transcriptomics, and experimental analysis, Michel Sadelain and colleagues ranked several LILRBs among the top 24 AML target candidates (14). LILRBs act as both immune checkpoint molecules and tumor sustaining factors but may not affect normal development (8). Thus, they have potential as attractive targets for cancer treatment.
Monocytic AML is a subtype of AML in which a majority of the leukemia cells are of the monocytic lineage. Extramedullary disease, including gum infiltrates and cutaneous and cerebrospinal fluid involvement, is common in monocytic AML (15). In agreement with the finding from Colovai and colleagues (16), we reported that LILRB4, a member of the LILRB family, is a marker for monocytic AML (17, 18). We further demonstrated that LILRB4 is more highly expressed on monocytic AML cells than on their normal counterparts and that LILRB4 expression inversely correlates with overall survival of AML patients (17, 18). LILRB4 (also known as CD85K, ILT3, LIR5, and HM18) has two extracellular Ig-like domains (D1 and D2) and three ITIMs. We have identified apolipoprotein E (ApoE) as an extracellular binding protein of LILRB4. ApoE binding is coupled with T-cell suppression and tumor infiltration through LILRB4-mediated downstream signaling in AML cells (18). Collectively, these findings show LILRB4, with restrictive and lower expression on normal monocytic cells, is a marker for monocytic AML with restrictive and lower expression on normal monocytic cells that inhibits immune activation and supports tumor invasiveness. Therefore, LILRB4 represents an attractive target for developing drugs to treat patients with monocytic AML.
In this study, we report an LILRB4-targeted humanized mAb, h128–3, that blocks LILRB4/APOE interaction in a competitive manner. This blocking antibody inhibits monocytic AML cell tissue infiltration and reverses T-cell suppression. In addition, h128–3 triggers ADCC- and ADCP-mediated AML cell killing. Treatment with h128–3 significantly reduced the AML tumor burden in various mouse models including PDX and syngeneic immunocompetent mouse models. These results suggest that LILRB4-neutralizing antibodies such as mAb h128–3 can be applied to anti-cancer therapeutic strategies.
Materials and Methods
Cell lines and human AML samples
HEK293F and CHO cell lines were obtained from Life Technologies (Carlsbad). Human monocytic AML cell lines (THP-1, MV4–11 and U937), mouse leukemia cell line C1498, and mouse macrophage cell line RAW264.7 were obtained from ATCC and maintained in a humidified atmosphere of 5% CO2 at 37°C, in media suggested by ATCC supplemented with fetal bovine serum (FBS) (HyClone) and 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies). Cell lines were not authenticated in the past year and cultured for fewer than 10 passages in indicated medium. All cell lines were routinely tested using a mycoplasma-contamination kit (R&D Systems). Primary human AML samples were obtained from the University of Texas Southwestern Medical Center (UTSW). Informed consent was obtained under a protocol reviewed and approved by the Institutional Review Board at UTSW. LILRB4 expressed samples were analyzed by flow cytometry.
Animals
C57BL/6 and NOD-SCID IL2Rγ null (NSG) mice were purchased from and maintained at the animal core facility of UTSW. For each experiment, the same-sex and age-matched (4–8 weeks) mice were used and randomly allocated to each group. All animal experiments were performed with the approval of the UTSW Institutional Animal Care and Use Committee (IACUC).
Generation of LILRB4 rabbit mAbs
Two New Zealand white rabbits were immunized subcutaneously with 0.5 mg recombinantly expressed human LILRB4 ECD protein (Sino Biological). After the initial immunization, animals were given boosters four times in a three-week interval. Serum titers were evaluated by indirect enzyme-linked immunosorbent assay (ELISA) and memory B cells were isolated after immunization was performed five times (RevMab Biosciences). A large panel of single memory B cells were collected and cultured for two weeks, and the supernatants were analyzed by ELISA (RevMab). Variable region genes from these positive single B cells were recovered by reverse transcription PCR (RT-PCR), using primers (RIgH-F: ATGGAGACTGGGCTGCGCTGGCTYC; RIgH-R: CCATTGGTGAGGGTGCCCGAG; RIgK-F: ATGGACACSAGGGCCCCCACTC; and RIgK-R: CAGAGTRCTGCTGAGGTTGTAGGTAC) that were specific to rabbit heavy and light chain variable regions. Two rounds of PCR were performed by incorporating overlapping sequences at the 3′ and 5′ ends allowing infusion cloning of the variable regions into vectors for expression of rabbit heavy and light chains. Heavy and light chain constructs were cotransfected into human embryonic kidney freestyle 293 (HEK293F) cells using transfection reagent PEI (Sigma) (19). After 7 days of expression, supernatants were harvested and antibodies were purified by affinity chromatography using protein A resin as we reported before (Repligen) (19). A panel of 26 purified rabbit monoclonal antibodies was generated and used for this study.
Chimeric receptor reporter assay
We constructed a stable chimeric receptor reporter cell system as previously described (20). One aim of this system was to test the ability of a ligand to bind to the ECD of LILRB4. Another aim of the system was to trigger the activation or inhibition of the chimerically fused intracellular domain of paired immunoglobulin-like receptor (PILR) β. This receptor signals through the adaptor DAP-12 to activate the NFAT promoter. If an agonist antibody binds the ECD and activates the chimeric signaling domain, an increase in GFP expression is observed. If an antagonist antibody binds the ECD and suppresses the chimeric signaling domain, a decrease in GFP expression is observed. A competition assay was used to screen LILRB4 blocking antibodies. Briefly, recombinant APOE protein (10 μg/ml) was precoated on 96-well plates at 37 °C for 3 hours. After washes with PBS twice, 2 × 104 LILRB4 reporter cells were seeded in each well. Meanwhile, indicated LILRB4 antibodies were added into culture media. After culture for 16 hours, the percentage of GFP+ reporter cells was measured by flow cytometry.
Epitope binning with Bio-layer Interferometry (BLI)
An Octet RED96 System, protein A biosensors, and kinetics buffer were purchased from ForteBio (Menlo Park). The epitope binning data was obtained by a BLI-based sandwich epitope binning assay performed on an 8-channel Octet RED96 instrument. First, antibodies (40 μg/mL) were loaded onto protein A biosensors for 4 minutes. The remaining Fc-binding sites on the biosensors were blocked with an irrelevant rabbit antibody (200 μg/ml) for 4 minutes, followed by soaking the biosensors in kinetics buffer for 10 seconds. The biosensors were then exposed to recombinant LILRB4 (25 μg/mL) for 4 minutes to saturate the binding site of the first antibody. Finally, the biosensors were exposed to the secondary antibodies (40 μg/mL) for 4 minutes to detect the binding. If no increased binding signal was observed with the second antibody over the binding signal of the first antibody, the pair of antibodies was classified in the same epitope bin (competitor). In contrast, if an increased binding signal was observed with the second antibody over the binding signal of the first antibody, the pair of antibodies was classified in different epitope bins (non-competitor). Bins are groups of antibodies that recognize the same binding site/area on an antigen. Protein A biosensors were reused for 10 times, and the surfaces were regenerated for 30 secones in 100 mM glycine (pH 2.6). Raw data were processed using ForteBio’s data analysis software 7.0.
Affinity measurement with BLI
For antibody affinity measurement, antibody (30 μg/mL) was loaded onto the protein G biosensors for 4 minutes. Following a short baseline in kinetics buffer, the loaded biosensors were exposed to a series of recombinant LILRB4 concentrations (0.1–200 nM) and background subtraction was used to correct for sensor drifting. All experiments were performed with shaking at 1,000 rpm. Background wavelength shifts were measured from reference biosensors that were loaded only with antibody. ForteBio’s data analysis software was used to fit the data to a 1:1 binding model to extract an association rate and dissociation rate. The Kd was calculated using the ratio koff/kon.
Humanization of rabbit mAb
Humanization of the LILRB4 antibody was based on a CDR-grafting strategy as described previously (21, 22). Briefly, CDRs in the heavy and light chains of the rabbit antibody were defined by a combination of three methods: Kabat, IMGT, and Paratome. The parental rabbit mAb and the most closely related human germline sequence were then aligned. Residues which are known not to be structurally critical and/or subjected to change during the in vivo maturation process were identified in the mutational lineage guided analysis and humanized (21). DNA encoding humanized VK and VH were synthesized (GenScript) (VH: EVQLLESGGGLVQPGGSLRLSCAASGIDFSNHYYMYWVRQAPGKGLEWIGSIFSGDSASTYYADSAKGRFTISRDNSKNTLYLQMNSLRAEDTAVYYCARGMSTNDWASDLWGQGTLVTVSS; VK: DIQMTQSPSSLSASVGDRVTITCQASESINSIYLAWYQQKPGKAPKLLIYRASTLASGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQSYDWGDVENTFGGGTKVEIK). The human IgG signal peptides and a Kozak sequence were engineered at the 5′ ends of the VK and VH sequences. The humanized VK and VH fragments were then cloned into human IgG1 CK and CH vectors separately. Expression, purification, and quantification of the humanized mAbs are the same as those for rabbit mAbs.
Generation of D1 and h128–3 Fab for structure work
The DNA encoding the D1 domain of LILRB4 was cloned into the vector pET21a (Novagen) with NdeI and XhoI restriction sites and then expressed in E. coli strain BL21 (DE3) (Novagen) (23). The bacteria were cultivated in LB medium containing the corresponding antibiotics (100 μg/ml ampicillin) in a shaker incubator at 37 °C. Expression was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the culture reached an OD600 of 0.8–1.0. Then, the culture was continued for 4–6 hours before harvest. Centrifuged cells were suspended in PBS buffer and disrupted using a homogenizer (JNBIO, China). The inclusion bodies of the recombinant proteins were purified and refolded as described, with some modifications (23). Briefly, aliquots of inclusion body were dropwise diluted in an agitating refolding buffer (100 mM Tris-HCl, 2 mM EDTA, 400 mM L-arginine, 0.5 mM oxidized glutathione, and 5 mM reduced glutathione, pH 8.0) for 8 hours at 4 °C. The refolded protein was concentrated and buffer-exchanged using an Amicon 8400 concentrator to the solution containing 20 mM Tris-HCl and 50 mM NaCl, pH 8.0. Subsequently the proteins were further purified by gel-filtration chromatography. The eligible peak fractionated proteins were concentrated for further study or crystallization. To produce the Fab, antibodies were concentrated to approximately 20 mg/mL and digested using papain (Pierce) protease at an antibody-to-papain ratio of 160:1 (w/w) at 37°C for 6 hours. The digestion mixture was loaded into a Protein A column (GE Healthcare) by applying the flow through mode to separate the Fab fragment with Fc region and undigested antibody. Fab fragments were collected, concentrated, and purified to homogeneity on a HiLoad® 16/600 Superdex 200 pg column (GE Healthcare).
Determination of h128–3-fab/D1 complex structure
D1 protein was mixed with purified Fab fragment of h128–3 at a molar ratio of 1.5:1, and incubated at 4°C overnight for complex formation. The mixture was then loaded onto a Superdex 75 10/300 GL column to purify the D1/Fab complex from any excess D1. Peak fractions corresponding to the complex were collected and concentrated for crystallization. D1/Fab complex crystals were grown by vapor diffusion in sitting drops. A total of 1 μl of complex protein solution at 5 mg/ml or 10 mg/ml was mixed with an equal volume of reservoir solution. The crystals obtained diffracted synchrotron radiation anisotropically to about 4.0 Å resolution. Finally, the better resolution of 3.0 Å was obtained using streak-seeding method with the collected complex proteins in the same reservoir solution at 4 °C for two weeks (PDB: 6K7O). X-ray diffraction data were collected at 100 K at the Shanghai Synchrotron Radiation Facility beamline BL17U and indexed, integrated and scaled with HKL2000. The complex structure was solved by the molecular replacement method using Phaser from the CCP4 program suite with the structures of LILRB4 (PDB: 3P2T) and the Fab (PDB: 4OQT) as the search models. Initial restrained rigid-body refinement was performed using REFMAC5. Initial manual model building was performed using COOT. Further refinement was performed using Phenix. Final statistics for data collection and structure refinement are presented in Supplementary Table S1.
Flow cytometry analysis
For analysis of human AML cells engraftment in NSG mice, a previously published protocol was followed (24). Cells were run on either Calibur or FACSAria for analysis. Original flow data were analyzed by Flowjo software. Binding of LILRB4 mAbs on AML cells was measured using a Guava easyCyte HT instrument based on the manufacturer’s instructions (Millipore). Briefly, 2 × 105 cells were dispensed in 100 μL aliquots and blocked with 100 μg/mL hIgG. LILRB4 mAbs (5 μg/mL) were then added for 1 hour on ice, followed by the addition of FITC-conjugated anti-rabbit or anti-human secondary IgG-F(ab)2 (Jackson ImmuneResearch Laboratories). After washing with PBS buffer, the cells were analyzed for fluorescence intensity. Irrelevant rabbit or human IgG was used as negative control.
Generation of LILRB4 mutants
LILRB4 mutants were generated using two rounds PCR overlapping method with LILRB4 wild type DNA construct as the template. The mutagenic primers were synthesized by Sigma (E54A-F: TGTCAGGGGACCCTGGCGGCTCGGGAGTACCGT; E54A-R: ACGGTACTCCCGAGCCGCCAGGGTCCCCTGACA; E54S-F: TGTCAGGGGACCCTGTCGGCTCGGGAGTACCGT; E54S-R: ACGGTACTCCCGAGCCGACAGGGTCCCCTGACA; E54R-F: TGTCAGGGGACCCTGAGGGCTCGGGAGTACCGT; E54R-R: ACGGTACTCCCGAGCCCTCAGGGTCCCCTGACA; R56E-F: GGGACCCTGGAGGCTGAGGAGTACCGTCTGGAT; R56E-R: ATCCAGACGGTACTCCTCAGCCTCCAGGGTCCC; R56A-F: GGGACCCTGGAGGCTGCGGAGTACCGTCTGGAT; R56A-R: ATCCAGACGGTACTCCGCAGCCTCCAGGGTCCC; R56Q-F: GGGACCCTGGAGGCTCAGGAGTACCGTCTGGAT; R56Q-R: ATCCAGACGGTACTCCTGAGCCTCCAGGGTCCC; P103T-F: TGTTACTATCGCAGCACTGTAGGCTGGTCACAG; P103T-R: CTGTGACCAGCCTACAGTGCTGCGATAGTAACA; P103R-F: TGTTACTATCGCAGCCGTGTAGGCTGGTCACAG; P103R-R: CTGTGACCAGCCTACACGGCTGCGATAGTAACA; P103S-F: TGTTACTATCGCAGCAGCGTAGGCTGGTCACAG; and P103S-R: CTGTGACCAGCCTACGCTGCTGCGATAGTAACA.), and the mutations were confirmed by DNA sequencing performed by GENEWIZ. The wild type and mutant fragments were then cloned into hIgG1 Fc-tag vector using infusion cloning method (Clontech). These LILRB4-Fc mutants were expressed by transient transfection of HEK293F cells and purified by protein A affinity chromatography.
Sequences alignment and phylogenetic analysis
D1 amino acid sequences of eleven LILR family members were analyzed. The accession numbers of proteins in GenBank are as follows: LILRB1, Q8NHL6; LILRB2, Q8N423; LILRB3, O75022; LILRB4, Q8NHJ6; LILRB5, O75023; LILRA1, O75019; LILRA2, Q8N149; LILRA3, Q8N6C8; LILRA4, P59901; LILRA5, A6NI73; LILRA6, Q6PI73. We defined the amino acid residues from position 27 to position 118 as D1. Multiple alignments were performed using ClustalX (Version 2.09) with eleven D1 sequences. A phylogenetic tree was generated using MEGA (Version 5.0).
ELISA binding assay
Corning 96-well EIA/RIA plates were coated overnight at 4°C with LILRB4 recombinant proteins (1 μg/mL) and blocked for 2 hours at 37°C with 5% non-fat milk. After washing with PBST 3 times, 100 μL of serial diluted LILRB4 antibodies were added and incubated for 45 minutes at 37°C. Subsequently, the plates were washed with PBST and incubated for 30 minutes with anti-rabbit or anti-human F(ab’)2 HRP-conjugated IgG (Jackson ImmunoResearch Laboratories). The immunoreactions were developed with TMB substrates (Sigma) and stopped by the addition of 2 M sulfuric acid before the plate was read at 450 nm.
Annexin-V/PI apoptosis assay
For analysis of LILRB4 antibody–induced apoptosis, 5 × 104 THP-1 cells were seeded in 4 replicates in 12-well plates. After 24 hours, antibodies were added to a final concentration of 20 μg/mL. Cells were treated with an irrelevant human IgG1 as isotype control. After 48 hours of incubation at 37°C in humidified air with 5% CO2, cells were collected, washed twice with PBS and resuspended in 150 μL of binding buffer. 5 μL of FITC-conjugated Annexin-V and PI (propidium Iodide) (BD Biosciences) was added to the cells, vortexed and incubated at RT in dark for 10 minutes. Apoptosis was measured by flow cytometry.
Leukemia cell and T cell coculture assay
In the coculture assay, human T cells (5 × 104 cells per well) isolated from health donor peripheral blood (Allcells) were placed in the lower chambers of a 96-well transwell plate. Leukemia cells were cultured in the upper chamber of transwell inserts (pore size, 3 μM, Thermo Fisher) in U-bottom 96 well-plate. Irradiated indicated leukemia cells (E:T ratio = 2:1) were added to the upper chambers and treated with indicated antibodies. After culture with anti-CD3/CD28-coated beads (Thermo Fisher) and 50 U/ml rhIL2 in the lower chambers for 5–7 days, representative T cells in lower chambers were photographed using an inverted microscope. T cells were stained with anti-CD3-APC, anti-CD4-PE, or anti-CD8-PE and analyzed by flow cytometry. Mouse IgG isotype was used as a negative control. Propidium Iodide (PI) was used as dead cell indicator.
Transwell migration assay
To measure the migration ability of AML cells, 2 × 105 THP-1 cells were seeded in the upper chamber (pore size, 8 μm, Thermo Fisher) and treated with antibodies (10 μg/ml). After culture for 20 hours, cells in the lower chamber were collected and counted.
Homing of leukemia cells
Cells (5 × 106 cells per mouse) were intravenously injected into NSG mice. Mice were treated with 10 mg/kg of anti-LILRB4 or control IgG1 immediately after injection of leukemia cells. Mice were sacrificed after 8 or 20 hours. Peripheral blood, bone marrow, liver, and spleen were harvested, and single-cell suspensions were examined by flow cytometry. CFSE, GFP, or anti-human CD45 were used to detect target human leukemia cells in indicated experiments. Numbers of leukemia cells in recipient liver, spleen, and bone marrow are reported as a ratio relative to cell numbers in peripheral blood. For normalization, the leukemia cells ratio in different organs treated with hIgG are normalized as 100%.
Human AML xenograft
Xenografts were performed essentially as described (18, 25). Briefly, 6–8-week-old NSG mice were used for transplantation. Human leukemia cells were resuspended in 200 μL PBS containing 1% FBS. Mice were given 1 × 106 human cultured leukemia cells or 5 to 10 × 106 human primary AML cells via tail-vein injection. 4 hours after transplantation, anti-LILRB4 or human IgG1 control was administrated by tail-vein injection. At indicated days after transplantation, the peripheral blood, bone marrow, spleen, and liver were assessed for the engraftment. Tumor growth was also monitored over time by luminescence.
Mouse AML allograft
Mouse AML allograft was performed as described before (18, 24). Briefly, 6–8 week-old wild-type C57BL/6 mice were used for transplantation. 1 × 106 mouse leukemia cells expressing human LILRB4 were resuspended in 200 μl PBS for subcutaneous implantation of each mouse. Mice were then given 10 mg/kg of anti-LILRB4 or control human IgG intravenously at day 7 after leukemia cell implantation and were treated every 3 days until euthanization. Tumor sizes were determined by caliper measure (width × width × length). For CD8+ T cell depletion, 10 mg/kg CD8 antibody (YTS 169.4.2) was intravenously injected at day 3 after leukemia cells implantation and were treated for additional two times every 3 days. To determine whether LILRB4 antibody treatment generates memory T cells, percentages of CD8+CD62L+ memory T cells in spleen were assessed by flow cytometry.
ADCC assay
Human buffy coats were obtained from heathy donors through Stanford Blood Center. 2.5×106 freshly isolated human PBMCs were used as effector cells and 5×104 THP-1 cells with stably transfected GFP were used as target cells in a 50:1 ratio. Human PBMCs, target THP-1-GFP cells, and increasing concentrations of antibodies were combined in 200 μl total in RPMI + 10% heat-inactivated FBS and 50 ng/ml of IL2 (R&D systems) in U-shaped 96-well plate and incubated for 20 hours at 37°C. Cells were washed and resuspended in 200 μl of staining buffer (BD Biosciences) containing 7-AAD. 150,000 cells were acquired by FACS Celesta and percentages of GFP-positive cells were measured. Cell cytotoxicity was calculated as: percent of cytotoxicity = 100 – ([T/NT] x100), where T and NT are the percentages of GFP+ cells treated with or without antibodies, respectively.
CDC assay
THP-1 cells were seeded in triplicates in 96 well plate at a density of 2 × 104 cells per well. Anti-LILRB4 or human IgG1 isotype control was added to a final concentration 20 μg/mL. After 30 minutes, normal human serum (Innovative) was added to a final concentration of 20%. After incubation for 4 hours at 37°C, cell supernatants were transferred to a 96-well plate to determine the amount of LDH released using LDH Cytotoxicity Assay Kit (Pierce).
ADCP assay
In vitro antibody-dependent cellular phagocytosis (ADCP) was performed with mouse macrophage cell line RAW 264.7 (mouse Macs) or human PBMC derived macrophages (human Macs) and analyzed by flow cytometry (26). Lipopolysaccharide (LPS) stimulated RAW 264.7 cells or human macrophages were stained with efluor 670 (Red) and seeded at a density of 4 × 104 cells per well and allowed to adhere overnight. Target THP-1 cells were stained with CellVue Jade (Green), pre-incubated with 20 μg/mL of anti-LILRB4 or human IgG1 isotype control for 30 minutes, and added at a 1:1 E/T ratio to the wells in triplicates. After incubation for 2–4 hours at 37°C, suspension THP-1 cells were washed away with PBS and adherent mouse macrophages or human macrophages were collected for flow cytometry analysis. ADCP percentage was calculated by double-positive macrophages/total macrophages.
Statistical analyses
Statistical analyses were performed with Prism 7.0 (GraphPad software). Statistical differences were determined to be significant at p < 0.05 using two-tailed Student t test and two-tailed Mann-Whitney log-rank test. In vitro data were presented as mean ± SEM. In all figures, * indicates p < 0.05; ** indicates p < 0.01.
Results
Generation and characterization of LILRB4 mAbs
To characterize the potential of LILRB4/APOE blockade as a therapeutic strategy for AML, we generated a panel of LILRB4 mAbs. The protocol for generating these mAbs is illustrated in Fig. 1A (21, 27, 28). Briefly, we serially immunized rabbits with LILRB4 extracellular domain (ECD) protein. The rabbit that exhibited the highest serum titer for LILRB4 binding in ELISA was selected for single memory B cell isolation and culture. Out of 400 LILRB4-specific single B cell clones screened, a total of 229 LILRB4 binders were generated (Supplementary Fig. S1A and S1B). We then cloned variable regions of heavy chain and light chain sequences from the top 26 hits with the highest binding activity. After recombinant expression in HEK293F cells as rabbit IgG1, we determined the binding ability by ELISA and found EC50 for 21 antibodies in the low nanomolar range (0.05–0.3 nM) (Fig. 1B). To group the mAbs by their binding epitopes, we performed a sandwich epitope binning assay with an Octet RED96 (29). A total of seven epitope bins (bin 1 to bin 7) were identified for the 21 high affinity binders (Fig. 1C). To further characterize the binding epitopes in each bin, we took representative antibodies from each bin and used ELISA to measure their binding to different domains of LILRB4 ECD: D1 (residues 27–118), D2 (residues 119–218), and SR (stalk region, residues 219–259). We used full-length LILRB4 ECD (residues 22–259) as a control (Fig. S2). The mAbs in three epitope bins bind to D1, the mAbs in one epitope bin to D2, and the mAbs in three epitope bins to SR (Fig. 1D). As expected, in a flow cytometry assay, mAbs from all epitope bins bind to THP-1 cells, a monocytic AML cell line expressing a high density of LILRB4. This result suggests these mAbs can bind to monocytic AML cells in physiological conditions (Fig. 1E).
Fig. 1. Generation and characterization of LILRB4 blocking mAbs.
(A) Single antigen-specific memory B cell isolation, culture, and cloning strategy used to generate LILRB4 rabbit mAbs. After isolation and culture of LILRB4-specific memory B cells from immunized rabbits, desired B cells were screened for binding to LILRB4 in ELISA. vH and vL genes were then cloned into rabbit IgG backbones and recombinant mAbs were produced using a transient HEK293F cell expression system. (B) EC50 of 26 LILRB4 rabbit mAbs. An irrelevant rabbit antibody (rIgG) was used as negative control. EC50 ≥ 1.0 nM showed as 1.0 nM. Two independent experiments were performed. (C) Node plot of the epitope bins of 21 LILRB4 rabbit mAbs determined by Octet RED96 using a classic sandwich epitope binning assay. (D) Binding of seven representative rabbit mAbs (from different bins) to ECD (residues 22–259), D1 (first Ig-like domain, residues 27–118), D2 (second Ig-like domain, residues 119–218) and SR (stalk region, residues 219–259) of LILRB4 determined by ELISA. (E) Binding of seven representative rabbit mAbs (from different bins) to LILRB4-expressing THP-1 cells determined by flow cytometry. Two independent experiments were performed. (F) Screening of LILRB4 blocking mAbs in chimeric receptor reporter assay. APOE2 was used as a functional ligand to activate LILRB4 reporter cells. The two LILRB4 blocking antibodies are shown in red. (G and H) Kinetics of 128–3 and 216–1 binding to LILRB4 were assessed using an Octet RED96. (I) Binding ability of 128–3 and 216–1 with LILRB4 determined by ELISA. (J) LILRB4 blocking efficacy of 128–3 and 216–1 was determined by reporter assay. Two independent experiments were performed. (K) Specificities of 128–3 and 216–1 were assessed by ELISA. (L) A combined KABAT/IMGT complementarity determining regions (CDR) graft strategy was used to humanize rabbit mAb 128–3. (M) Binding of humanized 128–3 (h128–3) to LILRB4 was determined by ELISA. (N) Affinity of humanized 128–3 (h128–3) to LILRB4 was determined by Octet RED96. (O) LILRB4 blocking efficacy of h128–3 was determined by reporter assay. Two independent experiments were performed.
Identification and humanization of a LILRB4 blocking mAb
To examine whether these LILRB4 mAbs block LILRB4 activation by APOE, a functional extracellular binding protein of LILRB4, we screened the 21 mAbs using a GFP-based LILRB4 chimeric receptor reporter assay (Fig. S3) (18). Two antibodies from the same epitope bin (bin 4) were found to neutralize APOE-mediated LILRB4 activation (Fig. 1F). Using an Octet RED96, we further assessed LILRB4 binding kinetics of these two mAbs, 128–3 and 216–1. Kd values for binding to LILRB4 are 2.9 nM for 128–3 and 14.5 nM for 216–1 (Fig. 1G and 1H). We selected 128–3 as the lead mAb based on its higher LILRB4 binding activity as measured by ELISA (Fig. 1I), higher affinity to LILRB4 as measured by the Octet RED96 system (Fig. 1G and 1H), and more potent LILRB4/APOE blocking efficacy as assessed in the chimeric receptor reporter assay (Fig. 1J). Nonspecific binding of a therapeutic antibody may cause severe adverse effects due to off target activity. The LILR family of receptors contains 11 members (B1–B5 and A1–A6) that are closely related phylogenetically, especially in the D1 domain (Supplementary Fig. S4). These receptors are expressed on a wide range of normal cells. To determine the specificity of 128–3, we next expressed full-length ECD proteins for all eleven LILR family receptors and tested their binding with 128–3 by ELISA. As shown in Fig. 1K, mAb 128–3 is specific to LILRB4 and does not cross react with other LILR family members.
For potential therapeutic development, we used a CDR grafting strategy to humanize 128–3 (h128–3), converting it into a human IgG1 subclass with a kappa light chain (Fig. 1L). Next, we measured the binding affinity of h128–3 to LILRB4 by Octet RED96 and ELISA. We also measured LILRB4 blocking efficacy using a chimeric receptor reporter assay. The EC50 for h128–3 binding to LILRB4 as measured in ELISA is 0.23 nM. This is less than three times of the EC50 of 0.08 nM for the parental 128–3 (Fig. 1M). Similarly, the Kd of h128–3 binding to LILRB4 as measured by Octet RED96, 3.5 nM, is comparable to the Kd of the parental 128–3, 2.9 nM (Fig. 1N). The IC50 for LILRB4 blocking activity for the humanized h128–3 is 0.24 nM and the IC50 for the parental 128–3 is 0.14 nM. These results suggest that h128–3 maintained the binding affinity and blocking activity of the parental rabbit mAb 128–3 (Fig. 1M–O).
Structural elucidation of h128–3/LILRB4 interaction
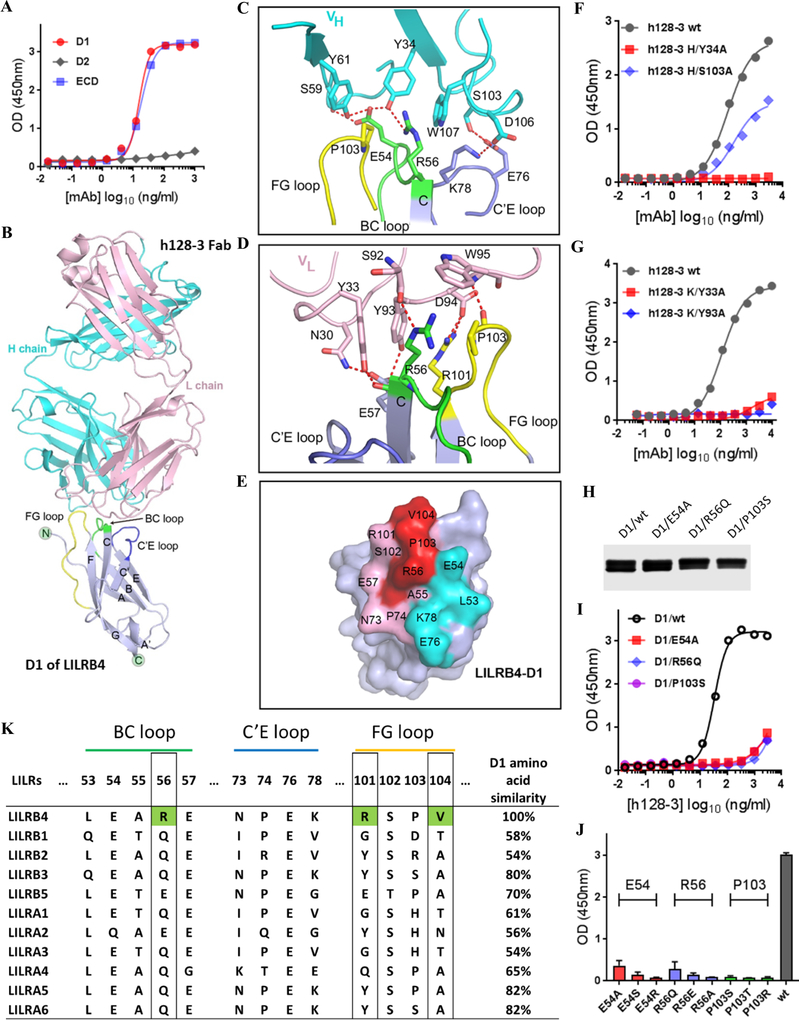
Studies of binding between h128–3 and the various ECD domains of LILRB4 showed that the antibody binds to the D1 domain of LILRB4 (Fig. 2A). Next, we sought to use X-ray crystallography to elucidate the molecular basis of this interaction and characterize the epitope on LILRB4 contributing to h128–3 binding. To achieve this objective, we expressed the D1 domain of LILRB4 as inclusion bodies in E. coli. From these inclusion bodies, we obtained soluble proteins by in vitro refolding. The D1/h128–3 Fab complex was subsequently prepared for crystal screening. The complex structure was determined by molecular replacement at a resolution of 3.0 Å (Supplementary Table S1). The overall structure reveals that h128–3 binds to the tip of the D1 domain (Fig. 2B). The mAb h128–3 utilizes both heavy (VH) and light (VL) chains to interact with LILRB4, involving five CDR loops in all h128–3 VH and VL regions except LCDR2 (Fig. 2C–D and Supplementary Table S2). In particular, h128–3 binds to the BC, C’E, and FG loops of D1. Residue E54 in the BC loop of D1 forms four hydrogen bonds, with residues both in HCDR1 (Y34) and HCDR2 (S59 and Y61) regions (Fig. 2C). Residue R56 in the BC loop interacts with residue Y34 in HCDR1 by forming one hydrogen bond (Fig. 2C). Furthermore, R56 contributes two hydrogen bonds, with residues Y33 in LCDR1 and S92 in LCDR3 (Fig. 2D). In addition, E76 and K78 in the C’E loop of D1 make contact with S103 and D106 in HCDR3 by forming two hydrogen bonds (Fig. 2C). Residues R101 and P103 in the FG loop of D1 form five hydrogen bonds, with residues Y93, D94, and W95 in LCDR3 (Fig. 2D). The binding surface in D1 of LILRB4 are shown in Fig. 2E. In summary, the three loops of D1 are targeted by h128–3 through a series of contacts including multiple hydrogen bond interactions.
Fig. 2. Decoding of the interaction of h128–3 with D1.
(A) Binding of h128–3 to D1, D2, and ECD of LILRB4 was determined by ELISA. (B) Overall crystal structure of D1/h128–3-Fab complex. D1 is shown in gray, the antibody h128–3 heavy (H) chain is shown in cyan and its light (L) chain shown in pink. The FG loop of D1 is shown in yellow, the BC loop of D1 is shown in green, and the C’E loop of D1 is shown in blue. (C) Detailed interaction of D1/h128–3-VH. (D) Detailed interaction of D1/h128–3-VL. Residues involved in the hydrogen bond interaction are shown as sticks and labeled. Hydrogen bonds are shown as red dashed lines. (E) The epitope residues in D1 are labeled by black characters. Residues contacted by the h128–3-Fab VH are colored cyan. Residues contacted by the h128–3-Fab VL are colored pink. Residues contacted by both chains are colored red. Binding of h128–3 heavy (F) and light (G) chain mutants to LILRB4 were performed by ELISA. (H) Generation and purification of D1 mutants, which were fused with human IgG1 Fc tag and expressed in HEK293F cells. (I) Binding of h128–3 to D1 mutants was determined by ELISA. (J) Binding of h128–3 with LILRB4 ECD mutants. Three identified critical amino acid residues that were mutated to three different types of amino acid residues. (K) Sequence alignment of h128–3 binding motif (BC loop, C’E loop, and FG loop) in all eleven LILR family members. LILRB4 unique amino acid residues R56, R101, and V104 are marked with green. Total amino acid similarities of D1 in percentages compared with LILRB4 (100%) are shown.
To confirm the role these key residues play in LILRB4 binding, we generated single alanine mutants of h128–3 (Y34A in HCDR1, S103A in HCDR3, Y33A in LCDR1, and Y93A in LCDR3). The mutant antibodies were tested by ELISA to assess the impact on LILRB4 binding. The single mutant variants Y34A, Y33A, and Y93A completely abolished the binding of h128–3 to LILRB4. Only S103A retained partial LILRB4 binding ability (Fig. 2F–G). We also generated single amino acid mutants in the LILRB4 D1 domain: E54A, R56Q, and P103S. Among LILRB family members, LILRB3 is the closest relative of LILRB4 based on D1 amino acid similarity. Therefore, the mutations were designed based on sequence alignment of D1 domains of LILRB4 and LILRB3. Where residues differ between LILRB4 and LILRB3, the LILRB4 amino acid residue was replaced by the corresponding residue in LILRB3. If the residue is conserved in LILRB4 and LILRB3, it was mutated to alanine. Consistent with predictions from analysis of the crystal structure of D1/h128–3 Fab complex, all three mutations in the LILRB4 D1 domain abolished binding to h128–3 (Fig. 2H–J).
Alignment of the h128–3 binding motif (BC loop, C’E loop, and FG loop) in all eleven LILR family members showed that this motif is unique to LILRB4: R56, R101, and V104 are LILRB4 specific amino acid residues (Fig. 2K). As expected, given the lack of cross reactivity of 128–3 to other LILR family members (Fig. 1K), crystal structure analysis and epitope mapping results further confirmed h128–3 as an LILRB4 specific mAb. In agreement with findings that the FG loop is critical for LILRB4 activation induced by APOE (18), our results indicate that h128–3 blocks ApoE-induced LILRB4 signaling activation in a competitive manner.
Reversal of T cell suppression by h128–3
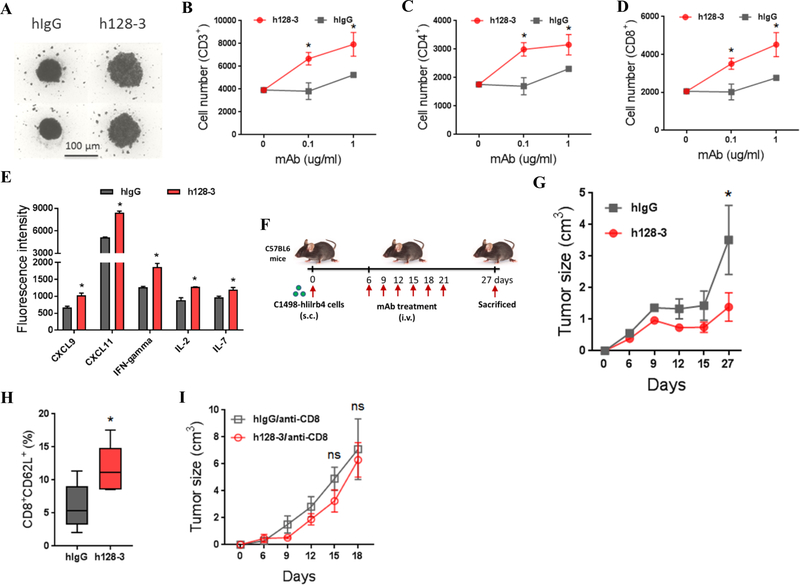
We previously demonstrated that LILRB4 expressed on monocytic AML cells suppresses T cell-mediated antitumor immunity through the APOE/LILRB4/SHP-2/NF-κB/uPAR/Arginase-1 axis (18). Arginase-1 is a key downstream effector of LILRB4/NF-kB/uPAR signaling and Arginase-1 can be secreted by AML cells to inhibit T cell activity. In this study, we sought to determine whether blocking LILRB4 signaling by mAb h128–3 reverses T-cell suppression. We cocultured human T cells with THP-1 cells and treated them with h128–3. hIgG served as a control. Treatment with h128–3 significantly increased T cell proliferation in these studies of cocultured T cells and THP-1 cells (Fig. 3A). The numbers of CD3+, CD4+, and CD8+ T cells were higher in the cocultures treated with mAb h128–3 (Fig. 3B–D). This result suggests that the T-cell suppressive ability of THP-1 cells was reversed by LILRB4 blockade. Moreover, in the human PBMC and AML THP-1 cell coculture assay, treatment with h128–3 increased the secretion of multiple T-cell proliferation and activation related cytokines including CXCL9, CXCL11, IFNγ, IL2, and IL7 (Fig. 3E).
Fig. 3. h128–3 reverses T-cell suppression in vitro and in vivo.
(A) Representative image of T cells in coculture assay. T cells isolated from healthy donors were incubated in the lower chamber of a 96-well transwell plate with irradiated THP-1 cells (E:T of 2:1) in the upper chamber separated by a membrane with 3 μm pores. After coculture with anti-CD3/CD28-coated beads and rhIL2 for 7 days, representative cells were photographed using an inverted microscope (scale bar, 100 μm). Two independent experiments were performed. T cells were stained with anti-CD3 (B), anti-CD4 (C), and anti-CD8 (D) and analyzed by flow cytometry. (E) Quantitative analysis of the cytokines in supernatants of human PBMC and THP-1 cells coculture assay. Human PBMC (1.5 × 106 cells/ml) and THP-1 cell (3 × 105 cells/ml) along with h128–3 or hIgG (20 μg/ml) were cocultured for 48 hours. The supernatants were then harvested and detected using RayBio G-Series human cytokine antibody array 1000 Kit. (F) Study design. C57BL/6 mice were subcutaneously implanted with human LILRB4 forced expressing mouse AML C1498 cells (C1498-hlilrb4) followed by treatment with h128–3, h128–3 along with anti-CD8, control human IgG (hIgG) or hIgG along with anti-CD8. Endpoints were assessed at day 27 after AML cells transplantation. (G) Tumor growth of subcutaneous C1498-hlilrb4-bearing mice (n=5) treated with h128–3 or hIgG. (H) Quantitation of CD8+CD62L+ memory T cells in subcutaneous C1498-hlilrb4-bearing mice treated with h128–3 or hIgG. (I) Tumor growth of subcutaneous C1498-hlilrb4-bearing mice (n=5) treated with h128–3 or hIgG in T cell depletion condition.
To further evaluate whether h128–3 reverses T-cell suppression in vivo, we next generated a human LILRB4 expressing mouse AML cell line, C1498-hlilrb4. We subcutaneously implanted C1498-hlilrb4 cells into C57BL/6 mice to establish a syngeneic immunocompetent mouse model. In this model, treatment with 10 mg/kg of h128–3 at day 6 after implantation of C1498-hlilrb4 cells significantly lowered the tumor burden and increased the percentage of CD8+CD62L+ memory T cells in the spleen, suggesting h128–3 reversed the T-cell suppression in vivo (Fig. 3F–H). Depletion of CD8+ T cells in this mouse model by treatment with CD8 antibodies almost completely eliminated the antitumor efficacy of h128–3 (Fig. 3I). Together, these results suggest that h128–3 restores T cell activity against tumors in vitro and in vivo.
Blocking tissue infiltration and inducing mobilization of AML cells by mAb h128–3
One of the characteristic features of monocytic AML is enhanced extramedullary infiltration of tumor cells (30). As we reported previously, LILRB4 guides leukemia cells to migrate to internal organs through the APOE/LILRB4/SHP-2/NF-κB/uPAR/Arginase-1 axis (18). Depletion of LILRB4 in AML cells significantly decreased their homing ability in vivo (Fig. 4A). In order to determine whether h128–3 inhibits AML cells migration, we performed a transwell migration assay using AML THP-1 cells. Treatment with h128–3 significantly decreased the migration of AML cells in vitro (Fig. 4B). To further investigate whether h128–3 inhibits AML cells tissue infiltration in vivo, we evaluated its efficacy for blocking AML cell homing in an NSG mouse model. AML MV4–11 cells were intravenously injected into NSG mice followed by immediate treatment with h128–3 or hIgG as a control. Treatment with h128–3 significantly decreased short-term (8 hours and 20 hours) homing of AML MV4–11 cells to bone marrow (BM), liver (LV), and spleen (SP) (Fig. 4C–D). Moreover, h128–3-mediated LILRB4 blockade showed significant inhibition of long-term (21 day) tissue infiltration of AML MV4–11 cells to bone marrow, liver, and spleen (Fig. 4E). Consistent with h128–3 blocking AML MV4–11 cells homing and tissue infiltration, whole animal bioluminescence imaging showed that treatment with h128–3 significantly blocked the establishment of AML THP-1 cells, delayed body weight loss, and prolonged survival of xenografted NSG mice (Fig. 4F–I). To assess whether blocking establishment of AML in NSG mice by h128–3 is dose-dependent, luciferase expressing THP-1 cells (THP-1-luc) were intravenously injected into NSG mice, followed on the same day by a single dose treatment with h128–3 of different doses (0.01 mg/kg, 0.1 mg/kg and 1 mg/kg). Bioluminescence imaging showed that h128–3 blocks engraftment of AML THP-1 cells in NSG mice in a dose-dependent manner. Treatment with 1 mg/kg h128–3 showed much higher therapeutic activity than did treatment with 0.01 mg/kg or 0.1 mg/kg (Fig. 4J). We also observed time dependence in h128–3 blocking of tissue infiltration by AML THP-1 cells tissue infiltration. Treatment with h128–3 at day 0 after injection of the AML cells showed the best efficacy in reducing AML cell tissue infiltration in NSG mice. Treatment at day 12 after injection of the AML cells exhibited minimal effect on AML cell tissue infiltration (Supplementary Fig. S5).
Fig. 4. h128–3 blocks AML cell migration and tissue infiltration.
(A) Comparison of the short-term (20 hours) tissue infiltration of wt or lilrb4-KO AML MV4–11 cells in NSG mice (n=4). The numbers of leukemia cells (GFP+) in bone marrow (BM), liver (LV), and spleen (SP) were determined by flow cytometry and normalized to number in peripheral blood (PB). Homing ratio of MV4–11cells in mice treated with hIgG was normalized to 100%. (B) Comparison of transwell migration abilities of THP-1 cells treated with h128–3 or hIgG. Two independent experiments were performed. Short-term homing abilities of CFSE-labeled MV4–11 cells that were injected into NSG mice followed by immediately treated with h128–3 or hIgG at 8 (C) or 20 (D) hours post-injection. (E) Long-term (21 days) tissue infiltration of THP-1 cells in NSG mice after treatment with h128–3 or hIgG. (F) Study design. NSG mice (n=5) were intravenously injected with luciferase expressing THP-1 cells (THP-1-luc) followed by immediate treatment with h128–3 or hIgG. Bioluminescence imaging (G), Survival curve (H) and body weight changes (I) of NSG mice treated with h128–3 or hIgG. (J) NSG mice were injected with 1×106 THP-1-luc cells followed immediately by treatment with 0.01 mg/kg, 0.1 mg/kg, or 1 mg/kg of h128–3 and monitored by bioluminescence imaging. (K) Study design. Primary AML cells from monocytic AML patients were injected into irradiated NSG mice. After 9 weeks of engraftment, mice were sacrificed and tissues (single cell suspension) with engrafted AML cells were then injected into other irradiated NSG mice followed by immediate h128–3 or hIgG treatment. Endpoints were assessed at day 21 after AML cells injection. Human AML cells in bone marrow, liver, spleen, and peripheral blood were analyzed by flow cytometry. (L) Percentages of human AML cells (CD45+LILRB4+) engrafted in indicated organs were analyzed by flow cytometry at day 21 post-transplant. (M) Study design. NSG mice (n=5 or 6) were intravenously injected with THP-1 cells followed by cytarabine (10 mg/kg) and h128–3 or hIgG (10 mg/kg) treatment at indicated time points. Endpoints were assessed at day 21 after THP-1 cells injection. Anti-human CD45 was used to detect human leukemia cells (THP-1 cells) in liver by flow cytometry. (N) Shown are percentages of human cells engrafted in liver at day 21 post-transplantation.
Next, we sought to assess whether the tissue infiltration blocking effects of h128–3 in AML THP-1 cells xenograft NSG mouse model could be reproduced with primary human monocytic AML cells. To this end, we first obtained primary AML cells from patients with monocytic AML (UT Southwestern Medical Center). Then, we intravenously transplanted these cells into irradiated NSG mice for 9 weeks. Mouse tissues along with engrafted human AML cells were then injected into other irradiated NSG mice followed by treatment with h128–3 or hIgG. The percentages of human CD45+LILRB4+ AML cells in bone marrow, liver, spleen, and peripheral blood (PB) of the injected NSG mice were measured by flow cytometry (Fig. 4K–L). Treatment with h128–3 significantly inhibited AML development in primary human monocytic AML-derived xenografts (Fig. 4L). Collectively, these results showed that h128–3 inhibits monocytic AML cells migration and tissue infiltration in vitro and in vivo.
Chemotherapy has limited capability to target cancer cells in niches. Based on the ability of LILRB4 to support homing, we hypothesize that blocking LILRB4 signaling enhances mobilization of those niche-homed AML cells into circulation. If this is the case, the LILRB4 blocking antibody would enhance anti-AML efficacy of chemotoxic drugs that kill tumor cells outside of their niches. To test the hypothesis, we performed h128–3 and cytarabine combination treatment in the THP-1 cells xenograft NSG mouse model. After transplantation of THP-1 cells into NSG mice, the mice were treated with cytarabine along with h128–3 or hIgG at day 6 or day 14 (Fig. 4M). Blocking LILRB4 by h128–3 significantly enhanced the anti-AML effects of cytarabine (Fig. 4N). These results suggest that h128–3 could be further developed as part of a combination therapy with chemotherapy drugs for AML treatment.
ADCC and ADCP mediated by mAb h128–3 contribute to tumor cell killing
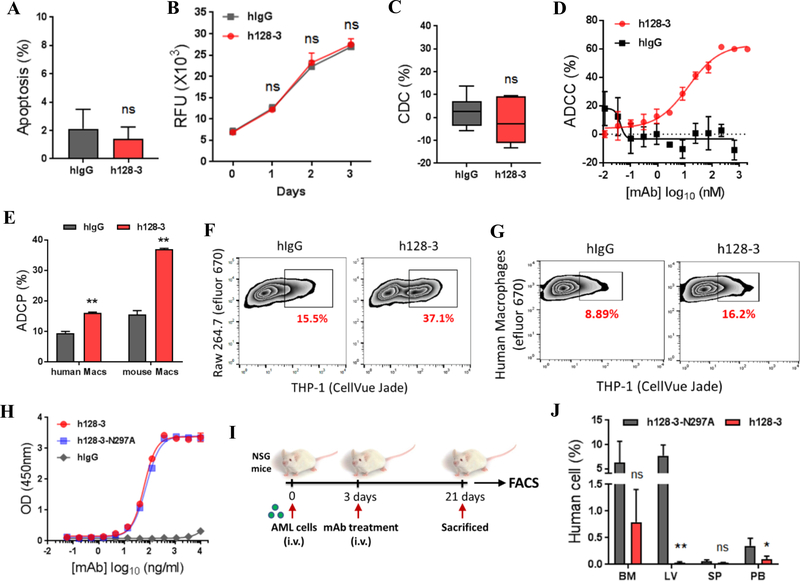
Fc-mediated immune functions such as antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement-dependent cytotoxicity (CDC) have been validated as modes of action of therapeutic monoclonal antibodies used in cancer therapy (31, 32). h128–3 has no effect on apoptosis or proliferation of AML THP-1 cells (Fig. 5A–B). To further investigate possible Fc-mediated mechanisms for anti-AML effects in vitro and in vivo, h128–3 was analyzed for CDC, ADCC, and ADCP activity using complement or effector cells. In an experiment using THP-1 cells as target cells and normal human serum as complement, no CDC activity of h128–3 was detected, even at a high antibody concentration of 20 μg/mL (Fig. 5C). We detected ADCC and ADCP activities for h128–3. AML THP-1 cells were specifically killed by human PBMCs in the presence of h128–3, in a dose-dependent manner with EC50 value of 13.5 nM (Fig. 5D). Moreover, h128–3 could direct both mouse macrophages (mouse Macs, RAW 264.7 cells stimulated with LPS) and human PBMC derived macrophages (human Macs, differentiated from CD14+ monocytes selected from human PBMC) to phagocytose AML THP-1 cells in a flow cytometry ADCP assay (Fig. 5E–G). To verify that Fc-mediated immune function is one of the mechanisms for anti-AML activity of h128–3, we generated Fc mutated h128–3 (h128–3-N297A), which is defective in ADCC and ADCP, by abolishing the binding of h128–3 to FcγRs on immune cells (33). As expected, h128–3 and h128–3-N297A showed the same binding affinity to LILRB4 as assessed by ELISA (Fig. 5H). Next, we sought to determine the contribution of h128–3 Fc-mediated anti-AML activity to leukemia cells killing in vivo. To accomplish this objective, we evaluated the activities of h128–3 and h128–3-N297A in AML THP-1 cells xenografted NSG mice (Fig. 5I). Treatment with 10 mg/kg of h128–3-N297A was correlated with significantly higher engraftment of AML THP-1 cells in NSG mice than was treatment with h128–3 (Fig. 5J). Taken together, these results suggest that h128–3 Fc-mediated immune functions contribute to anti-AML activity in vitro and in vivo.
Fig. 5. h128–3 triggers ADCC and ADCP.
(A) Comparison of the apoptosis of THP-1 cells induced by h128–3 or hIgG. Two independent experiments were performed. (B) Comparison of the effect on THP-1 cells proliferation treated with h128–3 or hIgG. Two independent experiments were performed. (C) Complement-dependent cytotoxicity (CDC) of h128–3 was assessed in a lactate dehydrogenase (LDH) assay using normal human serum as complement and THP-1 cell as target cell. Two independent experiments were performed. (D) Antibody dependent cellular cytotoxicity (ADCC) triggered by h128–3 or hIgG were assessed by flow cytometry. THP-1-GFP cells were used as target cells and fresh isolated PBMCs from healthy donors used as effector cells in a E:T ratio of 50:1. Two independent experiments were performed. (E-G) Antibody dependent phagocytosis (ADCP) triggered by h128–3 or hIgG was detected in flow cytometry assay using THP-1 cells as target cells and mouse macrophage cell line RAW 264.7 (mouse Macs) or human PBMC derived macrophages (human Macs) as effector cells. After incubation of h128–3 or hIgG together with target and effector cells for 2 hours at 37°C, adherent macrophages were collected and determined by flow cytometry. Phagocytosis percentage was calculated by double positive macrophages/total macrophages. Representative FACS profiles are shown. Three independent experiments were performed. (H) Comparison of the binding of wild-type (h128–3) and N297A mutated h128–3 (h128–3-N297A) to LILRB4 in ELISA. (I) Study design. NSG mice (n=5) were intravenously injected with AML THP-1 cells followed by treated with h128–3 or h128–3-N297A at day 3 after THP-1 cells injection. Endpoints were assessed at day 21 after THP-1 cells injection. Human cells (THP-1 cells) in bone marrow, liver, spleen, and peripheral blood were analyzed by flow cytometry. (J) Percentages of human AML cells engrafted in indicated organs were analyzed by flow cytometry at day 21 post-transplant.
Discussion
Despite an increased understanding of the biology of AML, antibody therapies directly targeting the antigens expressed on AML cells with appropriate specificity and functionality are not well established. For example, a CD33 antibody (gemtuzumab ozogamicin, CD33 antibody–drug conjugate) was in the clinic for more than twenty years and showed limited success in treating AML (7, 34). Therapies directed against novel targets are urgently needed for AML treatment. As we have reported, LILRB4 is a surface marker for monocytic AML. LILRB4 expression is primarily observed on normal monocytic cells but elevated on monocytic AML cells. Further, we have shown that LILRB4 activation by extracellular binding protein APOE inhibits T-cell activation and supports infiltration of leukemia cells (18). Together, these characteristics make LILRB4 a compelling target for the development of AML therapeutics.
Monoclonal antibodies, mAbs, have been validated as a highly effective drug modality for cancer therapy. These antibodies are particularly well suited for blocking ligand-receptor interaction driven disease mechanisms. In this study, we sought to generate LILRB4/APOE blocking mAbs and test whether these mAbs have anti-AML activity in vitro and in vivo. We generated a comprehensive panel of LILRB4 rabbit mAbs using a strategy optimized in this study. This strategy involved memory B cell isolation, culture, and cloning. The lead mAb 128–3 was selected based on its specificity to LILRB4 and potency of blocking LILRB4/APOE downstream signaling. This lead antibody was humanized. In both the human AML cell line and primary human AML cell xenograft NSG mouse model, the humanized antibody h128–3 exhibited potent anti-AML activity. Since NSG mice are immunodeficient and lack T cells, the anti-AML activity in the NSG mice of h128–3 is likely the result of inhibition of cancer cell migration and Fc-mediated immune effector functions such as ADCC and ADCP. LILRB4 contains two Ig-like motifs in its ECD and ITIM motifs in its intracellular domain, which are the hallmarks of immune checkpoint inhibitory receptors (35). We sought to determine the immune checkpoint inhibitory function of LILRB4. For studies addressing this question, we selected a human LILRB4 expressing mouse AML cell line allograft C57BL/6 mouse model. In this mouse model, T cells are critical for anti-AML activity. Depletion of T cells by CD8 antibody treatment reduced the anti-AML activity of h128–3. This result is consistent with the findings that the antitumor efficacy of other receptors targeted antibodies, such as Her2, CD47, PD-1, and CTLA-4, is T-cell dependent. Depletion of T cells sharply decreases the antitumor activity of these therapeutic antibodies (36–39).
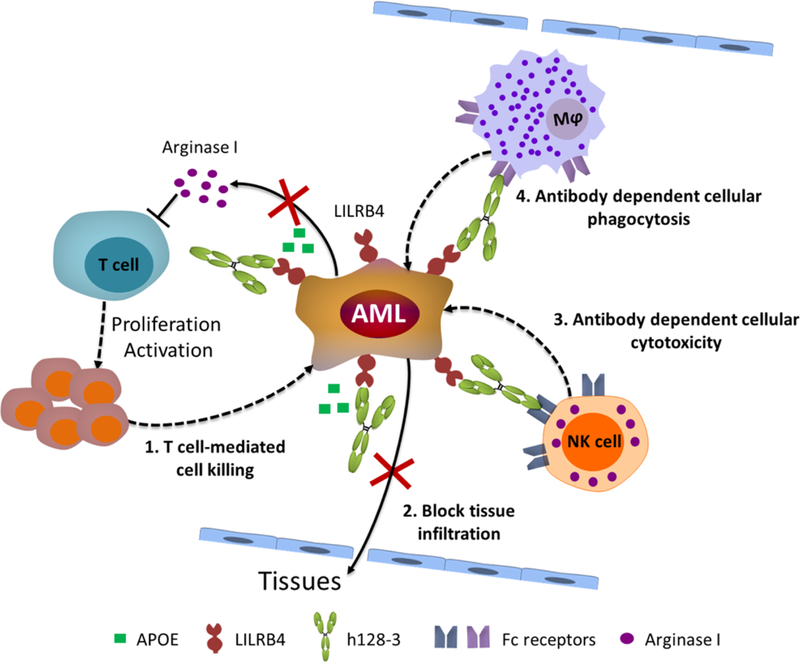
Our studies revealed that the anti-AML activity of the LILRB4-targeting antibody h128–3 possesses multiple mechanisms: 1) reversal of T-cell suppression mediated by LILRB4; 2) inhibition of AML cell tissue infiltration by blocking LILRB4 activation by APOE; 3) Fc-mediated ADCC and ADCP (Fig. 6). Fc-mediated immune functions are well-established modes of action for many cancer antibody therapies. In some cases, Fc-mediated immune effector functions are avoided to minimize the risk of depleting normal immune cells. For example, most of the currently approved immunomodulatory antibodies, such as pembrolizumab and nivolumab, are of the IgG4 isotype which has low or no binding to the Fc gamma receptors (FcγRs) that trigger ADCC or ADCP. LILRB4 is expressed on monocytic AML cells; normal monocytes and dendritic cells have lower expression of the receptor compared to monocytic AML cells (8). Therefore, Fc-mediated immune functions of h128–3 may be employed to enhance the anti-AML efficacy with potentially minimal side effects, due to the receptor copy number dependency of Fc-mediated effector functions.
Fig. 6. Multiple mechanisms of h128–3 contribute to anti-AML activity.
Leukocyte immunoglobulin-like receptors are categorized into two families of immunoregulatory receptors: LILRBs (B1–B5) and LILRAs (A1–A6). LILRBs contain cytoplasmic tails with immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that provide negative signals (8, 40). LILRAs have short cytoplasmic domains lacking signaling motifs. LILRAs transmit activating signals by linking to immunoreceptor tyrosine-based activation motifs (ITAMs) of the FcR gamma-chain (8, 40). LILRBs and LILRAs are expressed on a wide range of hematopoietic cell types such as macrophages, dendritic cells, NK cells, basophils and eosinophils (8, 40). Human genes encoding these receptors are found in a gene cluster at chromosomal region 19q13.4. Phylogenetic analysis showed that these receptors share a high degree of D1 sequences similarity. As all LILR family members are expressed on normal immune cells and hematopoietic cells, nonspecific binding of an LILRB4 therapeutic antibody may lead to serious adverse effects. In order to generate LILRB4-specific antibodies, we characterized a large panel of LILRB4 binding antibodies for their binding affinity to all eleven LILR family members made as Fc fusion proteins. The high LILRB4 specificity of h128–3 should increase the safety profile of the antibody as an AML therapy; this will need to be validated in clinical studies.
In summary, our study demonstrated that LILRB4 is a viable drug target for monocytic AML. The LILRB4-specific blocking antibody h128–3 exhibited anti-AML efficacy in both in vitro and in vivo models. Mechanistic studies revealed at least four modes of action for the anti-AML efficacy of h128–3: include inhibition of AML cell infiltration, stimulation of T-cell activation, ADCC and ADCP. Taken together, the results of this work open doors for development of LILRB4 targeting antibodies such as h128–3 for the treatment of monocytic AML.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Xuejun Fan, Dr. Wei Xiong and Ms. Hui Deng for the technical assistance and helpful advice. We thank Dr. Georgina T. Salazar for editing the manuscript.
Funding: This work was supported by the Cancer Prevention and Research Institute of Texas (RP140402, DP150056, RP180435, and RP150551), the Welch Foundation (AU-0042–20030616 and I-1834), and National Cancer Institute (1R01CA172268).
Footnotes
Disclosure of Potential Conflicts of Interests: The Board of Regents of the University of Texas System has filed patent applications with PCT Application Nos. PCT/US2016/020838, which covers LILRB antibodies and their uses in detecting and treating cancer, and PCT/US2017/044171, which covers the methods for identifying LILRB-blocking antibodies. Authors C.C.Z., M.D., Z.A., N.Z., and X.G. are listed as inventors of PCT/US2016/020838. Authors C.C.Z., Z.A., N.Z., M.D., J.K. and X.G. are listed as inventors of PCT/US2017/044171. Both patent applications have been exclusively licensed to Immune-Onc Therapeutics by the Board of Regents of the University of Texas System. Authors N.Z., C.C.Z., and Z.A. have a sponsored research agreement with Immune-Onc Therapeutics. Authors C.C.Z. and Z.A. are Scientific Advisory Board members with Immune-Onc Therapeutics, who also own equities. Authors T.H. and X.C.L. are employees and own equities of Immune-Onc Therapeutics.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD, Acute myeloid leukemia. New England Journal of Medicine 373, 1136–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A, Cancer statistics, 2016. CA: a cancer journal for clinicians 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Meyers J, Yu Y, Kaye JA, Davis KL, Medicare fee-for-service enrollees with primary acute myeloid leukemia: an analysis of treatment patterns, survival, and healthcare resource utilization and costs. Applied health economics and health policy 11, 275 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Curran EK, Godfrey J, Kline J, Mechanisms of Immune Tolerance in Leukemia and Lymphoma. Trends in immunology 38, 513–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas D, Majeti R, Optimizing next-generation AML therapy: activity of mutant IDH2 inhibitor AG-221 in preclinical models. Cancer discovery 7, 459–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz SE, Wilmot B, McWeeney S, Vellanki A, Local A, Benbatoul K, Folger P, Sheng S, Zhang H, Howell SB, CG′806, a first-in-class FLT3/BTK inhibitor, exhibits potent activity against AML patient samples with mutant or wild type FLT3, as well as other hematologic malignancy subtypes. (AACR, 2017). [Google Scholar]
- 7.Khan N, Hills RK, Virgo P, Couzens S, Clark N, Gilkes A, Richardson P, Knapper S, Grimwade D, Russell NH, Burnett AK, Freeman SD, Expression of CD33 is a predictive factor for effect of gemtuzumab ozogamicin at different doses in adult acute myeloid leukaemia. Leukemia 31, 1059–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang X, Kim J, Deng M, John S, Chen H, Wu G, Phan H, Zhang CC, Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle 15, 25–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirayasu K, Arase H, Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. Journal of human genetics 60, 703–708 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Carosella ED, Rouas-Freiss N, Roux DT, Moreau P, LeMaoult J, HLA-G: An Immune Checkpoint Molecule. Adv Immunol 127, 33–144 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Zheng J, Umikawa M, Cui C, Li J, Chen X, Zhang C, Hyunh H, Kang X, Silvany R, Wan X, Ye J, Canto AP, Chen SH, Wang HY, Ward ES, Zhang CC, Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature 485, 656–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang X, Lu Z, Cui C, Deng M, Fan Y, Dong B, Han X, Xie F, Tyner JW, Coligan JE, Collins RH, Xiao X, You MJ, Zhang CC, The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nat Cell Biol 17, 665–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, Satterthwaite A, Paietta E, Hunger SP, Willman CL, Melnick A, Loh ML, Jung JU, Coligan JE, Bolland S, Mak TW, Limnander A, Jumaa H, Reth M, Weiss A, Lowell CA, Muschen M, Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature 521, 357–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perna F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, Hendrickson RC, Brennan CW, Sadelain M, Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 32, 506–519 e505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohner H, Weisdorf DJ, Bloomfield CD, Acute Myeloid Leukemia. N Engl J Med 373, 1136–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Dobrowolska H, Gill KZ, Serban G, Ivan E, Li Q, Qiao P, Suciu‐Foca N, Savage D, Alobeid B, Bhagat G, Colovai A, Expression of immune inhibitory receptor ILT3 in acute myeloid leukemia with monocytic differentiation. Cytometry Part B: Clinical Cytometry 84, 21–29 (2013). [DOI] [PubMed] [Google Scholar]
- 17.John S, Chen H, Deng M, Gui X, Wu G, Chen W, Li Z, Zhang N, An Z, Zhang CC, A Novel Anti-LILRB4 CAR-T Cell for the Treatment of Monocytic AML. Molecular Therapy 26, 2487–2495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng M, Gui X, Kim J, Xie L, Chen W, Li Z, He L, Chen Y, Chen H, Luo W, Lu Z, Xie J, Churchill H, Xu Y, Zhou Z, Wu G, Yu C, John S, Hirayasu K, Nguyen N, Liu X, Huang F, Li L, Deng H, Tang H, Sadek AH, Zhang L, Huang T, Zou Y, Chen B, Zhu H, Arase H, Xia N, Jiang Y, Collins R, You MJ, Homsi J, Unni N, Lewis C, Chen G-Q, Fu Y-X, Liao XC, An Z, Zheng J, Zhang N, Zhang CC, LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng W, Li L, Xiong W, Fan X, Deng H, Bett AJ, Chen Z, Tang A, Cox KS, Joyce JG, Freed DC, Fu TM, Casimiro DR, Zhang N, Vora K, An Z, Efficient generation of monoclonal antibodies from single rhesus macaque antibody secreting cells. mAbs 7, 707–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng M, Lu Z, Zheng J, Wan X, Chen X, Hirayasu K, Sun H, Lam Y, Chen L, Wang Q, Song C, Huang N, Gao F, Jiang Y, Arase H, Zhang CC, A motif in LILRB2 critical for Angptl2 binding and activation. Blood 124, 924–935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Lee P, Ke Y, Zhang Y, Yu Q, Lee J, Li M, Song J, Chen J, Dai J, Couto FJ, An Z, Zhu W, Yu GL, A humanized anti-VEGF rabbit monoclonal antibody inhibits angiogenesis and blocks tumor growth in xenograft models. PloS one 5, e9072 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y-F, Ho M, Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: Rationale and examples mAbs 9, 419–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H, Mohammed F, Nam G, Chen Y, Qi J, Garner LI, Allen RL, Yan J, Willcox BE, Gao GF, Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance. The Journal of biological chemistry 286, 18013–18025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang X, Lu Z, Cui C, Deng M, Fan Y, Dong B, Han X, Xie F, Tyner JW, Coligan JE, Collins RH, Xiao X, You MJ, Zhang CC, The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nature cell biology 17, 665–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Xie J, Wu G, Shen J, Collins R, Chen W, Kang X, Luo M, Zou Y, Huang LJ-S, Amatruda JF, Slone T, Winick N, Scherer PE, Zhang CC, Fasting selectively blocks development of acute lymphoblastic leukemia via leptin-receptor upregulation. Nature medicine 23, 79–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gholamin S, Mitra SS, Feroze AH, Liu J, Kahn SA, Zhang M, Esparza R, Richard C, Ramaswamy V, Remke M, Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Science translational medicine 9, eaaf2968 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Seeber S, Ros F, Thorey I, Tiefenthaler G, Kaluza K, Lifke V, Fischer JA, Klostermann S, Endl J, Kopetzki E, Pashine A, Siewe B, Kaluza B, Platzer J, Offner S, A robust high throughput platform to generate functional recombinant monoclonal antibodies using rabbit B cells from peripheral blood. PloS one 9, e86184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freed DC, Tang Q, Tang A, Li F, He X, Huang Z, Meng W, Xia L, Finnefrock AC, Durr E, Espeseth AS, Casimiro DR, Zhang N, Shiver JW, Wang D, An Z, Fu TM, Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proceedings of the National Academy of Sciences 110, E4997–E5005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, Rajpal A, High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PloS one 9, e92451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Straus DJ, Mertelsmann R, Koziner B, McKenzie S, de Harven E, Arlin ZA, Kempin S, Broxmeyer H, Moore MA, Menendez-Botet CJ, Gee TS, Clarkson BD, The acute monocytic leukemias: multidisciplinary studies in 45 patients. Medicine 59, 409–425 (1980). [DOI] [PubMed] [Google Scholar]
- 31.Uchida J, Hamaguchi Y, Oliver JA, Ravetch JV, Poe JC, Haas KM, Tedder TF, The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. The Journal of experimental medicine 199, 1659–1669 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clynes RA, Towers TL, Presta LG, Ravetch JV, Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nature medicine 6, 443–446 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, Fan X, Deng H, Brezski RJ, Rycyzyn M, Jordan RE, Strohl WR, Zou Q, Zhang N, An Z, Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcγ receptors on macrophages. The Journal of Immunology 194, 4379–4386 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Jen EY, Ko CW, Lee JE, Del Valle PL, Aydanian A, Jewell C, Norsworthy KJ, Przepiorka D, Nie L, Liu J, Sheth CM, Shapiro M, Farrell AT, Pazdur R, FDA Approval: Gemtuzumab ozogamicin for the treatment of adults with newly-diagnosed CD33-positive acute myeloid leukemia. Clinical Cancer Research 24, 3242–3246 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Couzin-Frankel J, Cancer Immunotherapy. Science 342, 1432–1433 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Zhang X, Mortenson ED, Radkevich-Brown O, Wang Y, Fu YX, Cetuximab-mediated tumor regression depends on innate and adaptive immune responses. Molecular therapy : the journal of the American Society of Gene Therapy 21, 91–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S, Jiang Z, Mortenson ED, Deng L, Radkevich-Brown O, Yang X, Sattar H, Wang Y, Brown NK, Greene M, Liu Y, Tang J, Wang S, Fu YX, The therapeutic effect of anti-HER2/neu antibody depends on both innate and adaptive immunity. Cancer cell 18, 160–170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Pu Y, Cron K, Deng L, Kline J, Frazier WA, Xu H, Peng H, Fu YX, Xu MM, CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nature medicine 21, 1209–1215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer 12, 252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Touw W, Chen HM, Pan PY, Chen SH, LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol Immunother 66, 1079–1087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.