Abstract
Brain lobar volumes are heritable but genetic studies are limited. We performed genome-wide association studies of frontal, occipital, parietal and temporal lobe volumes in 16,016 individuals, and replicated our findings in 8,789 individuals. We identified six genetic loci associated with specific lobar volumes independent of intracranial volume. Two loci, associated with occipital (6q22.32) and temporal lobe volume (12q14.3), were previously reported to associate with intracranial and hippocampal volume, respectively. We identified four loci previously unknown to affect brain volumes: 3q24 for parietal lobe volume, and 1q22, 4p16.3 and 14q23.1 for occipital lobe volume. The associated variants were located in regions enriched for histone modifications (DAAM1 and THBS3), or close to genes causing Mendelian brain-related diseases (ZIC4 and FGFRL1). No genetic overlap between lobar volumes and neurological or psychiatric diseases was observed. Our findings reveal part of the complex genetics underlying brain development and suggest a role for regulatory regions in determining brain volumes.
Subject terms: Biomarkers, Genome-wide association studies, Neurology
Sven van der Lee et al. report a genome-wide association study for lobar brain volumes in more than 16,000 individuals, with replication in nearly 9,000. They find 6 loci associated with occipital, parietal and temporal lobe volume, of which two were previously reported to associate with intracranial and hippocampal volume.
Introduction
Four lobes of the human brain are distinguished and several diseases can partially be attributed to lobe-specific structural changes. Functions of the frontal brain lobe include reasoning, movement, social behavior, planning, parts of speech, and problem solving1; functions attributed to the parietal lobe include recognition and perception of stimuli2; functions attributed to the temporal lobe include memory and speech3; and lastly, visual input is mainly processed by the occipital lobe. Brain diseases with lobe-specific abnormalities include Alzheimer’s disease (in particular early onset), frontotemporal lobar degeneration4, temporal lobe epilepsy5, primary progressive aphasia, and cortical basilar ganglionic degeneration.
Environmental factors, such as smoking and hypertension, affect lobar brain volumes6, but previous studies have shown that genetic differences across individuals also contribute to variability in volumetric brain measures7,8. The estimated heritability of brain lobar volumes is high, ranging from 26% to 84% for the frontal lobe, from 32% to 74% for the occipital lobe, from 30% to 86% for the parietal lobe, and from 55% to 88% for the temporal lobe9–15. In addition, genetic analyses in families suggest that the lobes are determined by independent genetic factors15. The observation that brain lobes are highly and differentially heritable makes them compelling targets to unravel the genetic architecture of the brain. Recent large genome-wide association studies (GWAS) have efficiently identified associations between genetic determinants and volumetric brain measures16,17. However, to date no genetic variants influencing brain lobar volumes have been identified. GWAS of the four lobar volumes of the brain can contribute to our understanding of brain lobe development and may provide a biological link between brain lobar volumes and brain-related traits and diseases.
To identify genetic variants of influence on lobar brain volumes, we performed GWAS of four brain lobar volumes in 16,016 individuals and replicated our findings in a sample of 8,789 individuals. We identified six loci significantly associated with specific brain lobar volumes independent of intracranial volume. With this study, we shed light on common genetic variants determining human brain volume and allow for a deepened understanding of the genetic architecture of the brain lobes.
Results
In total, 16,016 individuals from 19 population-based or family-based cohort study and one case–control study were included in the current study. Additional information regarding the population characteristics, genotyping, and imaging methods are provided in the “Methods” section and in Supplementary Data 1–4.
Heritability of lobar brain volumes
Using a family-based approach, we found an age- and sex-adjusted heritability (h2) for occipital lobe of 50% (95% confidence interval (CI) 38–62%), for frontal lobe of 52% (95% CI 40–64%), for temporal lobe of 59% (95% CI 49–69%), and for parietal lobe of 59% (95% CI 49–69%) (all: p ≤ 1.9 × 10−19) (Supplementary Data 5). In comparison, the age- and sex-adjusted heritability estimate for total brain volume was 34% (95% CI 22–46%, p = 8.8 × 10−11).
Novel genetic associations with brain lobar volumes
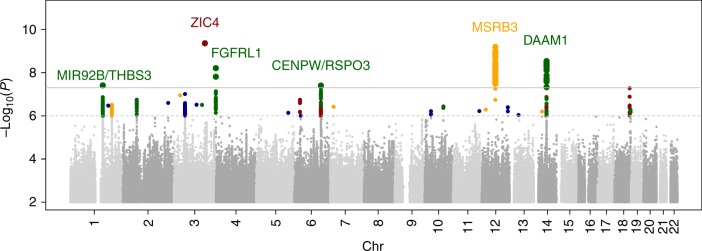
Our multi-ethnic meta-analysis (n = 16,016 individuals of which 15,269 were of European ancestry) identified significant associations between genotypes and brain lobar volumes in five independent loci, even though we adjusted for intracranial volume (Figs. 1 and 2, Table 1). The quantile–quantile plots did not show high genomic inflation (λGC ≤ 1.05) (Supplementary Fig. 1). Of these five loci, variants in one locus associated with temporal lobe volume, in one with parietal lobe volume, and in three with occipital lobe volume. The variant rs146354218 (12q14.3, pmulti-ethnic = 6.4 × 10−10) associated with temporal lobe volume and rs2279829 (3q24, pmulti-ethnic = 4.4 × 10−10) associated with parietal lobe volume. Three loci associated with occipital lobe volume: index variants rs147148763 (small indel GTTGT→G, 14q23.1, pmulti-ethnic = 2.9 × 10−9), rs74921869 (4p16.3, pmulti-ethnic = 6.2 × 10−9), and rs1337736 (6q22.32, pmulti-ethnic = 4.0 × 10−8). In the European ancestry-only meta-analysis, we found a significant association with occipital lobe volume in one additional independent locus (1q22, rs12411216, pEuropean ancestry-only = 3.9 × 10−8). In the multi-ethnic meta-analysis, this association was below the genome-wide significance threshold (pmulti-ethnic = 1.3 × 10−7). There was no significant heterogeneity observed for any of the six significant loci (Supplementary Figs. 2–7). The sensitivity meta-analysis including only the studies using the k-Nearest-Neighbor (kNN) algorithm for measuring lobar volumes showed similar results compared to the studies using other methods (Supplementary Figs. 2–7). The index variants of these total six loci were common (minor allele frequency ranging from 0.13 to 0.46) and associations with volume variations were between 0.48 and 0.95 cm3 per copy of the variant allele, explaining up to 0.27% of lobar volume variance per allele (Table 1). No variants were significantly associated with frontal lobe volume. All variants showing significant associations with brain lobar volumes are shown in Supplementary Data 6. Study-specific effects of all six significant loci are shown in Supplementary Figs. 2–7.
Fig. 1.
Common genetic variants associated with frontal, parietal, temporal and occipital lobe volume. ‘Manhattan’ plot displaying the association p value for each tested single-nucleotide polymorphism (SNP) (displayed as –log10 of the p value). Genome-wide significance threshold is shown with a line at p = 5 × 10−8 (solid black line) and also the suggestive threshold at p = 1 × 10−5 (dashed line). Dots represent SNPs, results of the four lobes are shown in a single figure, and the nearby gene is labeled. Above the suggestive threshold SNPs are colored by the associated lobe: yellow = temporal lobe, green = occipital lobe, red = parietal lobe, blue = frontal lobe
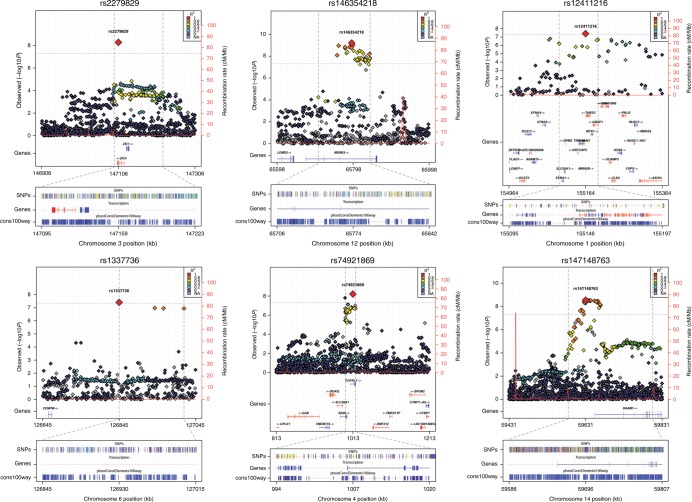
Fig. 2.
Regional view of the genome-wide significant loci. For each panel, zoomed in Manhattan plots (±kb from top single-nucleotide polymorphism (SNP)) are shown with gene models below (GENCODE version 19). Plots are zoomed in to highlight the genomic region that contains the index SNP and SNPs in linkage disequilibrium with the index SNP (R2 > 0.8). Each plot was made using the LocusTrack software (http://gump.qimr.edu.au/general/gabrieC/LocusTrack/)
Table 1.
Genetic variants at six loci significantly associated with lobar brain volumes
| Lobe | Annotation (nearby) | Gene | Rs-ID | Chr | Position | A1 | A2 | European ancestry meta-analysis | Multi-ethnic meta-analysis | Replication sample (n = 8,789) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frq | Effect | SE | p | Frq | Effect | SE | p | N | R2 (%) | Frq | Effect | SE | p | ||||||||
| Parietal | UTR3 | ZIC4 | rs2279829 | 3 | 147106319 | T | C | 0.21 | −0.92 | 0.16 | 5.3 × 10−9 | 0.21 | −0.95 | 0.15 | 4.4 × 10−10 | 16,015 | 0.24 | 0.22 | −0.63 | 0.18 | 6.0 × 10−4 |
| Temporal | Intronic | MSRB3 | rs146354218 | 12 | 65793942 | A | G | 0.37 | 0.69 | 0.11 | 1.2 × 10−9 | 0.37 | 0.69 | 0.11 | 6.4 × 10−10 | 15,789 | 0.24 | 0.37 | 0.54 | 0.14 | 6.4 × 10−5 |
| Occipital | Intergenic | DAAM1 (dist = 24302) | rs147148763a | 14 | 59631075 | G | GTTGT | 0.13 | −0.83 | 0.14 | 6.6 × 10−9 | 0.13 | −0.85 | 0.14 | 2.9 × 10−9 | 15,220 | 0.23 | 0.12 | −0.80 | 0.12 | 3.1 × 10−11 |
| Occipital | Intronic | FGFRL1 | rs74921869 | 4 | 1013382 | A | G | 0.2 | −0.84 | 0.14 | 5.9 × 10−9 | 0.2 | −0.82 | 0.14 | 6.2 × 10−9 | 12,424 | 0.27 | 0.19 | −0.22 | 0.10 | 3.0 × 10−2 |
| Occipital | Intergenic | CENPW (dist = 175626) | rs1337736 | 6 | 126845380 | A | G | 0.23 | −0.64 | 0.12 | 8.1 × 10−8 | 0.23 | −0.64 | 0.12 | 4.0 × 10−8 | 16,016 | 0.19 | 0.23 | −0.46 | 0.09 | 8.0 × 10−7 |
| Occipital | Upstream | MIR92B/THBS3 | rs12411216 | 1 | 155164480 | A | C | 0.46 | −0.52 | 0.10 | 3.9 × 10−8 | 0.46 | −0.49 | 0.09 | 1.4 × 10−7 | 16,016 | 0.17 | 0.45 | −0.19 | 0.08 | 1.6 × 10−2 |
The allele frequency (frq) and effect size are given for A1. Effect sizes are given in units of cm3 per effect allele. Results are provided for the discovery samples and the meta-analysis of all European ancestry and the multi-ethnic meta-analysis. The variance explained gives the percentage variance explained of a SNP16
A1 effect allele, A2 reference allele, Chr chromosome, Frq effect allele frequency, N number of individuals with genetic variant, p p value, Rs-ID reference SNP cluster ID, SE standard error
aIn the replication sample, another significant variant in high LD (R2 = 0.99) with this variant was used (rs76341705, p value = 4.8 × 10−9)
Abbreviations: effect allele (A1), reference allele (A2), chromosome (Chr), effect allele frequency (Frq), number of individuals with genetic variant (N), p-value (P), reference SNP cluster ID (Rs-ID), standard error (SE)
Notably, two genome-wide significant variants identified here, rs146354218 and rs2279827, were exclusively associated with the temporal lobe and parietal lobe, respectively (Supplementary Data 7). In contrast, rs147148763 and rs12411216 were not only significantly associated with occipital lobe volume but also appeared to be associated to some extent with parietal lobe volume (p = 2.5 × 10−6; variance explained = 0.15% and p = 2.4 × 10−5; variance explained = 0.11%, respectively). The other two variants also showed nominally significant associations with other lobar volumes.
Replication
Five out of the six index variants were available in the imputation reference panel of our replication sample (n = 8,789). Unfortunately, the haplotype reference consortium (HRC) reference panel does not contain insertions and deletions. Therefore, for replication we selected rs76341705, a variant that showed a comparable signal in the meta-analysis (prs147148763 = 2.9 × 10−9, vs. prs76341705 = 4.8 × 10−9), and in high linkage disequilibrium (LD) with the index variant (R2 = 0.99). We were able to replicate all these six variants at a nominal significance level (p values ranging from 3.0 × 10−2 to 8.0 × 10−7) with the same direction of effect as the discovery sample (Table 1, Supplementary Fig. 8).
Variance explained in lobar volumes by common variants
Based on the LD score regression single-nucleotide polymorphism (SNP)-based heritability analyses, common variants across the whole genome explained as much as 20.3% (95% CI 13.2–27.4%) of the variance in occipital lobe volume, 19.6% (95% CI 12.3–26.9%) of frontal lobe volume, 17.5% (95% CI 10.7–24.3%) of temporal lobe volume, and 17.9% (95% CI 11.7–24.1%) of parietal lobe volume (Supplementary Data 8). Common genetic variants account for 30–41% of the total heritability of brain lobar volumes (Supplementary Data 8).
Genetic overlap with other brain volumes and related diseases
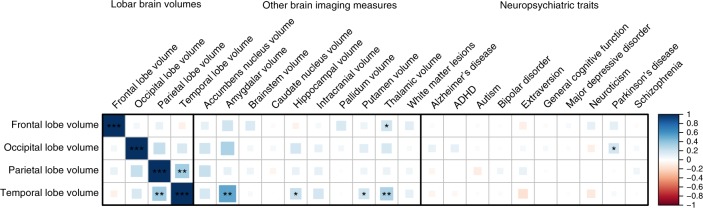
Although no top variant was significantly associated with the volume of multiple lobes, nominally significant correlation (rg) was observed between genetic components of the parietal and temporal lobe (rg = 0.35, p = 1.5 × 10−3), although this did not withstand correction for multiple testing (Fig. 3, Supplementary Data 9). Some suggestive correlation was observed between temporal and frontal lobe volume with genetic determinants of subcortical volumes; however, none survived multiple testing adjustments. When studying brain diseases, only occipital lobe volume showed a suggestive genetic correlation with Parkinson’s disease (rg = 0.18, p = 0.03). No significant genetic correlation was observed with any of the other tested neurological or psychiatric traits.
Fig. 3.
Genetic correlation between lobar brain volumes and other brain imaging measures and neuropsychiatric traits. Heatmap showing the genetic correlations estimates (rg) as calculated by linkage disequilibrium score regression. Larger blocks and darker colors present stronger correlations, with blue and red indicating positive and negative correlations, respectively. The strength of the significance levels are indicated by asterisks: *p < 0.05; **p < 1.3 × 10−2 (0.05/4), adjusted for the lobe count; ***p < 5.8 × 10−4 (0.05/86), adjusted for the number of correlations tested
Discussion
In our genome-wide association study of in up to 16,016 individuals, we identified 6 independent loci where variants had significant associations with brain lobar volumes, independent of intracranial volume. We were able to replicate these findings in a sample of 8,789 individuals. Four out of the six identified loci have not been linked to brain volume measures before; the other two loci are located in regions previously associated with brain volume measures (12q14.3 with hippocampal volume and 6q22.32 with intracranial volume). These new loci provide intriguing new insights into the genetics underlying brain lobar volumes.
We estimated that, after adjusting for intracranial volume, 17.5–20.3% of the variance in lobar volumes could be explained by common genetic variation. This forms 30–40% of the total heritability we estimated, suggesting a major contribution of common genetic variation in brain development. More genetic variants associated with brain volume may be discovered by increasing the sample sizes of genetic studies. An interesting observation is that we were able to replicate our findings using only gray matter volumes of each lobe, while the discovery studied the sum of gray and white matter. This difference might explain that not all loci replicated as strong as others and that differential effects might exist on gray and white matter volume effects of the genetic variants. Future studies will have to elucidate the biological mechanisms of the discovered associations.
Interestingly, the majority of the identified loci contained variants associated with occipital lobe volume, whereas the other brain lobes have more often been linked to disease outcomes. Yet, the heritability estimates for the occipital lobe do not exceed the heritability estimates of the other brain lobes. One possible explanation for this finding is the smaller volume of the occipital lobe compared to the other lobes, making it a more specific region—also in terms of the genetic architecture. It may also be explained by a less polygenic nature of the occipital lobe compared to the other lobes, allowing one to identify stronger associations for a single genetic variant.
Regarding the identified genome-wide significant loci, two identified loci have been previously associated with brain volume measurements. The locus 12q14.3 associated with temporal lobe volume in our study and was previously associated with hippocampal volume17. Our index variant rs146354218 (p = 6.4 × 10−10) is an intronic variant in the MSRB3 gene and lies 39 kilobases (kb) from the previously published rs61921502 variant associated with hippocampal volume18; however, the LD (R2 = 0.1, D’ = 1, p < 0.0001)19 is low. This previously published variant also showed some evidence of association with total temporal lobe volume (effect = 0.57 cm3, p = 6.5 × 10−4). Thus the 12q14.3 locus not only influences hippocampal volume but also seems to have a more generalized effect on the temporal lobe volume as would be expected by the genetic correlation between temporal lobe volume and hippocampal volume. The signal (rs1337736, p = 4.0 × 10−8) at 6q22.32 near to the gene CENPW (Centromere Protein W) is associated with occipital volume. This signal overlapped previously associated signals with intracranial volume16,17 and is further implicated in bone mineral density20, height21, waist-hip ratio22, and infant length23. The index variant associated with intracranial volume (rs11759026) and our top variant are in linkage equilibrium (R2 = 0.07, D’ = 1, p = 0.0002)19. We also found suggestive associations between rs11759026 and both frontal (−1.0 cm3, p = 6.3 × 10−5) and occipital lobar volume (−0.31 cm3, p = 6.6 × 10−3). Each locus was located in regions that are under epigenetic regulation in brain tissue (Supplementary Data 6) or close to genes or genomic loci associated with Mendelian brain-related diseases. The variant rs2279829 (3q24) is located in the 3’-untranslated region (UTR) of the Zic Family Member 4 (ZIC4) gene and close to the related ZIC1 gene. This variant localizes within enhancer sites in predominantly neurological cell types, among which the brain germinal matrix (Supplementary Data 6) and both ZIC4 and ZIC1 are expressed throughout the brain (Supplementary Fig. 9). Heterozygous deletions of ZIC1 and ZIC4 cause Dandy–Walker malformation24. Children with this malformation have no vermis, the part connecting the two cerebellar hemispheres24. Gain-of-function mutations in ZIC1 lead to coronal craniosynostosis and learning disability25. Variant rs147148763 was located 24 kb from the disheveled-associated activator of morphogenesis 1 (DAAM1). There is evidence for the most significant variants to localize within enhancer sites, as well as DNA-hypersensitivity sites in brain tissues. Also, genome-wide significant SNPs in the locus are expression quantitative trait loci (eQTLs) of DAAM1 in blood (Supplementary Data 6). Daam1 is a formin protein that has been linked to actin dynamics26, is regulated by RhoA27, and is expressed in the shafts of dendrites28. Expression patterns in brain development of animals further suggest a role in neuronal cell differentiation and movement29. Variant rs4647940 is located in the 3’-UTR of fibroblast growth factor receptor (FGFRL1) and is in LD with a missense variant in Alpha-L-iduronidase (IDUA) (rs3755955, R2 = 0.87) that was previously associated with bone mineral density (p = 5.0 × 10−15). Deletion of the 4p16.3 locus causes Wolf–Hirschhorn syndrome, a neurodevelopmental disorder characterized by mental retardation, craniofacial malformation, and defects in skeletal and heart development. Variant rs12411216 is located in an intron of MIR92B and THBS3, but the signal peak in this locus covers >20 genes. Promotor histone marks overlap the variant and it is an eQTL for multiple genes, both in a multitude of different tissues among which brain tissues (Supplementary Data 6). In summary, these findings link genes that cause Mendelian syndromes affecting cranial skeletal malformations, brain malformations, and intelligence with brain lobe volume in healthy individuals. One other interesting variant in tight LD with our index variant (rs4072037, R2 = 0.94) is a missense SNP in the MUC1 gene that decreased levels of blood magnesium concentrations30. It is not clear how decreased magnesium levels are involved in decreased occipital brain volume, but it is an interesting avenue to explore as magnesium is known to be important for neural transmissions31 and magnesium infusions have anti-convulsive effects and is still used to prevent convulsions in pre-eclampsia32.
Using genetic correlation analysis, we did not find a strong significant genetic correlation between most of the brain lobes, which suggests that the genetic basis of the brain lobes is largely independent15. We also did not find significant genetic overlap between lobar brain volumes and neurological and psychiatric disease outcomes. The most significant genetic correlation with brain lobar volume and diseases we observed was between occipital lobe volume and Parkinson’s disease (rg = 0.18, p = 0.03). However, this finding was not significant after multiple testing correction, leading us to report this finding with caution. The absence of significant genetic correlations between other brain lobes and clinical diseases could be due to true absence of a genetic overlap. However, other explanations can be put forward. First, it could also mean that our lobar volume GWAS and those for other diseases were still too underpowered to show significant genetic correlations. Second, the anatomical boundaries for the different lobes can be quite arbitrary and do not necessarily have to coincide with underlying gene function or biological processes leading to neurological or psychiatric disorders.
There are several limitations to our study. First, we have accepted differences in analytical methods of the magnetic resonance imaging (MRI) scans to allow for the largest sample size to be studied. This might have resulted in different effects over the studies. However, we did not observe significant heterogeneity after correcting for multiple testing for the six loci. In addition, a sensitivity analysis showed similar effects of the genome-wide significant variants for the studies using the kNN algorithm in comparison to the other studies. False negative findings due to differences in analytical methods of the MRI scans cannot be excluded. Second, a limitation of our study is that a different reference panel for imputation was used for the discovery and replication sample. This is due to the historic limited availability of the HRC reference panel at the initiation of this study. As the variants were well imputed (R2 > 0.5) in all studies, this is not expected to have influenced the results, although it is possible that additional variants may be discovered if the larger HRC reference panel would be used in future studies. Last, for the UK Biobank only gray matter parcellations were available to us. Despite this limitation, we were able to replicate our findings. This suggests that the identified variants have an effect on gray as well as white matter volumes.
In summary, brain lobar volumes are differentially heritable traits, which can in large part be explained by common genetic variation. We identified six loci where genotypes are associated with specific brain lobes, four of which have not been implicated in brain morphology before. These loci are compelling targets for functional research to identify the biology behind their genetic signals.
Methods
Study population
The study sample consisted of dementia- and stroke-free individuals with quantitative brain MRI and genome-wide genotypes from 19 population- and family-based cohort studies participating in the Cohorts of Heart and Aging Research in Genomic Epidemiology consortium and the case–control Alzheimer’s Disease Neuroimaging Initiative study. In total, 16,016 participants were included, 15,269 participants of European ancestry, 405 African Americans, 211 Chinese, and 131 Malay. We attempted to replicate our findings in 8,789 European ancestry individuals from the UK Biobank, an ongoing prospective population-based cohort study located in the United Kingdom. Descriptive statistics of all populations are provided in Supplementary Data 1. DNA from whole blood was extracted and genome-wide genotyping was performed using a range of commercially available genotyping arrays. Genotype imputations were performed in each discovery cohort using 1000 Genomes version 133 as reference and using the Haplotype Reference Consortium (HRC) version 1.1 in the replication cohort (Supplementary Data 2).
MRI methods
Three-dimensional T1-weighted brain MRI data were acquired by each cohort (Supplementary Data 1). Cohorts in the discovery sample segmented the T1-weighted images into supra-tentorial gray matter, white matter, and cerebrospinal fluid. The methods of image segmentation varied across study cohorts (Supplementary Methods). However, the majority used a previously described kNN algorithm, which was trained on six manually labeled atlases34, or in-house image-processing pipelines. In each study, MRI scans were performed and processed with automated protocols, without reference to clinical or genetic information. We studied the total volume (sum of white and gray matter and the left and right hemisphere) of the frontal, parietal, temporal, and occipital brain lobes, adjusted for intracranial volume. Descriptive information of the lobar volumes across the different studies is provided in Supplementary Data 3. Differences in average brain lobar volumes were accepted as differences in MRI acquisition, processing, segmentation, and demographics, which exist over cohorts. As a replication, we used the released volume measurements of 8,789 UK Biobank participants, extracted using the FreeSurfer software version 6.0, which obtains lobar volumes by adding up regions of interest volumes35,36. As only FreeSurfer gray matter volumes were available for this study sample, the replication sample volumes were smaller than the volumes in the discovery sample (Supplementary Data 3).
Estimation of heritability
The heritability of lobar brain volumes was estimated using family structure in the Framingham Heart Study (n = 2080), which constitutes a community-based cohort of non-demented individuals without evidence of significant brain injury (e.g., stroke or multiple sclerosis). In total, 619 extended families with a family size of 3.6 ± 6.6 individuals were included in the analyses. These families consisted of the following pairs of relatives: 316 parent–offspring, 1135 sibling, 340 avuncular, 1772 first cousin, and 826 second cousin pairs. We calculated additive genetic heritability without shared environmental effects (C) using a variance-components analysis under an AE model in SOLAR37, adjusted for age, age2, and sex.
GWAS of lobar volumes
Associations of imputed genotype dosages with lobar volumes were examined using linear regression analyses under an additive model. Associations were adjusted for age, age2, sex, the first four principal components to account for possible confounding due to population stratification, and study-specific covariates. Linear mixed models with estimated kinships were used for association analyses in cohorts with related samples. Details on the analysis methods used in each cohort are provided in Supplementary Data 1 and 2. Post-GWAS quality control (QC) was conducted using EasyQC38 and filtering. Genetic variants with a low imputation quality (R2 < 0.5), a minor allele count <10, and allelic or locational mismatching of SNPs with the reference panel were removed prior to the meta-analyses. The number of variants after filtering and the genomic inflation per study are provided in Supplementary Data 4. After QC, summary statistics were adjusted by the genomic control method in each of the participating cohorts39. We then performed two inverse-variance weighted fixed-effect meta-analyses in METAL39. First, we meta-analyzed all participants of European ancestry, then performed a multi-ethnic meta-analysis including African Americans (n = 405), Chinese (n = 211), and Malay (n = 131). After meta-analyses, genetic variants with a total sample size of <5000 were excluded. We performed conditional analysis on the index variants to determine whether there were multiple independent genome-wide significant variants in a locus using the Genome-Wide Complex Trait Analysis (GCTA) software (--cojo, --p-cojo)40,41. Genotypes in the Rotterdam-study (all 6291 individuals of the baseline cohort who were genotyped) were used as reference for this analysis. For loci with a genome-wide significant association (p < 5 × 10−8), we tested for heterogeneity using the I2 statistic39. In a sensitivity meta-analysis, we tested whether the studies using the kNN algorithm had similar effects of the genome-wide significant variants for the studies using the kNN-algorithm in comparison to the other studies. We also searched for candidate genes in the loci using publically available databases for differential expression of the SNPs (eQTL database in GTEx)42 and HaploReg, an online tool that summarizes the ENCODE database for epigenetic markings and proteins binding to DNA43.
Variance explained by common variants and genetic correlations
The variance explained by all SNPs, or SNP-based heritability, was calculated from summary statistics using LD score regression44. The percentage of variance explained by all SNPs was determined based on meta-analysis results using the LD Hub45. We used the same LD score regression44 to quantify the amount of genetic correlation between the four brain lobes and other brain-related traits and diseases, using summary statistics for meta-analyses of genetic studies of subcortical structures16, intracranial volume17, white matter hyperintensities46, general cognitive ability47, neuroticism48, schizophrenia49, attention-deficit/hyperactivity disorder50, autism51, major depressive disorder52, bipolar disorder¸49 Parkinson’s disease53, and Alzheimer’s disease54.
Ethical compliance
All participants, or their parents or guardians in the case of minors, provided written informed consent for study participation, the use of brain MRI data, and the use of their DNA for genetic research. Approval for the individual studies was obtained by the relevant local ethical committees and institutional review boards.
Statistics and reproducibility
Software used for the data analysis of this study: EasyQC (www.genepi-regensburg.de/easyqc), FreeSurfer (https://surfer.nmr.mgh.harvard.edu/), GCTA (http://cnsgenomics.com/software/gcta/), GenABEL (http://www.genabel.org), GTeX (https://gtexportal.org/home/), HaploReg (https://www.encodeproject.org/software/haploreg/), HASE (https://github.com/roshchupkin/hase), LD-hub (http://ldsc.broadinstitute.org/ldhub/), LD score regression (https://github.com/bulik/ldsc), mach2qtl (https://www.nitrc.org/projects/mach2qtl/), METAL (http://csg.sph.umich.edu/abecasis/metal/), Perl (https://www.perl.org/), PLINK (https://www.cog-genomics.org/plink2), R (https://www.r-project.org/), SNPTEST (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html), and SOLAR (http://www.sfbr.org).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, many investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Full acknowledgements are provided in the Supplementary Information.
Author contributions
(Co)wrote draft: S.J.v.d.L., M.J.K., N.A., H.H.H.A., C.D. Meta-analysis: S.J.v.d.L. Analyzed data: G.C., C.S., A.V.S., E.H., J.C.B., D.P.H., S.H., E.B.v.d.A., M. Bernard, L.R.Y., F.C., J.W.C., Y.S., S.L., L.Y., G.V.R., N.J., Q.Y., R.A.M., S.B., C.Y.C., P.M., W.Z., P.A.N., B.M.P., Z.P., P.M.T. Critical revision of manuscript: G.C., C.S., A.V.S., E.H., J.C.B., D.P.H., E.B.v.A., M. Bernard, L.R.Y., N.A., J.W.C., Y.S., O.L.L., S.L., J.v.d.G., L.Y., T.P., G.V.R., P.A., N.J., Q.Y., R.A.M., S.B., B.M., C.Y.C., P.M., W.J.N., M. Beekman, W.Z., P.A.N., M.W.V., H.S., Z.P., D.M.B., P.L.D.J., P.M.T., C.M.v.D., R.S., W.T.L., M.A.I., S.S. Contributed to sample collection: J.C.B., S.H., K.A., T.B.H., O.L.L., J.v.d.G., T.P., K.D.T., B.M., C.Y.C., K.R., D.v.H., T.Y.W., W.J.N., A.S.B., M. Beekman, P.A.N., C.C., L.L., B.M.P., M.K.I., M.W.V., H.S., Z.P., D.M.B., P.L.D.J., C.M.v.D., D.A.B., P.E.S., R.S., W.T.L., M.A.I., S.S., S.D., V.G. Secured funding: K.A., T.B.H., O.L.L., P.A., K.D.T., B.M., K.R., D.v.H., T.Y.W., W.J.N., A.S.B., C.C., L.L., B.M.P., M.K.I., H.S., D.M.B., P.L.d.J., C.M.v.D., D.A.B., P.E.S., R.S., W.T.L., M.A.I., S.S., S.D., V.G. Study supervision: H.H.H.A., C.D.
Data availability
The genome-wide summary statistics that support the findings of this study will be made available via the NHGRI-EBI GWAS Catalog website (https://www.ebi.ac.uk/gwas/downloads/summarystatistics) upon publication. Quantitative brain MRI and genotype data are available from the corresponding authors H.H.H.A. and C.D.C. upon reasonable request.
Code availability
No previously unreported custom computer code or mathematical algorithm was used to generate results central to the conclusions. The code is available upon request from the corresponding authors H.H.H.A. and C.D.C.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hieab H. H. Adams, Email: h.adams@erasmusmc.nl
Charles DeCarli, Email: cdecarli@ucdavis.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0537-9.
References
- 1.Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu. Rev. Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- 2.Culham JC, Valyear KF. Human parietal cortex in action. Curr. Opin. Neurobiol. 2006;16:205–212. doi: 10.1016/j.conb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Levy DA, Bayley PJ, Squire LR. The anatomy of semantic knowledge: Medial vs. lateral temporal lobe. Proc. Natl Acad. Sci. USA. 2004;101:6710–6715. doi: 10.1073/pnas.0401679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendez MF, Selwood A, Mastri AR, Frey WH., 2nd Pick’s disease versus Alzheimer’s disease: a comparison of clinical characteristics. Neurology. 1993;43:289–292. doi: 10.1212/WNL.43.2.289. [DOI] [PubMed] [Google Scholar]
- 5.de Lanerolle NC, et al. A retrospective analysis of hippocampal pathology in human temporal lobe epilepsy: evidence for distinctive patient subcategories. Epilepsia. 2003;44:677–687. doi: 10.1046/j.1528-1157.2003.32701.x. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasa RN, et al. Cardiovascular risk factors associated with smaller brain volumes in regions identified as early predictors of cognitive decline. Radiology. 2016;278:198–204. doi: 10.1148/radiol.2015142488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulshoff Pol HE, et al. Genetic contributions to human brain morphology and intelligence. J. Neurosci. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen W, et al. Distinct genetic influences on cortical and subcortical brain structures. Sci. Rep. 2016;6:32760. doi: 10.1038/srep32760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batouli SA, et al. Heritability of brain volumes in older adults: the Older Australian Twins Study. Neurobiol. Aging. 2014;35:937 e5–18. doi: 10.1016/j.neurobiolaging.2013.10.079. [DOI] [PubMed] [Google Scholar]
- 10.Baare WF, et al. Quantitative genetic modeling of variation in human brain morphology. Cereb. Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- 11.Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc. Natl Acad. Sci. USA. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmore JH, et al. Genetic and environmental contributions to neonatal brain structure: A twin study. Hum. Brain Mapp. 2010;31:1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon U, Fahim C, Perusse D, Evans AC. Lateralized genetic and environmental influences on human brain morphology of 8-year-old twins. Neuroimage. 2010;53:1117–1125. doi: 10.1016/j.neuroimage.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace GL, et al. A pediatric twin study of brain morphometry. J. Child Psychol. Psychiatry. 2006;47:987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- 15.DeStefano AL, et al. Bivariate heritability of total and regional brain volumes: the Framingham Study. Alzheimer Dis. Assoc. Disord. 2009;23:218–223. doi: 10.1097/WAD.0b013e31819cadd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams, H. H. et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 19,1569–1582 (2016). [DOI] [PMC free article] [PubMed]
- 18.Hibar DP, et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2011;43:1164. doi: 10.1038/ng1111-1164a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Valk RJP, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum. Mol. Genet. 2015;24:1155–1168. doi: 10.1093/hmg/ddu510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grinberg I, et al. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat. Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- 25.Twigg SR, et al. Gain-of-function mutations in ZIC1 are associated with coronal craniosynostosis and learning disability. Am. J. Hum. Genet. 2015;97:378–388. doi: 10.1016/j.ajhg.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/S0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 27.Matusek T, et al. The Drosophila formin DAAM regulates the tracheal cuticle pattern through organizing the actin cytoskeleton. Development. 2006;133:957–966. doi: 10.1242/dev.02266. [DOI] [PubMed] [Google Scholar]
- 28.Salomon SN, Haber M, Murai KK, Dunn RJ. Localization of the Diaphanous-related formin Daam1 to neuronal dendrites. Neurosci. Lett. 2008;447:62–67. doi: 10.1016/j.neulet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 29.Kida Y, Shiraishi T, Ogura T. Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Dev. Brain Res. 2004;153:143–150. doi: 10.1016/j.devbrainres.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Meyer Tamra E., Verwoert Germaine C., Hwang Shih-Jen, Glazer Nicole L., Smith Albert V., van Rooij Frank J. A., Ehret Georg B., Boerwinkle Eric, Felix Janine F., Leak Tennille S., Harris Tamara B., Yang Qiong, Dehghan Abbas, Aspelund Thor, Katz Ronit, Homuth Georg, Kocher Thomas, Rettig Rainer, Ried Janina S., Gieger Christian, Prucha Hanna, Pfeufer Arne, Meitinger Thomas, Coresh Josef, Hofman Albert, Sarnak Mark J., Chen Yii-Der Ida, Uitterlinden André G., Chakravarti Aravinda, Psaty Bruce M., van Duijn Cornelia M., Kao W. H. Linda, Witteman Jacqueline C. M., Gudnason Vilmundur, Siscovick David S., Fox Caroline S., Köttgen Anna. Genome-Wide Association Studies of Serum Magnesium, Potassium, and Sodium Concentrations Identify Six Loci Influencing Serum Magnesium Levels. PLoS Genetics. 2010;6(8):e1001045. doi: 10.1371/journal.pgen.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borges LF, Gucer G. Effect of magnesium on epileptic foci. Epilepsia. 1978;19:81–91. doi: 10.1111/j.1528-1157.1978.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 32.Pritchard JA, Pritchard SA. Standardized treatment of 154 consecutive cases of eclampsia. Am. J. Obstet. Gynecol. 1975;123:543–552. doi: 10.1016/0002-9378(75)90042-3. [DOI] [PubMed] [Google Scholar]
- 33.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrooman HA, et al. Multi-spectral brain tissue segmentation using automatically trained k-Nearest-Neighbor classification. Neuroimage. 2007;37:71–81. doi: 10.1016/j.neuroimage.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 36.Fischl B, et al. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 37.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler TW, et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium GT, et al. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Jie, Erzurumluoglu A. Mesut, Elsworth Benjamin L., Kemp John P., Howe Laurence, Haycock Philip C., Hemani Gibran, Tansey Katherine, Laurin Charles, Pourcain Beate St., Warrington Nicole M., Finucane Hilary K., Price Alkes L., Bulik-Sullivan Brendan K., Anttila Verneri, Paternoster Lavinia, Gaunt Tom R., Evans David M., Neale Benjamin M. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2016;33(2):272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhaaren BF, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ. Cardiovasc. Genet. 2015;8:398–409. doi: 10.1161/CIRCGENETICS.114.000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies G, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53 949) Mol. Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turley P, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell173, 1705.e16–1715.e16 (2018). [DOI] [PMC free article] [PubMed]
- 50.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang D, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome-wide summary statistics that support the findings of this study will be made available via the NHGRI-EBI GWAS Catalog website (https://www.ebi.ac.uk/gwas/downloads/summarystatistics) upon publication. Quantitative brain MRI and genotype data are available from the corresponding authors H.H.H.A. and C.D.C. upon reasonable request.
No previously unreported custom computer code or mathematical algorithm was used to generate results central to the conclusions. The code is available upon request from the corresponding authors H.H.H.A. and C.D.C.