Abstract
Although the overall five-year survival of patients with pancreatic ductal adenocarcinoma (PDAC) is dismal, there are survival differences between cases with clinically and pathologically indistinguishable characteristics, suggesting that there are uncharacterized properties that drive tumor progression. Recent mRNA sequencing studies reported gene-expression signatures that define PDAC molecular subtypes that correlate with differences in survival. We previously identified Keratin 17 (K17) as a negative prognostic biomarker in other cancer types. Here, we set out to determine if K17 is as accurate as molecular subtyping of PDAC to identify patients with the shortest survival. K17 mRNA was analyzed in two independent PDAC cohorts for discovery (n = 124) and validation (n = 145). Immunohistochemical localization and scoring of K17 immunohistochemistry (IHC) was performed in a third independent cohort (n = 74). Kaplan-Meier and Cox proportional-hazard regression models were analyzed to determine cancer specific survival differences in low vs. high mRNA K17 expressing cases. We established that K17 expression in PDACs defines the most aggressive form of the disease. By using Cox proportional hazard ratio, we found that increased expression of K17 at the IHC level is also associated with decreased survival of PDAC patients. Additionally, within PDACs of advanced stage and negative surgical margins, K17 at both mRNA and IHC level is sufficient to identify the subgroup with the shortest survival. These results identify K17 as a novel negative prognostic biomarker that could inform patient management decisions.
Subject terms: Pancreatic cancer, Prognostic markers
Introduction
The prognosis of patients with pancreatic ductal adenocarcinoma (PDAC), the most common form of pancreatic cancer, is mainly based on the assessment of the clinical and pathologic presentation at the time of diagnosis1,2. While tumor size, lymph node status, metastasis and surgical margins2 predict survival, patients with the same clinicopathologic characteristics can have different survival outcomes3, suggesting that other undetermined properties of PDAC may drive these differences.
Currently available biomarkers that guide the management of PDAC have limited ability to predict patient clinical course or survival and prior large-scale genomic studies demonstrated that common driver mutations or other mutations lack prognostic value4–7. CA 19-9 is the only FDA-approved PDAC biomarker used to determine resectability but is not an accurate predictor of outcome8–11. Thus, identification of new prognostic biomarkers to stratify patient subgroups with differential survival probabilities at baseline could be important to guide clinical decision making12. Furthermore, understanding the underlying molecular heterogeneity of PDAC may one day guide the development of novel therapeutic strategies.
We discovered that keratin 17 (K17) was a biomarker of aggressive cervical cancers through an unbiased proteomic screen13. Several additional studies, including ours, have identified K17 as a negative prognostic biomarker not only in cervical squamous cell carcinoma13 but also in endocervical adenocarcinoma14, epithelial ovarian carcinoma15, triple negative breast carcinoma16, gastric adenocarcinoma17, gallbladder adenocarcinoma18 and head and neck squamous cell carcinoma19. Recent RNA sequencing (RNA-Seq) studies20,21, including ours22, reported that mRNA expression defines molecular subtypes of PDAC including the relatively less aggressive “classical” subtype and the more aggressive “basal-like” subtype that is defined based on the expression of a 25-gene expression signature20,22. Embedded within this gene-expression signature, we identified the presence of K17 mRNA. Thus, two different and unbiased approaches, biomarker-based discovery by proteomics and molecular subtyping by RNA-Seq, point to the importance of K17 in PDAC.
Here, we show that K17 identifies PDAC patients with the shortest survival, even in those with the same advanced stage and negative margin status. As detected by immunohistochemistry (IHC), K17 offers a rapid and deployable clinical pathological-assessment technique to guide clinical management at the time of diagnosis. This also represents the first step in the development of a CLIA-certified assay that can be tested in clinical trials and implemented in pathology diagnostic laboratories for PDAC subtyping.
Results
Keratin 17 mRNA alone is as accurate as molecular subtyping to predict PDAC prognosis
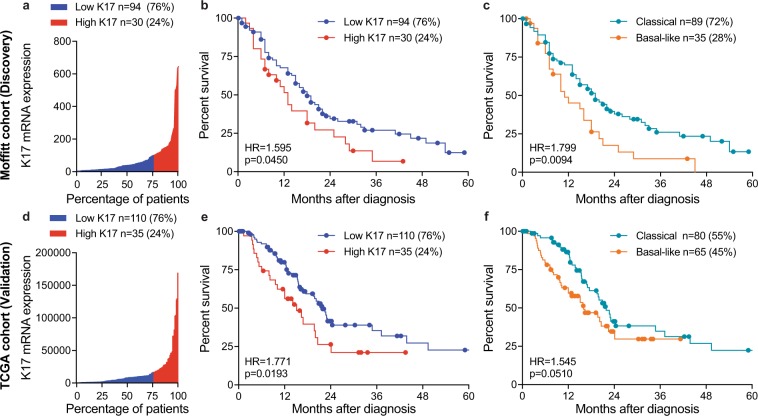
Based on our previous findings that K17 is a validated prognostic biomarker in other cancer types13–19, we set out to determine its prognostic value in PDAC. Using the Moffitt et al.22 patient dataset (n = 124) (Table 1), we first analyzed the distribution of K17 mRNA expression across cases. To define a threshold that provided maximal stratification of survival differences based on K17 mRNA, the maximum likelihood fit of a Cox proportional hazard model was used. 76% of the cases were classified as low K17 while 24% of cases were classified as high K17 (Fig. 1a). On average, K17 mRNA expression was 5-times higher in high versus low K17 cases.
Table 1.
Patient cohort demographics.
| Patient Cohorts | |||
|---|---|---|---|
| mRNA | IHC | ||
| MOFFITTa | TCGAb | SBU + UMASSc | |
| Months of follow-up, mean ± SD | 13 ± 14 | 14.1 ± 13.9 | 17.8 ± 15.2 |
| Age at diagnosis, mean ± SD | No information | 65.0 ± 11.1 | 65.5 ± 9.9 |
| Gender, number (%) | n = 112 | n = 145 | n = 74 |
| Female | 64 (57%) | 66 (46%) | 38 (51%) |
| Male | 48 (43%) | 79 (54%) | 36 (49%) |
| Histologic grade (G), number (%) | No Information | n = 145 | n = 74 |
| G1 + G2, Low and Moderate Grade | 76 (52%) | 42 (57%) | |
| G3, Poor Grade | 69 (48%) | 32 (43%) | |
| Surgical Margins (R), number (%) | n = 104 | n = 128 | n = 74 |
| R0, Negative margin | 66 (63%) | 80 (63%) | 52 (70%) |
| R1, Positive margin | 38 (37%) | 48 (38%) | 22 (30%) |
| Primary Tumord (T), number (%) | n = 109 | n = 144 | n = 74 |
| T1 + T2 | 20 (18%) | 21 (15%) | 15 (20%) |
| T3 + T4 | 89 (82%) | 123 (85%) | 59 (80%) |
| Lymph node statusd (N), number (%) | n = 111 | n = 144 | n = 74 |
| N0, No regional lymph node metastasis | 34 (31%) | 37 (26%) | 28 (38%) |
| N1, Regional lymph node metastasis | 77 (69%) | 107 (74%) | 46 (62%) |
| Cancer Staged, number (%) | n = 109 | n = 143 | n = 74 |
| I-IIA | 33 (30%) | 34 (24%) | 26 (35%) |
| IIB-IV | 76 (70%) | 109 (76%) | 48 (65%) |
| Molecular Subtype, number (%) | n = 124 | n = 145 | Not available |
| Classical | 89 (72%) | 80 (55%) | |
| Basal-like | 35 (28%) | 65 (44%) | |
| K17 status, number (%) | n = 124 | n = 145 | Not applicable |
| Low K17 | 94 (76%) | 110 (76%) | |
| High K17 | 30 (24%) | 35 (24%) | |
aMoffitt et al., Nature Genetics 2015.
bTCGA: The Cancer Genome Atlas, Cancer Cell 2017.
cSBU: Stony Brook University, UMASS: University of Massachusetts.
dStage classification per AJCC 7th edition.
Figure 1.
K17 mRNA is as accurate as molecular subtyping to predict prognosis of PDAC. (a) Water plot depicts K17 mRNA expression levels in Moffitt et al. cohort22. 76th percentile defined the cut-off to predict outcome by maximum likelihood fit of a Cox proportional hazard model, 76% of PDAC cases were found to be low K17 (blue) while 24% of cases were classified as high K17 (red). K17 mRNA ranged from 3.559 to 645.377 absolute fluorescence reads. (b) Kaplan-Meier curve depicting the overall survival for K17 of resected PDAC primary tumors from Moffitt et al. cohort22. Cox proportional model was used for analysis. Hazard ratios (HR) and p-values are shown. (c) Kaplan-Meier curve of overall survival analysis for mRNA molecular subtypes of resected PDAC primary tumors from Moffitt et al. cohort22. For analysis, Cox proportional model was used. Hazard ratios (HR) and p-values are shown. (d) Water plot depicts K17 mRNA expression levels in The Cancer Genome Atlas (TCGA) cohort23. 76th percentile defined the best cut-off to predict outcome by the maximum likelihood fit of a Cox proportional hazard model, 76% of PDAC cases where found to be low-K17 (blue) while 24% of cases were classified as high-K17 (red). K17 mRNA ranged from 75.39 to 170,437.66 RSEM reads. (e) Kaplan-Meier curve depicting the overall survival for K17 of resected PDAC primary tumors from TCGA cohort23. Cox proportional model was used for analysis. Hazard ratios (HR) and p-values are shown. (f) Kaplan-Meier curve of overall survival analysis for mRNA molecular subtypes of resected PDAC primary tumors from TCGA cohort23. Cox proportional model was used for analysis. Hazard ratios (HR) and p-values are shown.
To determine survival differences between low and high K17 cases, we performed a Kaplan- Meier (K-M) and Cox proportional-hazard regression model of survival and found that patients with high K17 PDACs had a median survival of 13 months, which was shorter than for patients with low K17 PDACs, who had a median survival of 18 months (HR = 1.6, p = 0.0450, CI = 0.935–2.723) (Fig. 1b). To determine the percentage of agreement between the Moffitt molecular subtype and K17 status in the same patient cohort, we performed a chi-square analysis of both K-M curves. The percentage of total agreement between K17 mRNA (Fig. 1b) and molecular subtype (Fig. 1c) K-M curves was 73.4. This showed that out of the 35 cases classified as basal-like subtype, 16 (46%) were also high K17. In addition, out of 89 cases classified as classical subtype, 75 (84%) were also low K17. Thus, the basal-like molecular subtype correlated with high K17 expression in a high proportion of cases. In comparison to the survival probabilities of molecular subtypes reported by Moffitt et al.22 (HR = 1.8, p = 0.0094, CI = 1.053–3.074) (Fig. 1c), the low versus high K17 classification were statistically similar (Fig. 1b).
To validate the accuracy of K17 mRNA as a negative prognostic biomarker for PDAC in an independent dataset, we employed The Cancer Genome Atlas (TCGA) PDAC database23 (n = 145; Table 1). Applying the same maximum likelihood fit of a Cox proportional hazard model threshold identified in the initial mRNA cohort22 (Fig. 1d), we found that high K17 mRNA was again a negative prognostic biomarker, with a median survival of 15 months for high K17 and 22 months for low K17 PDACs (HR = 1.8, p = 0.0193, CI = 1.006–3.119) (Fig. 1e). In comparison to basal-like versus classical molecular subtyping (HR = 1.5, p = 0.0510, CI = 0.968–2.467) (Fig. 1f), the negative prognostic accuracy of K17 mRNA status was once again statistically similar. The percentage of agreement by chi-square showed that 26 cases (40%) classified as basal-like subtype were also high K17 and 70 cases (88%) classified as classical subtype were low K17. The percentage of total agreement between K17 mRNA (Fig. 1e) and molecular subtype (Fig. 1f) K-M curves was 66.2. Together, these results indicate that high K17 mRNA status identifies an overlapping subset of patient cases and is as accurate as molecular subtyping in stratifying PDAC patients with the worst outcome in both the TCGA23 and the Moffitt et al.22 cohorts.
Immunohistochemical expression of K17
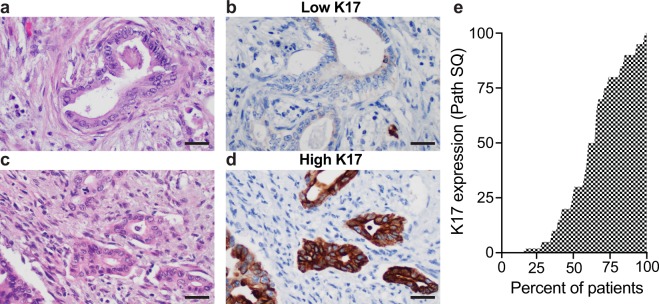
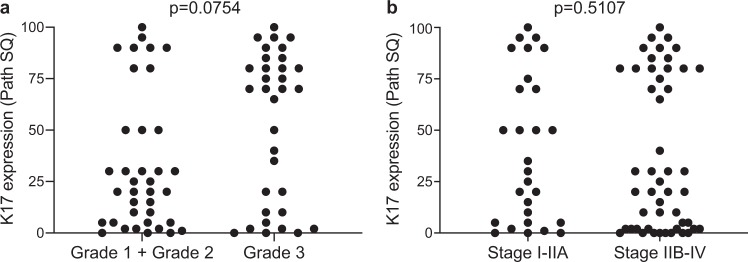
Based on virtual microdissection from bulk-tumor RNA-Seq22, we previously predicted that K17 mRNA is derived from tumor cells but not from other benign tumor cellular components. We used IHC to detect K17 at the protein level in 74 formalin-fixed paraffin-embedded (FFPE) PDAC tissue sections, each representing a unique patient (protein cohort; Table 1). K17 protein was identified in the tumor epithelial cells (Fig. 2a–d), in pancreatic intraepithelial neoplasia grade 2 and 3 (PanIN2/3), and was occasionally seen in centroacinar cells but was not detected in PanIN1, benign proliferative ducts, or in other cellular elements, substantially validating our previous in silico predictions22. To determine the level of expression of K17 by immunohistochemistry in PDAC, we used a semi-quantitative IHC scoring method, the PathSQ score, where the percentage of strong-positively stained tumoral cells on a scale of 0–100% was determined within a single representative histologic section from each case (Fig. 2e)13. The range of K17 staining was similar within the same grade or stage of the tumors (Fig. 3a,b). These results show that K17 IHC specifically identifies tumor cells.
Figure 2.
K17 immunohistochemistry in PDAC cases. (a–d) Representative images from two PDAC cases with similar histologic grade and immunohistochemical stains. Hematoxylin and eosin stained sections (a,c) and corresponding sections processed for K17 IHC (b,d). Note similar histologic features of low versus high-K17 expression. Scale bar = 20 μm. (e) Water plot depicts K17 IHC score levels in the IHC cohort.
Figure 3.
K17 IHC expression is independent of the histologic grade and tumor stage. (a) Graph illustrating K17 IHC expression for each case within the same grade category (grade 1 + grade 2 vs grade 3). Path SQ score ranges from 0 to 100% in both categories. P-value was calculated using the Mann Whitney test. (b) Graph showing expression of K17 IHC for each case within the same tumor stage category (stage I-IIA vs stage IIB-IV). Path SQ score ranges from 0 to 100% in both categories. P-value was calculated using the Mann Whitney test.
K17 is an independent negative prognostic biomarker at both mRNA and IHC levels and provides additional clinicopathological information
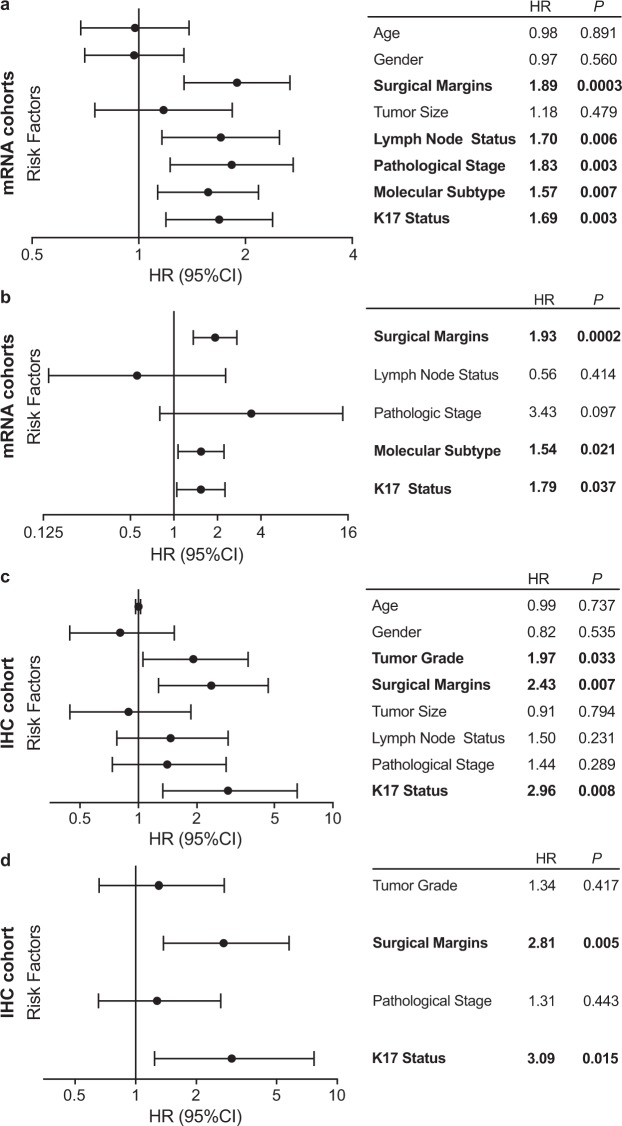
To evaluate if K17 status is independent of other clinicopathologic features, we performed univariate and multivariate analyses using Cox proportional hazard regression of individual risk factors in the combined mRNA cohort (Fig. 4a,b). K17 mRNA, molecular subtype and surgical margin status were independent negative prognostic biomarkers (Fig. 4b). To determine if K17 IHC for K17 was also prognostic, we used a Cox proportional hazard ratio and found that increased expression of K17 was associated with shorter survival of PDAC patients. (HR = 2.9, p = 0.008, CI = 1.3–6.5/100 units of Path SQ score) (Fig. 4c). Multivariate analysis also showed that K17 and surgical margin status were independent negative prognostic biomarkers (Fig. 4d).
Figure 4.
K17 is an independent negative prognostic biomarker, at both the mRNA and IHC (protein) levels. (a) Forest plot showing the univariate analysis using Cox proportional hazards regression for K17 mRNA as a binary variable and other PDAC risk factors from combined mRNA cohorts (Moffitt et al.22 and The Cancer Genome Atlas [TCGA])23. Surgical margin status, lymph node status, pathologic stage, molecular subtype and K17 status were all negative prognostic markers with significant p-values. (b) Forest plot showing the multivariate analysis from K17 mRNA as a binary variable and other risk factors, from combined mRNA cohorts. Surgical margins, molecular subtype and K17 showed significant p-values. (c) Forest plot showing the univariate analysis using Cox proportional hazards regression for K17 as a continuous variable and other PDAC risk factors from the IHC cohort. Tumor grade, surgical margins and K17 showed significant p-values. (d) Forest plot showing the multivariate analysis from K17 as a continuous variable and other risk factors, from IHC cohort. K17 and surgical margins show significant p-values.
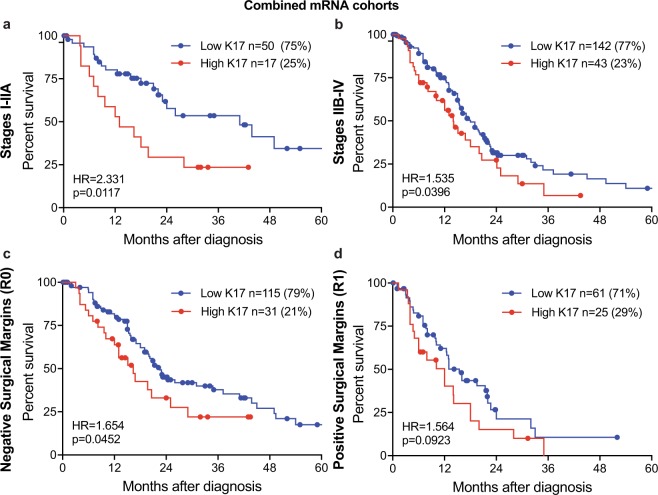
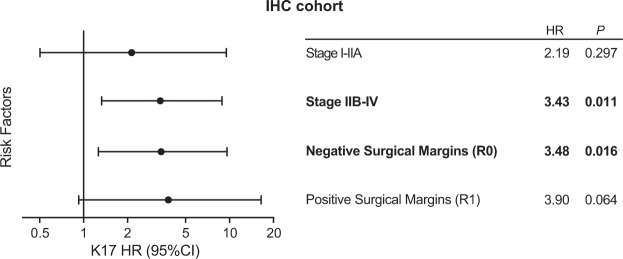
To determine if K17 provides additive prognostic value in combination with other clinical prognostic factors, we analyzed potential interactions in the mRNA cohorts. K17 mRNA stratified survival differences within patients with localized and advanced disease (Fig. 5a,b) and negative surgical margin status (Fig. 5c) identifying PDACs with the most aggressive clinical course. There was no statistical significance for positive margin status (Fig. 5d). For the IHC cohort, K17 also provided additional information within advanced stage and negative margin group (Fig. 6), as K17 correlated with shorter survival. However, neither K17 mRNA nor IHC data enhanced prognostic value within cases that had positive tumor margins (Figs 5d and 6). Of note, the prognostic significance of K17 was comparable when cases were staged according to 7th and 8th AJCC system.
Figure 5.
K17 predicts survival based on stage and surgical margins at the mRNA level. (a–d) Kaplan–Meier curves depicting the overall survival of the combined mRNA cohorts (Moffitt et al.22 and The Cancer Genome Atlas [TCGA])23 integrating K17 status and tumor stage (a: stage I-IIA, b: stage IIB-IV) and surgical margins (c: negative margins, d: positive margins). P-values were calculated using the log-rank test. Hazard ratios (HR) and p-values are shown.
Figure 6.
K17 provides additional prognostic value in advance stage and negative margin status groups at the protein level. Forest plot showing interaction of K17 IHC status and tumor stage and surgical margins. Hazard ratios (HR) and p-values are shown. Each HR is computed in subsets of the data.
In summary, K17 is an overall negative independent prognostic biomarker in PDAC and has additional prognostic value within advanced stage PDACs and for patients that had negative surgical margins of resection at both the mRNA and IHC levels.
Discussion
Here, we demonstrate that K17 is a negative and independent prognostic biomarker at both mRNA, as defined by RNA-Seq, and at the level of protein expression as evaluated by IHC. As a prognostic biomarker, K17 is as accurate as molecular subtyping to predict the patient population with the most aggressive form of the disease.
Currently, there are no methods deployed in clinical practice to predict the differential survival outcome of PDAC patients with indistinguishable clinical and pathological disease characteristics at the time of diagnosis. Through detection and analysis of K17 by IHC in tissues collected at the time of resection, we determined that among patients with the same advanced disease stage, patients with increased expression of K17 have shorter survival. Diagnosis at early stage and negative surgical margin are currently the best clinical factors associated with a higher probability of long-term survival24. Keratin 17 independently identifies a subset of patients with worst outcome within advanced stage and negative margin groups. Lower K17 tumor expression may therefore provide a new indicator of the potential PDAC patient long-term survivors. As an independent prognostic biomarker for PDACs, K17 could provide more accurate prediction of prognosis and potentially guide patient management decisions.
We recently identified two PDAC molecular subtypes defined as “classical” and “basal-like” by correlating tumor cell gene expression and patient survival22. This classification demonstrates that there are innate biological differences between tumors that can impact treatment decisions and responses. In the survival analysis presented here, molecular subtypes and K17 mRNA status displayed very similar overall survival curves. Although the correlation between molecular subtype and K17 in both cohorts was high, there are clear differences in the separation of the survival curves. There is not a one to one correlation between high-K17 and basal-like subtype and low-K17 and classical subtype, however, we demonstrate that by K17 alone, one of the 25 basal-like subtype genes, we are able to identify patients with the worst prognosis. These findings indicate that the determination of K17 mRNA expression status is as accurate as molecular subtyping as a biomarker of PDACs that have the most aggressive course. Interestingly, there have been reports that other components of the basal-like subtype signature also correlate with poor PDAC prognosis25–30. S100A227, AREG25, FAM83A29, GPR8728 and Slc2a127 have all been studied at the IHC level in PDAC tissues and have all been identified as independent prognostic markers, however, the prognostic value of these proteins has not been compared to the molecular subtypes. Further studies will be necessary to understand the role of these proteins in disease biology and response to treatment.
Importantly, recent preliminary results from the COMPASS trial31 suggest that the basal-like subtype is less responsive to first-line chemotherapy. Furthermore, GATA6 mRNA identification by in-situ hybridization, was identified as a surrogate biomarker of the classical subtype, and prior reports have indicated that patients with GATA6high tumors have better survival than those with GATA6low expression32. As expected, GATA6 expression is inversely correlated with that of K17 in both Moffitt et al.22 dataset and TCGA23 database and shows promise as a biomarker of the more favorable form of the disease, predicting survival and response to chemotherapy. The availability of more than one biomarker to subtype patients may be useful in improving prognostic accuracy and identification of the best candidates for novel targeted therapies. Although keratin 81(K81) has also been correlated with shorter survival in two different cohorts (HR = 1.036 and 1.730), K17 status (HR = 2.9) provides a higher HR value. It is also unknown whether K81 is prognostic in advanced stage PDAC or is impacted by surgical margin status33. Future work is required to continue understanding the molecular differences between the PDAC subtypes with the aim of improving treatment options.
Previous reports show that K17 mRNA is highly transcribed in PDAC34,35 and that at least 30% of cases can be detected by K17 IHC in neoplastic cells36,37. One of the most important advantages of IHC is that this method enables interrogation of biomarker expression at the histologic level while RNA-Seq or other methods based on tissue solubilization dilute cancer-specific biomarkers by components derived from other benign cellular elements, including stromal cells, inflammatory cells and benign ductal cells, acinar cells, islet cells, or other benign cellular elements. Thus, IHC enables the determination of K17 status, even in cases that have low tumor cellularity. IHC is also a clinically well established and deployable assay, widely used by pathology laboratories as a routine diagnostic test platform. Here, we demonstrate the prognostic value of K17 IHC test in a small cohort of PDAC patients. Future studies should validate these findings and evaluate the predictive value of K17 IHC testing in retrospective analyses using cohorts of PDAC cases involved in precision medicine testing, potentially including the Pancreatic Cancer Action Network “Know Your Tumor” initiative38, among others. Of note, less than 20% of PDAC patients are eligible for surgical resection but in many of those cases, the diagnosis is confirmed only by fine needle aspiration (FNA) biopsy. Thus, further studies are indicated to determine if immunocytochemical scoring of K17 status in FNA specimens could also be used to define PDAC K17 status in surgically unresectable PDACs, as a future prognostic strategy and to help guide decisions regarding selection of options for therapeutic intervention.
In summary, we demonstrate that K17 mRNA is as accurate as molecular subtyping to predict PDAC prognosis. By either RNA-Seq or IHC localization, K17 predicts survival, independent of tumor grade, stage, and margin status. In conclusion, IHC is an optimal platform to clinically deploy K17 as a prognostic biomarker for PDAC.
Methods
Patient demographics
Three independent patient cohorts were analyzed for K17 expression, of which two cohorts were analyzed for K17 mRNA sequencing and one for K17 immunohistochemical analysis (Table 1). For our K17 mRNA-1 cohort, we used the same cohort that was reported by Moffitt et al.22 (n = 124). For the K17 mRNA-2 cohort, we analyzed the TCGA23 database patient samples (n = 145). For the K17 IHC analysis, formalin-fixed paraffin-embedded (FFPE) surgical tissue blocks from consecutive PDAC cases from a five-year interval (2008–2012) were selected from the archival collections of the Departments of Pathology at Stony Brook Medicine (n = 56) and the University of Massachusetts (n = 18). The patient selection criteria were diagnosis of primary PDAC and patients over the age of 18 years old at the time of diagnosis. Patients with survival of less than a month after surgery or a diagnosis of cancer metastatic to the pancreas from other anatomic sites were excluded. Survival and adjuvant therapy data was obtained from the respective institution’s registry. All studies were performed in accordance with guidelines and regulations of the Stony Brook Medicine Institutional Review Board (IRB) protocol 94651-36 and University of Massachusetts IRB protocol H00015796. Patient written consent was waived by the IRB of both institutional IRBs because the research was restricted to the analysis of de-identified remnant waste surgical pathology specimens that were provided by each institution.
Patients were stratified based on the American Joint Committee on Cancer (AJCC) 7th edition staging criteria, as recorded in the original surgical pathology diagnostic reports. Tumor size categories were grouped as tumor limited to the pancreas (T1-T2) and tumor that extended beyond the pancreas (T3-T4); lymph node status was recorded as no regional metastases (N0) versus regional metastases (N1); cancer stage was grouped according to tumor limited to the pancreas with no regional or distant lymph node metastases (IA-IIA) versus tumor that extended beyond pancreas and/or was metastatic (IIB-IV). Histological grades were grouped into well and moderately differentiated (G1-G2) versus poorly differentiated (G3). Surgical margins were scored as negative (R0) versus positive (R1) for carcinoma. All analyses were performed in accordance with these criteria.
Bioinformatic analysis of K17 mRNA expression across databases
We screened the Moffitt et al.22 and the TCGA23 PDAC databases for K17 mRNA expression levels13. For the Moffitt et al.22 dataset, there were 124 cases with survival information. For the TCGA database, 145/150 cases had both RNA Sequencing data (RNASeq V2 RSEM) and survival data. K17 mRNA ranged from 3.559 to 645.377 absolute fluorescence reads in the Moffitt et al.22 dataset and from 75.39 to 170,437.66 RSEM reads in the TCGA database. Based on the maximum likelihood fit of a Cox proportional hazard model, we selected the 76th percentile of mRNA expression at which to split samples into K17 high or K17 low groups. This threshold was trained on the University of North Carolina22 cohort, and subsequently fixed before application to the TCGA23 cohort of cases.
K17 immunohistochemistry
All tumor sections were reviewed from each case to identify the single tissue block for immunohistochemical studies that contained the greatest total surface area of viable carcinoma. Exclusion criteria included cases where the total surface area of viable tumor was <1 cm2. An indirect immunoperoxidase method was used to identify the presence of K17 protein, as previously described13. Briefly, after incubation at 60 °C, slices were deparaffinized in xylene and rehydrated in alcohols. Antigen retrieval was performed in citrate buffer at 120 °C for 10 minutes in a decloaking chamber. Endogenous peroxidase was blocked by 3% hydrogen peroxide and sections were incubated overnight at 4 °C with: mouse monoclonal-[E3] anti-human K17 antibody (Abcam, Cambridge, MA). After primary antibody, biotinylated horse secondary antibodies (R.T.U. Vectastain ABC kit; Vector Laboratories, Burlingame, CA) were added. Development was done with 3, 3′ diaminobenzidine (DAB) (Dako, Carpinteria, CA) and counter-staining was done with hematoxylin. Negative controls were performed on all runs using an equivalent concentration of a subclass-matched immunoglobulin. K17 staining intensity was classified by one pathologist (K.R.S) based on a subjective assessment of absent (0), light (+1), or strong (+2) staining. The overall proportion of tumor cells with 2+ intensity staining (the PathSQ score) was determined by review of the representative immunostained histologic section from each case, blinded to corresponding clinical data.
Statistical analyses
The clinicopathologic features were categorized into meaningful groups, hence chi-square test and Fisher’s exact test were used to analyze associations between two categorical variables. The best cutoff point was chosen according to the lowest Akaike’s information criterion from a Cox proportional-hazard regression model to determine low versus high K17 mRNA expression. A data-driven cut off point classified patients into two groups: high K17 (24% of cases) and low K17 (76% of cases). The same cut off point was validated in the TCGA database which showed 24.8% of cases expression high K17 and 75.2% of cases expressing low K17. Overall survival was calculated from the date of diagnosis to the date of death and was estimated using the Kaplan–Meier method, using median or rate at specific time points with 95% confidence interval. Alive patients were censored at the last follow-up; all prognostic analyses were performed as described by Ballman39. Univariate analyses compared survival for K17 level (low vs. high), histological grade, tumor size, node status, tumor stage and surgical margins. To examine survival rates while adjusting for potential confounders, multivariate analyses were performed by Cox proportional hazards regression.
For the IHC analysis, K17 Path SQ score was used as a continuous variable in Cox proportional hazard model to determine the survival differences. Similarly, univariate and multivariate analyses compared survival for K17 level of expression, histological grade, tumor size, node status, tumor stage and surgical margins by Cox proportional hazards regression. Statistical significance was set at p-value ≤ 0.05 and analysis was done using SAS 9.4 (SAS Institute, Cary, NC, USA) and Graph pad prism 7 (Graph Pad Software, La Jolla, CA, USA).
Patent
Shroyer, K, Escobar-Hoyos, L.F. Keratin 17 as a Prognostic and Predictive Marker of Pancreatic Cancer. Stony Brook University Research Foundation. Filed 10/29/15. Pending Provisional Patent Application.
Acknowledgements
The authors thank The Cancer Genome Atlas (TCGA) and University of North Carolina for RNAseq data, the Stony Brook Cancer Center Biorepository for expert assistance to obtain tissue specimens (Dr. R. Kew), Dr. W. Hou for biostatistical support, and the Cancer Registries of the UMass Memorial Medical Center and Stony Brook University (X. Barzilay). This work was supported by the 2018 Pancreatic Cancer Action Network Translational Research Grant, grant number 18-65-SHRO (K.R.S.), NCI K99-R00 CA226342-01 (L.F.E.-H.), Pancreatic Cancer Action Network-AACR Pathway to Leadership Grant, grant number 17-70-25-ESCO (L.F.E.-H.) and a National Pancreas Foundation Grant (L.F.E-H). C.V.L. was partly supported by a postdoctoral fellowship awarded by the National Science Foundation’s workforce development program, the Alliance for Graduate Education in the Professoriate-Transformation grant (HRD-1311318), through the Stony Brook University Center for Inclusive Education, Stony Brook, NY, USA.
Author Contributions
Study concept and design: L.F.E.-H., K.R.S.; development of methodology: L.R.-P., L.F.E.-H., K.R.S.; acquisition of data: (managed patient samples): L.R.-P., A.A., L.F.E.-H.; analysis and interpretation of data: L.R.-P., S.B., E.A.V., C.V.L., C.-H.P., J.B., R.A.M., L.F.E.-H., K.R.S.; writing, editing/review, and intellectual content of the manuscript: L.R.-P., C.V.L., S.B., C.-H.P., R.A.M., L.F.E.-H., K.R.S.; study supervision: L.F.E.-H., K.R.S. All authors have read and approved the final manuscript.
Competing Interests
L.F. Escobar-Hoyos and K.R. Shroyer are consultants for KDx Diagnostics Inc. and OncoGenesis Inc.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kenneth R. Shroyer and Luisa F. Escobar-Hoyos jointly supervised this work.
Contributor Information
Kenneth R. Shroyer, Email: Kenneth.Shroyer@stonybrookmedicine.edu
Luisa F. Escobar-Hoyos, Email: Luisa.Escobarhoyos@stonybrook.edu
References
- 1.Paniccia A, et al. Characteristics of 10-Year Survivors of Pancreatic Ductal Adenocarcinoma. JAMA Surg. 2015;150:701–710. doi: 10.1001/jamasurg.2015.0668. [DOI] [PubMed] [Google Scholar]
- 2.Kamarajah SK, Burns WR, Frankel TL, Cho CS, Nathan H. Validation of the American Joint Commission on Cancer (AJCC) 8th Edition Staging System for Patients with Pancreatic Adenocarcinoma: A Surveillance, Epidemiology and End Results (SEER) Analysis. Ann Surg Oncol. 2017;24:2023–2030. doi: 10.1245/s10434-017-5810-x. [DOI] [PubMed] [Google Scholar]
- 3.Brennan MF, Kattan MW, Klimstra D, Conlon K. Prognostic nomogram for patients undergoing resection for adenocarcinoma of the pancreas. Ann Surg. 2004;240:293–298. doi: 10.1097/01.sla.0000133125.85489.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dal Molin M, et al. Very Long-term Survival Following Resection for Pancreatic Cancer Is Not Explained by Commonly Mutated Genes: Results of Whole-Exome Sequencing Analysis. Clin Cancer Res. 2015;21:1944–1950. doi: 10.1158/1078-0432.CCR-14-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirkin KA, Hollenbeak CS, Wong J. Prognostic impact of carbohydrate antigen 19-9 level at diagnosis in resected stage I-III pancreatic adenocarcinoma: a U.S. population study. J Gastrointest Oncol. 2017;8:778–788. doi: 10.21037/jgo.2017.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennon, A. M. & Goggins, M. Diagnostic and Therapeutic Response Markers. in Pancreatic Cancer, Vol. 28 (eds Neoptolemos, J. P., Urrutia, R., Abbruzzese, J. L. & Büchler, M. W.) 675–701 (Springer, New York, NY, 2010).
- 11.Scara S, Bottoni P, Scatena R. CA 19-9: Biochemical and Clinical Aspects. Adv Exp Med Biol. 2015;867:247–260. doi: 10.1007/978-94-017-7215-0_15. [DOI] [PubMed] [Google Scholar]
- 12.Biankin AV, Maitra A. Subtyping Pancreatic Cancer. Cancer Cell. 2015;28:411–413. doi: 10.1016/j.ccell.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Escobar-Hoyos LF, et al. Keratin 17 in premalignant and malignant squamous lesions of the cervix: proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker. Mod Pathol. 2014;27:621–630. doi: 10.1038/modpathol.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mockler D, et al. Keratin 17 Is a Prognostic Biomarker in Endocervical Glandular Neoplasia. Am J Clin Pathol. 2017;148:264–273. doi: 10.1093/ajcp/aqx077. [DOI] [PubMed] [Google Scholar]
- 15.Wang YF, et al. Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol. 2013;34:1685–1689. doi: 10.1007/s13277-013-0703-5. [DOI] [PubMed] [Google Scholar]
- 16.Merkin RD, et al. Keratin 17 is overexpressed and predicts poor survival in estrogen receptor-negative/human epidermal growth factor receptor-2-negative breast cancer. Hum Pathol. 2017;62:23–32. doi: 10.1016/j.humpath.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Ide M, et al. Keratin 17 expression correlates with tumor progression and poor prognosis in gastric adenocarcinoma. Ann Surg Oncol. 2012;19:3506–3514. doi: 10.1245/s10434-012-2437-9. [DOI] [PubMed] [Google Scholar]
- 18.Kim K, et al. Cytokeratin 17 Expression is Associated With Poor Prognosis in Gallbladder Adenocarcinoma. Appl Immunohistochem Mol Morphol. 2017;25:346–350. doi: 10.1097/PAI.0000000000000307. [DOI] [PubMed] [Google Scholar]
- 19.Regenbogen, E. et al. Elevated expression of keratin 17 in oropharyngeal squamous cell carcinoma is associated with decreased survival. Head Neck (2018). [DOI] [PubMed]
- 20.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 21.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell32, 185–203.e113 (2017). [DOI] [PMC free article] [PubMed]
- 24.Kim KS, Kwon J, Kim K, Chie EK. Impact of Resection Margin Distance on Survival of Pancreatic Cancer: A Systematic Review and Meta-Analysis. Cancer Res Treat. 2017;49:824–833. doi: 10.4143/crt.2016.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, et al. Expression of amphiregulin predicts poor outcome in patients with pancreatic ductal adenocarcinoma. Diagnostic pathology. 2016;11:60. doi: 10.1186/s13000-016-0512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizzi S, et al. Glucose transporter-1 expression and prognostic significance in pancreatic carcinogenesis. Histol Histopathol. 2009;24:175–185. doi: 10.14670/HH-24.175. [DOI] [PubMed] [Google Scholar]
- 27.Biankin AV, et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology. 2009;137(558–568):568.e551–511. doi: 10.1053/j.gastro.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, et al. Overexpression of G protein-coupled receptor GPR87 promotes pancreatic cancer aggressiveness and activates NF-κB signaling pathway. Molecular Cancer. 2017;16:61. doi: 10.1186/s12943-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, et al. FAM83A is amplified and promotes cancer stem cell-like traits and chemoresistance in pancreatic cancer. Oncogenesis. 2017;6:e300. doi: 10.1038/oncsis.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu XL, et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:33535. doi: 10.1038/srep33535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aung KL, et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res. 2018;24:1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinelli P, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut. 2017;66:1665–1676. doi: 10.1136/gutjnl-2015-311256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muckenhuber A, et al. Pancreatic Ductal Adenocarcinoma Subtyping Using the Biomarkers Hepatocyte Nuclear Factor-1A and Cytokeratin-81 Correlates with Outcome and Treatment Response. Clinical Cancer Research. 2018;24:351–359. doi: 10.1158/1078-0432.CCR-17-2180. [DOI] [PubMed] [Google Scholar]
- 34.Iacobuzio-Donahue CA, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–1094. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein NS, Bassi D. Cytokeratins 7, 17, and 20 reactivity in pancreatic and ampulla of vater adenocarcinomas. Percentage of positivity and distribution is affected by the cut-point threshold. Am J Clin Pathol. 2001;115:695–702. doi: 10.1309/1NCM-46QX-3B5T-7XHR. [DOI] [PubMed] [Google Scholar]
- 37.Real FX, Vila MR, Skoudy A, Ramaekers FC, Corominas JM. Intermediate filaments as differentiation markers of exocrine pancreas. II. Expression of cytokeratins of complex and stratified epithelia in normal pancreas and in pancreas cancer. Int J Cancer. 1993;54:720–727. doi: 10.1002/ijc.2910540503. [DOI] [PubMed] [Google Scholar]
- 38.Pishvaian MJ, et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clinical Cancer Research. 2018;24:5018–5027. doi: 10.1158/1078-0432.CCR-18-2645. [DOI] [PubMed] [Google Scholar]
- 39.Ballman KV. Biomarker: Predictive or Prognostic? J Clin Oncol. 2015;33:3968–3971. doi: 10.1200/JCO.2015.63.3651. [DOI] [PubMed] [Google Scholar]