Abstract
Hydrogen peroxide (H2O2) has a major function in host-microbial interactions. Although most studies have focused on the endogenous H2O2 produced by immune cells to kill microbes, bacteria can also produce H2O2. How microbial H2O2 influences the dynamics of host-microbial interactions is unclear. Here we show that H2O2 released by Streptococcus pneumoniae inhibits inflammasomes, key components of the innate immune system, contributing to the pathogen colonization of the host. We also show that the oral commensal H2O2-producing bacteria Streptococcus oralis can block inflammasome activation. This study uncovers an unexpected role of H2O2 in immune suppression and demonstrates how, through this mechanism, bacteria might restrain the immune system to co-exist with the host.
Subject terms: Inflammasome, Bacterial immune evasion
The functions of microbial hydrogen peroxide (H2O2) in host-pathogen interactions are unclear. Here, Erttmann and Gekara show that H2O2 released by Streptococcus pneumoniae inhibits inflammasomes, and thereby contributes to the pathogen’s ability to colonize the host.
Introduction
Hydrogen peroxide (H2O2) is a ubiquitous metabolic byproduct of aerobic unicellular and multicellular organisms1–4 that plays a major role in determining the outcome of host-microbial interactions5. Thus far, studies have focused on endogenous H2O2 produced by immune cells and its role in killing microbes or driving inflammatory processes1,5–8. However, H2O2 generation is not a preserve of eukaryotic host cells—many microbes do produce H2O21,4. How H2O2 produced by microbes affect the dynamic of host-microbial interactions, and in particular the ability of immune cells to respond to microbes, remains unknown.
Accumulation of H2O2 to high levels can be toxic to both the host and the microbe. To mitigate these undesired effects, most organisms are equipped with enzymes such as catalase to neutralize H2O22. However, some bacterial species lack catalase. One well-known example is S. pneumoniae. Consequently, a major hallmark of S. pneumoniae infection is massive production of H2O2, which can accumulate up to millimolars in in vitro cultures9,10. Considering that up to ∼108 cfu/mL of S. pneumoniae has been documented in infected lungs11, it is likely that the concentration of H2O2 in vivo, especially at the infection foci, is equally high hence can be detected in the breath of patients12.
While H2O2 production is responsible for autolysis of bacteria at stationary phase, generally, S. pneumoniae is highly resistant to H2O2 via mechanisms not fully understood13,14. This resistance allows S. pneumoniae not only to outcompete other bacteria from the host niche but likely also to withstand H2O2 produced by immune cells. But how H2O2 produced by bacteria afffects the anti-microbial response of immune cells is unresolved.
Microbes—especially those that have established a long-standing co-existence with their hosts have evolved diverse mechanisms to mitigate anti-microbial innate immune defenses13–15. S. pneumoniae is a habitant of the upper respiratory tract and the most common causative agent of community-acquired pneumonia, a leading cause of death worldwide16,17. Infections with S. pneumoniae possibly also promote invasion of the airways by other pathogens16,17.
How S. pneumoniae overcomes the innate immune system to persist in the host has remained unclear. Here we reveal that S. pneumoniae actively inhibits inflammasomes—key components of the innate immune system. This contributes to its ability to colonize the host but also renders immune cells unresponsive to other inflammasome stimuli during co-infections. We demonstrate that this phenomenon is due to S. pneumoniae-generated H2O2, which causes oxidative inactivation of inflammasomes. Further, we show that other H2O2-producing bacteria such as Streptococcus oralis, an oral commensal, similarly block inflammasomes. This study uncovers an unanticipated role of microbial H2O2 in innate immune suppression, and how this promotes host colonization by microbes.
Results
S. pneumoniae dampens inflammasome-dependent immunity via SpxB
The major source of H2O2 generated by S. pneumoniae is via its pyruvate oxidase (SpxB)9,18–20, which catalyzes the phosphate-dependent oxidation of pyruvate to form acetyl phosphate, CO2 and H2O2 (Fig. 1a). SpxB has diverse and counter-intuitive effects on the host and the bacteria. While responsible for spontaneous bacterial lysis19,21 and promotion of phagocytic uptake22, SpxB confers survival fitness to S. pneumoniae in vivo21 and contributes to resistance to oxidative stress9. Based on these divergent context-dependent effects of SpxB, we sought to investigate the impact of bacteria-derived H2O2 on the interaction between S. pneumoniae and a murine host.
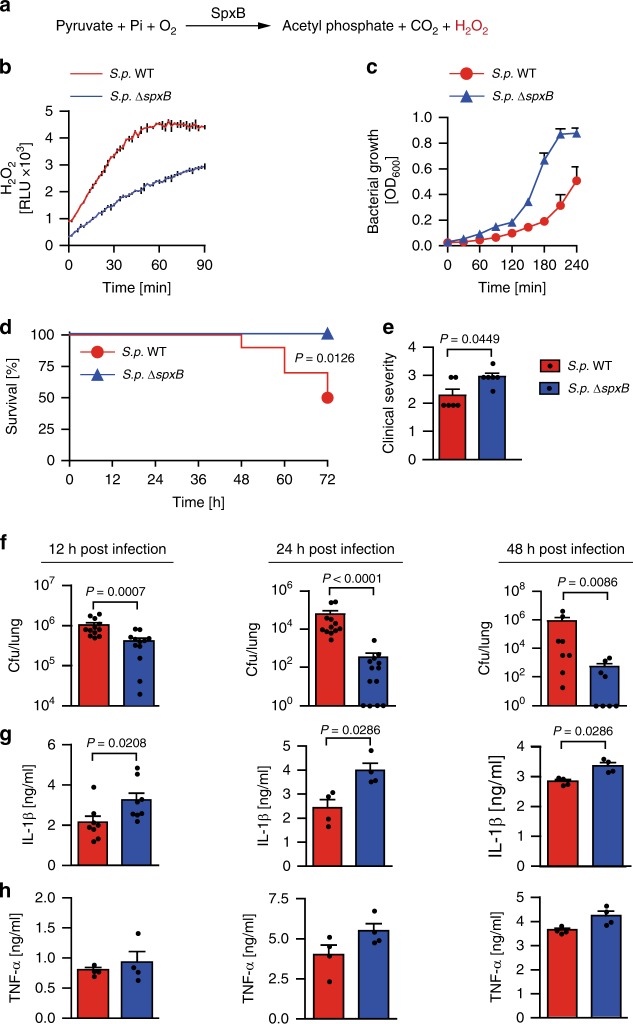
Fig. 1.
Pyruvate oxidase SpxB dampens inflammasome-dependent cytokine response promoting S. pneumoniae survival. a The pyruvate oxidase (SpxB)-mediated H2O2 generation reaction in S. pneumoniae. b H2O2 release by wild-type S. pneumoniae D39 (S.p. WT) and S.p.ΔspxB. c In vitro growth rate of S.p. WT (D39) and S.p. ΔspxB. b and c are representative of 3 independent experiments performed in triplicates; data shown as mean ± standard deviation (±s.d.). d Survival of WT mice after intranasal infection (1–2 × 107 cfu/mouse) with S.p. WT (D39) or S.p. ΔspxB analysed by the Kaplan–Meier method, with n = 10 animals per group. P value determined by Gehan-Breslow-Wilcoxon test. e Clinical severity of WT mice infected with S.p. WT (D39) or S.p. ΔspxB (1–2 × 107 cfu/mouse) for 6 h (n = 6 animals per group). P value determined by Mann Whitney test. f S.p. WT (D39) and S.p. ΔspxB counts (cfu) in the lungs of WT mice 12, 24 and 48 h after intranasal infection (1–2 × 107 cfu/mouse). g IL-1β and h TNF-α in the lung fluid of WT mice 12, 24 and 48 h after intranasal infection (1–2 × 107 cfu/mouse) with S.p. WT (D39) or S.p. ΔspxB. Results in f to h are from 2–3 independent experiments with a total of 4–12 animals per group; data shown as the mean ± standard error of the mean (±s.e.m.). P value determined by Mann Whitney test. Source data are provided as a Source Data file
Whereas the SpxB-deficient S. pneumoniae strain D39 (S.p. ΔspxB) that is highly defective in H2O2 production (Fig. 1b) exhibits a growth advantage over the wild-type strain in vitro (Fig. 1c), we find that upon intranasal inoculation in mice, S.p. ΔspxB causes severer clinical symptoms and is cleared faster without causing mortality (Fig. 1d–f). The severer symptoms and faster clearance of S.p. ΔspxB in vivo prompted us to investigate whether this mutant elicits a stronger anti-bacterial immune response. Indeed, although killed faster than wild-type S. pneumoniae D39 (S.p. WT) in vivo, S.p. ΔspxB elicits significantly higher levels of the inflammasome-dependent cytokine IL-1β (Fig. 1g). This is in contrast to the inflammasome-independent cytokine TNF-α, which, although showing a similar trend, is not significantly different between mice infected with wild-type or mutant bacteria (Fig. 1h). These results indicate that in the absence of SpxB, S. pneumoniae activates a stronger anti-bacterial immunity and in particular an inflammasome-dependent innate immune response.
S. pneumoniae blocks inflammasomes via SpxB-generated H2O2
Inflammasomes are intracellular multiprotein complexes that control the activation of Caspase-1, mediating pyroptotic cell death and the maturation of IL-1 family cytokines that are essential for optimal defence against pathogens including S. pneumoniae23–26. Generally, inflammasome activation involves two checkpoints: a priming step to induce the synthesis of pro-IL-1β and certain inflammasome components and a second step triggering the assembly and activation of inflammasome complexes24. To evaluate how S. pneumoniae modulates the second step of inflammasome activation, bone marrow-derived macrophages (BMDMs) were first primed with the TLR ligand lipopolysaccharide (LPS) and then infected with S. pneumoniae D39 for different durations. Thereafter, the magnitude and kinetics of Caspase-1 and IL-1β processing was compared with that by classical inflammasome activators. Here we show that although BMDMs infected with S. pneumoniae exhibit large amounts of pro-IL-1β and pro-Caspase-1, accumulation of the processed forms (IL-1β p17 and Casp-1 p20) is highly delayed and remains undetectable until 6 to 12 h post infection. Even at these delayed time points, the ratio of processed IL-1β and Caspase-1 to their precursors is very low (Supplementary Fig. 1a-c). This is in clear contrast to stimulation with ATP or nigericin (agonists of the NLRP3 inflammasome), Salmonella Typhimurium or Pseudomonas aeruginosa (activators of the NLRC4 inflammasome)27,28, which elicit robust processing of Caspase-1 and IL-1β within 30 to 60 min (Supplementary Fig. 1d–g). This demonstrates that S. pneumoniae does not directly activate inflammasomes at the early stage of infection and that the weak response detected at later time points is likely a secondary outcome of danger signals such as ATP from dead or stressed cells or by pneumolysin extruded from lysed bacteria29–32.
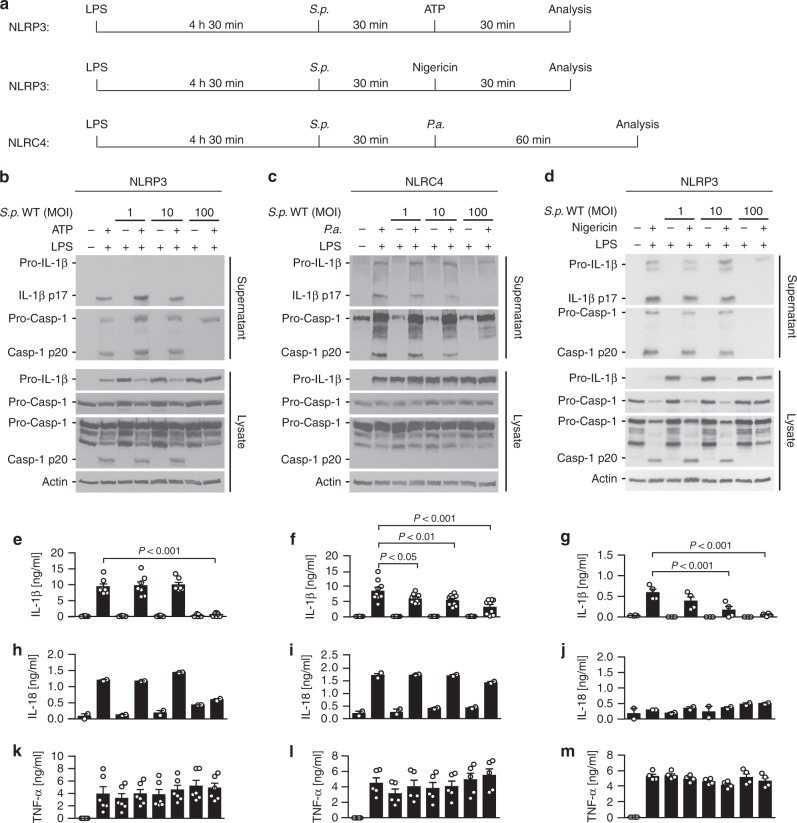
In view of these observations, we wondered whether such limited responses are due to inhibition of inflammasomes by S. pneumoniae. To directly test this idea, we pre-treated BMDMs with S.p. WT before challenging them with agonists for different inflammasomes including ATP, nigericin or P. aeruginosa (schematically illustration in Fig. 2a). S.p. WT dose-dependently inhibits the processing and secretion of Caspase-1, IL-1β and IL-18 (Fig. 2b–j). In contrast, TNFα is unaltered (Fig. 2k–m).
Fig. 2.
S. pneumoniae impairs NLRP3- and NLRC4-dependent inflammasome activation. a Schematic diagrams of experimental setups used in b to m. Immunoblots of Caspase-1 and IL-1β processing in LPS-primed BMDMs pre-treated with increasing S.p. WT (D39) doses (MOI 1, 10 or 100) for 30 min before stimulation with b ATP for 30 min, c P. aeruginosa (P.a., MOI 20) for 60 min or d nigericin for 30 min. Results are representative of at least 4 independent experiments. ELISA analysis of corresponding supernatants for e–g IL-1β, h–j IL-18 and k–m TNF-α. Results are obtained from at least two independent experiments; data are shown as mean ± s.e.m. P values determined by one-way ANOVA followed by Bonferroni post-test. Source data are provided as a Source Data file
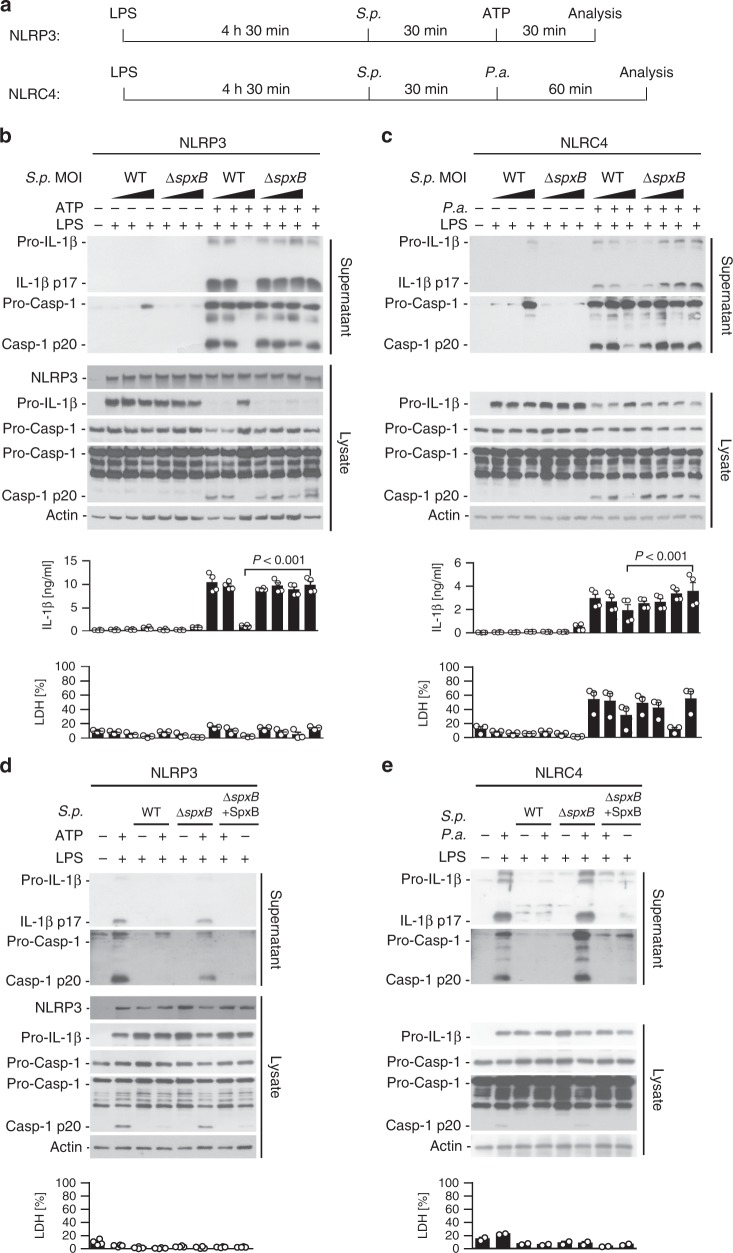
As S.p. ΔspxB induces elevated IL-1β levels in vivo (Fig. 1g), we tested whether S. pneumoniae blocks inflammasomes via SpxB. Indeed, inflammasome activation by ATP and P. aeruginosa is highly inhibited in the presence of S.p. WT (Fig. 3a–c) but not S.p. ΔspxB, except upon complementation with a plasmid carrying the spxB gene (Fig. 3d, e).
Fig. 3.
S. pneumoniae blocks multiple inflammasome pathways through its SpxB activity. a Schematic diagram of experimental setups used in (b–e). b, c Immunoblots of Caspase-1 and IL-1β in cell lysates and supernatants of LPS-primed BMDMs pre-treated with increasing MOIs (1, 10, 100) of S.p. WT (D39) or S.p. ΔspxB and then stimulated with agonists for NLRP3 (ATP, 5 mM, 30 min), or NLRC4 (P.a., MOI 20, 60 min). ELISA for IL-1β secretion and lactate dehydrogenase (LDH) assay in corresponding supernatants. The results are representative of four independent experiments. P values determined by one-way ANOVA followed by Bonferroni post-test. d, e Caspase-1 and IL-1β processing and LDH release of LPS-primed BMDMs pre-treated with S.p. WT (D39), S.p. ΔspxB or S.p. SpxB-complemented ΔspxB (ΔspxB + SpxB) (MOI 50) for 30 min before d stimulation with 5 mM ATP for 30 min or e infection with P.a. (MOI 20) for 60 min. The results are representative of four or two independent experiments, respectively. LDH release is depicted as the mean ± s.e.m. Source data are provided as a Source Data file
However, pre-infection of cells with S. pneumoniae does not inhibit the induction of Pro-IL-1β or TNFα by the TLR ligand LPS, thus ruling out priming as the target of S. pneumoniae-mediated inflammasome inhibition (Supplementary Fig. 2a–f). Further, LDH release and bacterial uptake assays show that the diminished inflammasome response is not due to loss of phagocytic activity or viability (Supplementary Fig. 2g–i). Notably, long-term infection of macrophages reveals that the SpxB mutant bacteria have an intrinsically increased capacity to activate the inflammasome when compared to WT bacteria (Supplementary Fig. 3). Together, these data demonstrate that S. pneumoniae actively blocks inflammasome activation.
Next, we sought to determine the bacterial factors responsible for inflammasome activation by S. pneumoniae. Pneumolysin (PLY) is an intracellular thiol-dependent pore forming toxin released into the environment upon bacterial lysis33,34. In view of previous studies29–32,35,36, we compared the ability of S.p. WT and ply-deficient S. pneumoniae (S.p. Δply) to induce inflammasome activation. As observed before, short (1 h) infection with S.p. WT or S.p. Δply fail to induce inflammasome activation. However, long-term (12 h) infection lead to a modest response by S.p. WT but not S.p. Δply, confirming the importance of PLY in S. pneumoniae-mediated inflammasome activation (Supplementary Fig. 4a). As a proxy to evaluate inflammasome activation by PLY and how this is affects by S. pneumoniae we used a highly purified listeriolysin O (LLO)—a homologous pore-forming toxin. LLO induces robust inflammasome activation that is inhibited by S.p. WT but not S.p. ΔspxB (Supplementary Fig. 4b–d). Similar to PLY, LLO is a thiol-dependent toxin hence highly sensitive to oxidation37. Therefore, we employed catalase, an H2O2-neutralizing enzyme, to reduce the accumulation of reactive oxygen species. Treatment with catalase enhances LLO-mediated inflammasome activation and increases the ability of S.p. WT to induce inflammasome activation (Supplementary Fig. 4e, f). Hence, we concluded that the restrained inflammasome response elicited by S. pneumoniae is likely due to oxidative inactivation of host signaling molecules as well as the bacterial inflammasome agonist PLY.
S. pneumoniae blocks the assembly of inflammasome complexes
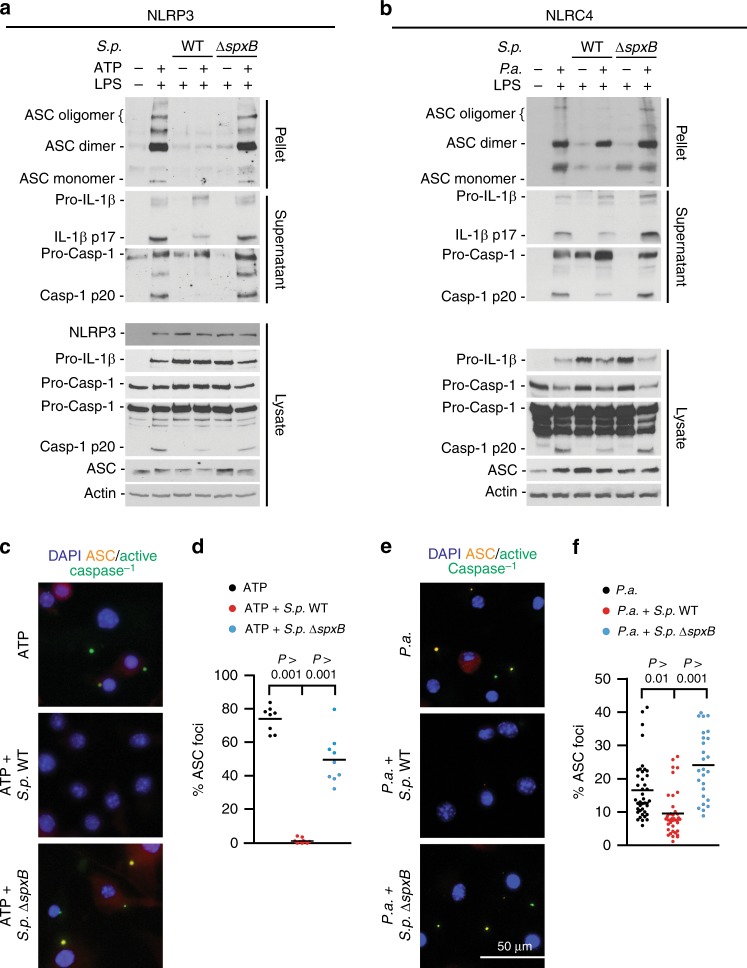
ASC oligomerization is a critical step in the assembly and activation of inflammasome complexes38,39. To test whether S. pneumoniae D39 affects this step, we employed differential centrifugation and chemical cross-linking40 to isolate and analyze ASC oligomers. LPS-primed BMDMs pre-treated with S.p. WT exhibit severely impaired ASC oligomerization and diminished processing of Caspase-1 and IL-1β in response to ATP or P. aeruginosa. This is in contrast to S.p. ΔspxB-pre-treated cells that do not exhibit inhibited ASC oligomerization (Fig. 4a, b), except upon complementation with the plasmid carrying the spxB gene (Supplementary Fig. 5a, b). To verify these findings, BMDMs were fluorescently labelled with the Caspase-1 fluorescent activity-based probe FLICA (FAM-YVAD-FMK) and an anti-ASC antibody and then analyzed microscopically for ASC/Caspase-1 complexes (specks). BMDMs pre-treated with WT S. pneumoniae D39 exhibit highly reduced ASC/Caspase-1 speck formation in response to ATP or P. aeruginosa (Fig. 4c–f). To determine whether inflammasome inhibition is a general feature of S. pneumoniae, we tested other S. pneumoniae strains including A66.1 (serotype 3), TIGR4 (serotype 4), and the unencapsulated strain R6. All such strains inhibit ASC oligomerization and processing of Caspase-1 and IL-1β (Supplementary Fig. 6a–d), thereby demonstrating that the observed inflammasome inhibition is not strain-specific.
Fig. 4.
Inflammasome inhibition by S. pneumoniae involves defects in ASC complex formation. a, b Immunoblot analyses of ASC oligomers or Caspase-1 and IL-1β processing in LPS-primed BMDMs pre-treated with S.p. WT or S.p. ΔspxB (D39, MOI 50, 30 min) before stimulation with a ATP for 30 min (NLRP3) or b P.a. (NLRC4, MOI 20) for 60 min. Results are representative of 3 independent experiments. c–f Microscopic visualisation of ASC/active Caspase-1 complexes (red/green; overlay yellow) in LPS-primed BMDMs pre-treated with S.p. WT or S.p. ΔspxB (D39, MOI 50) and subsequently stimulated with c, d ATP (5 mM, 30 min) or e, f P.a. (MOI 20, 60 min). Panels d and f depict corresponding percentages of cells in c and e, respectively, containing ASC/active Caspase-1 specks (determined by enumerating at least 100 cells per sample). Scale bar: 50 µm. Results in d and f are from three independent experiments. The data are shown as the mean ± s.e.m. P values determined by one-way ANOVA followed by Bonferroni post-test. Source data are provided as a Source Data file
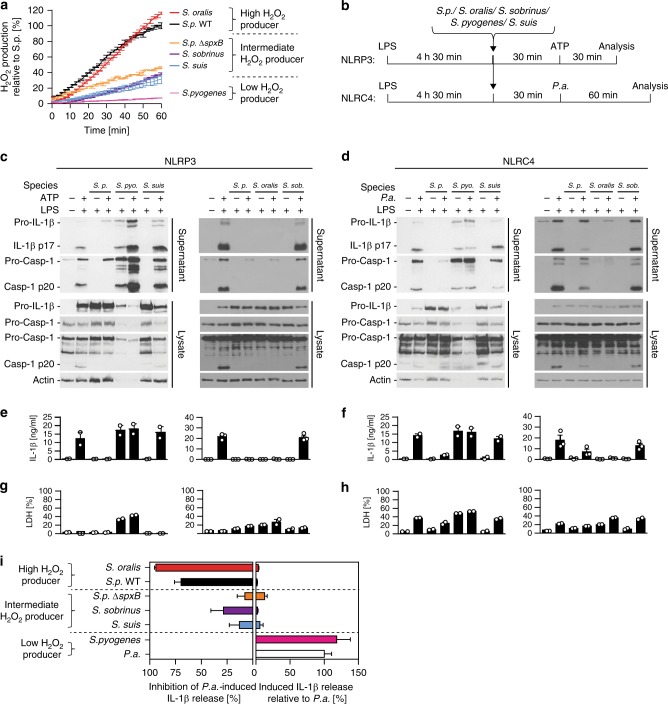
High H2O2-producing bacteria inhibit inflammasome activation
To further examine the generality of inflammasome inhibition by H2O2-producing bacteria, we tested other bacterial species including the oral commensals Streptococcus oralis and Streptococcus sobrinus, and the human and pig pathogens Streptococcus pyogenes and Streptococcus suis, respectively. By comparing the magnitude of H2O2 production, we can classify these bacteria into high, intermediate and low H2O2 producers (Fig. 5a). When tested on BMDMs, the high H2O2 producers S. pneumoniae and S. oralis strongly inhibit inflammasome activation. The intermediate H2O2 producers S.p. ΔspxB, S. sobrinus and S. suis neither inhibit nor induce inflammasome activation. In contrast, the low H2O2 producer S. pyogenes does not inhibit but rather induces inflammasomes (Fig. 5b–i). These data demonstrate that inflammasome inhibition by different bacterial species correlates with the strength of H2O2 release.
Fig. 5.
High H2O2-producing bacteria block inflammasome activation whereas low producers do not. a H2O2 production by S. oralis, S. pneumoniae ΔspxB (D39), S. sorbnius, S. suis, S. pyogenes relative to that by S. pneumoniae WT (D39) (100%). Data are representative of two independent experiments performed in duplicates or triplicates presented as the mean ± s.d. b Schematic diagrams of experimental setups used in (c–h). c, d Immunoblot analysis of Caspase-1 and IL-1β processing, e, f ELISA analysis of corresponding supernatants for IL-1β secretion and g, h LDH release of LPS-primed BMDMs pre-treated with S. pneumoniae D39 WT (S. p.), S. pyogenes (S. pyo.), S. suis, S. oralis or S. sobrinus (S. sob.) (MOI 40) for 30 min before c, e, g stimulation with ATP (5 mM) for 30 min or d, f, h infection with P. aeruginosa (MOI 20) for 60 min. Data in c and d are representative of two or three independent experiments, respectively. ELISA and LDH release data are from two or three independent experiments and are depicted as the mean ± s.e.m. i Relative inflammasome inhibition or activation by Streptococcus species presented as the mean ± s.e.m. Inhibition depicts decrease in P.a.-induced IL-1β release during co-infection with indicated bacteria. Activation depicts IL-1β release by indicated bacteria relative to that by P.a.. Data are presented as the mean ± s.e.m. of 2–9 experiments. Source data are provided as a Source Data file
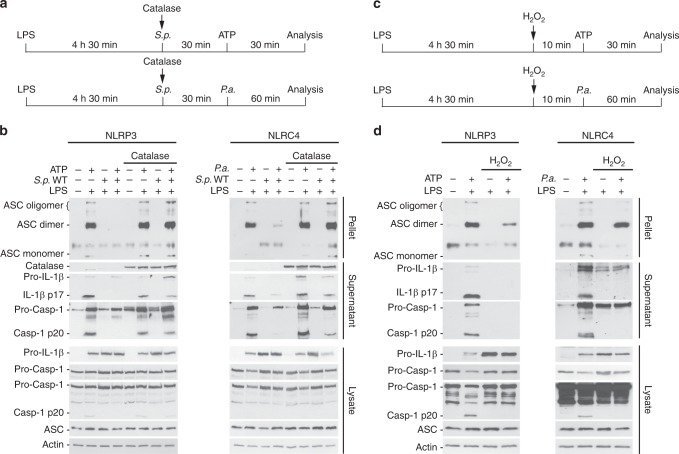
To verify whether the observed inflammasome inhibition is mediated by H2O2, we used catalase. Catalase promotes the in vitro growth of the high H2O2 producers WT S. pneumoniae and S. oralis, but not the intermediate or low H2O2 producers S.p. ΔspxB, S. sobrinus, S. suis or S. pyogenes (Supplementary Fig. 7a–c). Notably, catalase rescues inflammasome inhibition by S.p. WT (Fig. 6a, b and Supplementary Fig. 7d–o) and S. oralis, but has no effect on the IL-1β response by S. sobrinus or TNF-α production (Supplementary Fig. 7d–o). Moreover, S. pneumoniae is unable to block inflammasome activation in the presence of catalase in human PBMCs (Supplementary Fig. 8a–h). In contrast, inflammasome inhibition can be recapitulated by direct addition of H2O2 to BMDMs (Fig. 6c, d). Titration of the H2O2 amount reveals that 50–100 µM H2O2 are sufficient to inhibit Caspase -1 as well as Caspase -3 and Caspase -8 (Supplementary Fig 9a). In contrast to S.p. ΔspxB, S.p. WT and the S.p. ΔspxB carrying the SpxB complementing plasmid release similarly high amounts of H2O2 into the cell culture medium during infection (Supplementary Fig. 9b, c).
Fig. 6.
S. pneumoniae impairs inflammasome activation via SpxB-mediated H2O2 release. a Schematic diagrams of the experimental setups used in (b). b ASC oligomers, Caspase-1 and IL-1β processing in LPS-primed BMDMs pre-treated with S.p. WT (D39, MOI 100) in the presence of catalase (100 U mL−1) for 30 min before ATP stimulation (5 mM, 30 min) or P.a. infection (MOI 20, 60 min). c Schematic diagrams of the experimental setups used in d. d ASC oligomers, Caspase-1 and IL-1β processing in BMDMs pre-treated with 50 µM H2O2 for 10 min prior to stimulation with ATP (5 mM, 30 min) or infection with P.a. (60 min, MOI 20). The data in b and d are representative of three independent experiments. Source data are provided as a Source Data file
The above data unequivocally demonstrate that H2O2 is a potent inhibitor of inflammasomes. This led us to hypothesize that such inhibition of inflammasomes is due to oxidation of inflammasome components. Protein carbonylation is the most general marker of severe oxidative protein damage. It involves covalent adduction of carbonyl groups (e.g. aldehydes and ketones) on oxidised side chains of susceptible amino acid residues such as cysteine, proline, arginine, lysine and threonine41. Carbonylated proteins can be labelled with dinitrophenylhydrazine and detected using anti-dinitrophenol (DNP) antibodies. Immunoblotting of DNP-derivatized cell extracts reveals increased protein carbonylation in macrophages exposed to S.p. WT (Supplementary Fig. 10a, b), indicating that H2O2 production by S. pneumoniae causes oxidation of proteins in host cells. Given that oligomerization of ASC, a key inflammasome component, is impaired in the presence of S. pneumoniae or H2O2 (Fig. 4, Fig. 6, Supplementary Fig. 5 and Supplementary Fig 6), we next tested whether ASC is among those proteins oxidised by S. pneumoniae. Immunoprecipitation and immunoblot analyses of DNP-derivatized cell extracts reveal that ASC is carbonylated upon exposure to WT S. pneumoniae and that such oxidation is associated with defective Caspase-1 and IL-1β processing (Supplementary Fig. 10c). Based on the results, we conclude that H2O2-mediated inflammasome inhibition is due to the oxidative inactivation of ASC and most likely other components including the oxidation-sensitive cysteine-rich caspases42,43.
H2O2 blocks inflammasome-dependent bacterial clearance
Considering that S.p. ΔspxB elicits a stronger inflammasome response and is cleared faster from mice (Fig. 1), we investigated whether inflammasome activation contributes to enhanced clearance of S.p. ΔspxB using ASC−/− mice. At early time points, no ASC-dependent clearance of S.p. WT D39 is evident. However, as previously observed25,26, ASC−/− mice exhibit higher bacterial burden than wild-type mice 48 h after infection (Fig. 7a–c). ASC-dependent clearance of S.p. WT at later rather than early time points is consistent with its H2O2-mediated inhibitory effects on inflammasomes. Hence, the elicited inflammasome response is both low and delayed. In contrast to S.p. WT, ASC-dependent clearance of S.p. ΔspxB is observed as early as 12 and 24 h after infection (Fig. 7d–f). Noteworthy, although carrying a higher bacterial burden—consistent with the lack of inflammasome-dependent inflammation—the ASC−/− mice exhibit lower clinical severity and body temperature changes (Fig. 7g, h).
Fig. 7.
H2O2 release by S. pneumoniae suppresses inflammasome-dependent bacterial clearance. a–c S.p. WT (D39) burden (cfu) in the lungs of WT and ASC−/− mice a 12 h, b 24 h, and c 48 h after intranasal infection (1–2 × 107 cfu/mouse). (d–f) S.p. ΔspxB (D39) burden in the lungs of WT and ASC−/− mice d 12 h, e 24 h, and f 48 h post infection (1–2 × 107 cfu/mouse). g Clinical severity and h subcutaneous body temperatures of WT and ASC−/− mice after S.p. ΔspxB (D39) infection. Results in a to h are representative of 2 independent experiments with a total of 4–8 animals per group (1–2 × 107 cfu/mouse). i Bacterial burden, j clinical severity, and k subcutaneous body temperature of WT mice co-inoculated with S.p. WT (D39) and active or denatured catalase (500 U per mouse) for 24 h. Results are representative of three independent experiments with a total of 16 animals per group (1 × 107 cfu/mouse). l Bacterial burden, m clinical severity, and n subcutaneous body temperature of WT mice co-inoculated with S.p. ΔspxB (D39) and active or denatured catalase (500 U per mouse) for 24 h. Results are representative of two independent experiments with a total of 10 animals per group (1 × 107 cfu/mouse). The data in a to n are shown as mean ± s.e.m. a–f, i, l P values determined by Mann Whitney test. g, h, j, k, m, n P values determined by two-way ANOVA followed by Bonferroni post-test. o Proposed model of inflammasome inactivation by high H2O2-producing bacteria. Source data are provided as a Source Data file
In addition to metabolism and H2O2 production, SpxB also plays a role in other aspects of bacterial physiology9,44,45, which together with the modulation of inflammasomes may contribute to the in vivo attenuation of S.p. ΔspxB. Therefore, to specifically evaluate the contribution of H2O2 on the infection outcome, we asked whether in vivo neutralization of H2O2 by catalase could similarly increase S. pneumoniae-induced inflammation and whether this in turn could promote bacterial clearance. In agreement, when intranasally co-inoculated with catalase, S.p. WT elicits severer clinical symptoms and is cleared faster (Fig. 7i–k), while clearance of S.p. ΔspxB is unaltered (Fig. 7l–n). This independently confirms that H2O2 release by S. pneumoniae does inhibit innate immune activation and that this contributes to host colonization as schematically summarized in Fig. 7o.
Discussion
Here we report that in order to persist in the host S. pneumoniae interferes with a key arm of the innate immune response—inflammasome activation. We show that this feature is due to the ability of S. pneumoniae to release large quantities of hydrogen peroxide (H2O2) thereby causing oxidative inactivation of inflammasomes in immune cells. Further, we demonstrate that inflammasome inhibition is a common feature of other high H2O2-producing bacterial species. These findings are significant. Firstly, they uncover a new mechanism of innate immune subversion by bacteria. Secondly, they highlight an unexpected role of H2O2 in the negative regulation of inflammasomes.
Inflammasomes are vital players in the eradication of bacterial pathogens23,25,26. Correspondingly, recent studies also reveal that an increasing number of pathogens are equipped with mechanisms to circumvent inflammasome activation13,14,46. Such pathogens can overcome inflammasome-dependent host defenses either by evading receptor recognition or through active mechanisms of inflammasome suppression14. The avoidance strategy is a stealth approach whereby pathogens disguise their presence from the host for example by restricting the bioavailability of inflammasome stimuli47–50. Active suppression on the other hand involves targeted modulation of inflammasomes by virulence factors that interact with host cell proteins28,51–54. The active mechanisms of inflammasome suppression reported thus far seem to require an intimate host-pathogen contact to permit the delivery of inhibitory effectors into the host cells13,14,46. Arguably, such directed delivery of virulence factors is probably essential to ensure that immune suppression is not global but only limited to the infected and not the healthy bystander cells. In this study, we report a new mechanism of inflammasome suppression that does not require direct bacterial contact with or delivery of bacterial proteins into host cells. H2O2 is a diffusible small molecule that easily crosses biological membranes and can modulate the activity of different cellular proteins. We show that by releasing H2O2 into their surroundings, bacteria such as S. pneumoniae and S. oralis cause oxidative stress in host cells and that this results in suppression of different inflammasome pathways.
The demonstration that bacteria-derived H2O2 inhibits inflammasomes and that this is likely a common mechanism employed by diverse bacterial species to counter the innate immune system is somewhat unexpected. Previously, several studies have linked reactive oxygen species (ROS) with the activation of the NLRP3 inflammasome55–58. However, other studies have indicated that ROS do not trigger inflammasomes per se but support the priming phase of NLRP3 activation25,59–61. In fact, curiously and in agreement with the conclusions herein, it has been observed that genetic defects in ROS generation do not result in diminished inflammasome activation but rather in enhanced inflammasome activation25,61,62. On the other hand excessive generation of ROS and reactive nitrogen species by immune cells have been linked to diminished inflammasome activation25,43,63,64.
Oxidative stress is known to result in oxidative damage of many cellular proteins65,66. The data herein show that ASC oligomerization—a central node for many inflammasome pathways—is highly sensitive to oxidative inhibition. Therefore, as previously discussed6, although transient ROS generation might be essential for some steps such as priming, our findings support the view that oxidative stress is largely unfavourable for inflammasome activation.
Our understanding of host-microbe interactions has mostly been gleaned from studying monomicrobial infections. However, many infections such as those of the respiratory system are polymicrobial16,67. In these infections, one microbe predisposes the host to colonisation by others. S. pneumoniae possibly contributes to the aetiology of polymicrobial lung colonization16,17. The results here show that H2O2-mediated inflammasome inhibition not only enables bacteria to avoid invoking strong self-limiting inflammatory responses, but also to block the responsiveness of immune cells to other danger signals in co-infection setting. Thus, beyond demonstrating how S. pneumoniae and other bacteria subvert the innate immune system, this work opens up possibilities for further studies to better understand the aetiology of polymicrobial host colonization.
Finally, the demonstration that H2O2 inhibits inflammasomes advances the current knowledge beyond the conventional view of ROS as drivers of inflammation7,68 and highlights an underappreciated anti-inflammatory effect of ROS. The latter notion is consistent with increasing clinical and experimental evidence linking a reduction in intracellular ROS with hyper-inflammation61–63,69.
Methods
Ethics statement
Experiments involving humans were performed according to the recommendations of the local Research Ethics Committee of Umeå University (Regionala etikprövningsnämnden i Umeå), as approved in permit Dnr 2012-501-31M. Full informed consent was obtained from donors in compliance with the Declaration of Helsinki. Animal experiments were carried out according to the guidelines set out by the Umeå Regional Animal Ethic Committee (Umeå Regionala Djurförsöksetiska Nämnd), Approval no. A53-14.
Mice
All mice in this study were on C57BL/6 background. Asc−/− mice27 were obtained from Genentech, South San Francisco, USA. All mice were bred at the Umeå Transgene Core Facility and in vivo infection experiments were performed at the Umeå Centre for Comparative Biology. Mice were maintained under specific pathogen free conditions.
Animal infections
Age- and sex-matched adult mice (8–20 weeks old) were anaesthetized using isoflurane with the XGI-8 gas anaesthesia system (Caliper), inoculated intranasally with 30 μl of a suspension containing 1–2 × 107 cfu of S. pneumoniae strain D39 wild type or D39 ΔspxB and then suspended by their teeth for 10 min to enable the infection inoculum to descend into the lungs. At different time points after infection, mice were monitored for clinical symptoms such as weight loss and temperature. Clinical severity scoring was based on an arbitrary scale from 1 to 4 where 1 represented mice with mild but visible symptoms such as slowed activity whereas 4 represented those with highest morbidity, i.e. with a combination of hunchback posture, lethargy, loose fecal pellet, ruffled fur, >25% weight loss and difficulties in breathing and movement and hence had to be euthanized. Bacterial burden in the lung was determined by plating lung homogenates on blood agar plates to determine the cfu. For in vivo analysis of cytokines, the lungs were gently disrupted in PBS to release cells from the connective tissue. Lung flushings containing extracellular fluids and cell pellets were separated by centrifugation, and extracellular fluids were analysed by ELISA.
Bacterial strains and growth conditions
S. pneumoniae D39 wild-type and D39 ΔspxB strains were provided by Jetta Bijlsma45 (MSD animal health, Netherlands). TIGR4 Serotype 4 (BAA-334) and R6 Serotype 2 (BAA-255) were obtained from ATCC. The S. pneumoniae SpxB-complemented D39 ΔspxB + SpxB (P878 + pMU1328::300 + spxB: P1221) strain and the control strain D39 ΔspxB (D39 spxB::TnphoA(erm): P878), R6x and R6x ΔspxB (R6x spxB::TnphoA(erm): P1167)9,19 were from Jeffrey Weiser (NYU School of Medicine, New York, USA). S. pneumoniae D39 wild-type and D39 Δply strains were provided by Sven Hammerschmidt (Universtiy of Greifswald, Germany). Salmonella enterica serovar Typhimurium strain SL1344 was provided by Siegfried Weiss (Helmholtz Centre for Infection Research, Braunschweig, Germany). P. aeruginosa-Xen41 and S. pneumoniae-Xen10 strain A66.1 Serotype 3 were obtained from Caliper. Streptococcus pyogenes NZ131 (serotype M49) was provided by Victor Nizet (University of California, USA). Streptococcus suis (serotype 2) strain 10 was provided by Peter Valentin-Weigand (Tierärztliche Hochschule Hannover, Germany). Streptococcus oralis ATCC 35037 and Streptococcus sobrinus CCUG 21019 were provided by Jan Oscarsson (Umeå University, Umeå, Sweden). All bacterial strains used in this study were grown to exponential phase for infection experiments. Streptococcus species were incubated on blood agar plates overnight at 37 °C and 5% CO2 followed by inoculation and growth in Brain Heart Infusion broth at 37 °C and 5% CO2 for 2 1/2 h; S. Typhimurium and P. aeruginosa were incubated for 24 h at 37 °C on LB-agar (Lennox), then inoculated and grown overnight in Luria broth at 37 °C at 150 rpm to stationary phase followed by subculture in Luria broth at 37 °C at 150 rpm for 2 h to exponential phase.@@@
ROS measurements
H2O2 production by the different Streptococcus species was determined by adding 50 µM luminol and 1.2 U mL−1 horseradish peroxidase (HRP) to 100 µl of pre-warmed OptiMEM at 37 °C containing 1.25 × 105 bacteria. The H2O2 levels, expressed as relative light units (RLU), were measured every 2 min for 1.5 h at 37 °C using a Tecan Infinite M200 plate reader.
The H2O2 amounts produced by S.p. D39 WT, S.p. D39 ΔspxB, and S.p. D39 ΔspxB + SpxB were determined by the Fluorometric Hydrogen Peroxide Assay Kit from Sigma according to the manufacturers’ instructions. 1.5 × 106 BMDMs in 1 ml OptiMEM were infected with S.p. at MOI 1, 10, or 100 in the presence (or not) of 100 U mL−1 catalase for 1.5 h. Cell culture supernantants were collected, bacteria removed by centrifugation at 3000 × g for 2 min, and directly subjected to H2O2 quantification.
Cell culture, infection and stimulation
BMDM differentiation was performed by culturing bone marrow cells in 20% conditioned L-929 culture medium for 5 days. Human PBMCs were isolated from peripheral blood using Ficoll-Paque PLUS. To measure inflammasome responses, cells were primed with 500 ng mL−1 LPS for 4.5 h and then infected with the indicated Streptococcus species for 30 min at the indicated multiplicity of infection (MOI). Thereafter, cells were further stimulated with 5 mM ATP or 1 µM nigericin for 30 min, or infected with P. aeruginosa Xen41 at MOI 20 or S. Typhimurium SL1344 at MOI 10 for 60 min. In all cases, the bacteria were centrifuged onto BMDMs at 250 × g for 5 min to ensure comparable cell contact. 100 U mL−1 catalase (except where indicated otherwise) was added simultaneously with the indicated Streptococcus species to the cell culture medium, while 50 µM H2O2 was added 10 min before the addition of inflammasome −activators. For infection experiments, when cells were incubated for several hours (6–12 h), cells were infected for 60 min with S. pneumoniae, while for −further incubation the cell culture medium was replaced by medium containing 25 µg mL−1 gentamicin.
Calculation of bacterial uptake
LPS-primed BMDMs for 4.5 h were infected (or not) with S. pneumoniae WT at MOI 100 for 30 min. Cells were then infected with P. aeruginosa at MOI 20 for 1 h. Infections were synchronised by 5 min centrifugation at 250 × g. Cells were washed three times with PBS and then lysed in PBS containing 1% Triton X-100 and serial dilutions were plated on tetracycline-containing LA plates.
Cytokine ELISAs and LDH assay
The BD OptEIA Mouse IL-1β ELISA Set and BD OptEIA Human IL-1β ELISA Set (BD Biosciences), Mouse IL-18 ELISA Kit (MBL International), TNF alpha Human ELISA Kit (abcam) and Mouse TNF-α DuoSet (R&D Systems) were used according to the manufacturers’ instructions. LDH release was determined by CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega) according to the manufacturers’ instructions.
Immunoblotting analysis
Supernatants from BMDMs (1.5 × 106/well) maintained in serum-reduced medium (OptiMEM, Invitrogen) during stimulation were collected. Cell debris was removed by centrifugation, and proteins were precipitated by methanol-chloroform extraction. The precipitates were resuspended in 2 × Laemmli buffer, and the cells were directly lysed in 2 × Laemmli buffer. Proteins were separated on 13.5% SDS-PAGE gels and immunoblotted onto nitrocellulose membranes (Amersham). Membranes were blocked for 1 h in 1 × Roti Block (Roth) and subsequently incubated with different primary antibodies overnight. After incubation with HRP-labelled secondary antibodies, the proteins were detected using ECL substrate and X ray films. All uncropped Western blots are provided as part of the Source Data file.
Analysis of carbonylated proteins
Equal amounts of total protein lysates in 6% SDS in 20 mM Tris-HCl, pH 7.4, and 150 mM NaCl were treated with 5 µM 2,4-dinitrophenylhydrazine (DNPH) in 5% trifluoroacetic acid (TFA) for 15 min. Negative controls were treated with 5% TFA without DNPH. Cell lysates were neutralised by adding 2 M Tris, pH > 11, mixed with 2 × Laemmli buffer and analysed for DNP by immunoblotting. Alternatively, the cells were lysed in 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% deoxycholic acid, and 0.2% SDS containing a protease inhibitor cocktail (Roche). Protein A/G PLUS beads (Santa Cruz) with bound anti-ASC (or rat IgG control) were added to the lysates, and the mixture was incubated overnight at 4 °C. The immunoprecipitates were collected, washed, resuspended in 6% SDS in 20 mM Tris, pH 7.6, with 137 mM NaCl and heated for 5 min at 70 °C. The immunoprecipitated proteins were derivatized with 10 mM DNPH in 5% TFA for 15 min and neutralised using 2 M Tris, pH > 11, and buffered with 6 × Laemmli buffer. Then the proteins were analysed for DNP by immunoblotting.
ASC oligomerization
ASC oligomerization was assayed in LPS-primed cells treated with inflammasome activatiors. 3 × 106 cells were lysed in 20 mM Hepes, pH 7.5, 150 mM KCl, and 1% NP-40 with a protease inhibitor tablet (Roche) and sheared using a 21G × 2” needle. The cell lysates were centrifuged at 3300 × g for 10 min at 4 °C, and the pellets were washed twice with 1 ml of PBS. This was followed by incubation in 2 mM suberic acid bis(N-hydroxy succinimide ester) (DSS) for 30 min at RT. After centrifugation at 3300 × g for 10 min at 4 °C, the supernatants were removed and the resulting pellets were dissolved in 2 × Laemmli buffer. The proteins were separated on 10% SDS-PAGE gels and analysed by immunoblotting as described before.
Microscopy
For imaging of active Caspase-1, BMDMs were seeded on glass coverslips in 24-well plates at a density of 2.5 × 105 cells/well. LPS-primed cells were treated with FAM-YVAD-FMK FLICA reagent in DMSO or DMSO only for 15 min, followed by inflammasome activation. Afterwards, the cells were washed with PBS, fixed in 4% (w/v) paraformaldehyde and incubated with the rat anti-ASC (1:500) primary antibody in blocking buffer (3% (w/v) BSA and 0.1% (w/v) saponin) for 30 min. After washing, the cells were incubated with an AlexaFluor 546-conjugated secondary antibody (1:250) for 30 min, stained with DAPI before mounting and imaged using a Nikon Eclipse C1 Plus Eclipse confocal microscope. To quantify Caspase-1 activity, random image sections with at least 100 cells were counted for each condition.
Reagents and antibodies
Adenosine 5′-triphosphate disodium salt hydrate (ATP), nigericin sodium salt, bovine catalase (C-40), gentamicin, suberic acid bis(N-hydroxy succinimide ester) (DSS), 2,4-dinitrophenylhydrazine (DNPH) and monoclonal anti-β-Actin (clone AC-74, #A2228; dilution 1:10,000) were purchased from Sigma-Aldrich. Catalase for in vivo experiments was from Worthington Biochemical Corporation. Lipopolysaccharide (LPS-EB, TLR4/TLR2 ligand) was from InvivoGen. Listeriolysin O (LLO) was purified from supernatants of non‐pathogenic Listeria innocua hyperexpressing listeriolysin O as described previously70. H2O2 was from Merck-Millipore. Rat anti-mouse Caspase-1 p20 (Clone 4B4.2.1; used dilution 1:2,000) and rat anti-mouse ASC (CARD5) (Clone 8E4.1; for immunoblotting: dilution 1:2,000, for immunoprecipitation: 2 µg per 500 µg cell lysate, for immunofluorescence staining: dilution 1:500) were obtained from Genentech, San Francisco USA. Goat anti-IL-1β was from R&D Systems (#AF-401-MA; dilution 1:2,500). NLRP3 rabbit monoclonal antibody (D4D8T) was from Cell Signaling Technology (#15101; dilution 1:1,000). Polyclonal rabbit anti-dinitrophenol (DNP) was from Abcam (#AB6306; dilution 1:1,000). Anti-HRP-labelled secondary anti-mouse and anti-rabbit were from GE Healthcare Amersham (#NA931; dilution 1:10,000) and Cell Signaling Technology (#7074; dilution 1:10,000), respectively. The anti-HRP-labelled secondary anti-goat and anti-rat antibodies (#sc-2020 and sc-2006; dilution 1:10,000) and normal rat-IgG control (#sc-2026; used for immunoprecipitation: 2 µg per 500 µg cell lysate) were from Santa Cruz Biotechnology. The AlexaFluor 546 goat anti-rat IgG secondary antibody was from Life Technologies (#A11081; dilution 1:250). ECL substrates and X-ray films were purchased from GE Healthcare Amersham (Hyperfilm ECL). FAM-YVAD-FMK FLICA (dilution 1:150) was from ImmunoChemistry Technologies. Ficoll-Paque PLUS was from GE Healthcare.
Statistical information
The data in the text and Figures are expressed as the mean with the standard error of the mean (±s.e.m.) or if representative measurements of technical replicates are shown as mean with standard deviation (±s.d.). Statistical comparisons were done using either a one–way ANOVA or two-way ANOVA with Bonferroni post-test or Mann Whitney test. P < 0.05 was considered as statistically significant. The survival rates were analysed by the Kaplan-Meier method and P values were determined by Gehan-Breslow-Wilcoxon test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to Jetta J. E. Bijlsma of the University of Groningen, Groningen, Netherlands for providing D39 ΔspxB strain, and to Jeffrey Weiser, New York University School of Medicine, USA for the S. pneumoniae SpxB-complemented strain (D39 ΔspxB + SpxB) and others. We thank, Victor Nizet (University of California, USA) for Streptococcus pyogenes NZ131 (serotype M49), Peter Valentin-Weigand (Tierärztliche Hochschule Hannover, Germany) for Streptococcus suis serotype 2 strain 10 and Jan Oscarsson (Umeå University, Sweden) for Streptococcus oralis ATCC 35037 and Streptococcus sobrinus CCUG 21019. This work was supported by funding from the Laboratory for Molecular Infection Medicine Sweden (MIMS). This work was supported by Umeå University, Stockholm University and Swedish Research Council grants (reference numbers 2015-02857 and 2016-00890) to N.O.G.
Author contributions
N.O.G. conceived, designed and supervised the study. S.F.E. conceived, designed and performed experiments. S.F.E. and N.O.G. prepared the manuscript.
Data availability
All data are available in the manuscript and its Supplementary Information files. Raw source data for Figs. 1–7 and Supplementary Figs. 1–10 are presented in the Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information: Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41467-019-11169-x.
References
- 1.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Zamocky M, Furtmuller PG, Obinger C. Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 2008;10:1527–1548. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid. Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubartelli A. Redox control of NLRP3 inflammasome activation in health and disease. J. Leukoc. Biol. 2012;92:951–958. doi: 10.1189/jlb.0512265. [DOI] [PubMed] [Google Scholar]
- 7.Varga G, Gattorno M, Foell D, Rubartelli A. Redox distress and genetic defects conspire in systemic autoinflammatory diseases. Nat. Rev. Rheuma. 2015;11:670–680. doi: 10.1038/nrrheum.2015.105. [DOI] [PubMed] [Google Scholar]
- 8.Lambeth JD, Neish AS. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu. Rev. Pathol. 2014;9:119–145. doi: 10.1146/annurev-pathol-012513-104651. [DOI] [PubMed] [Google Scholar]
- 9.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J. Bacteriol. 2003;185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overweg K, et al. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect. Immun. 2000;68:4604–4610. doi: 10.1128/IAI.68.8.4604-4610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piroth L, et al. Development of a new experimental model of penicillin-resistant Streptococcus pneumoniae pneumonia and amoxicillin treatment by reproducing human pharmacokinetics. Antimicrob. Agents Chemother. 1999;43:2484–2492. doi: 10.1128/AAC.43.10.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majewska E, et al. Elevated exhalation of hydrogen peroxide and thiobarbituric acid reactive substances in patients with community acquired pneumonia. Respir. Med. 2004;98:669–676. doi: 10.1016/j.rmed.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Ulland TK, Ferguson PJ, Sutterwala FS. Evasion of inflammasome activation by microbial pathogens. J. Clin. Investig. 2015;125:469–477. doi: 10.1172/JCI75254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin S, Brodsky IE. The inflammasome: learning from bacterial evasion strategies. Semin. Immunol. 2015;27:102–110. doi: 10.1016/j.smim.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 16.Cilloniz C, et al. Microbial aetiology of community-acquired pneumonia and its relation to severity. Thorax. 2011;66:340–346. doi: 10.1136/thx.2010.143982. [DOI] [PubMed] [Google Scholar]
- 17.van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374:1543–1556. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 18.Ramos-Montanez S, et al. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol. Microbiol. 2008;67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- 19.Spellerberg B, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 1996;19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 20.Pesakhov S, et al. Effect of hydrogen peroxide production and the Fenton reaction on membrane composition of Streptococcus pneumoniae. Biochim Biophys. Acta. 2007;1768:590–597. doi: 10.1016/j.bbamem.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 2007;189:6532–6539. doi: 10.1128/JB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Syk A, et al. Emergence of hypervirulent mutants resistant to early clearance during systemic serotype 1 pneumococcal infection in mice and humans. J. Infect. Dis. 2014;210:4–13. doi: 10.1093/infdis/jiu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Moltke J, Ayres JS, Kofoed EM, Chavarria-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev. Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 24.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Erttmann SF, et al. Loss of the DNA damage repair kinase ATM impairs inflammasome-dependent anti-bacterial innate immunity. Immunity. 2016;45:106–118. doi: 10.1016/j.immuni.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Fang R, et al. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J. Immunol. 2011;187:4890–4899. doi: 10.4049/jimmunol.1100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 28.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martner A, Dahlgren C, Paton JC, Wold AE. Pneumolysin released during Streptococcus pneumoniae autolysis is a potent activator of intracellular oxygen radical production in neutrophils. Infect. Immun. 2008;76:4079–4087. doi: 10.1128/IAI.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemon JK, Weiser JN. Degradation products of the extracellular pathogen Streptococcus pneumoniae access the cytosol via its pore-forming toxin. MBio. 2015;6:e02110–e02114. doi: 10.1128/mBio.02110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witzenrath M, et al. The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. J. Immunol. 2011;187:434–440. doi: 10.4049/jimmunol.1003143. [DOI] [PubMed] [Google Scholar]
- 32.Harvey RM, et al. The impact of pneumolysin on the macrophage response to Streptococcus pneumoniae is strain-dependent. PLoS ONE. 2014;9:e103625. doi: 10.1371/journal.pone.0103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alouf JE. Cholesterol-binding cytolytic protein toxins. Int J. Med. Microbiol. 2000;290:351–356. doi: 10.1016/S1438-4221(00)80039-9. [DOI] [PubMed] [Google Scholar]
- 34.Palmer M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon. 2001;39:1681–1689. doi: 10.1016/S0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 35.Shoma S, et al. Critical involvement of pneumolysin in production of interleukin-1alpha and caspase-1-dependent cytokines in infection with Streptococcus pneumoniae in vitro: a novel function of pneumolysin in caspase-1 activation. Infect. Immun. 2008;76:1547–1557. doi: 10.1128/IAI.01269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNeela EA, et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog. 2010;6:e1001191. doi: 10.1371/journal.ppat.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark RA. Oxidative inactivation of pneumolysin by the myeloperoxidase system and stimulated human neutrophils. J. Immunol. 1986;136:4617–4622. [PubMed] [Google Scholar]
- 38.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalle-Donne I, et al. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell Mol. Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borutaite V, Brown GC. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Lett. 2001;500:114–118. doi: 10.1016/S0014-5793(01)02593-5. [DOI] [PubMed] [Google Scholar]
- 43.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat. Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 44.Echlin H, et al. Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLoS Pathog. 2016;12:e1005951. doi: 10.1371/journal.ppat.1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho SM, et al. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PLoS ONE. 2013;8:e68277. doi: 10.1371/journal.pone.0068277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taxman DJ, Huang MT, Ting JP. Inflammasome inhibition as a pathogenic stealth mechanism. Cell Host Microbe. 2010;8:7–11. doi: 10.1016/j.chom.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodsky IE, et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wynosky-Dolfi MA, et al. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J. Exp. Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Lopez A, Rosales-Reyes R, Alpuche-Aranda CM, Ortiz-Navarrete V. Salmonella downregulates Nod-like receptor family CARD domain containing protein 4 expression to promote its survival in B cells by preventing inflammasome activation and cell death. J. Immunol. 2013;190:1201–1209. doi: 10.4049/jimmunol.1200415. [DOI] [PubMed] [Google Scholar]
- 50.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell. Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LaRock CN, Cookson BT. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe. 2012;12:799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schotte P, et al. Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1beta. J. Biol. Chem. 2004;279:25134–25142. doi: 10.1074/jbc.M401245200. [DOI] [PubMed] [Google Scholar]
- 53.Galle M, et al. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J. Cell Mol. Med. 2008;12:1767–1776. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Master SS, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cruz CM, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 57.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 58.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauernfeind F, et al. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 2011;187:613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juliana C, et al. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J. Biol. Chem. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Veerdonk FL, et al. Reactive oxygen species-independent activation of the IL-1beta inflammasome in cells from patients with chronic granulomatous disease. Proc. Natl Acad. Sci. USA. 2010;107:3030–3033. doi: 10.1073/pnas.0914795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meissner F, et al. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–1573. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao K, et al. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 2013;23:201–212. doi: 10.1038/cr.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernandez-Cuellar E, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J. Immunol. 2012;189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 65.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J. Biol. Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai Z, Yan LJ. Protein oxidative modifications: beneficial roles in disease and health. J. Biochem. Pharmacol. Res. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]
- 67.Brogden KA, Guthmiller JM, Taylor CE. Human polymicrobial infections. Lancet. 2005;365:253–255. doi: 10.1016/S0140-6736(05)70155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinon F. Signaling by ROS drives inflammasome activation. Eur. J. Immunol. 2010;40:616–619. doi: 10.1002/eji.200940168. [DOI] [PubMed] [Google Scholar]
- 69.Winter S, Hultqvist Hopkins M, Laulund F, Holmdahl R. A reduction in intracellular reactive oxygen species due to a mutation in NCF4 promotes autoimmune arthritis in mice. Antioxid. Redox Signal. 2016;25:983–996. doi: 10.1089/ars.2016.6675. [DOI] [PubMed] [Google Scholar]
- 70.Darji A, et al. Hyperexpression of listeriolysin in the nonpathogenic species Listeria innocua and high yield purification. J. Biotechnol. 1995;43:205–212. doi: 10.1016/0168-1656(95)00138-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and its Supplementary Information files. Raw source data for Figs. 1–7 and Supplementary Figs. 1–10 are presented in the Source Data file.