Abstract
DNA damaging chemotherapies are successful in cancer therapy, however, the damage can be reversed by DNA repair mechanisms that may be up-regulated in cancer cells. We hypothesized that inhibiting RAD51, a protein involved in homologous recombination DNA repair, would block DNA repair and restore the effectiveness of DNA damaging chemotherapy. We used phage-display to generate a novel synthetic antibody fragment that bound human RAD51 with high affinity (KD = 8.1 nM) and inhibited RAD51 ssDNA binding in vitro. As RAD51 is an intracellular target, we created a corresponding intrabody fragment that caused a strong growth inhibitory phenotype on human cells in culture. We then used a novel cell-penetrating peptide “iPTD” fusion to generate a therapeutically relevant antibody fragment that effectively entered living cells and enhanced the cell-killing effect of a DNA alkylating agent. The iPTD may be similarly useful as a cell-penetrating peptide for other antibody fragments and open the door to numerous intracellular targets previously off-limits in living cells.
Subject terms: Protein delivery, Antibody fragment therapy
Introduction
Chemotherapy is a predominant strategy for treating cancer. With the exception of targeted therapies, most chemotherapy works on the premise that drugs causing DNA damage kill rapidly dividing cancer cells better than their normal counterparts. Unfortunately, chemotherapy often fails to eliminate cancer cells due to innate or acquired drug resistance mechanisms that prevent drug activity1,2. Further, normal DNA repair processes can counteract DNA damage-based chemotherapies3, and this mechanism of chemo-resistance is further compounded in cells that have misregulated DNA repair4,5.
Homologous recombination (HR) is a well-conserved and fundamentally important DNA repair pathway that uses homologous undamaged sister chromatids as an error-free template to restore genetic information. HR is typically responsible for double-strand break (DSB) repair, however, the HR machinery is also used for a “last-resort” process to overcome a wide variety of potentially lethal DNA modifications that may be encountered by the DNA replication fork. Various DNA lesions from base modifications and bulky adducts to intra- and inter-strand cross-links may all lead to single-ended DSBs, which if left un-repaired can lead to cell death. This is in contrast to other major DNA repair mechanisms, such as base excision repair, mismatch repair, and non-homologous end joining, which have a comparatively narrow spectrum of target lesions. The broad functions of HR proteins is highlighted by the participation of HR mechanisms in the repair of all major toxic lesions induced by cancer treatments with the exception of anti-metabolites3. HR proteins are therefore central to repairing chemotherapy induced DNA damage.
RAD51 is an essential6 HR recombinase conserved in nature7 that conventionally functions in two important homology-directed DNA repair steps. First, following resection of DSB lesions, RAD51 uses DNA binding8, ATP-binding, and ATPase activities9,10 to form a nucleoprotein filament. Next, the DNA-RAD51 nucleofilament promotes strand invasion11,12 of a homologous DNA duplex to form the characteristic D-loop DNA structure necessary for recombination. These activities are carried out in conjunction with and mediated by several protein-protein interactions13–16. Beyond this classic HR role, RAD51 is also necessary for replication fork reversal and chromatin remodeling17 in S-phase to address a variety of damage-induced lesions encountered during DNA replication18,19. Further, RAD51 is involved in replication fork maintenance by protecting nascent DNA strands from nuclease degradation during replication20–23.
The central activities of RAD51 in DNA repair and replication point toward a role in cancer and cancer therapy chemoresistance. Indeed, RAD51 overexpression is observed in immortal human cancer cell lines24,25 and in breast cancer26, prostate cancer27, pancreatic cancer28, non-small cell lung carcinoma29, and leukemia primary cancer cells30. In addition, RAD51 hyperactivity is thought to contribute fundamentally to the genesis of genome instability and cancer, and RAD51 overexpression leading to hyper-recombination is implicated in malignant transformation31,32. These associated phenotypes are therapeutically relevant, as RAD51 overexpression increases cellular resistance to radiation and chemotherapeutic drugs33–35. Additional connections to cancer have placed RAD51 as a possible biomarker36 and prognostic indicator37. Taken together, RAD51 is a potential therapeutic target that may be particularly effective for patients undergoing chemotherapy.
Several early studies showed success in inhibiting RAD51 for cancer treatments. Depletion of RAD51 by antisense RNA attenuates radiotherapy resistance38,39 and intensifies killing of immortal HeLa cells by cisplatin40. Similarly, targeted RAD51 inhibition using ribozyme treatments increases radiosensitivity41. These studies laid the foundation for targeting RAD51 with several small-molecule inhibitors. Those most extensively studied include DIDS42,43, B0244,45, IBR246, Halenaquinone47, and RI-1/RI-248,49. Together they inhibit various RAD51 activities, including homologous strand pairing and exchange, D-loop formation, ssDNA binding, and dsDNA binding50. Despite limited success in potentiating chemotherapeutic agents, most of these small molecules are limited by poor specificity, instability, and cellular toxicity leading to side-effects in patients. As a result, most RAD51 small molecule inhibitors to date have been limited to in vitro and research purposes51.
With shortcomings for small-molecule chemotherapeutics in treating RAD51-associated cancer, we hypothesized that an entirely different class of drugs might be successful. Therapeutic antibodies are prominent anti-cancer drugs52 that can offer several advantages over small-molecules, including tighter target binding, improved specificity, and longer in vivo half-lives53. However, due to their large size, antibodies and antibody fragments do not effectively enter living cells, and are normally limited to extracellular and cell-surface targets54. In order to inhibit RAD51 with an antibody, we fused a cell-penetration peptide (CPP) called “iPTD” to a RAD51 inhibitory antigen-binding fragment (Fab). The resulting Fab-iPTD was able to penetrate living cells and enhance the cell-killing activity of a DNA alkylating agent.
Results
Generation of a human antigen-binding fragment (Fab) against human RAD51
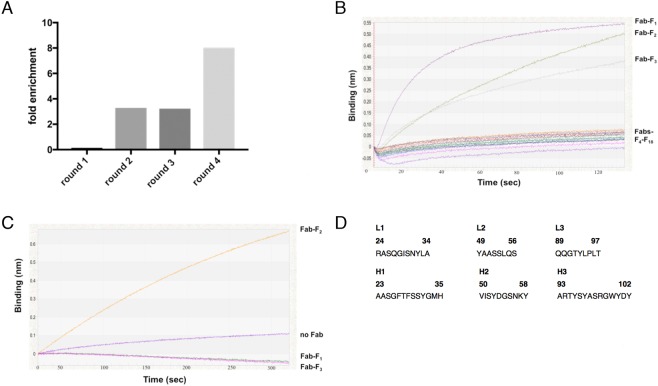
Recombinant human RAD51 was purified from E. coli (Fig. S3) and used as the target for phage display selection of Fab fragments (Fig. 1). We used a novel human IgG1 synthetic Fab phage library containing >1 × 1010 members, referred to as Library S55. Library S was designed with canonical CDRs to structurally complement randomized CDRL3 and CDRH355. Phage display selections were performed against surface-immobilized recombinant RAD51 and a modest enrichment of target binding phage over negative BSA controls was observed after four rounds of selection (Fig. 1A). The pool of Fab-encoding sequences from the fourth round of selection was sub-cloned from phage for small-scale Fab expression in E. coli. Eighteen Fab lysates were screened for Fab expression through successful loading (i.e. immobilization) onto Protein A biosensors using biolayer interferometry (BLI). Of three lysates with loaded Fab (Fig. 1B), only one subsequently bound RAD51 at a 500 nM screening concentration (Fig. 1C). This clone, Fab-F2, was sequenced (Fig. 1D) and purified to near homogeneity (Fig. S1).
Figure 1.
Selection of a Fab binding to RAD51. (A) Four rounds of phage display selection were performed and fold enrichment was calculated as eluted phage from target wells containing immobilized RAD51 divided by BSA negative control wells. (B) E. coli lysates from 18 random clones over-expressing Fab were sampled in parallel using OctetRED384 biolayer interferometry. The presence of expressed Fab is detected by binding to Protein A biosensors which causes an increase in optical thickness or binding (nm) over time. (C) Fabs immobilized to protein L biosensors are transferred to wells containing 500 nM RAD51 and the resulting association curves are shown. (D) Fab-F2 CDR sequences. The Kabat scheme was used for numbering amino acids and the CDR regions shown were defined according to North et al.91.
Kinetics and specificity of Fab-F2 binding to RAD51
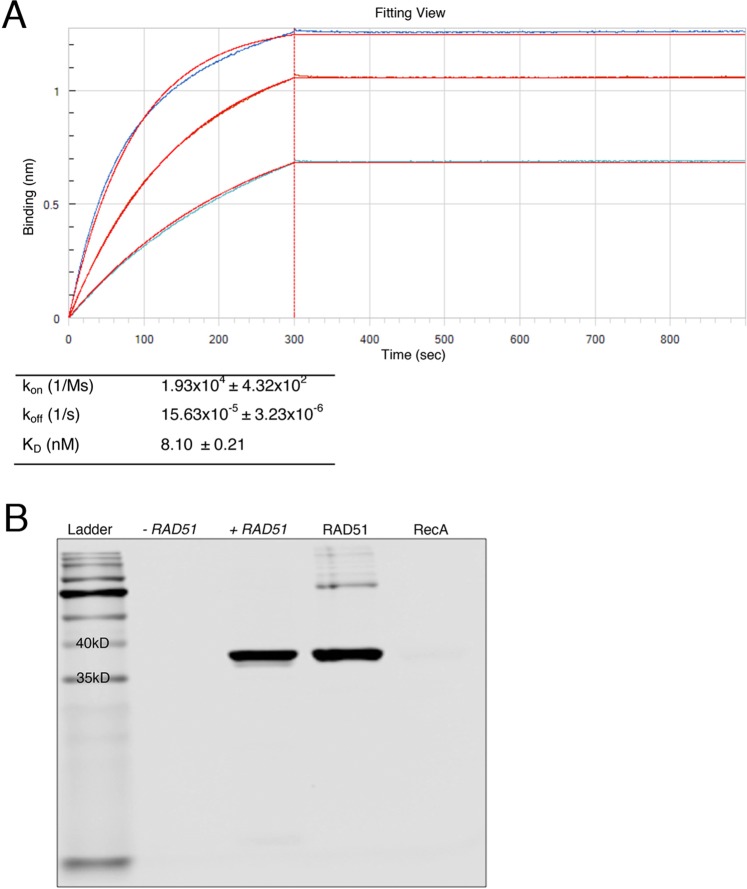
BLI was used to determine kinetics of Fab-F2 binding to RAD51 (Fig. 2A). Protein L biosensors were used to immobilize Fab-F2 and three concentrations of RAD51 were used to measure the association and dissociation phases of the interaction. Binding curves were fit globally to a 1:1 binding model, which exhibited a tight fit and yielded an on-rate (kon) of 1.93 × 104 1/Ms ± 4.32 × 102, an off-rate (koff)of 15.63 × 10−5 1/s ± 3.23 × 10−6, and a dissociation constant (KD) of 8.10 nM ± 0.21.
Figure 2.
Kinetics and specificity of Fab-F2. (A) Kinetic parameters for the binding of Fab-F2 to purified RAD51 were determined using biolayer interferometry. Equal amounts of Fab-F2 were immobilized on Protein L biosensors which were immersed in parallel to a range of RAD51 concentrations and then buffer alone to yield association and dissociation curves (blue traces), respectively. Global curve-fitting employing a 1:1 Langmuir binding model (red traces) using reference-well subtracted data was used to determine the kinetic values in the table. (B) A Western blot using IRDye 800CW-labeled Fab-F2 was used to detect expression of plasmid-based RAD51 from E coli lysates with (+) or without (−) induction by IPTG. Equal amounts of purified RAD51 and the homologue RecA were used to verify size and specificity, respectively. Minor aggregation of purified RAD51 and/or RAD51-DNA impurities was observed (upper bands). The lower ladder band indicates the gel dye-front.
E. coli RecA is a structural and functional homologue of RAD51 that shares 51% sequence similarity over its core-domain and a conserved homologous recombination function in DNA strand pairing and exchange56,57. To test binding specificity, Western analysis using Fab-F2 against equal amounts of purified RAD51 and RecA was performed. Fab-F2 was able to detect RAD51 but not RecA (Fig. 2B). Further specificity was observed through the detection of a single band in E. coli whole cell lysates containing plasmid-expressed RAD51 versus the control expression plasmid (Fig. 2B).
Fab-F2 inhibits RAD51 DNA binding but not ATPase activity in vitro
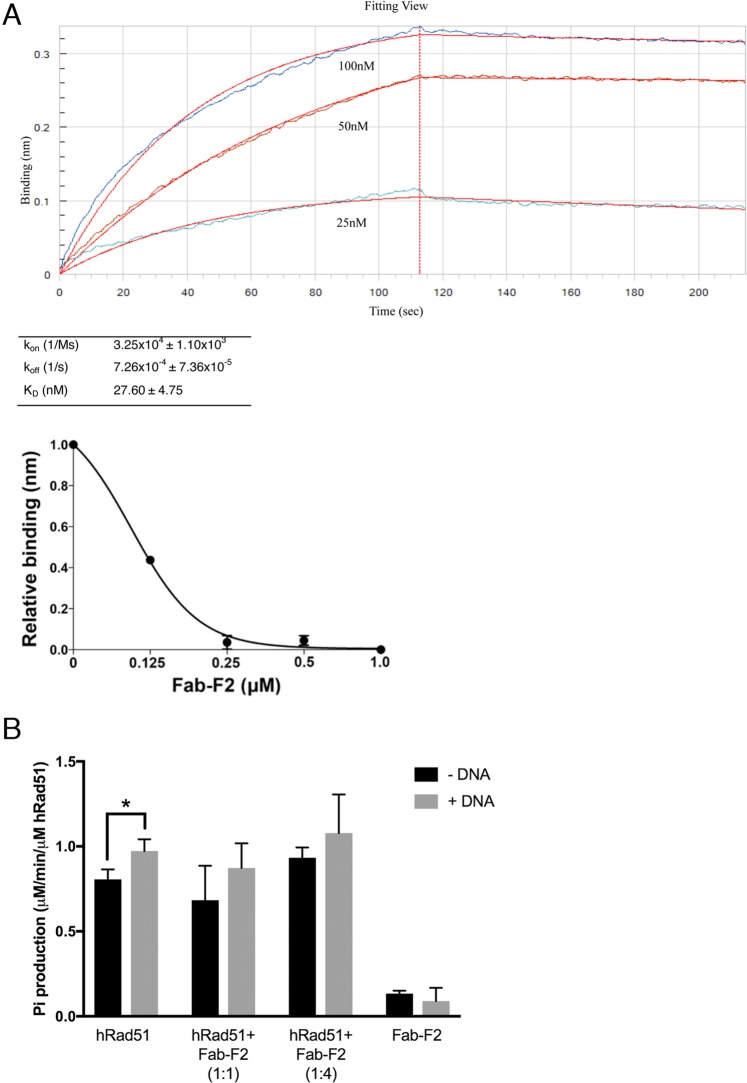
RAD51 posses several activities for DNA repair. In its role in HR, RAD51 binding to ssDNA is an integral part of DNA nucleoprotein filament formation, which is essential for the creation of a pre-synaptic filament/complex58–60. ssDNA binding by RAD51 is also a requirement for its role in protecting the DNA replication fork22. To test Fab-F2 inhibition of RAD51 ssDNA binding, we established a BLI DNA-binding assay. We first determined the KD of the RAD51-ssDNA interaction by immobilizing 5′-biotinylated poly-deoxythymidine (oligo(dT)36) onto a streptavidin biosensor and measuring RAD51 binding (Fig. 3A). The ssDNA-RAD51 KD was 27.60 ± 4.75 nM, which is similar to the previously reported KD for the interaction of RAD51 with oligo(dT)5061. To measure Fab-F2 inhibition of the ssDNA-RAD51 interaction, we repeated BLI in a similar manner, except with pre-incubation of 500 nM RAD51 with various concentrations of Fab-F2. This enabled measurement of RAD51 ssDNA binding inhibition by subtraction of binding signals without Fab-F2. Using this approach, an IC50 for the Fab-F2 was calculated to be less than 125 nM (Fig. 3A).
Figure 3.
Inhibition of RAD51 activities in vitro by Fab-F2. (A) Biolayer interferometry was used to assay the inhibition of RAD51 ssDNA binding by Fab-F2. First, the kinetics of RAD51 binding to DNA was determined (upper panel) using streptavidin-immobilized 5′-biotinylated oligo(dT)36 biosensors which were immersed in parallel to a range of RAD51 concentrations and then buffer alone to yield association and dissociation curves (blue traces), respectively. Global curve-fitting employing a 1:1 Langmuir binding model (red traces) using reference-well subtracted data was used to determine the kinetic values in the table. In the lower panel, the experiment was repeated using 0.5 μM RAD51 so that maximal binding at equilibrium in the absence of Fab-F2 (expressed as 1.0) could be plotted relative to binding in the presence of a range of Fab-F2 concentrations. Error bars indicate standard deviation from three independent experiments. (B) The effect of Fab-F2 on ATP hydrolysis by RAD51 was measured with a malachite green assay that enables spectrophotometric detection of free phosphates. The free inorganic phosphates produced by RAD51 (μM/min/μM RAD51) from ATP were determined in reactions containing RAD51 or Fab-F2 alone and in combination at the indicated ratios in the presence and absence of ssDNA. Error bars represent standard deviation from at least three independent measurements. *p-value 0.034.
RAD51 binds ATP to promote its own stable nucleofilament formation10,62. Following strand invasion and homology searching functions, RAD51 then hydrolyzes ATP through its ATPase domain, causing its release from the synapse11,12,63. To investigate whether Fab-F2 inhibits RAD51 ATPase activity, we monitored the release of inorganic phosphate by ATP hydrolysis over time using a malachite green assay64. The lower than anticipated yet statistically significant stimulation of ATPase activity in the presence of ssDNA may have been due to possible residual nucleic acid in the RAD51 preparations, as indicated in Fig. 2B. Nonetheless, the RAD51 ATPase activity was not inhibited by Fab-F2 to any extent in the presence or absence of ssDNA (Fig. 3B).
Intracellular expression of a Fab-F2 scFv fragment inhibits HEK293T cell growth
To determine the potential for Fab-F2 to affect RAD51 function in cells, we sub-cloned the light and heavy chain variable regions of Fab-F2 as a single chain variable fragment (scFv) that could be expressed inside cells. An scFv is encoded by a single gene that brings together the antigen-binding variable heavy and light chains with a flexible poly-linker65,66. To improve intracellular function, the scFv construct was fused to a fragment crystallizable (Fc) domain to enhance stability and a nuclear localization signal to promote nuclear uptake67. A cell import tag (iPTD, see below) was also added to streamline with prospective recombinant Fab-F2-iPTD. The construct was then cloned into the mammalian constitutive expression plasmid pcDNA and transiently transfected into HEK293T cells. scFv-Fc expression was confirmed by Western analysis of cell lysates (Fig. S2).
To test scFv-F2-Fc-iPTD on RAD51 DNA repair function in cells, we used the alkylating agent methylmethane sulfonate (MMS) to induce DNA damage and replication fork impairment68,69. The mutation of genes involved in homologous recombination DNA repair, such as RAD51, cause sensitivity to the alkylating agent MMS70. In addition, we expected MMS-induced replication fork impairment would impinge upon the role of RAD51 in HR-independent DNA replication fork fidelity22. Lastly, MMS induces RAD51 foci formation71 and is relevant to our overall goal in treating cancer with Fab-F2 because alkylating agents are commonly used in chemotherapy3.
We used a clonogenic survival assay to test transiently transfected HEK293T cells for colony growth as quantified by light microscopy72. Cells were seeded at 200 per well and treated with MMS the next day. Following culture for seven days, cells were then stained in order to count viable colony growth as a measure of tolerance to MMS. The pcDNA-SCFV-F2-FC-IPTD construct had a strong effect in preventing HEK293T cells to form colonies, independent of MMS treatment (Fig. 4).
Figure 4.
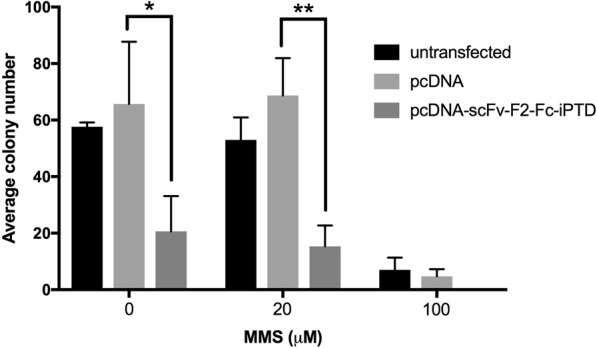
Cells in culture are sensitive to an scFv-Fc intracellular antibody based on Fab-F2. A clonogenic assay was used to test the effect of an intracellular-expressed Fab-F2 derivative. HEK293T cells were transiently transfected with a pcDNA-SCFV-F2-FC-IPTD expression plasmid and exposed to methyl methanesulfonate (MMS). Cells were cultured for 7 days post MMS treatment and colonies were stained with 0.3% crystal violent and enumerated by light microscopy. Error bars indicate standard deviation from three independent measurements. *p-value = 0.037 **p-value = 0.0036.
Fusion of a novel intracellular protein delivery domain, iPTD, with Fab-F2 does not block RAD51 binding
We fused a novel intracellular protein delivery domain iPTD to Fab-F2 for cell internalization. The iPTD (Patent No. WO 2014005219 A1) consists of a 35 amino acid sequence (MALGPCMLLLLLLLGLRLPGVWAPPRRRRRRRRR) that enhances interaction with the cell membrane and incorporates features from each major CPP class: cationic, hydrophobic, and amphipathic73. A design feature of the iPTD is enhanced retrograde transport that, similar to immunotoxins and intracellular pathogens, is an efficient and common mechanim relevant to almost any cell type74,75. The iPTD also contains a cleavage inhibition sequence (CIS) that enables retention of iPTD-fused cargo with retrotranslocon post cell membrane interactions to facilitate internalization
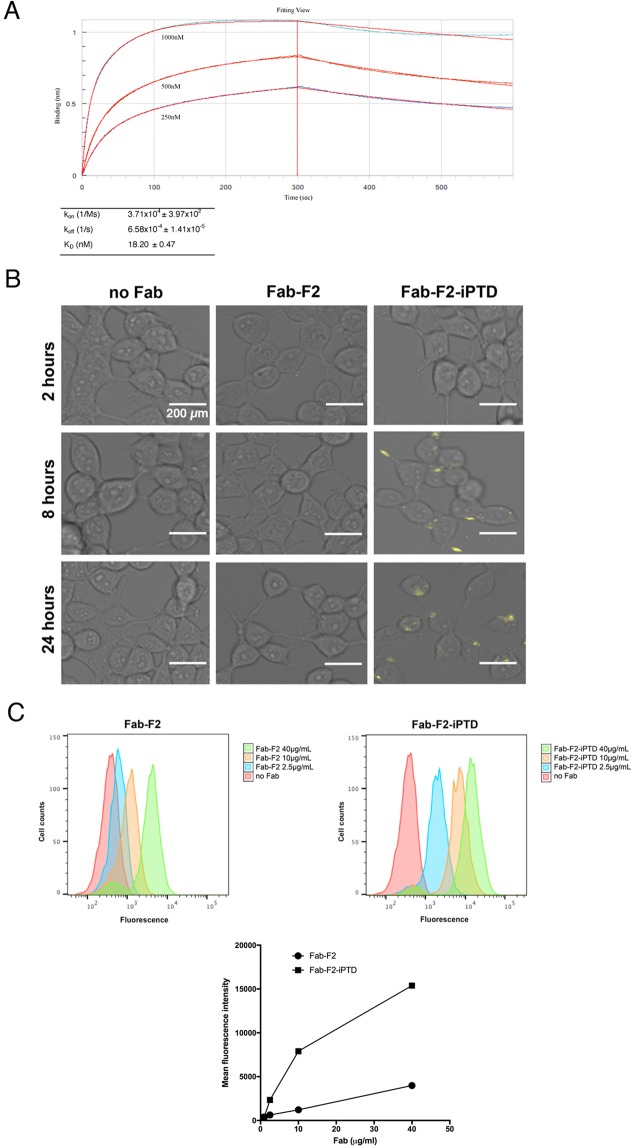
The iPTD-encoding sequence was cloned in-frame to the heavy chain C-terminus of Fab-F2. This construct, Fab-F2-iPTD, was expressed and purified from E. coli with yield and purity similar to Fab-F2 (Fig. S1). To verify that the iPTD did not interfere with RAD51 binding, we determined kinetics of Fab-F2-iPTD binding to RAD51. The iPTD had a modest effect on the Fab-F2 interaction with RAD51, with on- and off-rates lowered by approximately 2-fold (Fig. 5A vs. Figure 2A). The KD for Fab-F2-iPTD was 18.20 nM as compared to 8.10 nM for Fab-F2.
Figure 5.
A cell-penetrating Fab-F2-iPTD binds to RAD51. (A) Kinetic parameters for the binding of Fab-F2-iPTD to purified RAD51 were determined using biolayer interferometry. Equal amounts of Fab-F2-iPTD were immobilized on Protein L biosensors which were immersed in parallel to a range of RAD51 concentrations and then buffer alone to yield association and dissociation curves (blue traces), respectively. Global curve-fitting employing a 1:1 Langmuir binding model (red traces) using reference-well subtracted data was used to determine the kinetic values in the table. (B) HEK293T cells were incubated with 40 µM 800CW-labeled Fab (yellow) for the indicated time points and fluorescent microscopy was used to visualize cellular localization. Image contrast and brightness adjustments were performed equally across all panels. Bars indicate 200 µm. (C) Flow cytometry was used to quantify relative Fab internalization. The mean fluorescence intensity was measured for HEK293T cells treated with 800CW-labeled Fab for 24 hours (upper panels) and then plotted for comparison (bottom panel). Error bars, while not visible, were added to indicate standard deviation from three independent measurements.
Fab-F2-iPTD exhibits enhanced internalization into HEK293T cells
To compare the internalization of Fab-F2-iPTD versus Fab-F2 in HEK293T cells, we labeled Fab-F2 and Fab-F2-iPTD with the near-infrared dye IRDye®800CW, and analyzed cellular localization by fluorescent microscopy. A fluorescent signal could be observed for Fab-F2-iPTD after only eight hours following addition to the cell culture media, with predominant staining at the plasma membrane (Figs 5B and S4). After 24 hours, Fab-F2-iPTD entered cells and exhibited intracellular foci with little to no staining at the plasma membrane. No corresponding fluorescent signal for Fab-F2 was observed. To quantify the relative uptake of Fab-F2-iPTD and Fab-F2, HEK293T cells treated with Fab at the 24-hour time-point were measured by flow cytometry to detect cell-bound fluorescence. Over all concentrations, cells treated with Fab-F2-iPTD exhibited peak shifts greater than cells treated with Fab-F2 (Fig. 5C). The difference was most obvious in a plot of mean fluorescence intensity versus concentration (Fig. 5C), where the Fab-F2-iPTD signal sharply increases at lower concentrations. In contrast, Fab-F2 fluorescence increased linearly in a manner typical of background binding and/or non-specific uptake.
Fab-F2-iPTD increases the sensitivity of HEK293T cells to MMS
We tested Fab-F2-iPTD intracellular function by measuring its ability to enhance the activity of MMS in the clonogenic survival assay. HEK293T cells were treated with purified Fab-F2 or Fab-F2-iPTD in the presence and absence of MMS, and the resulting cell colonies were counted after 7 days. We observed no direct inhibition of clonogenicity for either Fab treatment lacking MMS. However, following MMS treatment, cells exposed to Fab-F2-iPTD at 40 μM and 10 μM caused a significant reduction in colony formation in comparison to Fab-F2 at 40 μM, which had no measurable effect on MMS-induced cell death (Fig. 6).
Figure 6.
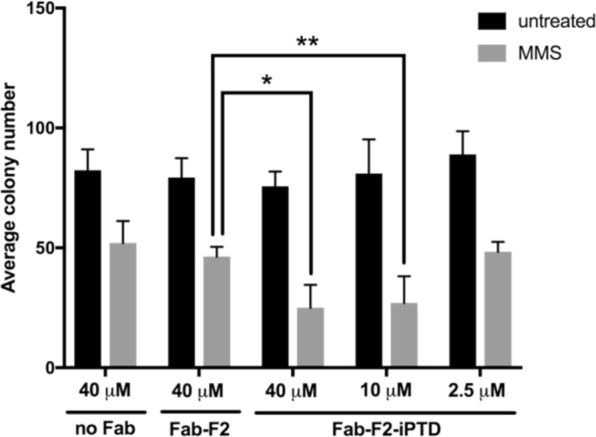
Fab-F2-iPTD increases cell sensitivity to MMS. A clonogenic assay was used to test the effect of adding purified Fab-F2 or -F2-iPTD protein to HEK293T cells treated with MMS at the indicated concentrations. Cells were cultured for seven days post treatment and colonies were then stained with 0.3% crystal violent and enumerated by light microscopy. Error bars represent standard deviation from three independent measurements. No statistical difference exists between untreated samples. *p-value = 0.023 **p-value = 0.042.
Materials and Methods
Plasmids
The coding sequence of Homo sapiens RAD51 was a gift from Richard Fishel at Ohio State University. The coding sequence for residues 21–339 of RAD51 was inserted between the NcoI and XhoI sites of plasmid pET28a (Novagen). The expressed RAD51 protein contained 8 extra amino acid residues (Leu-Glu-His-His-His-His-His-His) at the C-terminus.
To create pCW-Fab-F2 and -iPTD, Fab fragments were PCR amplified from phagemid clones using primers TGS157 (5′-TCCAGATGACCCAGTCCCCGAGCTCCCTG) and TGS160 (5′-CAAATCTTGTGACAAAACTCACACGGGTGGTTCGCACCACCACCACCACCACTGAG). The Fab DNA sequences were cloned by Gibson Assembly76 into a modified pCW-LIC (Addgene plasmid #26098) plasmid containing the 5′ portion of the Fab light chain followed by a SacI restriction site, and a 3′ portion containing an XhoI restriction site followed by a poly-His-encoding sequence or a poly-His-encoding sequence preceded by the iPTD encoding sequence (5′-ATGGCC TTGGGCCCTTGCATGTTGTTGTTGTTGTTGTTGTTGGGTTTGCGCCTGCCGGGTGTTTGGGCGCCGCCGCGTCGCCGCCGCCGTCGTCGCCGTCGT).
The gene sequence encoding scFv-F2-iPTD was synthesized in a pUC57 plasmid and subcloned directly (via restriction digestion with KpnI and BamHI) from pUC57 and ligated into the KpnI-BamHI linearized vector of an antibody-expressing pcDNA3.1(+) plasmid containing the Fc-NLS domain. The NLS domain is from the nuclear localization sequence (EGMLANLVEQNISVRRRQGVSIGRLHKQRKPDRRKRSRPYKAKRQ) at the C-terminal end of ubiquitin carboxyl-terminal hydrolase BAP1.
RAD51 purification
E. coli Rosetta(DE3) (Novagen) cultures harbouring pET28a-RAD51 were grown at 37 °C until an OD600nm of 1.2, and were then induced with 0.25 mM IPTG for four hours. Cells were harvested by centrifugation and pellets were resuspended in binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 25 mM imidazole at pH 7.4). Following sonication lysates were clarified by centrifugation at 12,000 g for 15 minutes. RAD51 was precipitated in a final concentration of 0.35 g/mL ammonium sulfate. Following centrifugation at 12,000 g for 15 minutes, the protein pellet was dissolved in binding buffer and passed through a DE52 anion exchange column (Sigma-Aldrich) to remove DNA. A HiTrap Ni-chelating column (GE Healthcare) was used as a final purification step per manufacturer’s instructions and eluted with 500 mM imidazole. Fractions containing RAD51 protein were dialyzed with PBS. Protein concentrations were determined using PierceTM BCA Protein Assay Kit.
Phage display
Phage display selection with synthetic Fab Library S as described in55 was performed by immobilizing 5 μg/mL of purified RAD51 on Nunc MaxiSorp Immunoplates, and 1013 total phages each selection round were incubated with RAD51 for 2 hours. Unbound phages were removed with eight washes of PBS and target-bound phages were eluted with acid. Enrichment of target-binding phages was calculated each round by titering the number of phage eluted from RAD51 wells divided by the BSA negative control wells performed in parallel.
Fab purification
Fabs were purified as described in55.
Small-scale lysate preparation
Soluble protein was extracted from small-scale E. coli cultures (<5 mL) using B-PER Bacterial Protein Extraction Reagent (ThermoScientific) per manufacturer’s instructions employing a 10 minute room temperature incubation with gentle rotation. The lysate supernatant was separated from insoluble material by centrifugation at 12,000 g for 5 minutes at 4 °C.
Western blot
HEK293T whole-cell lysates were prepared with SDS sample buffer followed by incubation at 95 °C for 10 minutes. Proteins were separated with 12% SDS-PAGE and blotted onto nitrocellulose. Following one hour blocking with LiCor Odyssey Blocking Buffer, the membrane was incubated for one hour in 1:15,000 anti-hIgG 800CW in Odyssey Blocking Buffer. The membrane was then rinsed with PBS and imaged using a LiCor Odyssey-CLx with auto-exposure settings.
Biolayer interferometry
The OctetRED384 (ForteBio Inc) was used for label-free measurements of binding as detected on biosensors as a function of optical thickness (nm) versus time as described in55. All assay steps were performed at 25 °C with 1000 RPM stirring in tilted-bottom 384-well plates (ForteBio Inc) containing 80 μL sample volume. PBS (pH 7.4) containing 0.1% Tween 20 and 10 mg/mL bovine serum albumin (BSA) was used as the kinetic assay buffer. For kinetic determination, approximately 0.5 nm optical thickness Fab was loaded onto Protein L biosensors. Next, an association phase was performed over a range of RAD51 concentrations for 2–3 minutes. Biosensors were then moved to kinetics buffer alone to measure the rate of dissociation. Association and dissociation rates and dissociation constants were calculated using ForteBio Data Analysis version 7.1 curve-fitting software with a 1:1 Langmuir binding model. A reference well with buffer alone was subtracted from all values to account for sensor drift.
To calculate the IC50 for Fab inhibition of RAD51 DNA binding, 1 nm optical thickness of 5′-biotinylated oligo(dT)36 was loaded onto a streptavidin biosensor from a 1 μM solution. Association to 0.5 μM RAD51 in the absence of Fab-F2 was performed to yield maximal binding at equilibrium, from which the relative binding at equilibrium of parallel wells containing Fab-F2 could be subtracted.
ATPase assay
2 μM RAD51 was incubated at 37 °C with Fab-F2 in 100 μL of 50 mM HEPES buffer (pH 7.4) containing 1 mM MgCl2, 45 mM NaCl, 3% glycerol, 0.6 mM 2-mercaptoethanol, 1 mM dithiothreitol, 30 μM EDTA, and 0.1 mg/mL bovine serum albumin (BSA), in the presence of 20 μM oligo(dT)36. After 10 minutes, the reaction was initiated by addition of 50 mM ATP. The 20 μl reaction was quenched after 20 minutes with 30 μl of 100 mM EDTA. The release of inorganic phosphate by ATP hydrolysis was measured with the malachite green assay described in77 modified from64.
Protein labeling
500 ug of Fab or anti-IgG antibody was was dissolved in 600 μL of PBS buffer at room temperature and mixed with 1.35 μL of 10 mg/mL IRDye800CW for 2 hours followed by 16 hours overnight at 4 °C in the absence of light. Free IRDye800CW was removed from the protein conjugates using Zeba® Desalting Spin Columns (Thermo Scientific) per manufacturer’s instructions. IRDye800CW/Fab ratio and Fab concentration were determined by measuring the absorbance of the conjugate at 280 nm and 780 nm using a UV-Vis spectrophotometer, and calculated per the IRDye 800CW Protein Labeling Kit.
Cell culture and transfection
Mammalian cell cultures were maintained at 37 °C with 5% CO2. HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium. Cells were passaged to 10–20% confluency for general maintenance once they reached 80–90%. pcDNA-SCFV-FC was transfected into HEK293T cells using Lipofectamine 2000® Reagent (Invitrogen) per manufacturer’s instructions. Cells were cultured for 48 hours and then collected for experimentation. A portion of the sample was lysed for Western analysis to confirm the presence of scFv-Fc expression.
Clonogenic survival assay
Clonogenic survival assay was used to test the sensitivity of HEK293T cells to increasing doses of a DNA-damaging agent in the presence of Fab or scFv-Fc72. HEK293T cells were trypsinized and reseeded in a 6-well tissue culture plate at 200 cells/well. Following overnight culture, cells were treated with indicated concentrations of MMS alone, or in combination with the indicated amount of Fab-F2 or Fab-F2-iPTD. For the scFv-Fc, cells transfected with pcDNA-SCFV-FC were seeded in a 6-well tissue culture plate at 200 cells/well. Following overnight culture, MMS was added at indicated concentrations. After 7 days, cells were fixed and stained using staining solution (0.3% crystal violet, 50% methanol in PBS). Colonies were counted using light microscopy (EVOS® FL Cell Imaging System, ThermoFisher Scientific). Cells treated with PBS and DMSO, or empty vector transfected cells were used as negative controls.
Fluorescence imaging and flow cytometry
Fab internalization into HEK293T cells was analyzed using fluorescent microscopy (EVOS® FL Cell Imaging System, Cy7 light box, ThermoFisher Scientific). Cells were seeded in a 48-well plate at 5 × 104 cells/well, and IRDye800CW dye-labeled Fab was added to the culture medium as indicated. For imaging, cells in each well were washed with 300 µL PBS. For quantitative analysis of fluorescent Fab uptake, cells were detached from wells with TripLE Express (ThermoFisher Scientific), washed twice with PBS, and the internalized fluorescence signal was measured using a Gallios Flow Cytometer (Beckman Coulter, Inc.). The IRDye800CW dye was excited with the 640 nm laser and fluorescence emission was monitored using a 755 LP filter. Five thousand cells were measured for each sample. Untreated cells were used to set the gate on live cells and mean fluorescence intensity for the gated cells was reported.
Statistical analysis
P-values were determined using an unpaired t test using SPSS 16.0 (SPSS, USA).
Discussion
In this study we used phage-display to generate a novel synthetic Fab targeting human RAD51. Fab-F2 bound RAD51 with high-affinity (KD = 8.10 nM) and inhibited RAD51 ssDNA binding in vitro. The corresponding intracellular scFv caused growth impairment for HEK293T cells transfected with an scFv-Fc intrabody construct. To enable crossing of the cell membrane, we fused Fab-F2 to a novel cell-penetrating peptide, iPTD. Fab-F2-iPTD was easily purified and retained strong binding (KD = 18.20 nM) to RAD51. As compared with Fab-F2, Fab-F2-iPTD was readily imported into HEK293T cells and exerted an intracellular phenotype related to RAD51 inhibition by enhancing the cell killing of the DNA alkylating agent MMS. To our knowledge, this is the first report of a completely RAD51-specific inhibitory antibody fragment. Furthermore, the iPTD fusion Fab opens the opportunity for its use as a viable companion drug to enhance DNA-damage based chemotherapies, and to help treat cancers prone to chemotherapy resistance via homologous recombination mechanisms.
While our study was in progress, another group reported a cell-penetrating antibody with affinity for human RAD5178. They investigated a characteristic autoantibody (3E10) from the autoimmune disease systemic lupus erythematosus, and found that it possesses dual specificity to bind RAD51 in addition to its well-known binding of DNA79. Significantly, the anti-DNA component is absolutely required for cell-penetration and 3E10 scFv fragments with DNA-binding mutations were unable to penetrate cells and inhibit RAD5178. The authors suggest that 3E10 DNA binding is non-toxic, however, it remains to be seen how its connection to a human disease may lead to success in clinical trials. The KD for 3E10 binding RAD51 was 388 nM, and 612 nM for a corresponding scFv antibody fragment, which is over 20-fold weaker as compared with the affinity of Fab-F2-iPTD for RAD51 (18.2 nM) reported in this study. Despite these limitations, 3E10 has attracted interest in clinical development and underscores the significance of our Fab-F2-iPTD.
Cell penetrating peptides are of growing therapeutic interest as they not only enable cellular import, but also facilitate cargo delivery into difficult to penetrate tissues such as tumors, and even allow crossing of the blood-brain barrier73,80. Well over 1000 unique CPPs have been experimentally tested to date81, and have been used to transport a wide variety of drugs and macromolecules including proteins, nucleic acids, and lipids82,83. However, relatively few CPPs reported to date have been used for antibodies and antibody fragments (e.g.)84–86. One reason may be because most CPPs are chemically conjugated and site-directed modifications are more difficult with large antibodies versus small molecules, leading to undesired functional compromises. For example, a study using an scFv chemically coupled to a well-characterized TAT CPP87 led to severely reduced tumor targeting performance as compared to the corresponding unconjugated antibody86. Our iPTD CPP is a genetic fusion and is therefore translated along with the antibody fragment in a 1:1 site-specific manner. We expect the iPTD will be useful for other antibody fragments selected from our synthetic library based on the 4D5 human IgG1 Fab framework, and may also be successful with other antibodies sharing the same scaffold88.
Our overall goal was to create a therapeutic antibody that inhibits the intracellular protein RAD51. To demonstrate that F2 antibody fragments have the potential to disrupt RAD51 function, we first tested an intracellular antibody based on Fab-F2 and found a strong growth inhibitory phenotype in the absence of exogenous DNA damage treatment. We suspect this phenotype is associated with the more recently identified role for RAD51 in protecting nascent DNA strands during replication22,23,89. The phenotype was not observed with treatments using purified Fab-F2 or Fab-F2-iPTD, possibly due to the transient nature of these treatments as compared with constitutive expression of the intrabody and/or the presence of an NLS in the transfected construct. Nonetheless, the phenotype exerted by the scFv-Fc expression construct could be a valuable research tool to study DNA damage-independent processes of RAD51 by reverse genetics. Such reagents are especially useful because RAD51 deletion mutants cause embryonic lethality6,90.
Supplementary information
Acknowledgements
This work was made possible through Western Diversification funding #12939.
Author Contributions
L.P., Y.F. and C.R.G. conceived the idea. C.R.G. supervised the entire study. L.P. and Y.F. conducted the experiments and analyzed the results with R.G. K.L. and L.L. created the iPTD. Y.L. provided the expression plasmid and supervised protein purification and ATPase experiments. J.F.D. supervised cell culture and flow cytometry experiments. L.P. wrote the manuscript with additions from C.R.G., K.L. and L.L. All authors made manuscript revisions.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
L.L. and K.L. have financial interests. L.L. and K.L. are employees of Iprogen Biotech Inc. They provided the pcDNA-SCFV-FC plasmid for this study. Iprogen Biotech has a patent for the iPTD peptide technology. Patent information: Applicant: IPROGEN BIOTECH INC. Inventors: LEE, Keun Ho; LIN, Leo Yen-Cheng; WANG, Aikun International Application Number: PCT/CA20 13/0006 14. Status of Application: International Publication Date 9 January 2014 (09.01.2014) International Publication Number: WO 2014/005219 Al. Specific aspect of manuscript covered in patent application: The manuscript used the iPTD amino acid sequence from the patent.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Landon Pastushok and Yongpeng Fu contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47600-y.
References
- 1.Alfarouk, K. O. et al. Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell International15 (2015). [DOI] [PMC free article] [PubMed]
- 2.Mellor HR, Callaghan R. Resistance to chemotherapy in cancer: A complex and integrated cellular response. Pharmacology. 2008;81:275–300. doi: 10.1159/000115967. [DOI] [PubMed] [Google Scholar]
- 3.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 4.Broustas CG, Lieberman HB. DNA Damage Response Genes and the Development of Cancer Metastasis. Radiat. Res. 2014;181:111–130. doi: 10.1667/RR13515.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nature Reviews Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 6.Tsuzuki T, et al. Targeted disruption of the RAD51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Semidominant mutations in the yeast RAD51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 1996;16:4782–9. doi: 10.1128/MCB.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namsaraev EA, Berg P. Binding of RAD51p to DNA. Interaction of RAD51p with single and double- stranded DNA. J. Biol. Chem. 1998;273:6177–6182. doi: 10.1074/jbc.273.11.6177. [DOI] [PubMed] [Google Scholar]
- 9.Li X, et al. RAD51 and Rad54 ATPase activities are both required to modulate RAD51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung CW, Fortin GS, Peterson SE, Symington LS. The RAD51-K191R ATPase-Defective Mutant Is Impaired for Presynaptic Filament Formation. Mol. Cell. Biol. 2006;26:9544–9554. doi: 10.1128/MCB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51. protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 12.Baumann P, Benson FE, West SC. Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/S0092-8674(00)81394-X. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama T, Kantake N. Dynamic Regulatory Interactions of RAD51, Rad52, and Replication Protein-A in Recombination Intermediates. J. Mol. Biol. 2009;390:45–55. doi: 10.1016/j.jmb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human RAD51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raschle M, Van Komen S, Chi P, Ellenberger T, Sung P. Multiple interactions with the RAD51 recombinase govern the homologous recombination function of Rad54. J. Biol. Chem. 2004;279:51973–51980. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- 17.Falbo KB, et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zellweger Ralph, Dalcher Damian, Mutreja Karun, Berti Matteo, Schmid Jonas A., Herrador Raquel, Vindigni Alessandro, Lopes Massimo. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. The Journal of Cell Biology. 2015;208(5):563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature Reviews Molecular Cell Biology. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 20.Quinet A, Lemaçon D, Vindigni A. Replication Fork Reversal: Players and Guardians. Molecular Cell. 2017;68:830–833. doi: 10.1016/j.molcel.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. RAD51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol. Cell. Biol. 1997;17:7151–7158. doi: 10.1128/MCB.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raderschall E, et al. Elevated levels of RAD51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- 26.Maacke H, et al. Over-expression of wild-type RAD51 correlates with histological grading of invasive ductal breast cancer. Int. J. Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::AID-IJC11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Mitra A, et al. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009;55:696–704. doi: 10.1111/j.1365-2559.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maacke H, et al. DNA repair and recombination factor RAD51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 29.Qiao G-B, et al. High-level expression of RAD51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br. J. Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christodoulopoulos G, et al. Chlorambucil induction of HsRAD51 in B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 1999;5:2178–2184. [PubMed] [Google Scholar]
- 31.Klein HL. The consequences of RAD51 overexpression for normal and tumor cells. DNA Repair. 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flygare J, et al. Effects of HsRAD51 overexpression on cell proliferation, cell cycle progression, and apoptosis. Exp. Cell Res. 2001;268:61–69. doi: 10.1006/excr.2001.5265. [DOI] [PubMed] [Google Scholar]
- 33.Vispé S, Cazaux C, Lesca C, Defais M. Overexpression of RAD51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannay, J. A. F. et al. RAD51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol. Cancer Ther (2007). [DOI] [PubMed]
- 35.Brown, E. T. & Holt, J. T. RAD51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol. Carcinog (2009). [DOI] [PMC free article] [PubMed]
- 36.Nagathihalli NS, Nagaraju G. RAD51 as a potential biomarker and therapeutic target for pancreatic cancer. Biochimica et Biophysica Acta - Reviews on. Cancer. 2011;1816:209–218. doi: 10.1016/j.bbcan.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, et al. Overexpression of RAD51 Predicts Poor Prognosis in Colorectal Cancer: Our Experience with 54 Patients. PLoS One. 2017;12:e0167868. doi: 10.1371/journal.pone.0167868. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Taki T, et al. Antisense inhibition of the RAD51 enhances radiosensitivity. Biochem. Biophys. Res. Commun. 1996;223:434–438. doi: 10.1006/bbrc.1996.0911. [DOI] [PubMed] [Google Scholar]
- 39.Ohnishi T, Taki T, Hiraga S, Arita N, Morita T. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem. Biophys. Res. Commun. 1998;245:319–324. doi: 10.1006/bbrc.1998.8440. [DOI] [PubMed] [Google Scholar]
- 40.Ito M, et al. RAD51 siRNA delivered by HVJ envelope vector enhances the anti-cancer effect of cisplatin. J. Gene Med. 2005;7:1044–1052. doi: 10.1002/jgm.753. [DOI] [PubMed] [Google Scholar]
- 41.Collis SJ, et al. Ribozyme minigene-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res. 2001;29:1534–8. doi: 10.1093/nar/29.7.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang, F. & Mazin, A. V. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 43.Ishida T, et al. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Res. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Fei, Motlekar Nuzhat A., Burgwin Chelsea M., Napper Andrew D., Diamond Scott L., Mazin Alexander V. Identification of Specific Inhibitors of Human RAD51 Recombinase Using High-Throughput Screening. ACS Chemical Biology. 2011;6(6):628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang F, Mazina OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J. Med. Chem. 2012;55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 46.Zhu J, et al. A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol. Med. 2013;5:353–365. doi: 10.1002/emmm.201201760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takaku M, et al. Halenaquinone, a chemical compound that specifically inhibits the secondary DNA binding of RAD51. Genes to Cells. 2011;16:427–436. doi: 10.1111/j.1365-2443.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- 48.Budke B, et al. RI-1: A chemical inhibitor of RAD51 that disrupts Homologous recombination in human cells. Nucleic Acids Res. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budke B, et al. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring michael acceptor reactivity. J. Med. Chem. 2013;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hengel SR, Spies MA, Spies MA. Small-Molecule Inhibitors Targeting DNA Repair and DNA Repair Deficiency in Research and Cancer Therapy. Cell Chem. Biol. 2017;24:1101–1119. doi: 10.1016/j.chembiol.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward A, Khanna KK, Wiegmans AP. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treatment Reviews. 2015;41:35–45. doi: 10.1016/j.ctrv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 53.Glassman PM, Balthasar JP. Mechanistic considerations for the use of monoclonal antibodies for cancer therapy. Cancer Biol. Med. 2014;11:20–33. doi: 10.7497/j.issn.2095-3941.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strome SE, Sausville EA, Mann D. A Mechanistic Perspective of Monoclonal Antibodies in Cancer Therapy Beyond Target-Related Effects. Oncologist. 2007;12:1084–1095. doi: 10.1634/theoncologist.12-9-1084. [DOI] [PubMed] [Google Scholar]
- 55.Vellalore Maruthachalam, B. et al. A Single-Framework Synthetic Antibody Library Containing a Combination of Canonical and Variable Complementarity Determining Regions. Chembiochem (2017). [DOI] [PubMed]
- 56.Shinohara A, Ogawa H, Ogawa T. RAD51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-K. [DOI] [PubMed] [Google Scholar]
- 57.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 1998;23:247–251. doi: 10.1016/S0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 58.Sung P, Krejci L, Van Komen S, Sehorn MG. RAD51 Recombinase and Recombination Mediators. Journal of Biological Chemistry. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 59.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human RAD51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 60.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and RAD51 on DNA. Proc. Natl. Acad. Sci. USA. 2001;98:8419–24. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein. III. Modulation of DNA binding by adenosine nucleotides. J. Biol. Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 62.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human RAD51 recombinase function. DNA Repair (Amst). 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 63.Kiianitsa K, Solinger JA, Heyer W-D. Terminal association of Rad54 protein with the RAD51-dsDNA filament. Proc. Natl. Acad. Sci. USA. 2006;103:9767–72. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Itaya, K. & Ui, M. A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta. (1966). [DOI] [PubMed]
- 65.Huston JS, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bird RE, Walker BW. Single chain antibody variable regions. Trends Biotechnol. 1991;9:132–137. doi: 10.1016/0167-7799(91)90044-I. [DOI] [PubMed] [Google Scholar]
- 67.Strube RW, Chen SY. Characterization of anti-cyclin E single-chain Fv antibodies and intrabodies in breast cancer cells: Enhanced intracellular stability of novel sFv-Fc intrabodies. J. Immunol. Methods. 2002;263:149–167. doi: 10.1016/S0022-1759(02)00035-2. [DOI] [PubMed] [Google Scholar]
- 68.Lundin C, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tercero JA, Diffley JFX. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 70.Prakash L, Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–35. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian RAD51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA. 1995;92:2298–302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Essers J, et al. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/S0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 73.Guidotti G, Brambilla L, Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017;38:406–424. doi: 10.1016/j.tips.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 74.Hampton Randolph Y, Sommer Thomas. Finding the will and the way of ERAD substrate retrotranslocation. Current Opinion in Cell Biology. 2012;24(4):460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Römisch, K. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. (1999). [DOI] [PubMed]
- 76.Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods (2009). [DOI] [PubMed]
- 77.Wu, Y., Qian, X., He, Y., Moya, I. A. & Luo, Y. Crystal structure of an ATPase-active form of RAD51 homolog from Methanococcus voltae: Insights into potassium dependence. J. Biol. Chem. (2005). [DOI] [PubMed]
- 78.Turchick, A., Hegan, D. C., Jensen, R. B. & Glazer, P. M. A cell-penetrating antibody inhibits human RAD51 via direct binding. Nucleic Acids Res. 1–18 (2017). [DOI] [PMC free article] [PubMed]
- 79.Zack DJ, Stempniak M, Wong AL, Taylor C, Weisbart RH. Mechanisms of cellular penetration and nuclear localization of an anti-double strand DNA autoantibody. J. Imunol. 1996;157:2082–8. [PubMed] [Google Scholar]
- 80.Razpotnik, R., Novak, N., Curin Šerbec, V. & Rajcevic, U. Targeting malignant brain tumors with antibodies. Frontiers in Immunology8 (2017). [DOI] [PMC free article] [PubMed]
- 81.Agrawal P, et al. CPPsite 2.0: A repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016;44:D1098–D1103. doi: 10.1093/nar/gkv1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raucher D, Ryu JS. Cell-penetrating peptides: Strategies for anticancer treatment. Trends Mol. Med. 2015;21:560–570. doi: 10.1016/j.molmed.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 83.Borrelli, A., Tornesello, A. L., Tornesello, M. L. & Buonaguro, F. M. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules23 (2018). [DOI] [PMC free article] [PubMed]
- 84.Ali SA, et al. A cell internalizing antibody targeting capsid protein (p24) inhibits the replication of HIV-1 in T cells lines and PBMCS: A proof of concept study. PLoS One. 2016;11:1–17. doi: 10.1371/journal.pone.0145986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Y, Lou D, Burkett J, Kohler H. Chemical engineering of cell penetrating antibodies. J. Immunol. Methods. 2001;254:137–45. doi: 10.1016/S0022-1759(01)00410-0. [DOI] [PubMed] [Google Scholar]
- 86.Niesner U, et al. Quantitation of the tumor-targeting properties of antibody fragments conjugated to cell-permeating HIV-1 TAT peptides. Bioconjug. Chem. 2002;13:729–736. doi: 10.1021/bc025517+. [DOI] [PubMed] [Google Scholar]
- 87.Vivès E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 88.Fellouse FA, et al. High-throughput Generation of Synthetic Antibodies from Highly Functional Minimalist Phage-displayed Libraries. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 89.Costanzo V. Brca2, RAD51 and Mre11: Performing balancing acts on replication forks. DNA Repair (Amst). 2011;10:1060–1065. doi: 10.1016/j.dnarep.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 90.Lim DS, Hasty P. A mutation in mouse RAD51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/MCB.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.North B, Lehmann A, Dunbrack RL. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.