Targeted modification of bacterial genomes is critical for genetic analysis of microorganisms. Allelic exchange is a technique that relies on homologous recombination to replace native loci with engineered sequences. However, current allelic-exchange vectors often enable only weak selection for successful homologous recombination. We developed a suite of new allelic-exchange vectors, pTOX, which were validated in several medically important proteobacteria. They encode visible nonfluorescent chromoproteins that enable easy identification of colonies bearing integrated vectors and permit stringent selection for the second step of homologous recombination. We demonstrate the utility of these vectors by using them to investigate the effect of inactivation of Serratia marcescens peptidoglycan amidohydrolases on β-lactam antibiotic resistance.
KEYWORDS: allelic exchange, Serratia marcescens, type VI toxin, AmilCP gene, ampC, ampD, antibiotic resistance, beta-lactamase, sacB, toxin-antitoxin
ABSTRACT
Despite the advent of new techniques for genetic engineering of bacteria, allelic exchange through homologous recombination remains an important tool for genetic analysis. Currently, sacB-based vector systems are often used for allelic exchange, but counterselection escape, which prevents isolation of cells with the desired mutation, occasionally limits their utility. To circumvent this, we engineered a series of “pTOX” allelic-exchange vectors. Each plasmid encodes one of a set of inducible toxins, chosen for their potential utility in a wide range of medically important proteobacteria. A codon-optimized rhaS transcriptional activator with a strong synthetic ribosome-binding site enables tight toxin induction even in organisms lacking an endogenous rhamnose regulon. Expression of the gene encoding blue AmilCP or magenta TsPurple nonfluorescent chromoprotein facilitates monitoring of successful single- and double-crossover events using these vectors. The versatility of these vectors was demonstrated by deleting genes in Serratia marcescens, Escherichia coli O157:H7, Enterobacter cloacae, and Shigella flexneri. Finally, pTOX was used to characterize the impact of disruption of all combinations of the 3 paralogous S. marcescens peptidoglycan amidohydrolases on chromosomal ampC β-lactamase activity and the corresponding β-lactam antibiotic resistance. Mutation of multiple amidohydrolases was necessary for high-level ampC derepression and β-lactam resistance. These data suggest why β-lactam resistance may emerge during treatment less frequently in S. marcescens than in other AmpC-producing pathogens, like E. cloacae. Collectively, our findings suggest that the pTOX vectors should be broadly useful for genetic engineering of Gram-negative bacteria.
IMPORTANCE Targeted modification of bacterial genomes is critical for genetic analysis of microorganisms. Allelic exchange is a technique that relies on homologous recombination to replace native loci with engineered sequences. However, current allelic-exchange vectors often enable only weak selection for successful homologous recombination. We developed a suite of new allelic-exchange vectors, pTOX, which were validated in several medically important proteobacteria. They encode visible nonfluorescent chromoproteins that enable easy identification of colonies bearing integrated vectors and permit stringent selection for the second step of homologous recombination. We demonstrate the utility of these vectors by using them to investigate the effect of inactivation of Serratia marcescens peptidoglycan amidohydrolases on β-lactam antibiotic resistance.
INTRODUCTION
The ever-increasing availability of bacterial genome sequence data has driven the demand for widely applicable and facile techniques enabling site-specific targeted mutagenesis. In general, such techniques can be divided into those that rely on exogenous enzymes versus those that depend exclusively on endogenous enzymes. Examples of methods in the former category are those utilizing the Lambda red recombinase (recombineering) (1), those employing clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 systems (2), or a combination of the two (3, 4). These systems can be fast and reliable but often require organism-specific modifications, rely on efficient transformation, and can leave genetic scars or result in off-target mutations.
In contrast, allelic exchange utilizes endogenous homologous-recombination enzymes to facilitate the replacement of a native genomic region with a foreign sequence of interest. It is a versatile technique that can routinely yield mutations ranging from kilobase scale deletions or insertions to the generation of precise point mutations. The early allelic-exchange vectors resulted in antibiotic-marked strains (5, 6); subsequent advances using counterselectable cassettes allowed the generation of truly scarless, unmarked mutant strains. However, many genes used in counterselection strategies (e.g., rpsL, pheS, thyA, and ccdB) require a specific host genotype, limiting their widespread utility (7). Background-independent counterselection strategies utilizing tetAR (8), sacB (9), or a combination of the two (10) are valuable but often require considerable optimization. Moreover, counterselection escape, where the integrated allelic-exchange vector remains lodged in the genome, preventing isolation of the desired mutant, may occasionally occur with such schemes despite optimization (9). This has been a key technical obstacle limiting wider use of allelic exchange.
Recently, a powerful negative-selection system using inducible toxins derived from toxin-antitoxin systems or from type VI secreted effector toxins was developed for use with recombineering (11). This system allowed high-stringency negative selection in clinical Escherichia coli and Salmonella isolates. Here, we repurpose these toxins for use in allelic exchange and engineer a counterselection escape surveillance system using visible chromoproteins. We demonstrate the utility of these new allelic-exchange vectors, designated “pTOX,” in multiple medically important proteobacteria. These vectors were used to systematically delete all combinations of the three peptidoglycan (PG) hydrolases in Serratia marcescens to characterize their contributions to β-lactam antibiotic resistance.
RESULTS
Engineering and testing of pTOX vectors.
The motivation for this work arose from our difficulties adapting common genetic tools for use in S. marcescens, a member of the Enterobacteriaceae that is a common cause of health care-associated urinary tract infections, pneumonia, and bacteremia (12). While chemical transformation and electrotransformation are possible in many S. marcescens strains (13, 14), they are often cumbersome and inefficient, which reduces the utility of Lambda red recombinase- and CRISPR/Cas9-based systems for genetic manipulation. Because of this, we sought to construct a conjugatable allelic-exchange vector for S. marcescens that would be widely useful.
Our set of new vectors (the pTOX vectors) is derived from pDS132, a sacB-based suicide plasmid that contains the conditional (π-dependent) R6K origin of replication (15). The sacB cassette was replaced with a rhamnose-inducible toxin obtained from the pSLC vector series (11) (Fig. 1A). Reasoning that a given toxin would be most useful in a strain that did not encode a chromosomal copy of that same toxin (and presumably the corresponding antitoxin or immunity protein, which could detoxify it), we performed protein BLAST analyses using the pSLC series toxins and identified a minimal set of three toxins (encoded by yhaV, mqsR, and tse2), at least one of which should be effective in the majority of medically important proteobacteria (see Fig. S1 in the supplemental material). Additional steps in the construction of this set of vectors included (i) introduction of a codon-optimized rhaS transcriptional activator (16) with a strong synthetic ribosome-binding site (17) to enable use of the well-characterized and stringent rhamnose-inducible system for toxin activation (18), even in strains that lack a rhamnose regulon; (ii) introduction of a strong forward transcriptional terminator upstream of the multiple-cloning site, minimizing read-through into the multiple-cloning site, thus facilitating the manipulation of toxin genes; and (iii) introduction of a greatly expanded polylinker region (19) (Fig. 1A, bottom) to facilitate insertion of new sequences into the vectors. Two versions of this set of plasmids, encoding either chloramphenicol or gentamicin resistance, were created (see Table S1 in the supplemental material). All molecular cloning was performed in the presence of glucose, which inhibits toxin production through catabolite repression.
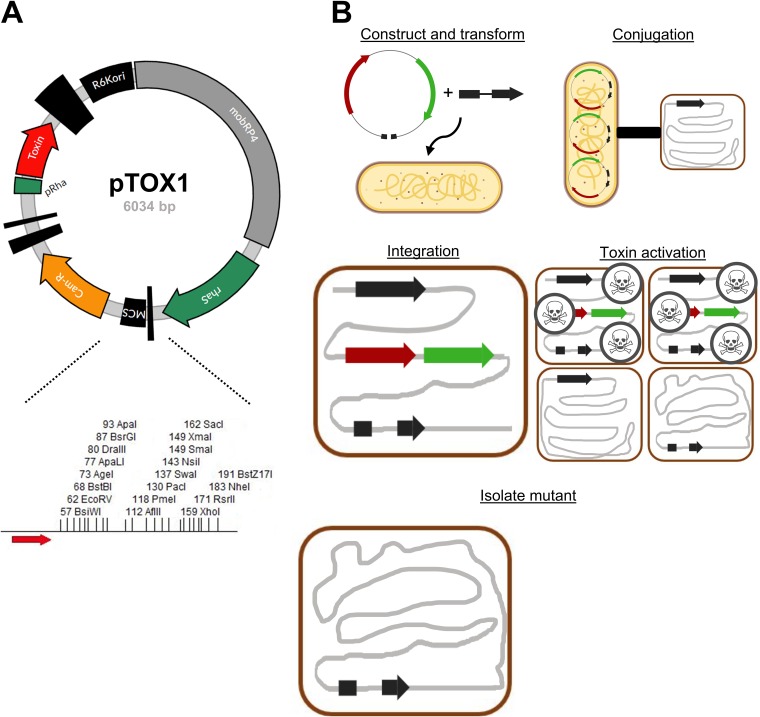
FIG 1.
Allelic exchange with pTOX. (A) (Top) Map of plasmid pTOX1. R6Kori, R6K origin of replication; mobRP4, mobilization region from the RP4 conjugative plasmid; rhaS, rhamnose transcriptional activator gene; MCS, multiple-cloning site; Cam-R, chloramphenicol resistance cassette; pRha, rhamnose promoter. The vertical black bars of various widths represent terminators. (Bottom) Expanded polylinker with restriction sites unique to pTOX1 (yhaV), shown. Red arrow, forward transcriptional terminator. (B) pTOX workflow. (Step 1) The desired allele is inserted into the MCS using isothermal assembly and transformed into donor E. coli (yellow bacillus). (Step 2) Conjugation is performed between the donor E. coli and the organism of interest (red coccobacillus). (Step 3) pTOX integrates into the appropriate chromosomal locus. (Step 4) Merodiploids are isolated, and toxin is induced. (Step 5) The desired clone is identified by colony PCR.
The utility of each of the three toxins was validated in S. marcescens ATCC 13880, which lacks rhaS and endogenous versions of the 3 toxins. First, 500 to 750 bp of the upstream and downstream regions homologous to the targeted chromosomal locus was inserted into the pTOX multiple-cloning site (see Materials and Methods for more detail) (Fig. 1B shows a schematic). Next, conjugation was used to introduce pTOX derivatives into S. marcescens. Single-crossover merodiploids were selected with the appropriate antibiotic. To assess the utility of the heterologous rhaS, we then compared the growth of merodiploids to that of wild-type S. marcescens in either glucose- or rhamnose-containing medium. Toxin-containing merodiploids grown in glucose-containing medium grew indistinguishably from the wild type, while growth in rhamnose-containing medium was undetectable (Fig. 2). These observations reveal that yhaV, mqsR, and tse2 enable robust growth inhibition in S. marcescens and that the exogenous rhaS is sufficient for stringent control of their expression.
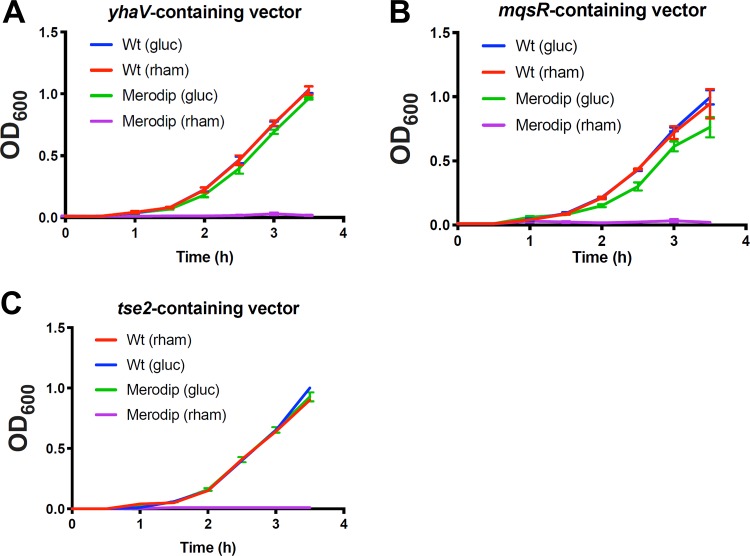
FIG 2.
Induction of specific bacterial toxins inhibit S. marcescens growth. S. marcescens wild type (Wt) or merodiploid (Merodip) bacteria harboring the indicated pTOX-carrying toxin were diluted from exponential-phase growth in LB into either 2% (wt/vol) glucose (gluc) or rhamnose-containing (rham) LB and incubated with agitation at 37°C. Note that the Wt (gluc) curve is obscured by the Wt (rham) curve in panel A and the error bars in panel C are smaller than the line for all but Merodip (gluc). Means and SEM are depicted for at least 3 independently generated merodiploids.
A limitation of the sacB counterselection system is the occasional outgrowth of merodiploids that either have a mutated sacB gene or have acquired resistance to its product (9). Such counterselection escape can confound isolation of double-crossover events. To assess whether counterselection escape also confounds yhaV-, mqsR-, or tse2-based selection, we randomly selected 23 colonies representing putative double crossovers (based on growth in the presence of rhamnose) from 3 independent experiments for each of the three toxin vectors. All 207 colonies screened were chloramphenicol sensitive and lacked pTOX vector sequences by PCR (see Fig. S2 in the supplemental material for a representative result). These observations suggest that selection mediated by the 3 toxins is potent and that the frequency of counterselection escape is very low.
Utility of pTOX vectors in diverse pathogens.
To investigate the versatility of the pTOX vectors, we tested their capacities to mediate diverse allele replacements, beginning with the S. marcescens hexS locus. S. marcescens ATCC 13880, like many isolates of this opportunistic pathogen, produces the red prodigiosin pigment; however, production is robust only at reduced temperatures, due to relief of repression mediated by the negative regulator hexS (20). A pTOX derivative carrying sequences flanking hexS was used to delete this regulator from the S. marcescens chromosome, resulting in prodigiosin hyperproduction even at 37°C (Fig. 3A). Subsequently, we replaced more than 20 loci in S. marcescens using pTOX1, pTOX2, and pTOX3 (see Table S1). All the attempts were successful, though, as with other allelic-exchange methods, the ratio of wild-type to mutant double crossovers can vary from balanced to skewed.
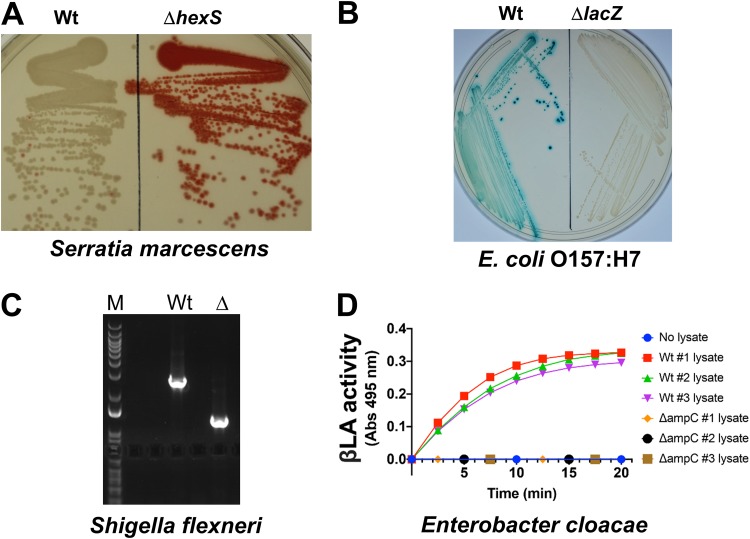
FIG 3.
pTOX for genomic modification in multiple pathogens. (A) S. marcescens colony coloration in Wt (left) and ΔhexS (right) bacteria grown at 37°C for 1 day. HexS inhibits expression of the red prodigiosin characteristic of S. marcescens. (B) E. coli O157:H7 colony coloration in Wt (left) and ΔlacZ (right) bacteria grown on X-Gal-containing medium. Blue-green colony color indicates lactose fermentation. (C) S. flexneri colony PCR and results of 1% agarose gel electrophoresis demonstrating deletion of ipgH from an S. flexneri virulence plasmid. M, marker; Wt, wild type; Δ, ΔipgH. (D) E. cloacae β-lactamase (βLA) activity in total clarified sonicate from 3 Wt double-crossover colonies and 3 ΔampC colonies induced with 50 μg/ml clavulanate prior to harvesting. The sonicates were incubated with nitrocefin, a chromogenic cephalosporin substrate that absorbs at 495 nm when hydrolyzed. An OD of 1 corresponds to 31 nmol hydrolyzed nitrocefin.
We also tested the utility of the pTOX vectors in 3 additional Gram-negative pathogens. E. coli O157:H7 (also known as enterohemorrhagic E. coli [EHEC]) is an important cause of foodborne diarrhea, as well as a systemic microangiopathy that can lead to hemolysis and renal failure. A pTOX3 derivative was used to delete lacZ, which produces a β-galactosidase that enables wild-type EHEC to ferment lactose. As seen in Fig. 3B, deletion of EHEC lacZ yielded colonies that were white on agar containing the chromogenic lactose analog 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Derivatives of pTOX3 were also used to replace nearly 20 additional loci in EHEC.
Shigella flexneri is an increasingly antibiotic-resistant cause of dysentery. In S. flexneri, most secreted virulence proteins (effectors) are encoded by a large, unstable virulence plasmid. Recombineering is useful in performing single gene deletions on the plasmid, but multiple gene deletions leave identical scar sequences that can enable undesired recombination within the plasmid. pTOX3 was used to delete the virulence plasmid ipgH locus (Fig. 3C), as well as chromosomal loci.
Finally, pTOX was efficacious in Enterobacter cloacae, an opportunistic hospital-associated pathogen associated with urinary tract and bloodstream infections. E. cloacae, like S. marcescens, possesses an inducible chromosomally encoded β-lactamase, AmpC, that hydrolyzes most β-lactam antibiotics. A pTOX3 derivative was used to delete E. cloacae ampC. Colonies harboring the ampC deletion exhibited no detectable β-lactamase activity, whereas colonies that reverted to wild type (ampC+) did (Fig. 3D). Collectively, these observations suggest that pTOX may be widely useful in Gram-negative bacteria, particularly those for which other methods are difficult or unavailable.
Chromoproteins facilitate visual detection of pTOX transconjugants.
Conjugation efficiency can vary between species and strains. For organisms like S. marcescens, in which conjugation can be inefficient, we incorporated an additional module coding for the AmilCP protein into the pTOX vectors (Fig. 4A). AmilCP is a nonfluorescent blue chromoprotein derived from the Acropora millepora coral; we sought to use its blue coloration as an additional method to discriminate wild-type colonies from transconjugants. With this aim, multiple combinations of promoters and ribosomal binding sites were tested to identify those that provided coloration sufficient for discrimination without special equipment.
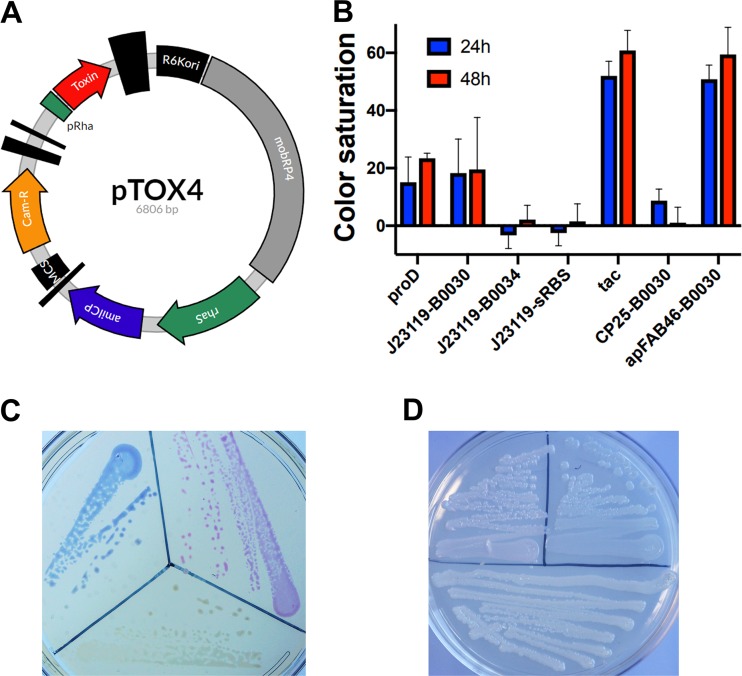
FIG 4.
A chromoprotein module facilitates monitoring of conjugation. (A) Plasmid map of pTOX4. R6Kori, the R6K origin of replication; mobRP4, the mobilization region from the RP4 conjugative plasmid; rhaS encodes the rhamnose transcriptional activator; amilCP, AmilCP gene, encoding the blue AmilCP chromoprotein; MCS, multiple-cloning site; Cam-R, chloramphenicol resistance cassette; pRha, rhamnose promoter. The vertical black bars of various widths represent terminators. (B) The tac promoter and apFAB46-B0030 allow optimal AmilCP gene expression. Shown is the relative color saturation at 24 h and 48 h of pTOX4-containing colonies with various promoters and ribosome-binding sites (RBS) (described in more detail in Materials and Methods). (C) Depiction of donor E. coli containing (clockwise from bottom) pTOX without chromoprotein, with the tac-AmilCP gene, and with the apFAB46-B0030-TsPurple gene after 24 h at 37°C. (D) E. cloacae pTOX merodiploids (clockwise from bottom) without chromoprotein, with the tac-AmilCP gene, and with the apFAB46-B0030-TsPurple gene after 24 h at 37°C and an additional 24 h at 25°C.
The series of AmilCP gene modules were first tested in E. coli donors, where we found that the tac promoter (21) or the apFAB46 promoter (22) offered the deepest-blue coloration (Fig. 4B; see Fig. S3A in the supplemental material). This level of AmilCP gene expression did not incur a detectable fitness cost (see Fig. S3B); however, several strategies to increase colony coloration further (e.g., increasing the AmilCP gene copy number) led to toxicity and were not pursued. pTOX vectors containing the AmilCP gene driven by the tac promoter and a pTOX vector expressing the magenta TsPurple gene chromoprotein driven by apFAB46 were created (Fig. 4C) (23). Both AmilCP and TsPurple were visible in restreaked merodiploid colonies after 24 to 48 h of incubation (Fig. 4D), though coloration was not as saturated as when they were expressed from the pTOX plasmids (which have medium-copy-number origins). Therefore, the pTOX chromoprotein modules may prove useful for monitoring the success of single and double crossovers, particularly in organisms with inefficient conjugation.
Application of the pTOX vectors to study inducible antibiotic resistance.
To further investigate the utility of the pTOX suite, we used the vectors to characterize the role of the S. marcescens PG amidohydrolases in inducible β-lactam resistance mediated by the AmpC β-lactamase. The PG component of the bacterial cell wall consists of a repeated disaccharide polymer linked through peptide cross-links. The peptidoglycan amidohydrolases facilitate remodeling of the cell wall by catalyzing hydrolysis of the amide bond linking the polysaccharide to the peptide component, generating muropeptide breakdown products that can subsequently be recycled in the cytoplasm (24). When the classical cytoplasmic PG amidohydrolase gene, ampD, becomes saturated with the substrate in the setting of catastrophic remodeling precipitated by β-lactam antibiotics, such as penicillins and cephalosporins, the accumulation of muropeptides leads to ampC derepression. The associated β-lactam resistance enables subsequent restoration of PG homeostasis (25).
In E. cloacae and Citrobacter freundii, expression of ampC at basal levels is sufficient for clinical resistance to penicillins and early-generation cephalosporins. After exposure to β-lactams and the resulting accumulation of muropeptide breakdown products, transcriptional upregulation can lead to transient intermediate resistance to late-generation cephalosporins, such as ceftriaxone. Under conditions where there is selection for high-level cephalosporin resistance (i.e., in patients who are subjected to prolonged cephalosporin treatment), mutation of the ampD amidohydrolase can occur. This leads to a constitutive increase in cytoplasmic muropeptide that is sufficient for high-level derepression of ampC and resistance to ceftriaxone (26, 27). However, it is unclear whether the insights gained from studies of E. cloacae and C. freundii can be generalized to all AmpC-producing organisms, because the pathway to full derepression may be more complicated in organisms with multiple amidohydrolases. For example, in Pseudomonas aeruginosa, full derepression of ampC requires inactivation of additional ampD paralogues (28), while in Yersinia enterocolitica, deletion of all three ampD paralogues does not result in obvious clinical resistance (29).
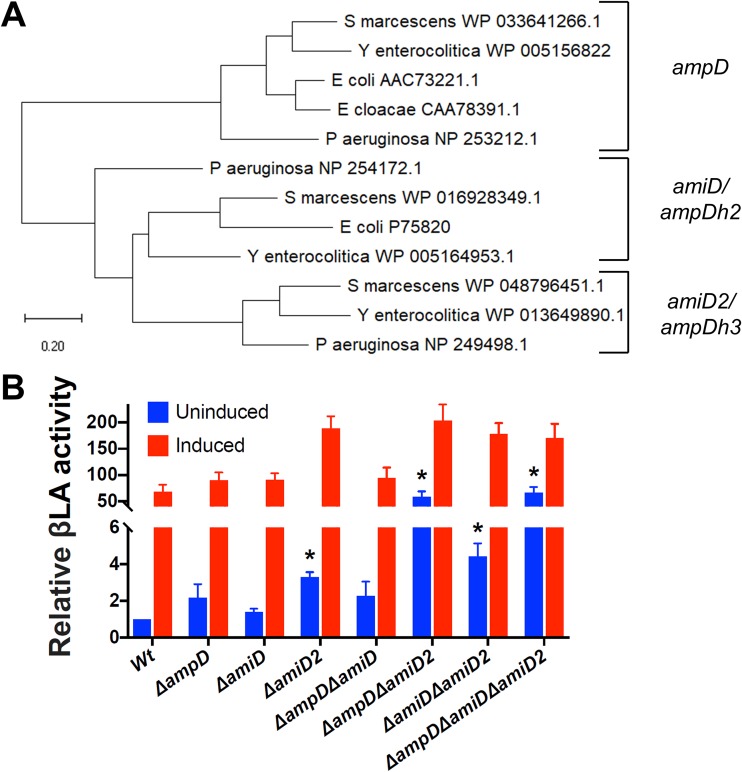
Systematic investigation of the contribution of S. marcescens PG amidohydrolases to ampC derepression and the resulting β-lactam resistance has not been performed. We found that S. marcescens encodes 3 PG amidohydrolases, which by sequence homology (30) we denote ampD (WP_033641266.1), amiD (WP_016928349.1), and amiD2 (WP_048796451.1) (Fig. 5A; see Fig. S4 in the supplemental material). Creation of pTOX derivatives targeting each of the S. marcescens PG amidohydrolases allowed the rapid generation of all combinations of single, double, and triple mutants (see Fig. S5 in the supplemental material). We found that, of the single mutants, only the ΔamiD2 mutant had a significant increase in basal AmpC activity (Fig. 5B); however, this corresponded to only a 2-fold increase in cephalosporin MICs (Table 1). In contrast, the ΔampD ΔamiD2 double mutant had a more than 50-fold increase in AmpC activity, which resulted in an 8-fold increase in the ceftriaxone MIC and a 4-fold increase in the cefepime MIC. The triple mutant exhibited no further increase in AmpC activity or in MICs. Under Clinical and Laboratory Standards Institute (CLSI) breakpoints, the ΔampD ΔamiD2 double mutant and the triple mutant, with MICs of 2 μg/ml, would be considered to have “intermediate” resistance to ceftriaxone but to still be fully susceptible to ceftazidime and cefepime. In comparison, inactivation of the single E. cloacae ampD was reported to result in a ceftriaxone MIC of 32 μg/ml (from a baseline of 0.5 μg/ml) (26).
FIG 5.
S. marcescens peptidoglycan amidohydrolase deletions lead to differential derepression of ampC. (A) Phylogenetic analysis performed using the maximum-likelihood method and a JTT matrix-based model in MEGA X (30). An unrooted tree with the lowest log likelihood (−4,913) is shown. (B) Clarified sonicates from the indicated strains were incubated with equal amounts of nitrocefin, a chromogenic cephalosporin β-lactam, and absorbance was measured in the kinetic mode for 10 min. The slope of the line from the first 5 data points was used to calculate β-lactamase (βLA) activity, which was then normalized to the Wt, which corresponds to 31.2 nmol nitrocefin/min/mg total protein. Measurements are shown without preinduction and with induction with 4 μg/ml cefoxitin for 2 h prior to harvesting. The data represent the means and SEM of the results of 4 independent experiments. Comparisons were made between all uninduced mutants and the Wt and between each induced sample and its uninduced control. *, P < 0.05 after performance of the Bonferroni correction. All the induced samples were also significantly different from the corresponding uninduced samples, except for the ΔampD ΔamiD and ΔampD ΔamiD2 mutants and the triple mutant (these asterisks are not shown for clarity).
TABLE 1.
MICs for amidohydrolase mutantsa
| Strain | MIC (μg/ml) |
|||
|---|---|---|---|---|
| FOX | CTX | CAZ | FEP | |
| S. marcescens ATCC 13880 | 16 | 0.13 | 0.25 | 0.06 |
| S. marcescens ATCC 13880 ΔampD | 16 | 0.13 | 0.25 | 0.03 |
| S. marcescens ATCC 13880 ΔamiD | 16 | 0.13 | 0.25 | 0.13 |
| S. marcescens ATCC 13880 ΔamiD2 | 16 | 0.25 | 0.50 | 0.13 |
| S. marcescens ATCC 13880 ΔampD ΔamiD | 16 | 0.50 | 0.25 | 0.13 |
| S. marcescens ATCC 13880 ΔampD ΔamiD2 | 16 | 2.00 | 0.50 | 0.25 |
| S. marcescens ATCC 13880 ΔamiD ΔamiD2 | 16 | 0.13 | 0.25 | 0.06 |
| S. marcescens ATCC 13880 ΔampD ΔamiD ΔamiD2 | 16 | 2.00 | 0.50 | 0.25 |
MICs were calculated using broth microdilution according to CLSI guidelines. FOX, cefoxitin; CTX, ceftriaxone; CAZ, ceftazidime; FEP, cefepime.
DISCUSSION
Here, we demonstrate the utility of pTOX in the clinically important Enterobacteriaceae S. marcescens, E. coli O157:H7, S. flexneri, and E. cloacae. We anticipate that pTOX may be even more widely useful, as we were purposeful in installing toxins absent from the genomes of a diverse array of pathogenic proteobacteria. These toxins have been used to facilitate recombineering (11), and the use of inducible toxins for allelic exchange promises to be a broadly generalizable approach, as systems have recently also been described for Vibrio and Aeromonas species (31), as well as for the archaeon Pyrococcus yayanosii (32).
The pTOX vectors contain an expanded multiple-cloning site, multiple antibiotic resistance cassettes, and chromoprotein modules that facilitate monitoring of crossover events. The utility of the pTOX vectors and all 3 of the toxins they encode was demonstrated through creation of multiple deletions in 4 different pathogens, including S. flexneri, an organism in which allele exchange has been difficult. These vectors are being deposited at Addgene to facilitate their distribution. Besides their utility for engineering Gram-negative organisms in research laboratories, the vectors may also be useful in the context of undergraduate education.
S. marcescens, along with E. cloacae, C. freundii, Klebsiella aerogenes, and Morganella morganii, is a member of a group of pathogenic Enterobacteriaceae with the potential for high-level, inducible expression of AmpC, which in some cases has been linked to resistance to almost all penicillins and cephalosporins (33). Original reports of cephalosporin failure in E. cloacae (34) engendered the practice of using ultra-broad-spectrum antibiotics (such as cefepime or carbapenems, which are resistant to AmpC hydrolysis) in the treatment of serious infections by pathogens with the potential for AmpC overexpression. However, this approach has untoward consequences, including increasing infections with carbapenem-resistant Enterobacteriaceae (35).
It is not clear if routine use of cefepime and carbapenems is warranted for all organisms with inducible AmpC expression. A recent review (36) emphasized that, except for E. cloacae, the data on ceftriaxone failure for pathogens with inducible AmpC are sparse. What data do exist emphasize that true on-treatment emergence of β-lactam resistance is probably rare, at least in S. marcescens and in M. morganii (37). In vitro experiments also hint at important heterogeneity among these pathogens; in this setting, the development of spontaneous cephalosporin resistance has been reported to be nearly 100-fold lower in S. marcescens than in E. cloacae and C. freundii and 10-fold lower still in M. morganii (38).
Our observations suggest that ultra-broad-spectrum antibiotics may not be necessary for treatment of S. marcescens infections. We used the pTOX vectors to investigate the roles of the 3 peptidoglycan amidohydrolases of S. marcescens in inducible β-lactam antibiotic resistance. We found that deletion of a single amidohydrolase locus had a minimal effect on cephalosporin MICs and that even the absence of all 3 amidohydrolase loci did not consistently render S. marcescens resistant to this class of antibiotics, although the triple mutant and the ΔampD ΔamiD2 double mutant did exhibit intermediate resistance to ceftriaxone. Thus, the effects of amidohydrolase deletion in S. marcescens differ from those in C. freundii and E. cloacae, in which resistance arises following the loss of a single amidohydrolase. Importantly, though current CLSI breakpoints would classify the ΔampD ΔamiD2 double mutant as having intermediate resistance to ceftriaxone, there is no evidence of increased clinical failure in this range (39). This is important, since ceftriaxone is less expensive, has more convenient dosing intervals, and is a narrower-spectrum agent than cefepime or carbapenems. Further work with additional S. marcescens isolates to clarify the generalizability of our findings is warranted.
MATERIALS AND METHODS
pTOX construction.
The DNA components of the pTOX series were obtained from pDS132 (15), the pSLC recombineering series (11) was a gift from Swaine Chen (Addgene plasmid no. 73194), pON.mCherry (21) was a gift from Howard Shuman (Addgene plasmid no. 84821), E. coli strain TP997 (40) was a gift from Anthony Poteete (Addgene plasmid no. 13055), and direct gene synthesis was performed by Integrated DNA Technologies; the components were assembled using Gibson or HiFi assembly (New England BioLabs) unless otherwise stated. All restriction enzymes were obtained from New England BioLabs, and all PCRs were performed with primers from Integrated DNA Technologies and Q5 polymerase (New England BioLabs). All cloning steps were performed in π-carrying hosts (either E. coli DH5αpir [41] for propagation or E. coli MFD-π [42] for conjugation) under catabolite repression in LB containing the appropriate antibiotic and 2% (wt/vol) glucose.
pSLC toxin vectors were first linearized with primers prJL1 and prJL2 and joined with the fragment obtained from pDS132 with primers prJL3 and prJL4 (Table 2 lists all the primers used in this study). mobRP4 (the mobilization region from the RP4 conjugative plasmid) from pDS132 was subsequently amplified with primers prJL5 and prJL6 and assembled with the prior vectors cut with NheI. The chloramphenicol resistance cassette from pON.mCherry was then amplified with primers prJL7 and prJL8 and inserted into the prior vectors digested with ClaI and BglII. The π-dependent origin from pDS132 was next isolated by SmaI digestion and inserted into the prior vectors linearized with prJL9 and prJL10. An S. marcescens codon-optimized rhaS gene (with the original primary protein sequence obtained from EHEC WP_000217135.1) and promoter (see Text S1 in the supplemental material for the sequences of all directly synthesized DNA fragments used in this study) were obtained by direct synthesis and assembled into the prior vectors linearized with prJL11 and prJL12. The expanded polylinker (19) with the forward transcriptional terminator BBa_B1002 (IGEM) was obtained by direct synthesis (sequence 2) and inserted into the prior vectors linearized with primers prJL13 and prJL14. The artificial ribosome-binding site was generated using the online calculator derived as described previously (17), synthesized as described above (sequence 3), and assembled into the prior vectors linearized with prJL15 and prJL16 to generate pTOX1 (containing yhaV), pTOX2 (containing mqsR), and pTOX3 (containing tse2). See Table S1 for all the plasmids used in this work. For insertion of the AmilCP gene or the TsPurple gene, the above-mentioned vectors were cut with SbfI, and sequence 4 and sequence 5 were inserted. For replacement of the chloramphenicol resistance cassette with one encoding gentamicin resistance, the appropriate vector was linearized with prJL17 and prJL18 and assembled with the cassette amplified from strain TP997 (using prJL19 and prJL20). See Text S2 in the supplemental material for a description of a general scheme for replacement of the antibiotic resistance cassette. Q5 GC enhancer (New England BioLabs) was used for amplification of mobRP4 and tse2.
TABLE 2.
Primers used in this study
Insertion of homology targeting regions.
pTOX vectors were cut with SmaI, and the relevant homologous regions were assembled after being amplified with prJL21, prJL22, prJL23, and prJL24 (for S. marcescens hexS); prAW1, prAW2, prAW3, and prAW4 (for EHEC lacZ); prCJK1, prCJK2, prCJK3, and prCJK4 (for S. flexneri ipgH); prJL25, prJL26, prJL27, and prJL28 (for E. cloacae ampC); prJL29, prJL30, prJL31, and prJL32 (for S. marcescens ampD); prJL33, prJL34, prJL35, and prJL36 (for S. marcescens amiD); and prJL37, prJL38, prJL39, and prJL40 (for S. marcescens amiD2). Note that some of the overlap regions in the above-mentioned primers correspond to an early version of pTOX with the original pDS132 polylinker.
Allelic exchange with pTOX.
On day 1, the appropriate upstream and downstream sequences from the targeted pathogen were amplified from genomic DNA (gDNA) in separate PCRs. After column purification of the resulting PCR product (Denville), the products were assembled with pTOX previously gel purified after restriction digestion of the polylinker and electroporated into an E. coli strain that could serve as a donor in conjugation. Throughout this work, we routinely used the diaminopimelic acid (DAP) auxotroph MFD-π (42) as the pTOX donor strain. Unless otherwise specified, all subsequent steps were performed in the presence of 2% glucose to avoid premature toxin induction. On day 2, colony PCR was performed on single MFD-π transformant colonies to confirm the appropriate insert size. On day 3, conjugation was performed between MFD-π bearing pTOX and the pathogen of interest. Optimizing the conjugation was crucial. For example, we found that conjugation was efficient at 4 to 8 h at 37°C with a 3:1 (vol/vol) ratio of MFD-π to the pathogen for EHEC, E. cloacae, and S. flexneri, but S. marcescens had markedly better efficiency when conjugated overnight at 30°C using a 50-fold excess volume of an early-logarithmic-phase growth culture of MFD-π. Exconjugants were isolated on appropriate antibiotics. On day 4, a single exconjugant colony was resuspended in 2 ml of LB containing glucose (but no selective antibiotic). This culture was incubated at 37°C with agitation to an optical density at 600 nm (OD600) of 0.2 and then washed twice with M9 salts (Sigma) with 2% (wt/vol) rhamnose before being plated on the M9-rhamnose agar described below. A short preliminary outgrowth in broth without selection minimized the possibility of the culture becoming dominated with a single double-crossover rhamnose-resistant clone. On day 5, the desired mutants could be identified with colony PCR on the resulting double-crossover colonies. The selection was stringent, and in this manner, individual colonies could frequently be isolated from a plate inoculated with the undiluted washed culture from before, but 10−1 and 10−2 dilutions should also be plated.
For the experiments shown in Fig. S2, primers prJL51 and prJL52 were used; their amplicons consisted of a small intergenic region that was largely replaced when the expanded polylinker was inserted.
AmilCP gene coloration optimization.
pTOX derivatives with different promoters and ribosome-binding sites to drive the AmilCP gene were created by assembling SbfI-cut pTOX1 with the AmilCP gene (sequence 4) amplified with prJL59 and either prJL41 (for J23119-B0030), prJL42 (for J23119-B0034), prJL43 (for CP25-B0030), or prJL44 (for apFAB46-B0030). The J23119 promoter and B0030 and B0034 ribosome-binding site sequences were obtained from IGEM. The insulated proD promoter (43) was amplified from pSB3C5-proD-B0032-E0051 (which was a gift from Joseph Davis and Robert Sauer; Addgene plasmid no. 107241) with prJL47 and prJL48, it was fused by splicing by overhang extension (SOE) PCR with the AmilCP gene coding sequence obtained from PGR-Blue (44) (which was a gift from Nathan Reyna; Addgene plasmid no. 68374) using prJL49 and prJL50, and after XbaI digestion of the product, it was ligated with XbaI-cut pTOX1. The J23119–synthetic ribosome-binding site (17) was amplified from sequence 6 with prJL45 and prJL46 and assembled in an SbfI-cut vector, and the AmilCP gene was amplified with prJL49 and prJL50 as described for proD above.
E. coli DH5αpir was transformed with the appropriate AmilCP gene-containing plasmid. Single colonies were grown in overnight cultures, diluted 1:100, and then back diluted once in logarithmic-phase growth to enable spot streaking onto solid agar at the same optical density. Digital images were taken at 24 h and 48 h, and saturation was obtained by splitting the resulting image into an “HSB stack” in ImageJ. The peak saturation was subsequently obtained using the “measure” function and then normalized by subtracting the peak saturation of the resulting spots from that of spots of E. coli DH5αpir carrying pTOX1 without the AmilCP gene. The resulting values represented the means and standard errors of the mean (SEM) of this procedure done on 3 different days.
β-Lactamase assay.
Overnight cultures of the indicated strains were back diluted 1:100 (vol/vol) into fresh medium and grown for an additional 2 h. The bacteria were then pelleted, washed twice in phosphate-buffered saline, and then flash frozen in liquid nitrogen. On the day of the assay, the pellets were thawed at 37°C and then subjected to a single round of sonication on ice (Sonic Dismembranator 60; Fisher Scientific; setting 8; 5 s). The lysates were clarified by centrifugation at a relative centrifugal force (rcf) of 20,000 for 60 min at 4°C. Total protein was quantitated by fluorometry using a Qbit protein assay kit (Thermo Fisher). β-Lactamase activity was determined by the addition of 7.8 nmol nitrocefin to either 250 ng or 1,000 ng of total protein. To facilitate accurate quantitation, 250 ng was used for all cefoxitin-induced S. marcescens samples and also for the ΔampD ΔamiD2 double mutant and the triple mutant; 500 ng was used with the E. cloacae lysates depicted in Fig. 3. Immediately after the addition of nitrocefin with a multichannel pipettor, absorbance was read kinetically at 495 nm every 5 min in a Synergy HT plate reader (BioTek).
MIC determination.
MICs were determined for the indicated S. marcescens isolates by broth microdilution according to CLSI guidelines by the method of Wiegand et al. (45). Briefly, overnight cultures were back diluted in cation-adjusted Mueller-Hinton broth, allowed to grow for 2 h, and adjusted to a final inoculum of 5 × 105 CFU per ml before being applied to wells with the appropriate antibiotic concentration. Results were read after 20 h of incubation at 37°C. The results shown in Table 1 represent the modes from 3 independent experiments.
Materials and strains.
Unless otherwise specified, all materials were purchased from Sigma. When appropriate, media were supplemented with 200 μg/ml streptomycin, 5 μg/ml gentamicin, and 20 μg/ml chloramphenicol for all E. coli, E. cloacae, and S. flexneri isolates. S. marcescens exconjugants were isolated on 100 μg/ml chloramphenicol. DAP was used at a final concentration of 0.3 mM, X-Gal at 60 μg/ml, and glucose at 2% (wt/vol) in all propagation steps with pTOX vectors. When the outgrown single crossovers were washed, rhamnose was used at 2% (wt/vol) in M9 salts. The resulting washed bacteria were plated on M9 agar supplemented with 0.2% (wt/vol) Casamino Acids, 0.5 mM MgSO4, 0.1 mM CaCl2, 25 μM iron chloride in 50 μM citric acid, the appropriate antibiotic, and rhamnose. Rhamnose at a final concentration of 0.2% to 2% facilitated good toxin induction in the organisms we tested; there was no obvious correlation with the concentration of rhamnose used, but it may be prudent to optimize this in new organisms. The S. marcescens ATCC 13880 isolate used throughout this work is a spontaneous mutant selected on streptomycin. E. cloacae was obtained from ATCC (isolate 13047). EHEC was isolate EDL933. S. flexneri was strain 2457T.
Miscellaneous analyses.
All figures and statistical analyses were prepared in Prism 8 (GraphPad). The growth curves in Fig. S3 were generated using Bioscreen C (Growth Curves USA). The plasmid maps were generated with Angular Plasmid and ApE (for the polylinker inset in Fig. 1).
Data availability.
The GenBank accession numbers that correspond to the annotated sequences of the pTOX vectors are as follows: pTOX1-yhaV, MK972837; pTOX12-tse2, MK972838; pTOX11-mqsR, MK972839; pTOX10-yhaV, MK972840; pTOX9-tse2-tsPurple, MK972841; pTOX8-mqsR-tsPurple, MK972842; pTOX7-yhaV-tsPurple, MK972843; pTOX6-tse2-amilCP, MK972844; pTOX5-mqsR-amilCP, MK972845; pTOX4-yhaV-amilCP, MK972846; pTOX3-tse2, MK972847; and pTOX2-mqsR, MK972848.
Supplementary Material
ACKNOWLEDGMENTS
J.E.L. was supported by grant T32AI007061 and by a Harvard Catalyst Medical Research Investigator Training fellowship, A.R.W. by grant T32AI132120, and M.K.W. by grant R01 AI-042347 and the Howard Hughes Medical Institute.
We thank the other members of our group for many productive conversations informing the design of pTOX and for comments on the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00990-19.
REFERENCES
- 1.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106:1.16.1–1.16.39. doi: 10.1002/0471142727.mb0116s78. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2015. Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol 81:5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisch CR, Prather KLJ. 2015. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci Rep 5:15096. doi: 10.1038/srep15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66:4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maloy SR, Nunn WD. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol 145:1110–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X-T, Thomason LC, Sawitzke JA, Costantino N, Court DL. 2013. Positive and negative selection using the tetA-sacB cassette: recombineering and P1 transduction in Escherichia coli. Nucleic Acids Res 41:e204. doi: 10.1093/nar/gkt1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khetrapal V, Mehershahi K, Rafee S, Chen S, Lim CL, Chen SL. 2015. A set of powerful negative selection systems for unmodified Enterobacteriaceae. Nucleic Acids Res 43:e83. doi: 10.1093/nar/gkv248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Rear J, Alberti L, Harshey RM. 1992. Mutations that impair swarming motility in Serratia marcescens 274 include but are not limited to those affecting chemotaxis or flagellar function. J Bacteriol 174:6125–6137. doi: 10.1128/jb.174.19.6125-6137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid JD, Stoufer SD, Ogrydziak DM. 1982. Efficient transformation of Serratia marcescens with pBR322 plasmid DNA. Gene 17:107–112. doi: 10.1016/0378-1119(82)90106-8. [DOI] [PubMed] [Google Scholar]
- 15.Philippe N, Alcaraz J-P, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Kelly CL, Liu Z, Yoshihara A, Jenkinson SF, Wormald MR, Otero J, Estévez A, Kato A, Marqvorsen MHS, Fleet GWJ, Estévez RJ, Izumori K, Heap JT. 2016. Synthetic chemical inducers and genetic decoupling enable orthogonal control of the rhaBAD promoter. ACS Synth Biol 5:1136–1145. doi: 10.1021/acssynbio.6b00030. [DOI] [PubMed] [Google Scholar]
- 17.Espah Borujeni A, Channarasappa AS, Salis HM. 2014. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res 42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW. 2006. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 40:355–364. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- 19.Latynski US, Valentovich LN. 2014. DNA tuner: a computer program for the construction of polylinker sequences of molecular vectors. Proc Belarusian State Univ Ser Physiol Biochem Mol Biol Sci 9:148–153. [Google Scholar]
- 20.Tanikawa T, Nakagawa Y, Matsuyama T. 2006. Transcriptional downregulator hexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol Immunol 50:587–596. doi: 10.1111/j.1348-0421.2006.tb03833.x. [DOI] [PubMed] [Google Scholar]
- 21.Gebhardt MJ, Jacobson RK, Shuman HA. 2017. Seeing red; the development of pON.mCherry, a broad-host range constitutive expression plasmid for Gram-negative bacteria. PLoS One 12:e0173116. doi: 10.1371/journal.pone.0173116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. 2013. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A 110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liljeruhm J, Funk SK, Tietscher S, Edlund AD, Jamal S, Wistrand-Yuen P, Dyrhage K, Gynnå A, Ivermark K, Lövgren J, Törnblom V, Virtanen A, Lundin ER, Wistrand-Yuen E, Forster AC. 2018. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J Biol Eng 12:8. doi: 10.1186/s13036-018-0100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera I, Molina R, Lee M, Mobashery S, Hermoso JA. 2016. Orthologous and paralogous AmpD peptidoglycan amidases from Gram-negative bacteria. Microb Drug Resist 22:470–476. doi: 10.1089/mdr.2016.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JW, Fisher JF, Mobashery S. 2013. Bacterial cell-wall recycling. Ann N Y Acad Sci 1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guérin F, Isnard C, Cattoir V, Giard JC. 2015. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother 59:7753–7761. doi: 10.1128/AAC.01729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/aac.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moya B, Juan C, Albertí S, Pérez JL, Oliver A. 2008. Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Wang X, Chen Y, Hao H, Li X, Liang J, Duan R, Li C, Zhang J, Shao S, Jing H. 2016. Three Yersinia enterocolitica AmpD homologs participate in the multi-step regulation of chromosomal cephalosporinase, AmpC. Front Microbiol 7:1282. doi: 10.3389/fmicb.2016.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiles TJ, Wall ES, Schlomann BH, Hay EA, Parthasarathy R, Guillemin K. 2018. Modernized tools for streamlined genetic manipulation and comparative study of wild and diverse proteobacterial lineages. mBio 9:e01877-18. doi: 10.1128/mBio.01877-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Q, Li Z, Chen R, Ma X, Xiao X, Xu J. 2018. Induction of a toxin-antitoxin gene cassette under high hydrostatic pressure enables markerless gene disruption in the hyperthermophilic archaeon Pyrococcus yayanosii. Appl Environ Microbiol 85:e02662-18. doi: 10.1128/AEM.02662-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. 1991. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med 115:585–590. doi: 10.7326/0003-4819-115-8-585. [DOI] [PubMed] [Google Scholar]
- 35.Chiotos K, Tamma PD, Flett KB, Naumann M, Karandikar MV, Bilker WB, Zaoutis T, Han JH. 2017. Multicenter study of the risk factors for colonization or infection with carbapenem-resistant enterobacteriaceae in children. Antimicrob Agents Chemother 61:e01440-17. doi: 10.1128/AAC.01440-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamma PD, Doi Y, Bonomo RA, Johnson JK, Simner PJ, Antibacterial Resistance Leadership Group . 2019. A primer on AmpC beta-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin Infect Dis 6:ciz173. doi: 10.1093/cid/ciz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi S-H, Lee JE, Park SJ, Choi S-H, Lee S-O, Jeong J-Y, Kim M-N, Woo JH, Kim YS. 2008. Emergence of antibiotic resistance during therapy for infections caused by Enterobacteriaceae producing AmpC beta-lactamase: implications for antibiotic use. Antimicrob Agents Chemother 52:995–1000. doi: 10.1128/AAC.01083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohlmann R, Bähr T, Gatermann SG. 2018. Species-specific mutation rates for ampC derepression in Enterobacterales with chromosomally encoded inducible AmpC β-lactamase. J Antimicrob Chemother 73:1530–1536. doi: 10.1093/jac/dky084. [DOI] [PubMed] [Google Scholar]
- 39.Tamma PD, Pierce VM, Cosgrove SE, Lautenbach E, Harris A, Rayapati D, Han JH. 2018. Can the ceftriaxone breakpoints be increased without compromising patient outcomes? Open Forum Infect Dis 5:ofy139. doi: 10.1093/ofid/ofy139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poteete AR, Rosadini C, St Pierre C. 2006. Gentamicin and other cassettes for chromosomal gene replacement in Escherichia coli. Biotechniques 41:261–264. doi: 10.2144/000112242. [DOI] [PubMed] [Google Scholar]
- 41.Platt R, Drescher C, Park SK, Phillips GJ. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12–23. doi: 10.1006/plas.1999.1433. [DOI] [PubMed] [Google Scholar]
- 42.Ferrieres L, Hemery G, Nham T, Guerout A-M, Mazel D, Beloin C, Ghigo J-M. 2010. Silent mischief: bacteriophage mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis JH, Rubin AJ, Sauer RT. 2011. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradshaw JC, Gongola AB, Reyna NS. 2016. Rapid verification of terminators using the pGR-Blue plasmid and Golden Gate assembly. J Vis Exp 110. doi: 10.3791/54064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand I, Hilpert K, Hancock R. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GenBank accession numbers that correspond to the annotated sequences of the pTOX vectors are as follows: pTOX1-yhaV, MK972837; pTOX12-tse2, MK972838; pTOX11-mqsR, MK972839; pTOX10-yhaV, MK972840; pTOX9-tse2-tsPurple, MK972841; pTOX8-mqsR-tsPurple, MK972842; pTOX7-yhaV-tsPurple, MK972843; pTOX6-tse2-amilCP, MK972844; pTOX5-mqsR-amilCP, MK972845; pTOX4-yhaV-amilCP, MK972846; pTOX3-tse2, MK972847; and pTOX2-mqsR, MK972848.