Abstract
The global spread and diversification of multidrug-resistant Gram-negative (MRGN) bacteria poses major challenges to healthcare. In particular, carbapenem-resistant Klebsiella pneumoniae strains have been frequently identified in infections and hospital-wide outbreaks. The most frequently underlying resistance gene (blaKPC) has been spreading over the last decade in the health care setting. blaKPC seems to have rapidly diversified and has been found in various species and on different plasmid types. To review the progress and dynamics of this diversification, all currently available KPC plasmids in the NCBI database were analysed in this work. Plasmids were grouped into 257 different representative KPC plasmids, of which 79.4% could be clearly assigned to incompatibility (Inc) group or groups. In almost half of all representative plasmids, the KPC gene is located on Tn4401 variants, emphasizing the importance of this transposon type for the transmission of KPC genes to other plasmids. The transposons also seem to be responsible for the occurrence of altered or uncommon fused plasmid types probably due to incomplete transposition. Moreover, many KPC plasmids contain genes that encode proteins promoting recombinant processes and mutagenesis; in consequence accelerating the diversification of KPC genes and other colocalized resistance genes.
Subject terms: Computational biology and bioinformatics, Clinical microbiology
Introduction
The global spread and diversification of multidrug-resistant Gram-negative (MRGN) bacteria with plasmid-localized β-lactamases poses major challenges to healthcare. β-lactamases in general are ancient enzymes1 and most likely developed in the environment2. The transmission to pathogenic bacteria might result from increasing industrial antimicrobial pollution of natural habitats3. It has been demonstrated that sublethal concentrations promote stable, high-level resistance at low fitness costs4. Highly selective urban and hospital environments promote the diversification and exchange of resistance genes, particularly on conjugable or mobilizable plasmids, which are additionally associated with transposons5. In particular, carbapenem-resistant Klebsiella pneumoniae strains have been frequently associated with infections and hospital-wide outbreaks6,7. The most frequent underlying resistance gene, blaKPC, is mainly located on a plasmid. KPC-2 and KPC-3 are predominantly identified (32 alleles are stored at NCBI BioProject 313047 as of January 2019)8.
KPC has been reported to be often associated with so-called pkpQIL-like plasmids, mostly due to an infamous nationwide outbreak with highly carbapenem-resistant K. pneumoniae in Israel in 2006, which led to a detailed sequence analysis of this plasmid9. Furthermore, researchers in other countries, such as the USA10, the UK11, Greece12, Italy12 and Taiwan13 have identified pkpQIL plasmids. In addition to pkpQIL-plasmids, many other KPC-bearing plasmid types have been identified14–16. The localization of KPC-β-lactamases on a transposon seems to be mainly responsible for different KPC-plasmids, with Tn4401 being the most prominent transposon type for KPC-217. Moreover, KPC-carrying plasmids often contain other genes for antibiotic resistance, virulence or detoxification as well as genes related to plasmid stability and longevity10,11.
The result of this transposon-directed transfer can continuously lead to different rearranged and newly mixed KPC plasmid types and thus to novelty. The aim of this study was to illustrate the result of this transposon-directed transfer by analysing the distribution of KPC and its association with different plasmid backbones.
The characterization of these KPC plasmids is of particular interest not only for outbreak control, but also for epidemiological surveillance of antibiotic resistance, because resistance plasmids of Gram-negatives usually carry multiple resistance genes and are exchanged between species18.
In order to avoid an overestimation of frequently described and deposited plasmids (e.g. pKpQIL type), a comparison based only on representative sequences was needed. However, similarity matching or clustering utilizing more conserved sequences (e.g., such as genomic DNA) could not be used because there is no consistent core genome between plasmid types. While a conserved ‘mosaic-like’ backbone exists within a plasmid type19,20 they cannot be easily utilized for a general overview across all KPC plasmid types. Furthermore, it is unclear how to deal with the mixed core backbone in fusion plasmids or plasmids with Inc-colocalization to achieve a reliable bioinformatic characterization of plasmid types.
To analyse the genetic heterogeneity of all KPC plasmids currently available at NCBI, a plasmid clustering approach tailored to this problem was applied.
Results and Discussion
Plasmid clustering into representative sequences
In total, we identified 759 blaKPC-bearing sequences in the NCBI database (May 2019). After selecting only circular plasmid sequence entries, 435 blaKPC-bearing plasmid sequences with sizes ranging from 7,995 bp (Accession Number KC609322) to 447,095 bp (Accession Number CP029436) were investigated.
Because plasmids usually contain a variable number of transposons and insertion sequences, they cannot be grouped properly with conventional genome analysis or binning tools (e.g. based on gene presence or absence). Furthermore, there is no consistent ‘core genome’ or a universal typing method that can be applied across different types of plasmids21. Moreover, mobile genetic elements can be large and could make up a significant proportion of the plasmids, creating close phylogenetic relation if a distance-based method is applied. As such, we used a clustering-based approach via psi-cd-hit.pl (detailed description in the Method section) to separate the plasmids into 257 representative KPC-plasmids (from here simply referred to as ‘representatives’). The clustering results can be found in the Supplementary Material Table S1, Section 2. We adjusted the psi-cd-hit.pl setting to consider certain plasmid properties for this large set of plasmid types. For example, many genes are similar for different plasmid types due to selective advantages (e.g. conjugation, toxin/antitoxin systems, antibiotic resistance). Therefore, only high scoring pairs (HSPs) covering 10% of the query were accepted, to avoid spurious clustering based solely on a similar gene content. This means that an HSP contains a minimal gene arrangement due to its length. In addition, the total length of all HSPs must cover at least 60% of the longer sequence. This setting was used to consider the occurrence of fusion plasmids. Many smaller KPC plasmids below 100 kbp would otherwise not be considered, because they match to fusion plasmids of 200 kbp or more. Only the representative sequence of each cluster (as labelled via psi-cd-hit.pl) was used for further analysis.
However, some limitations must be noted. Assembling plasmids remains difficult, especially for short reads and large amounts of repetitive sequences, as found in transposon or insertion sequences. This can lead to unfinished plasmid contigs or uncommon plasmid chimeras due to an incorrectly resolved assembly graph, which can be usually resolved with long-read sequencing technologies22. In our case, 63.4% of all 257 representatives (retrieved from GenBank) were sequenced using either PacBio (Pacific Biosciences) or Nanopore (Oxford Nanopore Technologies) technology, which should be assembled to current standards (e.g. hybrid assembly via Unicycler23; if long and short reads are available). Only circular sequences entries were used (as specified in the GenBank record), which should remove most incomplete sequences. Furthermore, only the representative sequence of each cluster is investigated. Plasmids with only a few missing genes (e.g. by deletion, gene loss or incomplete) would still be clustered to the representative sequence. Since most of the data available for KPC-plasmids were related to clinical samples, there was a bias favouring clinical pathogens.
All representatives were used to analyse genes of interest such as β-lactamases, toxin systems or other elements such as Inc groups and transposon regions. We arranged the representative plasmids based on the identified transposon types and summarized the extracted information in Fig. 1 (detailed results for each representative plasmid are given in Supplementary Material Table S1, Section 1). The species and the isolation date were extracted only from the representative plasmids. Information on microbial origins of other plasmid members of a cluster was not considered.
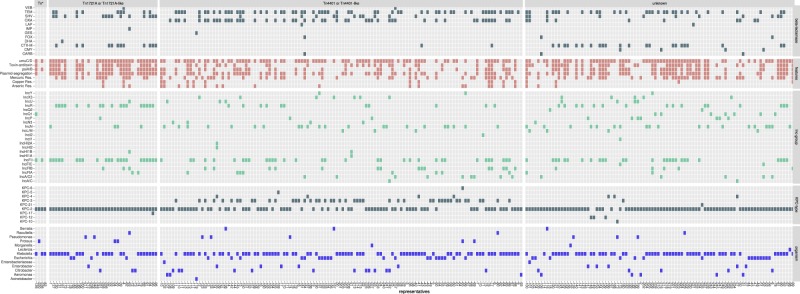
Figure 1.
Schematic presentation of the gene content and organism distribution for each representative KPC plasmid ordered by the identified transposon element. Each column corresponds to a representative plasmid. Each column is numbered based on the entry ‘representative number’ in the Supplementary Table S1, Section 1. Enterobacteriaceae corresponds to unclassified Enterobacteriaceae bacterium.
Incompatibility groups
In total, we identified Inc groups in 204 representatives using the PlasmidFinder database24. 128 out of 257 (49.8%) representatives could be assigned to a single specific Inc group (Fig. 2), with IncN (n = 30) being the most prevalent group, followed by IncFII (n = 29), IncR (n = 11) and IncA/C2 (n = 10). Multiple Inc group colocalizations were identified in 76 out of 257 (29.6%) representatives, with IncFII & IncR (n = 25) and IncFIB & IncFII (n = 12) occurring most frequently (for more details see Supplementary Material Table S1, Section 1).
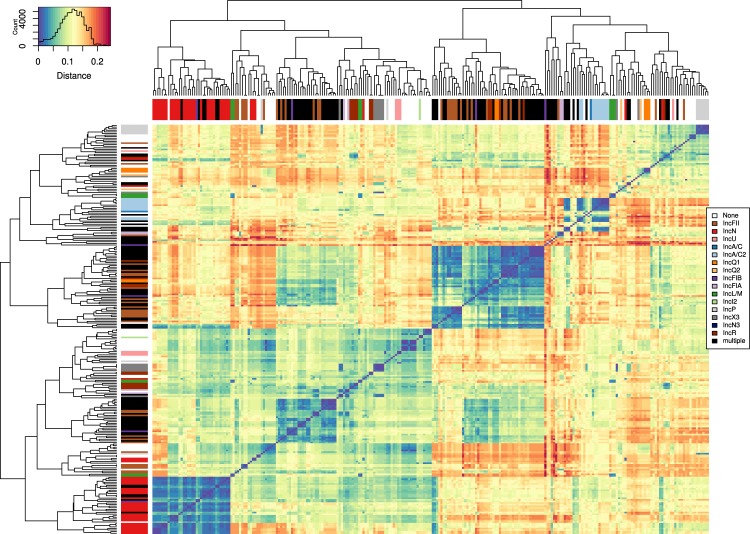
Figure 2.
Heatmap diagram of the distances between the individual 257 representative plasmids plotted using the heatmap.2 function in R. The blue/red gradient represents the estimated Mash distance (legend at the top left, reflects the amount of data corresponding to each colour/distance). Dark blue indicates closely related representatives with similar gene content but different gene arrangements. On the X and Y axes, the 257 representatives were arranged based on their distance to each other, and the branches are annotated with colours that correspond to the different plasmid incompatibility groups (legend on the right).
The similarity among all 257 representatives was assessed by estimating the distance between them using Mash25 (Fig. 2, detailed heat map in Supplementary Material Fig. S1). In order to identify representative plasmids, minimal gene ordering was required for clustering (see section ‘Plasmid clustering into representative sequences’), due to minimal HSP size. Thus, a small distance now reflects either (a) similar gene content but different order or (b) that representative sequence is partially or fully represented in a larger fusion/hybrid plasmid. Although the distance-based approach cannot be used to group representative plasmids in a tree like representation (or generally different types of plasmids), it provides an overview of the sequence content similarity between the representative plasmids as a heatmap. For the sake of clarity, the representatives were sorted based on distance in the figure. These sorted groups still show high similarities to various other distinct groups, due to the previously raised points about plasmid types with shared genes or mobile elements. The heatmap was useful to illustrate the behaviour of overall plasmid similarity between different Inc groups and Inc colocalizations.
Transposons
We identified in 165 of the 257 representatives the following transposon types Tn4401-like (n = 124), Tn1721-like (n = 37) and Tn6367-like (n = 4). Only matches with a minimal sequence coverage of 60% were considered. In ten representatives, the same transposon type (Tn4401-like or Tn1721-like) was present twice (representative number 7, 15, 23, 26, 95, 111, 113, 134, 160 and 190, detailed coverage and hit information in the Supplementary Material Table S1, Section 1). The majority (n = 72) of the remaining 92 representatives contained several transposon-related genes (retrieved from GenBank entries), such as tnpR or tnpA, which were mainly associated with Tn3 or Tn3-like structures, suggesting the presence of other active transposons. Transposon-specific genes did not appear to be restricted to a particular Inc group.
The high transposon frequency in the representative plasmids indicates that the often-described transposon-mediated transposition of KPC-β-lactamases remains the primary mechanism for the spread of the blaKPC gene to other plasmids. Especially Tn4401 or its variants were identified in 48.2% (124/257) of all representatives. The oldest described KPC-2 found in K. pneumoniae that was isolated in 1997 and was located on an IncN-plasmid within the Tn4401b transposon26,27. Tn4401 is a Tn3-class transposon that can mobilize blaKPC-2 genes under laboratory conditions at a frequency of 4.4 × 10−6 per recipient cell28. Tn1721A-like structures carrying blaKPC were identified in representative plasmids originating from Japan, Taiwan and China29. Tn1721A-like transposons seems to be preferable located on IncFII/IncR colocalized KPC-plasmids (12 of 37 representatives).
Resistance gene alleles
The KPC-2 was the most abundant allele (76.3%; 196/257) in all representatives; the KPC-3 allele was present in 18.3% (47/257). Other KPC variants identified were KPC-4 (n = 7), -5 (n = 1), -6 (n = 1), -10 (n = 1) -12 (n = 3), -17 (n = 1), and -21 (n = 1). We identified 273 KPC genes in all 257 representative plasmids due to KPC colocalizations like KPC-17 and KPC-2 in plasmid pF77_1 (Accession Number CP026137).
Other β-lactamases were present in 55.6% of all representative KPC plasmid sequences (143/257); in some cases, they were localized on integron structures as described previously for OXA-β-lactamases (class D)30. All detected β-lactamase combinations for the representatives can be found in the Supplementary Material Table S1, Section 1. The TEM β-lactamases (Ambler class A) (TEM-1 > TEM-150 > TEM-12 > TEM-40) most frequently accompanied the KPC alleles (37.7%; 97/257). TEM-12 is known as an extended spectrum β-lactamase (ESBL)31 hydrolysing 3rd generation cephalosporins and TEM-40 as a β-lactamase inhibitor resistant variant32. The second most common β-lactamases found in the KPC plasmids were various OXA variants (Ambler class D) (22.6%; 58/257), which do not belong to the carbapenemase-rich ‘Acinetobacter cluster’2. The identified variants were the narrow-spectrum β-lactamases OXA-1, -9, -10, and the ESBL OXA-129.
Other Ambler class A β-lactamases found less frequently were ESBLs of the CTX-M group (14.0%; 36/257) and SHV variants (14.4%; 37/257), which might exhibit narrow or ESBL phenotypes depending on the variant. Occasionally, β-lactamases such as the GES (ESBL), IMP (Carbapenemase), VEB (ESBL), the CARB or LAP (narrow spectrum β-lactamases), or the Ambler class C cephalosporinases CMY, DHA or FOX were identified.
Other plasmid features
Most representatives contained various types of genes that explain their successful spread and persistence. For example, in 48.6% stbB, parA and/or parM genes coding for plasmid stability proteins were found, both of which ensure inheritance to daughter cells during cell division. In addition, 38.5% of all representatives utilized a toxin/antitoxin system (usually a type II toxin-antitoxin system such as ccdA, higA, maze or yefM), which ensures high plasmid stability within the cells.
The psiB gene encoding a plasmid SOS inhibition protein B could be identified in 38.9% of all representatives. The PsiB protein is produced in the recipient during conjugation and suppresses an SOS DNA repair response by inhibiting all activities of the RecA protein33. Additionally, most of the plasmid groups (67.3%) contained an umuDC operon that encodes the error-prone DNA polymerase. This suggests that the presence of these genes potentially promotes the generation of allelic variants through point mutations for KPCs, but also for the other accompanying β-lactamases.
Presumably, resistance plasmids in general originated from the environment and could still carry typical environmental resistance genes34,35. The corresponding genes for mercury resistance (mer)36, were found in 34.2% (88/257) of all representatives, such as mercury reductase merA37 or other genes like merB, merC, merP, merR, merT. Some plasmids also carried resistance genes to arsenic (ars) (9.3%; 24/257)38 and copper (cop) (6.6%; 17/257)39. A specific distribution pattern for the heavy-metals resistance genes could not be correlated to the Inc groups or transposons within the analysed KPC plasmids. The exception is the apparent presence of mercury resistance, which is commonly found in environmental samples40–42.
Fusion plasmids or hybrid plasmids
Resistance plasmids have a highly heterogeneous sequence content, and mosaic plasmids consisting of two fused plasmids are not uncommon (e.g. IS26-mediated)43,44. As already mentioned, a large number of representatives (29.6%) have several Inc groups.
It is also interesting that in 13 representatives several KPC genes were present. Ten of these plasmids have a duplicate of the same transposon type (nine representatives with a Tn4401-like duplicate, one with a Tn1721A-like duplicate). This indicates that a transposition event caused these fusion plasmids. One might suggest that these entries are assembler chimeras; however, the plasmid pMNCRE44_5 (Accession Number CP010881.1, representative number 15) was sequenced using high-throughput (Illumina) and long-read sequencing technologies (PacBio) that usually resolves repeat regions commonly found in plasmids. In addition, the results were confirmed via PFGE45. During the transposition between the donor and acceptor plasmids guided by Tn3, a temporary fusion plasmid is formed in which the transposon is replicated46. Usually, this fusion plasmid is resolved by the resolvase. In the case of pMNCRE44_5, the plasmid seems to be not resolved; thus, it consists of two plasmids bearing two KPC-carrying transposons. Plasmid pMNCRE44_5 has a total size of 116,803 bp with two segments: one 56,475 bp segment with IncFIA and another 46,317 bp segment with IncX3. Both segments are connected to each other via Tn4401-like elements on both sides. Therefore, new types of KPC resistance plasmids may also arise due to incomplete or incorrect transpositions. However, as we are not aware of the complete assembly methodology of the representatives, we refrain from further speculation, as in some cases assembly chimeras are still likely.
Conclusions
Plasmid classification, typing or even grouping across different types remains difficult. Most resistance plasmids contain plenty of non-conserved and variable genes or gene cassettes and other genetic elements10,47. Gene rearrangements, horizontal gene transfer, and higher mutation rates additionally drive the diversification of plasmids48. There are several cgMLST typing schemes based on the specific plasmid backbone of an Inc group (https://pubmlst.org/plasmid/), but these genes are usually not conserved across different types of plasmids49,50. Furthermore, identifying the most suitable core backbone remains challenging for certain plasmid types20. In addition, essential bacterial genes that would be suitable as a consistent core genome are favourably chromosomally localized21. Considering all these limitations when analysing a large number of different plasmids, we applied a cluster-based approach to limit the influence of common plasmids. This should provide a less biased overview of the current distribution of KPC and its various plasmid backbones. This approach was intended and tested for the KPC plasmids and is not universally applicable to other plasmids. The results could show how diverse the KPC carrying plasmids already are: they cover at least 22 known Inc groups and 20.6% of the plasmids could not be assigned to any Inc group. The use of two or more functioning replicons is also widespread, reducing the incompatibility effects to allow for a wider host range51. These Inc colocalizations leads to numerous problems when investigating the dynamics and epidemiology of such resistance plasmids. For example, Inc group classification alone for epidemiological purposes would have insufficient resolution or discrimination for multi-replicon plasmids and would not extend to new plasmids either. Long-read sequencing technologies increasingly generate more complete plasmid sequences, but new analysis tools have to be developed which are specifically tailored for the genetic variability of the plasmids. This is where classic ‘backbone’ analyses of core genetic loci are limited52.
In total, approximately 1/3 of the analysed representative sequences were either fusion plasmids or hybrid plasmids containing two or more Inc groups. The currently understood modifications of the plasmid backbone via vertical and horizontal gene transfer make it already difficult to trace back the origin of a particular resistance gene50, but the potential fusion of plasmids like pMNCRE44_5 (IncFIA and IncX3, Accession CP010881) makes epidemiological investigations even more difficult. The consequence of fused or cointegrated plasmids for public health is difficult to estimate. Assigning a similar trend for other Gram-negative resistance plasmids, PCR-based detection methods may no longer be suitable for trustworthy clinical epidemiology. Whole genome sequencing offers many benefits in this regard but there are still some limitations hampering a comprehensive use in clinical routines or surveillance. For example, the development of automated pipelines is complex and dynamic as more and more bioinformatic tools become available, especially for long-read sequencing technologies. Standardized quality control and data interpretation requires additional bioinformatics know-how and infrastructure that supports data analysis, transfer and long term storage53. The general increase in sequencing activity54 and different assembly quality might have biased the presented results, but the recent increased use of long-read sequencing technologies and new bioinformatic methods will provide more insights to better understand the overall plasmid dynamics in the future.
Moreover, knowledge of endemic spreading of a particular KPC plasmid might improve empirical therapy, as accessory resistance genes or mechanisms might be localized on the plasmid interfering with the recommended therapy.
Materials and Methods
Data extraction and clustering
The KPC-2 sequence (Accession Number NC_019161.1) was used for a nucleotide-nucleotide BLAST search in the NCBI database (expected threshold 10E-70), resulting in 759 sequence hits (in May 2019). This list of sequences was analysed by a custom-made workflow (see ‘deposition’ section and Fig. 3). The workflow downloaded the respective GenBank entries from NCBI and filtered, clustered and annotated them. The downloaded gene bank entries were filtered based on their stored information. Only circular entries with a plasmid flag were included for further analysis. The sequences were clustered with psi-cd-hit.pl to identify representative sequences55,56. The following settings for psi-cd-hit.pl were used: -aL 0.6 -prog blastn -c 0.6 -g 1 -s “-evalue 10E-100 -max_target_seqs 100000 -qcov_hsp_perc 10 -max_hsps 10”. Only the representative sequence of each cluster (as labelled via psi-cd-hit.pl) was used for further analysis.
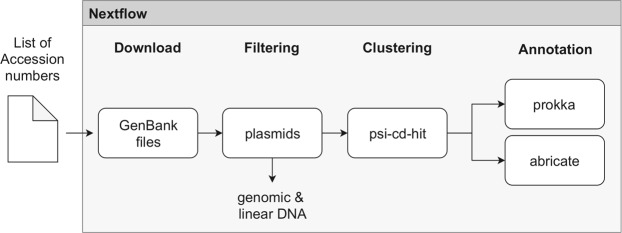
Figure 3.
Schematic illustration of the workflow for the clustering and annotation of the KPC-plasmids in Nextflow. Workflow is stored at https://github.com/replikation/plasmid_analysis.
Annotation
The representative sequences were annotated with Prokka57 and abricate (https://github.com/tseemann/abricate). Abricate was used with three databases. The NCBI BioProject 3130476 database for the identification of resistance genes. The PlasmidFinder database24 for identifying Inc groups and a custom transposon sequence set. Three transposon types were used for identification: Tn4401 (Accession Number NC021660 and CP004367.2)58, Tn1772A (Accession Number KF826292)29 and Tn6367/Tn6296 (Accession Number MF156709.1 and MF156712)59.
Only resistance genes with a coverage of >80% and >75% identity (proportion of exact nucleotide matches) were accepted. All identified β-lactamase genes have an identity of at least 98%. Inc groups have to cover at least 95% with >75% identity in order to be considered. Only transposon hits with a coverage of >60% are considered. All transposon hits have at least 97% identity. The coverage information of each transposon hit is included in Supplementary Material Table S1, Section 1. All representative sequences were analysed with Prokka, so the annotations of all sequences are based on the same settings. The results were used to identify additional plasmid features such as toxin/antitoxin systems or mercury resistance. Where appropriate, isolation dates have been taken from GenBank entries or related publications. Otherwise, the date of submission was used. Metadata such as organism or occurrence country were also extracted (see Supplementary Material Table S1, Section 1).
The β-lactamase genes in all representatives were subsequently confirmed with abricate with the databases NCBI BioProject 3130476, CARD60, ARG-ANNOT61 and ResFinder62 (not part of the workflow). We observed no differences for the group designations (e.g. KPC, TEM, SHV). We only observed a few re-occurring allelic differences for certain TEM or SHV β-lactamases, e.g. TEM-1 (in NCBI), TEM-1A_1 (in ResFinder), TEM-122 (in ARG-ANNOT), TEM-122 (in CARD).
Plasmid clustering
To cluster the 257 representative KPC plasmids, the distance between each one was determined using ‘Mash V.2.1.1’25, which estimates genome distances based on shared k-mer samples using the MinHash algorithm. First, ‘sketches’ were created for the 257 representatives and stored in a ‘sketch database’. Then, each plasmid was compared to the ‘sketch database’ to calculate the distance between each. The calculated distance table was converted into a matrix via R and then plotted with ‘gplots’ (https://cran.r-project.org/web/packages/gplots/index.html).
Deposition
The whole workflow is illustrated in Fig. 3, written in Nextflow63 (www.nextflow.io) and stored at https://github.com/replication/plasmid_analysis. For reproducibility, each step in the workflow runs in a docker container. Each container uses a defined program version, which is stored in a docker image either on dockerhub.com or biocontainers.pro. The metadata of all representative sequences, the cluster results and all 759 Accession Numbers can be found in Supplementary Material Table S1, Sections 1, 2 and 3.
Supplementary information
Acknowledgements
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) BR 5692/1-1 and supported by Grants from the Federal Ministry of Education and Research (Germany), Grant Numbers 13GW0096D and 01KI1501. The bioinformatics support of the BMBF-funded project ‘Bielefeld-Gießen Center for Microbial Bioinformatics; BiGi (Grant Number 031A533)’ within the German Network for Bioinformatics Infrastructure (de.NBI) is gratefully acknowledged.
Author Contributions
C.B. conceived, designed and coordinated the study and drafted the manuscript. C.B. wrote the workflow in Nextflow with Docker integration. C.B. and A.V. collected and cured the sequence data. C.B. performed the analysis. C.B. and A.S. performed sequence clustering and image rendering. C.B. and S.L. collected the sequence meta data. The works were supervised by M.W.P. and O.M., who critically edited the manuscript for clinical and diagnostic considerations. D.W., B.M., A.V. and J.K. have critically edited the methodological part of the manuscript and incorporated their bioinformatic considerations. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47758-5.
References
- 1.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Brandt C, et al. In silico serine β-lactamases analysis reveals a huge potential resistome in environmental and pathogenic species. Scientific Reports. 2017;7:43232. doi: 10.1038/srep43232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environmental Pollution. 2009;157:2893–2902. doi: 10.1016/j.envpol.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 4.Wistrand-Yuen E, et al. Evolution of high-level resistance during low-level antibiotic exposure. Nature Communications. 2018;9:1599. doi: 10.1038/s41467-018-04059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2018;65:34–44. doi: 10.1139/cjm-2018-0275. [DOI] [PubMed] [Google Scholar]
- 6.Kanamori H, et al. A Prolonged Outbreak of KPC-3-Producing Enterobacter cloacae and Klebsiella pneumoniae Driven by Multiple Mechanisms of Resistance Transmission at a Large Academic Burn Center. Antimicrobial Agents and Chemotherapy. 2017;61:e01516–16. doi: 10.1128/AAC.00912-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos AC, et al. Outbreak of Klebsiella pneumoniae carbapenemase–producing K pneumoniae: A systematic review. American Journal of Infection Control. 2016;44:1374–1380. doi: 10.1016/j.ajic.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Stoesser N, et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep. 2017;7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. Complete Nucleotide Sequence of KPC-3-Encoding Plasmid pKpQIL in the Epidemic Klebsiella pneumoniae Sequence Type 258. Antimicrob Agents Chemother. 2010;54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, et al. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob. Agents Chemother. 2014;58:2871–2877. doi: 10.1128/AAC.00120-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doumith M, et al. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J. Antimicrob. Chemother. 2017;72:2241–2248. doi: 10.1093/jac/dkx141. [DOI] [PubMed] [Google Scholar]
- 12.Papagiannitsis CC, et al. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J. Antimicrob. Chemother. 2016;71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 13.Tang H-J, et al. Identification of the first imported KPC-3 Klebsiella pneumoniae from the USA to Taiwan. Int. J. Antimicrob. Agents. 2014;44:431–435. doi: 10.1016/j.ijantimicag.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Wang, Y.-C. et al. Co-carriage of distinct blaKPC-2 and blaOXA-48 plasmids in a single ST11 carbapenem-resistant Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother., 10.1128/AAC.02282-18 (2019). [DOI] [PMC free article] [PubMed]
- 15.Papagiannitsis, C. C. et al. IncC blaKPC-2-positive plasmid characterized from ST648 Escherichia coli. J Glob Antimicrob Resist, 10.1016/j.jgar.2019.05.001 (2019). [DOI] [PubMed]
- 16.Fu, P. et al. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int. J. Antimicrob. Agents, 10.1016/j.ijantimicag.2019.03.014 (2019). [DOI] [PubMed]
- 17.Cheruvanky A, et al. Enhanced Klebsiella pneumoniae Carbapenemase Expression from a Novel Tn4401 Deletion. Antimicrob. Agents Chemother. 2017;61:e00025–17. doi: 10.1128/AAC.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-López R, et al. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 2006;30:942–966. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- 20.Hancock, S. J. et al. Identification of IncA/C Plasmid Replication and Maintenance Genes and Development of a Plasmid Multilocus Sequence Typing Scheme. Antimicrob. Agents Chemother. 61 (2017). [DOI] [PMC free article] [PubMed]
- 21.Tazzyman SJ, Bonhoeffer S. Why There Are No Essential Genes on Plasmids. Mol Biol Evol. 2015;32:3079–3088. doi: 10.1093/molbev/msu293. [DOI] [PubMed] [Google Scholar]
- 22.Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS. Long reads: their purpose and place. Hum. Mol. Genet. 2018;27:R234–R241. doi: 10.1093/hmg/ddy177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Computational Biology. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondov BD, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biology. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eilertson, B., Chen, L., Chavda, K. D. & Kreiswirth, B. N. Genomic Characterization of Two KPC-Producing Klebsiella Isolates Collected in 1997 in New York City. Antimicrob. Agents Chemother. 61 (2017). [DOI] [PMC free article] [PubMed]
- 27.Cuzon G, et al. Wide Dissemination of Pseudomonas aeruginosa Producing β-Lactamase blaKPC-2 Gene in Colombia. Antimicrob Agents Chemother. 2011;55:5350–5353. doi: 10.1128/AAC.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuzon G, Naas T, Nordmann P. Functional Characterization of Tn4401, a Tn3-Based Transposon Involved in blaKPC Gene Mobilization. Antimicrob. Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, Y. et al. Translocation of Carbapenemase Gene blaKPC-2 both Internal and External to Transposons Occurs via Novel Structures of Tn1721 and Exhibits Distinct Movement Patterns. Antimicrob. Agents Chemother. 61 (2017). [DOI] [PMC free article] [PubMed]
- 30.Ramirez MS, Parenteau TR, Centron D, Tolmasky ME. Functional characterization of Tn1331 gene cassettes. J Antimicrob Chemother. 2008;62:669–673. doi: 10.1093/jac/dkn279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford PA, Cherubin CE, Idemyor V, Rasmussen BA, Bush K. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing beta-lactamases in a single isolate. Antimicrob. Agents Chemother. 1994;38:761–766. doi: 10.1128/AAC.38.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blazquez J, Baquero MR, Canton R, Alos I, Baquero F. Characterization of a new TEM-type beta-lactamase resistant to clavulanate, sulbactam, and tazobactam in a clinical isolate of Escherichia coli. Antimicrob. Agents Chemother. 1993;37:2059–2063. doi: 10.1128/AAC.37.10.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell. 2009;36:121–130. doi: 10.1016/j.molcel.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 35.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebert CA, Wireman J, Smith T, Summers AO. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 1997;63:1066–1076. doi: 10.1128/aem.63.3.1066-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Møller AK, et al. Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine. FEMS Microbiol. Ecol. 2014;87:52–63. doi: 10.1111/1574-6941.12189. [DOI] [PubMed] [Google Scholar]
- 38.Cervantes C, Ji G, Ramírez JL, Silver S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 1994;15:355–367. doi: 10.1111/j.1574-6976.1994.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 39.Cooksey DA. Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 1994;14:381–386. doi: 10.1111/j.1574-6976.1994.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 40.Nascimento AMA, Chartone-Souza E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet. Mol. Res. 2003;2:92–101. [PubMed] [Google Scholar]
- 41.Smit E, Wolters A, van Elsas JD. Self-transmissible mercury resistance plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl. Environ. Microbiol. 1998;64:1210–1219. doi: 10.1128/aem.64.4.1210-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborn AM, Bruce KD, Strike P, Ritchie DA. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 43.Wong MH-Y, Chan EW-C, Chen S. IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Antimicrob. Chemother. 2017;72:2750–2754. doi: 10.1093/jac/dkx238. [DOI] [PubMed] [Google Scholar]
- 44.Papagiannitsis, C. C., Kutilova, I., Medvecky, M., Hrabak, J. & Dolejska, M. Characterization of the Complete Nucleotide Sequences of IncA/C2 Plasmids Carrying In809-Like Integrons from Enterobacteriaceae Isolates of Wildlife Origin. Antimicrob. Agents Chemother. 61 (2017). [DOI] [PMC free article] [PubMed]
- 45.Hargreaves ML, et al. Clonal Dissemination of Enterobacter cloacae Harboring blaKPC-3 in the Upper Midwestern United States. Antimicrob. Agents Chemother. 2015;59:7723–7734. doi: 10.1128/AAC.01291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolas, E. et al. The Tn3-family of Replicative Transposons. Microbiol Spectr3 (2015). [DOI] [PubMed]
- 47.Matsumura Y, et al. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:3034–3038. doi: 10.1093/jac/dky303. [DOI] [PubMed] [Google Scholar]
- 48.Fondi M, et al. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol. Biol. 2010;10:59. doi: 10.1186/1471-2148-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fricke WF, et al. Comparative Genomics of the IncA/C Multidrug Resistance Plasmid Family. J Bacteriol. 2009;191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sen D, Brown CJ, Top EM, Sullivan J. Inferring the Evolutionary History of IncP-1 Plasmids Despite Incongruence among Backbone Gene Trees. Mol Biol Evol. 2013;30:154–166. doi: 10.1093/molbev/mss210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orlek, A. et al. Plasmid Classification in an Era of Whole-Genome Sequencing: Application in Studies of Antibiotic Resistance Epidemiology. Front. Microbiol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 53.Kwong JC, McCallum N, Sintchenko V, Howden BP. Whole genome sequencing in clinical and public health microbiology. Pathology. 2015;47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 57.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 58.Pérez-Chaparro PJ, et al. Complete nucleotide sequences of two blaKPC-2-bearing IncN Plasmids isolated from sequence type 442 Klebsiella pneumoniae clinical strains four years apart. Antimicrob. Agents Chemother. 2014;58:2958–2960. doi: 10.1128/AAC.02341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, et al. Dissemination of KPC-2-Encoding IncX6 Plasmids Among Multiple Enterobacteriaceae Species in a Single Chinese Hospital. Front Microbiol. 2018;9:478. doi: 10.3389/fmicb.2018.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta SK, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Tommaso P, et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.