Abstract
Importance
Genetic testing of hereditary cancer using comprehensive gene panels can identify patients with more than one pathogenic mutation in high and/or moderate-risk-associated cancer genes. This phenomenon is known as multilocus inherited neoplasia alleles syndrome (MINAS), which has been potentially linked to more severe clinical manifestations.
Objective
To determine the prevalence and clinical features of MINAS in a large cohort of adult patients with hereditary cancer homogeneously tested with the same gene panel.
Patients and methods
A cohort of 1023 unrelated patients with suspicion of hereditary cancer was screened using a validated panel including up to 135 genes associated with hereditary cancer and phakomatoses.
Results
Thirteen (1.37%) patients harbouring two pathogenic mutations in dominant cancer-predisposing genes were identified, representing 5.7% (13/226) of patients with pathogenic mutations. Most (10/13) of these cases presented clinical manifestations associated with only one of the mutations identified. One case showed mutations in MEN1 and MLH1 and developed tumours associated with both cancer syndromes. Interestingly, three of the double mutants had a young age of onset or severe breast cancer phenotype and carried mutations in moderate to low-risk DNA damage repair-associated genes; two of them presented biallelic inactivation of CHEK2. We included these two patients for the sake of their clinical interest although we are aware that they do not exactly fulfil the definition of MINAS since both mutations are in the same gene.
Conclusions and relevance
Genetic analysis of a broad cancer gene panel identified the largest series of patients with MINAS described in a single study. Overall, our data do not support the existence of more severe manifestations in double mutants at the time of diagnosis although they do confirm previous evidence of severe phenotype in biallelic CHEK2 and other DNA repair cancer-predisposing genes.
Keywords: multilocus inherited neoplasia alleles syndrome, genetic testing, cancer syndromes, gene panel
Introduction
Hereditary cancer syndromes account for 5%–10% of all patients with cancer.1 Patients with these syndromes are carriers of pathogenic mutations in high or moderate-penetrance genes and are at risk of developing cancer at an early age as well as multiple synchronous or metachronous tumours. It is important to identify these patients because they will require specialised, long-term care and both they and their families can benefit from clinical follow-up appropriate to their risk, together with proper reproductive choices. One of the challenges in genetic counselling of these disorders is dealing with clinical heterogeneity and overlapping clinical manifestations. The phenotype variability could be explained by many factors, in isolation or in combination, such as incomplete penetrance, allelic and genetic heterogeneity, existence of genetic modifiers, environmental factors and stochastic events.2 3 Genetic diagnosis of these conditions has evolved over the last decade thanks to the introduction of next-generation sequencing, a cost-effective solution in terms of cost and time for the simultaneous sequencing of multiple genes. These new approaches for sequencing have led to the development of gene panels that contain clearly defined high-penetrance genes and moderate or even low-penetrance genes, arbitrary defined with a relative risk below 4 (moderate) or 2 (low). The use of these genes in the clinical setting is a matter of discussion and some clinical geneticists and genetic counsellors are reluctant to screen them for clinical purposes due to the uncertainty of changing medical management in carriers and in non-carriers of mutations in these genes because clinical utility has not yet been clearly established.4 Moreover, the use of large gene panels can lead to unexpected and complex findings, for example, to identify patients with more than one pathogenic mutation in genes implicated in different cancer syndromes.5–7 The term ‘MINAS’ (multilocus inherited neoplasia alleles syndrome) was coined by Whitworth and colleagues8 in a JAMA Oncology review in which the authors presented their experience (five cases) and the literature review (82 cases) of patients with cancer with two pathogenic mutations in hereditary cancer genes. No clear conclusion was reached and a database was created to record such cases (http://databases.lovd.nl/shared/diseases/04296); as of July 2018 it contained only 40 entries, all but one from the Whitworth group. Very recently, the same group presented data from a series of 460 patients with two or more tumours identifying two additional cases of MINAS.9
The authors highlighted that data gathered in the literature presented inherent ascertainment bias in relation to the genes and patients analysed. Most of the cases studied in the prepanel era were patients with breast and ovarian or colorectal cancer, in which only a few suspected genes were analysed, hence most examples with double mutations are patients with two germline BRCA1 and BRCA2 mutations or with constitutional mismatch repair deficiency syndrome, the latter with a clearly defined severe phenotype.10–13 In addition, homozygosity for the founder c.1100delC CHEK2 mutation has been associated with high breast cancer risk relative to heterozygous carriers.14
We report the genotype and phenotype of the largest unbiased MINAS cohort of patients with hereditary cancer analysed with a comprehensive gene panel that includes almost all hereditary cancer genes described in the literature.
Materials and methods
Our study population is a cohort of 1023 unrelated adult patients with clinical suspicion of hereditary cancer, visited in the Genetic Counselling Unit of the Catalan Institute of Oncology. Institutional review board approval was obtained for panel testing. For genetic testing, we used our validated custom I2HCP gene panel (containing 122–135 genes, depending on the version used).15 Variant classification was conducted under American College of Medical Genetics and Genomics guidelines.16 When possible, cosegregation analysis was performed. A brief description of the whole cohort is depicted in online supplementary table 1 and online supplementary figure 1 (comprehensive analysis is under publication, Feliubadaló et al, submitted manuscript). From our cohort of 1023 patients, 16% had multiple tumours. Of them, four patients are part of our MINAS series. For the purposes of this study, only patients with more than one pathogenic/likely pathogenic mutation (hereafter, ‘mutation’) in dominant cancer-predisposing genes are presented (online supplementary table 2). All the mutations reported here were confirmed by Sanger sequencing. All patients underwent a tiered-binned informed consent process. Twenty-four of our 135 panel genes were denominated CORE genes (Feliubadaló et al, submitted manuscript). Pathogenic variants in all these genes were returned to patients—including those with an unrelated phenotype, unless patients did not consent for the CORE panel analysis. The remaining genes of the panel were considered as research genes and are used for research purposes and the results are then explained to families in this scenario where cosegregation analysis is requested.
jmedgenet-2018-105700supp001.docx (78.8KB, docx)
jmedgenet-2018-105700supp002.jpg (774.6KB, jpg)
Results
We identified 13 unrelated patients carrying more than one mutation (1.37%) (table 1, figures 1 and 2), representing 5.75% of patients with pathogenic mutations in our study population (the overall mutation detection yield is 22.14%). The referral criteria for each are summarised in online supplementary table 1.
Table 1.
Description of patients with MINAS identified
| ID | Phenotype | Tumour/condition | Age Dx | Current age (β)/death (£) | Mutation A* | Mutation B* |
| Fam-1 | AFAP | Colorectal polyposis | 76 | 77 (£) | APC [c.423-3T>A; p.(Arg141Serfs*8)] | BRCA1 [c.1961delA; p.(Lys654Serfs*47)] |
| Fam-2 (proband) |
MEN1 | Neuroendocrine tumour | 41 | 45 (β) | MLH1 [c.244A>G; p.(Thr82Ala)] | MEN1 (c.784-9G>A; p.[Lys267Valfs*28,Arg280Serfs*2]) |
| Pituitary adenoma | 36 | |||||
| Parathyroid | 39 | |||||
| Hyperplasia | 29 | |||||
| Hepatic haemangiomas | ||||||
| Fam-2 (sister) |
HNPCC, MEN1 | Uterine carcinoma | 41 | 52 (β) | MLH1 [c.244A>G; p.(Thr82Ala)] | MEN1 (c.784-9G>A; p.[Lys267Valfs*28,Arg280Serfs*2]) |
| Colorectal cancer | 48 | |||||
| Haemangiomas | 45 | |||||
| Hyperparathyroidism | 45 | |||||
| Fam-3 | HBOC | Ovarian cancer | 45 | 51 (β) | BRCA1 [c.607C>T; p.(Arg1203*)] | TP53 [c.659A>G; p.(Tyr220Cys)] |
| Fam-4 | Tuberous sclerosis | Subcutaneous benign tumours Epilepsy |
6 | 28 (β) | TSC2 [c.5227C>T; p.(Arg1743Trp)] | RAD51D [c.694C>T; p.(Arg232*)] |
| Fam-5 | Reed’s syndrome | Cutaneous leiomyomas | 40 | 47 (β) | FH (c.905-2A>G; p.?) | BARD1 [c.157delT; p.(Cys53Valfs*5)] |
| Fam-6 | AFAP | Colorectal polyposis Colorectal cancer |
52 53 |
55 (β) | APC [c.5826_5829del; p.(Asp1942Glufs*27)] | EXO1 [c.1900C>T; p.(Arg634*)] |
| Fam-7 (proband) |
HBOC | Ovarian cancer | 51 | 52 (β) | BRCA1 [c.2309C>A; p.(Ser770*)] | XPA [c.553C>T; p.(Gln185*)] |
| Fam-7 (sister) |
HBOC | Ovarian cancer | 37 | 38 (β) | BRCA1 [c.2309C>A; p.(Ser770*)] | XPA [c.553C>T; p.(Gln185*)] |
| Fam-8 | HBOC Birt-Hogg-Dubé |
Ovarian cancer Pneumothorax (×5) |
66 33 |
68 (β) | FLCN [c.346C>T;p.(Gln116*)] | ERCC3 [c.325C>T;p.(Arg109*)] |
| Fam-9 | HBOC | Breast cancer Pancreatic cancer |
54 59 |
59 (β) | PALB2 [c.3256 C>T; p.(Arg1086*) | ATM (c.3802delG;p.Val1268*) |
| Fam-10† | HBOC | Bilateral breast cancer | 35 | 37 (β) | CHEK2 (c.433C>T; p.(Arg145Trp)] | CHEK2 [c.470T>C; p.(Ile157Thr)] |
| Fam-11† | HBOC | Breast cancer | 42 | 42 (£) | CHEK2 (whole gene deletion) | CHEK2 [c.499G>A; p.(Gly167Arg)] |
| Fam-12 | HBOC | Breast cancer | 35 | 38 (β) | ATM [c.3712_3716del; p.(Leu1238Lysfs*6)] | FANCA (c.2602-1G>C; p.?) |
| Fam-13 | HBOC | Ovarian cancer | 49 | 70 (β) | SDHB [c.505C>T; p.(Gln169*)] | FANCA [c.3558dupG; p.(Arg1187Glufs*28)] |
*Cell shadow code: dark grey: high-risk genes, light grey: moderate to low-risk genes (see online supplementary table 1).
†These patients are compound heterozygous for mutations at the same time, therefore they do not strictly fulfil the MINAS first definition.
AFAP, attenuated familial adenomatous polyposis; HBOC, hereditary breast-ovarian cancer; HNPCC, hereditary non-polyposis colorectal cancer; MEN1, multiple endocrine neoplasia type 1; MINAS, multilocus inherited neoplasia alleles syndrome.
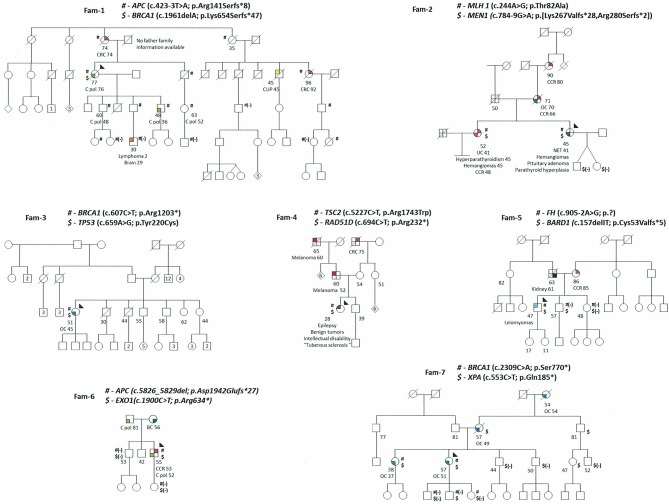
Figure 1.
Pedigrees of MINAS patients. Filled quarters of symbols indicate affected patients (each color denotes a specific type of tumor). Current age, age at death and age at diagnosis, when available, are also detailed. Proband is marked by an arrow, carrier status was studied in available relatives, and those carrying the variant are shown with the variant symbol (#,$) and if genotyped and not carriers a (-) is under the mutation symbol. A number inside a symbol denotes the number of siblings condensed in the symbol. Brain C (light orange), BC: breast cancer (emerald), C pol: colon polyposis (light green), CRC: colorectal cancer (red), CUP: carcinoma of unknown primary (yellow), Kidney Cancer (black), Leiomyomas (light blue), Lymphoma (orange), Melanoma (brown), NET: neuroendocrine tumor (dark green), OC: ovarian cancer (blue), Tuberous sclerosis (purple), UC: uterine carcinoma (pink).
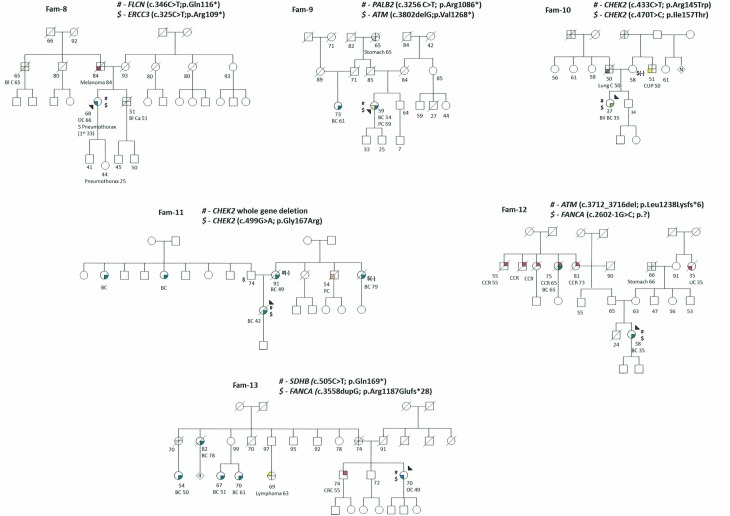
Figure 2.
Pedigrees of MINAS patients. Filled quarters of symbols indicate affected patients (each color denotes a specific type of tumor). Current age, age at death and age at diagnosis, when available, are also detailed. Proband is marked by an arrow, carrier status was studied in available relatives, and those carrying the variant are shown with the variant symbol (#,$) and if genotyped and not carriers a (-) is under the mutation symbol. A number inside a symbol denotes the number of siblings condensed in the symbol. Bl C: bladder cancer (light yellow), BC: breast cancer (emerald), Bil BC: bilateral breast cancer (emerald), CRC: colorectal cancer (red), CUP: carcinoma of unknown primary (yellow), Kidney Cancer (black), LC: lung cancer (grey), Lymphoma (orange), Melanoma (brown), OC: ovarian cancer (blue), PC: pancreas cancer (light orange), Stomach cancer (light grey), Tuberous sclerosis (purple), UC: uterine carcinoma (pink).
Nine patients had one high-risk gene mutation (Fam-1 to Fam-9); all showed an association with the proband’s clinical phenotype. The second mutation in these families did not translate into recognised clinical manifestations, except for Fam-2, in which two sisters carry two high-risk mutations; one presented a phenotype consistent with the clinical features of both cancer syndromes (MLH1 and MEN1), whereas the other showed only MEN1 clinical traits. In Fam-7, two sisters with double mutations (BRCA1 and XPA) presented ovarian cancer at ages 37 and 51, respectively. It should be noted that in two additional patients the second mutation was in a high-risk gene (BRCA1 and TP53), whereas in the remainder the mutations were in genes associated with moderate or low risk of breast, ovarian or colorectal cancer (RAD51D, XPA, BARD1, EXO1, ATM and ERCC3).
Three cases (Fam-10 to Fam-12) with clinical suspicion of hereditary breast-ovarian cancer harbour two mutations in moderate to low breast cancer risk genes: one patient with mutations in ATM and FANCA genes and two patients with biallelic mutations in CHEK2. Two of these patients were diagnosed with young-onset breast cancer (35 years) and the third was diagnosed with metastatic breast cancer at 42 years and had a poor outcome; none had a family history of breast cancer.
The last patient was diagnosed with ovarian cancer before the age of 50 and presented mutations in SDHB and FANCA, which are difficult to associate with the observed phenotype (Fam-13).
Discussion
The term MINAS was introduced with a view to discerning whether carriers of pathogenic mutations in more than one dominant hereditary cancer gene have specific clinical characteristics or are associated with a more severe phenotype. In our series, 15 patients from 13 families are carriers of two pathogenic mutations in dominant hereditary cancer genes. The most common situation was the presence of a mutation in a high-risk gene associated with the proband’s cancer phenotype and a second mutation without current clinical manifestations in the proband or the family. A mixed clinical presentation was only observed in one family where one of carriers of MEN1 and MLH1 mutations presented clinical traits of the two hereditary cancer conditions. Interestingly, in three cases of early-onset breast cancer the proband carried two pathogenic mutations in moderate to low-risk genes, suggesting an additive effect of these two mutations. This hypothesis merits further exploration and is additionally supported by Dutch population data for the analysis of the founder c.1100delC CHEK2 mutation.14 It is important to note that the two patients who were compound heterozygotes for CHEK2 mutations do not fulfil the strict definition of MINAS made formerly since both mutations are in the same gene, but we really believe that the fact of observing a severe phenotype in the three instances with mutations in moderate to low cancer risk genes makes it worth highlighting as well as being documented together in the MINAS open database. Notably, we highlight the identification of mutations in known high-risk cancer-associated genes (such as BRCA1, TP53 or RAD51C) that, in this context of double mutations, behave as low-penetrance pathogenic variants with no personal or family cancer history. There are different possible reasons for this, such as young age of the proband, incomplete penetrance, a de novo mutation in the proband, genetic mosaicism, lower risk than expected for the specific mutation identified or incomplete/missing family information. Hopefully, these can be clarified with cosegregation data, functional analysis or tumour profiling. In such situations, genetic counselling, clinical surveillance and cascade testing should be offered since these mutations are in genes of clearly known clinical utility.
In conclusion, further analysis and prospective follow-up of these patients is needed to improve our knowledge of the clinical relevance and consequences of MINAS. Of potential clinical and scientific interest is the putative relation of double mutations in moderate to low cancer risk genes with a severe clinical phenotype in early onset of cancer. As suggested by Whitworth, sharing genetic and clinical data and the continuous clinical update of these patients is crucial. To this end, all our cases have been submitted to the open database created by Whitworth (https://databases.lovd.nl/shared/diseases/04296).
Acknowledgments
We thank the participating patients and families and all members of the Genetic Counselling Unit and Genetic Diagnostic Unit involved in the Hereditary Cancer Program at the Catalan Institute of Oncology (ICO-IDIBELL). We thank CERCA Program/Generalitat de Catalunya for institutional support.
Footnotes
Contributors: Planning the project: CL, JB, JBG, GC, AS and JdV. Preparation of samples for further study: OC. Analysis of the mutations found in the sequencing and checking by Sanger: JdV, PR, LF, SG, GV, ET and CL. Correlation of the findings with the clinic of the individuals studied and their families: AS, JdV, LF, ÈGG, SG, ÀI, ET, MN, JBG, MP, JB and CL. Genetic counselling: ÈGG, ÀV, ÀI, MN and AS. Global analysis of the results and preparation of the manuscript: AS, JdV, ÈGG, JB, LF, MP and CL.
Funding: Contract grant sponsor: Supported by the Carlos III National Health Institute and Ministerio de Educación y Ciencia funded by FEDER funds–a way to build Europe (PI16/00563, PI16/01363, SAF2015-68016-R and CIBERONC); the Government of Catalonia (Pla estratègic de recerca i innovació en salut (PERIS), 2017SGR1282 and 2017SGR496); and the scientific foundation Asociación Española Contra el Cáncer.
Competing interests: None declared.
Ethics approval: Comité de Ética de Investigación Clínica del Hospital Universitari de Bellvitge.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Obtained.
References
- 1. Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med 2008;359:2143–53. 10.1056/NEJMra0802968 [DOI] [PubMed] [Google Scholar]
- 2. Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet 2013;132:1077–130. 10.1007/s00439-013-1331-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 2004;4:665–76. 10.1038/nrc1431 [DOI] [PubMed] [Google Scholar]
- 4. Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, Lipkin SM, Syngal S, Wollins DS, Lindor NM. American Society of clinical oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol 2015;33:3660–7. 10.1200/JCO.2015.63.0996 [DOI] [PubMed] [Google Scholar]
- 5. Thompson ER, Rowley SM, Li N, McInerny S, Devereux L, Wong-Brown MW, Trainer AH, Mitchell G, Scott RJ, James PA, Campbell IG. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol 2016;34:1455–9. 10.1200/JCO.2015.63.7454 [DOI] [PubMed] [Google Scholar]
- 6. Balmaña J, Digiovanni L, Gaddam P, Walsh MF, Joseph V, Stadler ZK, Nathanson KL, Garber JE, Couch FJ, Offit K, Robson ME, Domchek SM. Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing. J Clin Oncol 2016;34:4071–8. 10.1200/JCO.2016.68.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, Hallberg E, Moore R, Thomas A, Lilyquist J, Feng B, McFarland R, Pesaran T, Huether R, LaDuca H, Chao EC, Goldgar DE, Dolinsky JS. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol 2017;3:1190 10.1001/jamaoncol.2017.0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitworth J, Skytte A-B, Sunde L, Lim DH, Arends MJ, Happerfield L, Frayling IM, van Minkelen R, Woodward ER, Tischkowitz MD, Maher ER. Multilocus inherited neoplasia alleles syndrome. JAMA Oncol 2016;2:373 10.1001/jamaoncol.2015.4771 [DOI] [PubMed] [Google Scholar]
- 9. Whitworth J, Smith PS, Martin J-E, West H, Luchetti A, Rodger F, Clark G, Carss K, Stephens J, Stirrups K, Penkett C, Mapeta R, Ashford S, Megy K, Shakeel H, Ahmed M, Adlard J, Barwell J, Brewer C, Casey RT, Armstrong R, Cole T, Evans DG, Fostira F, Greenhalgh L, Hanson H, Henderson A, Hoffman J, Izatt L, Kumar A, Kwong A, Lalloo F, Ong KR, Paterson J, Park S-M, Chen-Shtoyerman R, Searle C, Side L, Skytte A-B, Snape K, Woodward ER, Tischkowitz MD, Maher ER, Aitman T, Alachkar H, Ali S, Allen L, Allsup D, Ambegaonkar G, Anderson J, Antrobus R, Armstrong R, Arno G, Arumugakani G, Ashford S, Astle W, Attwood A, Austin S, Bacchelli C, Bakchoul T, Bariana TK, Baxendale H, Bennett D, Bethune C, Bibi S, Bitner-Glindzicz M, Bleda M, Boggard H, Bolton-Maggs P, Booth C, Bradley JR, Brady A, Brown M, Browning M, Bryson C, Burns S, Calleja P, Canham N, Carmichael J, Carss K, Caulfield M, Chalmers E, Chandra A, Chinnery P, Chitre M, Church C, Clement E, Clements-Brod N, Clowes V, Coghlan G, Collins P, Cookson V, Cooper N, Corris P, Creaser-Myers A, DaCosta R, Daugherty L, Davies S, Davis J, De Vries M, Deegan P, Deevi SVV, Deshpande C, Devlin L, Dewhurst E, Dixon P, Doffinger R, Dormand N, Drewe E, Edgar D, Egner W, Erber WN, Erwood M, Erwood M, Everington T, Favier R, Firth H, Fletcher D, Flinter F, Frary A, Freson K, Furie B, Furnell A, Gale D, Gardham A, Gattens M, Ghali N, Ghataorhe PK, Ghurye R, Gibbs S, Gilmour K, Gissen P, Goddard S, Gomez K, Gordins P, Graf S, Gräf S, Greene D, Greenhalgh A, Greinacher A, Grigoriadou S, Grozeva D, Hackett S, Hadinnapola C, Hague R, Haimel M, Halmagyi C, Hammerton T, Hart D, Hayman G, Heemskerk JWM, Henderson R, Hensiek A, Henskens Y, Herwadkar A, Holden S, Holder M, Holder S, Hu F, Huis in’t Veld A, Huissoon A, Humbert M, Hurst J, James R, Jolles S, Josifova D, Kazmi R, Keeling D, Kelleher P, Kelly AM, Kennedy F, Kiely D, Kingston N, Koziell A, Krishnakumar D, Kuijpers TW, Kuijpers T, Kumararatne D, Kurian M, Laffan MA, Lambert MP, Allen HL, Lango-Allen H, Lawrie A, Lear S, Lees M, Lentaigne C, Liesner R, Linger R, Longhurst H, Lorenzo L, Louka E, Machado R, Ross RM, MacLaren R, Maher E, Maimaris J, Mangles S, Manson A, Mapeta R, Markus HS, Martin J, Masati L, Mathias M, Matser V, Maw A, McDermott E, McJannet C, Meacham S, Meehan S, Megy K, Mehta S, Michaelides M, Millar CM, Moledina S, Moore A, Morrell N, Mumford A, Murng S, Murphy E, Nejentsev S, Noorani S, Nurden P, Oksenhendler E, Othman S, Ouwehand WH, Ouwehand WH, Papadia S, Park S-M, Parker A, Pasi J, Patch C, Paterson J, Payne J, Peacock A, Peerlinck K, Penkett CJ, Pepke-Zaba J, Perry D, Perry DJ, Pollock V, Polwarth G, Ponsford M, Qasim W, Quinti I, Rankin S, Rankin J, Raymond FL, Rayner-Matthews P, Rehnstrom K, Reid E, Rhodes CJ, Richards M, Richardson S, Richter A, Roberts I, Rondina M, Rosser E, Roughley C, Roy N, Rue-Albrecht K, Samarghitean C, Sanchis-Juan A, Sandford R, Santra S, Sargur R, Savic S, Schotte G, Schulman S, Schulze H, Scott R, Scully M, Seneviratne S, Sewell C, Shamardina O, Shipley D, Simeoni I, Sivapalaratnam S, Smith KGC, Sohal A, Southgate L, Staines S, Staples E, Stark H, Stauss H, Stein P, Stephens J, Stirrups K, Stock S, Suntharalingam J, Talks K, Tan Y, Thachil J, Thaventhiran J, Thomas E, Thomas M, Thompson D, Thrasher A, Tischkowitz M, Titterton C, Toh C-H, Toshner M, Treacy C, Trembath R, Tuna S, Turek W, Turro E, Van Geet C, Veltman M, Vogt J, von Ziegenweldt J, Vonk Noordegraaf A, Wakeling E, Wanjiku I, Warner TQ, Wassmer E, Watkins H, Watt C, Webster ndrew, Welch S, Westbury S, Wharton J, Whitehorn D, Wilkins M, Willcocks L, Williamson C, Woods G, Woods G, Wort J, Yeatman N, Yong P, Young T, Yu P. Comprehensive cancer-predisposition gene testing in an adult multiple primary tumor series shows a broad range of deleterious variants and atypical tumor phenotypes. Am J Hum Genet 2018;103:1–16. 10.1016/j.ajhg.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, Krejdovsky J, DeLeonardis K, Sedgwick K, Soltis K, Roa B, Wenstrup RJ, Hartman AR. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015;121:25–33. 10.1002/cncr.29010 [DOI] [PubMed] [Google Scholar]
- 11. Silva-Smith R, Sussman DA. Co-occurrence of Lynch syndrome and juvenile polyposis syndrome confirmed by multigene panel testing. Fam Cancer 2018;17:87–90. 10.1007/s10689-017-0012-z [DOI] [PubMed] [Google Scholar]
- 12. Rebbeck TR, Friebel TM, Mitra N, Wan F, Chen S, Andrulis IL, Apostolou P, Arnold N, Arun BK, Barrowdale D, Benitez J, Berger R, Berthet P, Borg A, Buys SS, Caldes T, Carter J, Chiquette J, Claes KB, Couch FJ, Cybulski C, Daly MB, de la Hoya M, Diez O, Domchek SM, Nathanson KL, Durda K, Ellis S, Evans DG, Foretova L, Friedman E, Frost D, Ganz PA, Garber J, Glendon G, Godwin AK, Greene MH, Gronwald J, Hahnen E, Hallberg E, Hamann U, Hansen TV, Imyanitov EN, Isaacs C, Jakubowska A, Janavicius R, Jaworska-Bieniek K, John EM, Karlan BY, Kaufman B, Investigators K, Kwong A, Laitman Y, Lasset C, Lazaro C, Lester J, Loman N, Lubinski J, Manoukian S, Mitchell G, Montagna M, Neuhausen SL, Nevanlinna H, Niederacher D, Nussbaum RL, Offit K, Olah E, Olopade OI, Park SK, Piedmonte M, Radice P, Rappaport-Fuerhauser C, Rookus MA, Seynaeve C, Simard J, Singer CF, Soucy P, Southey M, Stoppa-Lyonnet D, Sukiennicki G, Szabo CI, Tancredi M, Teixeira MR, Teo SH, Terry MB, Thomassen M, Tihomirova L, Tischkowitz M, Toland AE, Toloczko-Grabarek A, Tung N, van Rensburg EJ, Villano D, Wang-Gohrke S, Wappenschmidt B, Weitzel JN, Zidan J, Zorn KK, McGuffog L, Easton D, Chenevix-Trench G, Antoniou AC, Ramus SJ. EMBRACE HEBON. Inheritance of deleterious mutations at both BRCA1 and BRCA2 in an international sample of 32,295 women. Breast Cancer Res 2016;18:112 10.1186/s13058-016-0768-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. katharina W, Kratz CP, Vasen HFA, Caron O, Colas C, Entz-Werle N, Gerdes AM, Goldberg Y, Ilencikova D, Muleris M, Duval A, Lavoine N, Ruiz-Ponte C, Slavc I, Burkhardt B, Brugieres L. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: Suggestions of the European consortium ‘Care for CMMRD’ (C4CMMRD). J Med Genet 2014;51:355–65. [DOI] [PubMed] [Google Scholar]
- 14. Adank MA, Jonker MA, Kluijt I, van Mil SE, Oldenburg RA, Mooi WJ, Hogervorst FB, van den Ouweland AM, Gille JJ, Schmidt MK, van der Vaart AW, Meijers-Heijboer H, Waisfisz Q. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. J Med Genet 2011;48:860–3. 10.1136/jmedgenet-2011-100380 [DOI] [PubMed] [Google Scholar]
- 15. Castellanos E, Gel B, Rosas I, Tornero E, Santín S, Pluvinet R, Velasco J, Sumoy L, Del Valle J, Perucho M, Blanco I, Navarro M, Brunet J, Pineda M, Feliubadaló L, Capellá G, Lázaro C, Serra E. A comprehensive custom panel design for routine hereditary cancer testing: preserving control, improving diagnostics and revealing a complex variation landscape. Sci Rep 2017;7:39348 10.1038/srep39348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–23. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2018-105700supp001.docx (78.8KB, docx)
jmedgenet-2018-105700supp002.jpg (774.6KB, jpg)