Abstract
Glioblastoma is the most lethal brain cancer in adults, with no known cure. This cancer is characterized by a pronounced genetic heterogeneity, but aberrant activation of receptor tyrosine kinase signaling is among the most frequent molecular alterations in glioblastoma. Somatic mutations of fibroblast growth factor receptors (FGFRs) are rare in these cancers, but many studies have documented that signaling through FGFRs impacts glioblastoma progression and patient survival. Small-molecule inhibitors of FGFR tyrosine kinases are currently being trialed, underlining the therapeutic potential of blocking this signaling pathway. Nevertheless, a comprehensive overview of the state of the art of the literature on FGFRs in glioblastoma is lacking. Here, we review the evidence for the biological functions of FGFRs in glioblastoma, as well as pharmacological approaches to targeting these receptors.
Keywords: FGFR, review, malignant glioma, brain cancer, astrocytoma, fibroblast growth factor
1. Introduction
Fibroblast growth factors (FGFs) were first isolated from bovine brain extracts in 1939 and characterized by their ability to induce proliferation of fibroblasts [1]. It took another 50 years to discover and clone the first of their cognate receptors [2]. FGFRs control many biological functions, including cell proliferation, survival, and cytoskeletal regulation (for review, see [3]). FGFR signaling is important during embryonal development of the CNS, and as a survival mechanism for adult neurons and astrocytes [4,5,6]. Furthermore, FGFR signaling was found to promote self-renewal and fate specification of neural stem cells [7].
In many cancers, FGFR aberrations have been implicated in tumor development and progression [8,9], and include FGFR overexpression, amplification, mutations, splicing isoform variations, and FGFR translocations [10,11]. While FGFR genomic alterations have been identified in many solid tissue cancers, such events remain rare in glioblastoma (GBM) (Table 1) [12]. Nonetheless, FGFR expression changes in astrocytes can lead to malignant transformation and GBM progression due to the activation of mitogenic, migratory, and antiapoptotic responses [13,14,15]. Of note, fusions between FGFR and TACC (transforming acidic coiled-coil containing proteins) genes were shown to be oncogenic in GBM [16], and occur in about 3% of GBM patients [12]. Whole-genome analyses of patient samples have revealed that the number of FGFR mutations and amplifications are generally very low in GBM (FGFR1: 51/3068 samples, FGFR2: 12/2662; FGFR3: 16/2887; FGFR4: 9/2456; cancer.sanger.ac.uk; [17]). Not only are oncogenic mutations in FGFRs rare in GBM, the lack of FGFR passenger mutations (i.e., mutations not providing a survival benefit) suggests that these are selected against, and that the maintenance of dynamic FGFR signaling is important for the development and/or progression of GBM.
Table 1.
Common FGFR genomic aberration in solid tumors. FGF signaling deregulation is involved in the development of many different human cancers. Four FGFR genomic alterations are represented in this table: gene amplification, point mutations, chromosomal translocations, and FGFR splicing isoforms. Each FGFR alteration is linked with the most significant cancers that contain those alterations. The role of the majority of the discovered point mutations in FGFR is unknown in cancer. Adapted from [10,18].
| Gene | Gene Amplifications | Point Mutations | Chromosomal Translocations | Splice Variants |
|---|---|---|---|---|
| FGFR1 | Breast, ovarian, bladder, and lung cancer | Majority of cancers. Example: Melanoma | Stem cell leukemia/lymphoma (SCLL), GBM | IIIc: small cell lung carcinoma Iβ: breast cancer and GBM |
| FGFR2 | Breast, gastric, lung cancer | Majority of cancers. Example: Endometrial carcinoma | IIIb: breast, endometrial, cervical, lung, pancreatic and colorectal cancer IIIc: prostate cancers |
|
| FGFR3 | Bladder cancer | Majority of cancers. Example: bladder cancer | GBM, T-cell lymphoma and bladder | IIIc: bladder cancer |
| FGFR4 | Colorectal cancer | Majority of cancers. Example: metastatic breast cancer and rhabdomyosarcoma |
We hypothesize that the neurodevelopmental and cell survival functions of this signaling pathway are at least partly conserved in GBM. Thus, dynamic FGFR signaling needs to be maintained for the survival of GBM cells, and therefore evolutionary pressure selects against both activating and inactivating mutations. Here, we review the current literature on FGFRs in GBM, and the evidence for differential functions of individual FGFRs in brain tumor progression.
2. FGFR Structure
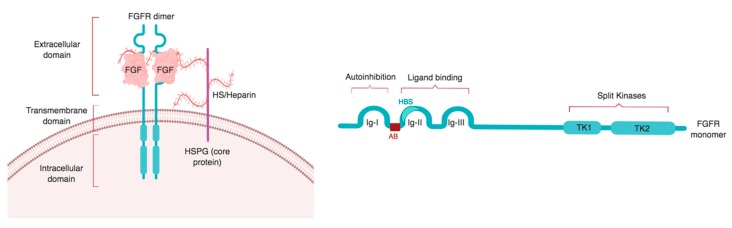
There are four known FGFRs, FGFR1–4, which are membrane-bound receptor tyrosine kinases (RTKs). A fifth member of the FGFR family, FGFRL1, is lacking a transmembrane domain and is therefore soluble. FGFRL1 acts as an antagonist to FGFR signaling [19,20]. Structurally, FGFR1–4 consist of three different domains: an extracellular ligand binding domain, a transmembrane domain, and an intracellular domain that interacts with cytoplasmic molecules and transduces FGFR signaling [8,21,22] (Figure 1).
Figure 1.
Domain structure of FGFRs: an extracellular domain containing ligand binding site is followed by a single transmembrane domain, and an intracellular domain containing split tyrosine kinases. Left panel: organization of the FGF–FGFR complex at the cell surface. The FGF–FGFR complex is stabilized by a heparin/HS chain of the HS proteoglycan (HSPG). Right panel: The extracellular domain of the receptor is composed of three Ig-like domains: Ig-I, Ig-II, and Ig-III. Ig-I has autoinhibitory capacity while Ig-II and Ig-III form the ligand binding domain. Ig-II contains the heparin/HS binding site (HBS) and is separated from Ig-I by an acid box (AB). The cytoplasmic domain is formed by two tyrosine kinases: tyrosine kinase 1 (TK1) and tyrosine kinase 2 (TK2). Image created with biorender.com.
The extracellular domain can bind FGF ligands, heparan sulfate (HS), and extracellular matrix molecules, which can act as a scaffold to enable receptor binding of specific FGFs. It is divided into three immunoglobulin-like (Ig) loops: Ig-I, Ig-II, and Ig-III (also called D1, D2, and D3) [23]. Ig-I is linked to Ig-II by a stretch of 30 acidic residues called the acid box, a unique region of FGFRs [10,24]. Ig-I and the acid box have receptor auto-inhibitory functions [25,26,27] while the Ig-II and Ig-III subdomains form the ligand binding site of the receptor [3,15,18,28,29]. Ig-II contains the heparin/HS binding region and FGF binding activity site, while the junction between Ig-II and Ig-III controls heparin and FGF affinity [21,30,31,32,33] (Figure 1).
Multiple FGFR isoforms are generated by alternative splicing of the region encoding for the extracellular domain. This modifies the affinity and sensitivity of the receptors for different FGF ligands [34,35]. Thus, an array of FGFR isoforms is created that can fine-tune the response of cells to the large number of potential FGF ligands available in their specific environment. FGF sensitivity is further modified by co-receptors, such as Klotho family members, which are required for binding of endocrine FGFs [36,37,38].
Alternatively-spliced β isoforms of FGFR1 or FGFR2 are produced by the exclusion of the Ig-I domain, which is encoded by exon 3. Due to the auto-inhibitory function of the Ig-I domain, the β isoform has considerably higher affinity for FGFs, and is oncogenic [39,40]. Retention of the Ig-I domain creates FGFR α isoforms [28].
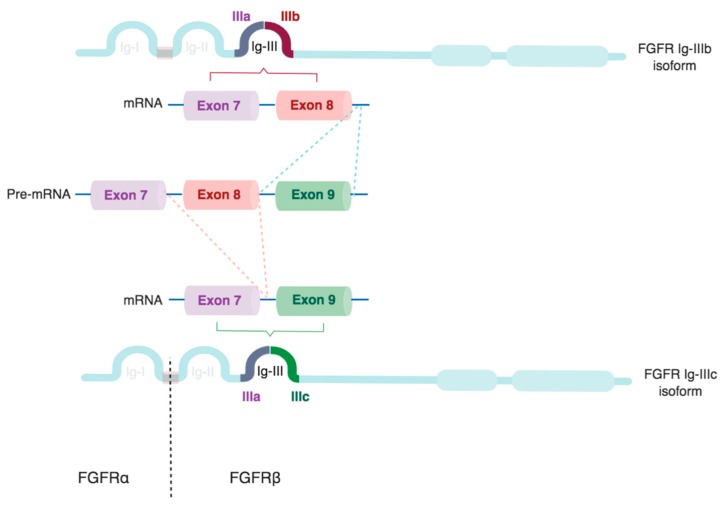
Additionally, alternative splicing generates two isoforms of the Ig-III domain, known as Ig-IIIb and Ig-IIIc, in FGFR1–3, but not FGFR4 [10,41,42,43]. Ig-IIIb and Ig-IIIc are generated by exon skipping, and are encoded by exons 8 and 9, respectively (Figure 2). By contrast, exon 7, encoding Ig-IIIa, is present in all splice variants. Different splice-regulatory proteins have been identified that control the splicing of Ig-IIIb, such as regulatory RNA-binding protein (RBP), and the epithelial splicing regulatory proteins (ESRP1/2) [32,40,44].
Figure 2.
Schematic representation of FGFR splice isoforms. The Ig-III domain of FGFR1–3 is encoded by exons 7–9. Exon 7 encodes Ig-IIIa, which consists of the N-terminal half of the Ig-III loop. The C-terminal half is formed by the IIIb or IIIc sequence, which is generated by the selective inclusion of exons 8 or 9, respectively. Truncation of the Ig-I loop creates FGFRβ isoforms (dotted line), while the full-length receptor is termed FGFRα. Image created with biorender.com.
The expression of FGFR splice variants is tissue-dependent. For example, the Ig-IIIb isoform is more prevalent in epithelial tissues, while Ig-IIIc is preferentially expressed in mesenchymal ones [22,24,32]. Switching of epithelial and mesenchymal isoforms occurs during epithelial–mesenchymal transition, which is known as the IIIb/IIIc switch. Hence, FGFR isoform expression is also related to tissue plasticity, and changes during tissue growth, proliferation, and remodeling [45].
The FGFR transmembrane domain is crucial for transferring the signal from the extracellular to the intracellular domain, the latter consisting of a juxtamembrane domain, two tyrosine kinase domains, and the C-terminal tail [10,32,46,47]. FGFR domains are highly conserved among receptors, and the tyrosine kinase domain shares the highest homology. The Ig-III domain is also highly conserved, especially between FGFR1 and FGFR2 [48].
The binding of FGF ligands to HSPGs causes FGFR dimerization and activation in the -COOH receptor tail of the cytoplasmic tyrosine residues by phosphorylation [49,50]. For instance, autophosphorylation of FGFR1 tyrosine (Y) residues occurs in three steps. Firstly, phosphorylation of Y653 leads to a 50–100-fold increase of the catalytic core activation of the intracellular domain. Secondly, Y583, Y463, Y766, and Y585 sites are consequently phosphorylated, and finally, the second tyrosine kinase domain phosphorylation increases the tyrosine kinase activity 10-fold. This sequence of autophosphorylation follows a specific and controlled order that, if deregulated, can induce malfunction of the pathway [49].
Different models have been proposed for FGFR dimerization depending on the ligand–heparin–receptor complex, specificity between the ligand and the receptor, and the heparin length required for the binding [21,24]. In the first model, HS increases the association between the receptor and the high affinity binding site of the ligand, forming a ternary complex (1:1:1 FGF–HS–FGFR) that then interacts with a second receptor, inducing FGFR dimerization through the FGF low affinity binding site (2:1:1 FGFR–HS–FGF). On the other hand, the symmetrical model suggests that two individual ternary complexes are formed. The FGFR dimerization will then occur by FGFR–FGFR direct interaction, FGF ligand interaction, or by HS–HS link (2:2:2 FGFR–HS–FGF). In this model, HS enhances FGFR dimerization, but it is not crucial. Finally, according to the asymmetric model, HS attaches to two FGF–FGFR complexes, binding both FGFs but only one of the receptors (2:1:2 FGFR–HS–FGF) [21]. Therefore, although different models have been suggested, more research is needed to clarify the stoichiometry of FGFR dimerization [21,24,51].
3. FGFR Signaling Cascade
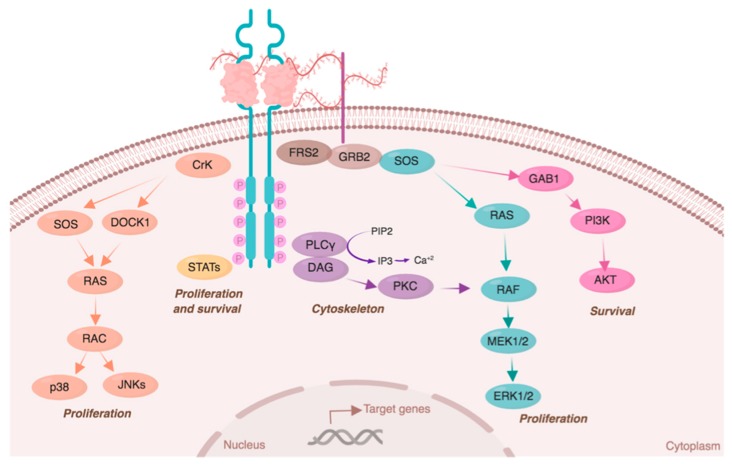
FGF–FGFR stimulates cell signaling pathways related to cell proliferation, survival, cytoskeletal regulation, and FGFR degradation [10]. Cell proliferation is mainly induced by RAC/JNK and RAS–MAPK signaling pathways [3]. RAC kinases can be activated by the transient phosphorylation of CRK, which simultaneously stimulates RAC phosphorylation trough DOCK1 or SOS/RAS [10]. RAC kinases promote proliferation by the activation of JNK and p38. Alternatively, the RAS/RAF/MEK/ERK signaling pathway can be activated by the FRS2–GRB2–SOS–SHP2 complex assembly or byPKC activation through PLC phosphorylation [18,22].
Cell survival is mainly promoted by phosphorylation of PI3K/AKT signaling through the FRS2–GRB2–GAB1 complex. Finally, FGFRs are also implicated in cytoskeletal regulation, as PLC phosphorylation leads to the hydrolysis of PIP2 into IP3, inducing calcium release [18] (Figure 3).
Figure 3.
FGFR signaling pathway. After ligand binding, FGFRs dimerize and activate multiple signal transduction pathways. Each pathway induces the expression of specific target genes related to cell proliferation (STATs, RAS/p38/JNKs, and RAS/MAPK/ERK), survival (STATs and PI3K/AKT), and cytoskeleton regulation (PLC/Ca2+). Kinases are color-coded according to their specific signaling pathway. Image created with biorender.com.
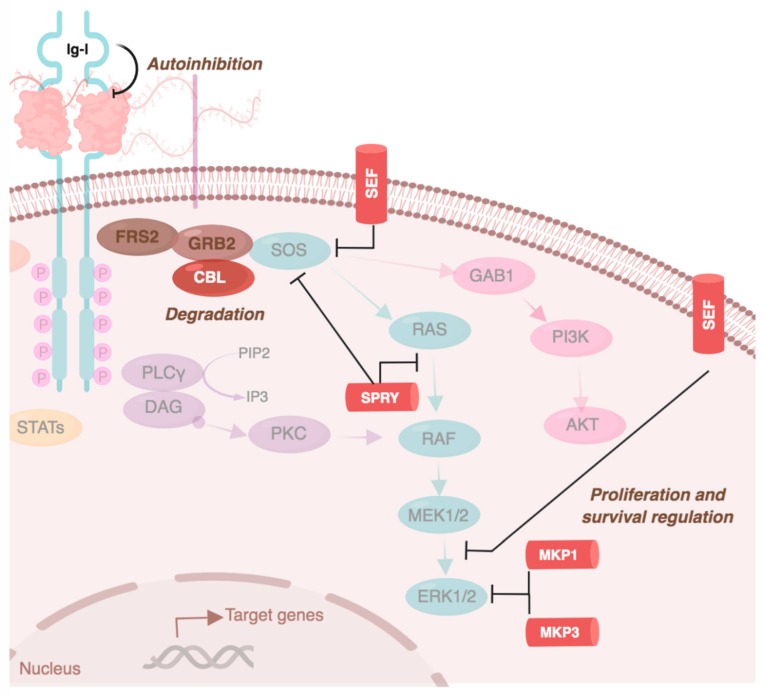
Because FGFR signaling acts upon many biological functions, a regulatory system that controls its timing, spread, and balances its activation is required. This is important as the activation of the signaling cascade depends on FGFR expression and localization to the cell membrane. Therefore, receptor availability depends on the balance between its recycling and degradation rate, which differ among receptors. One of these regulatory systems is FGFR internalization or constitutive endocytosis. FGFR synthesis occurs at a higher level than its internalization. However, after ligand-binding, FGFR internalization from the plasma membrane accelerates [52]. FGFR internalization is primarily mediated by clathrin-dependent endocytosis and requires the SRC–FRS2 complex [53]. The internalization rate depends on the receptor type—FGFR1 has the highest internalization rate and FGFR3 the lowest. Endocytosis of activated FGFRs involves detachment from the SRC complex [54]. FGFRs can then re-translocate to the cytosol, mitochondria, nucleus (to directly regulate gene expression), or to the endosomal compartment for receptor degradation [52]. The latter requires interaction between the FRS2–GRB complex and CBL, and is receptor-independent [22] (Figure 4). Indeed, FGFR1 has more ubiquitination sites than FGFR4, so its degradation rate is likely higher [10].
Figure 4.
FGFR signaling pathway regulation. FGFR signaling is negatively regulated, partly by CBL (inducing FGFR degradation after receptor internalization), by SEF, SPRY, MKP1, and MPK3 (which negatively regulate proliferation and survival related pathways). FGFRs can also regulate their own activation due to the autoinhibitory function of Ig-I. Image created with biorender.com.
Other regulatory systems of FGFR signaling are the negative regulators SEF, SPRY1/SPRY4, and MKP1/MKP3. The activation of cell proliferation is counterbalanced by SEF, which negatively regulates ERK and AKT activation [18]. Similarly, SPRY1/SPRY4 reduce proliferation by directly interacting with RAS/RAF kinases or by blocking the FRS2–GRB2–SOS–SHP2 complex. MKP1 and MKP3 also attenuate FGFR signaling by dephosphorylating MAPK and ERK [10] (Figure 4).
FGF signaling is also negatively regulated by the autoinhibitory (Ig-I) domain of the receptors. This is controlled by the electrostatic interactions between the negatively charged acid box with the highly basic heparin binding site in Ig-II [25]. This complex blocks the heparin–FGF binding, minimizing FGFR activation. The auto-inhibitory capacity is crucial for the modulation of the pathway, as the high amount of HSPGs from the cell surface and the extracellular matrix increases the probabilities of FGF–heparin binding and activation of the RTK cascade [24].
Other factors involved in FGFR pathway regulation are the ligand affinity for the receptor and the ligand amount and availability. Extracellular FGFs are protected and stored by HS proteoglycans. Heparanases are directly involved in FGF signaling regulation, as they cleave the HS chain, thus releasing FGFs in the vicinity of cells. Depending on the cell type and the growth factor released, heparanases are therefore involved in cell growth, differentiation, or stemness maintenance [55]. Likewise, sufficient amounts of ligand and heparin/HSPGs are necessary for stabilizing FGFR dimerization. The necessary ligand concentration is dependent on the ligand-binding affinity of the FGFRs, which depends on the FGFR splice isoforms [3,8,42]. For example, FGF2 activates both FGFR1 IIIb and IIIc isoforms, while it has a higher affinity for the isoform IIIc in FGFR2 and FGFR3 [42].
4. Crosstalk between FGFRs and Other Cell Surface Molecules
FGFRs can be modulated independently of their ligands by integral cell membrane proteins, such as G-protein-coupled receptors (GPCRs), cell adhesion molecules (CAMs), and other RTKs, which play a crucial role in the induction of specific cell responses and fate during development and cancer [56]. GPCRs can transactivate FGFRs by promoting the activation of matrix metalloproteinases, resulting in cleavage of FGFs, or by directly interacting with FGFRs [57]. GPCR-mediated FGFR1 transactivation is associated with neuronal differentiation, neurite growth, and synaptic plasticity [56]. FGFR1 modulation by GPCRs (e.g., CB1A, 5-HT1A, and mAChR) involves the activation of the SRC-ERK1/2 pathway [57,58,59]. In C6 glioma cells, crosstalk between the mu-Opioid receptor and FGFR1 was shown to activate this signaling cascade, but the specific mechanism is not yet completely understood [60].
FGFR activity is also modulated by CAMs, cell surface proteins that regulate cell–cell interactions and motility. Importantly, the FGFR acid box region is required for the CAM/FGFR interaction [61], hence the FGFR β isoforms cannot be transactivated by CAMs. CAMs of the integrin, cadherin, and immunoglobulin (e.g., NCAM and L1-CAM) superfamilies signal through FGFRs to induce neurite outgrowth, cell survival, and oncogenesis [61,62,63,64,65,66]. Integrins signaling can activate cell proliferation, survival, and invasion [67], and integrin α6 [68] and α7 [69] have been linked to GBM cancer stem cells (GSCs). A recent study suggested that integrin α6 regulates the expression of FGFR1 through ZEB1 and YAP transcription factors [70]. N-cadherin stabilizes FGFR1 and decreases its internalization, thus promoting invasion in breast cancer cells, and N-Cadherin/FGFR crosstalk promotes neurite outgrowth [71]. Of note, the stem cell transcription factor ZEB1 regulates N-Cadherin expression, which is associated with EMT and invasion. It is tempting to speculate that N-Cadherin/FGFR1 interactions could constitute a positive feedback loop in GSCs through the activation of ZEB1 and subsequent induction of N-Cadherin and FGFR1 expression [70,72] (see also Section 5.1). NCAMs physically associate with FGFRs and inhibit the high-affinity binding between these receptors and their canonical ligands [56,73]. Furthermore, polysialic acid-NCAM (PSA-NCAM) has been described as a marker of GBM patient prognosis [74]. This study showed that a targeted expression of PSA-NCAM in C6 glioma cells resulted in increased levels of Olig2, a transcription factor associated with GSCs [75]. While it remains unclear whether this was the result of FGFR transactivation by PSA-NCAM, we have recently shown that OLIG2 can be induced by FGFR1 signaling [72]. Furthermore, the L1-CAM/FGFR1/Anosmin-1 complex regulates neurite branching [76,77,78] and L1-CAM-mediated FGFR1 transactivation induces glioma cell proliferation and motility [79].
Crosstalk between FGFRs and other RTKs, such as EPHs and PDGFRs, has been identified in Y2H screens and endothelial cells [80,81]. FGFR/RTKs form a heterocomplex in which the tyrosine kinase domain of the FGFR is phosphorylated by the other receptor [56]. EPHA4 transactivated FGFR1 in the U251 glioma cell line, promoting cell growth and migration, and EPHA4 expression is increased in glioma [82]. Less is known about potential crosstalk between other RTKs and FGFRs in GBM. In summary, FGFR activity can be modulated non-canonically by other cell surface proteins, resulting in the activation of intracellular signaling pathways and cell responses associated with FGFR signaling.
5. Expression and Functions of FGFRs in Glioblastoma
Gene expression analysis of TCGA data (GBM 540) revealed profound heterogeneity of FGFR1–4 expression across GBM patients [72]. Below, we discuss the evidence for the functions of individual FGFRs in glioma.
5.1. FGFR1
Yamaguchi et al. found that expression of FGFR1 increases with WHO grade in astrocytomas [39], and increased FGFR1 levels in GBM are not due to amplification of the FGFR1 gene [83].
In addition to the increased expression of FGFR1 in malignant gliomas, the ratio of alternatively spliced FGFR1 α/β isoforms changes with progression to more aggressive brain cancers. While FGFR1 α is the predominant isoform in normal brain and low-grade gliomas, high-grade gliomas show a shift towards the expression of FGFR1 β [13,39]. Loss of the FGFR1 α exon increases the receptor–ligand affinity [28], thus, changes in alternative splicing may contribute to GBM malignancy by increasing the sensitivity of tumor cells to FGFs present in their environment.
Functionally, FGFR1 expression in malignant glioma has been associated with increased migration of cancer cells [82]. In this study, a high expression of EPHA4 in glioma cells was found to potentiate FGF2–FGFR1 signaling and promoted cell growth and migration through the AKT/MAPK and RAC1/CDC42 pathways, respectively. Data from our lab support that FGFR1 loss results in reduced tumor invasion in vivo (Jimenez-Pascual and Siebzehnrubl, unpublished observation).
Loilome and colleagues identified FGFR1 as a potential transducer of FGF2 effects on glioma cell proliferation [84], but whether other FGFRs also contribute was not directly tested. Nevertheless, the pharmacological inhibition of FGFR signaling significantly reduced tumor cell growth in a range of established and patient-derived glioma lines.
The malignancy-promoting effects of FGFR1 were further demonstrated in a study that found FGFR1 signaling promoting radioresistance in glioma cell lines through PLC1γ and HIF1α [85]. FGFR1 expression is regulated by the stem-cell associated transcription factor ZEB1 [70], suggesting that FGFR1 may be associated with GBM cancer stem cells. We recently performed a comprehensive analysis of the functions of FGFR1-3 in GBM and found that FGFR1 indeed is preferentially expressed on GSCs, where it regulates the expression of the critical stem cell transcription factors SOX2, OLIG2, and ZEB1, thereby promoting tumorigenicity in vivo [72]. In summary, FGFR1 is a key regulator of tumor growth, invasion, therapy resistance, and cancer stemness in malignant glioma.
5.2. FGFR2
While FGFR1 is mainly expressed on neurons [6], FGFR2 is the primary FGFR on astrocytes [5]. In contrast to FGFR1, FGFR2 expression decreases with glioma grade [43]. Reduced expression of FGFR2, as well as its IIIb and IIIc isoforms, is associated with a higher tumor grade and poorer survival in glioma patients [43]. Tumors with a higher expression of FGFR2 showed significantly less proliferation, as identified by Ki-67 staining, but whether there is a direct link between FGFR2 signaling and slowing or exiting the cell cycle remains unclear. By contrast, experimental tumors derived from in vivo implantation of C6 glioma cells exhibited decreased tumor growth after inhibition of FGFR2 signaling by a dominant negative construct [86].
Our recent analysis of cell-surface FGFR expression patterns in GBM stem cell lines indicates that FGFR2 is nevertheless highly prevalent on GBM cells in vitro [72], but it remains to be tested whether FGFR2 loss results in increased proliferation and/or tumorigenicity. Loss of FGFR2 is associated with a loss of Chr. 10q, which in and of itself carries an unfavorable prognosis [87]. It is therefore conceivable that FGFR2 loss is not causally linked to reduced patient survival, and further work is needed to clarify the functional relevance of FGFR2 signaling in GBM.
5.3. FGFR3
In a small subset of GBM patients, fusion of the FGFR3 and TACC3 genes generates an oncogenic FGFR3 form [16]. In rare cases, fusion between FGFR1 and TACC1 can occur as well [88]. In FGFR3–TACC3, the FGFR tyrosine kinase domain is fused to the TACC coiled-coil domain, resulting in constitutive activation of the fused receptor. Small-molecule FGFR inhibitors were effective at blocking tumor growth where FGFR–TACC fusion occurred, indicating that the fused receptor is causally linked to tumor development. Overall, FGFR–TACC fusions are found in ~3% of gliomas and are mutually exclusive with EGFR amplifications. Recently, it has been shown that FGFR–TACC fusions affect cell metabolism, activating oxidative phosphorylation and mitochondrial activity [89]. Of note, we have recently found that GSCs preferentially utilize oxidative phosphorylation and mitochondrial respiration [90], and therefore it would be interesting to investigate whether FGFR–TACC fusions also affect stemness pathways in GBM.
Whether FGFR3 has specific functions that differ from FGFR1 or -2 in GBM, and/or whether signaling through FGFR3 activates specific downstream signaling pathways remains unclear. Of note, global transcriptomic analysis of TCGA and CGGA datasets found increased expression of FGFR3 in the classical and neural subtypes of GBM [91]. Gene ontology analysis showed an association of FGFR3 expression with biological processes of cell differentiation in this study.
A recent study investigating gene expression using single-cell RNA-Seq in GBM patients found that FGFR3 expression is five-fold higher in invasive GBM cells compared to the tumor core [92]. Indeed, FGFR3 was the second highest differentially expressed gene between invasive and tumor core GBM cells. While this suggests that FGFR3 may be functionally associated with tumor invasion, whether FGFR3 signaling is driving GBM invasion remains to be shown.
5.4. FGFR4
Very little evidence exists of the expression of FGFR4 in GBM. An early study found increased expression of the FGFR4 protein, but not mRNA, with increasing grade in astrocytoma [93]. Another study demonstrated the expression of FGFR4 across different GBM cell lines [84]. We recently investigated FGFR protein expression in GBM cells [72]. In our study, FGFR4 was not detectable by western blot in primary patient-derived GBM cells, and analysis of GBM patient data in the TCGA dataset showed heterogeneous, but overall low expression of FGFR4. Moreover, we could not find differences in survival when stratifying patients for FGFR4 high or low expression. More research is needed to fully characterize whether FGFR4 is expressed on subsets of GBM cells, and whether it is functional in these cancers.
6. FGFRs as Therapeutic Targets in GBM
Oncogenic FGFR signaling promotes malignancy in many cancers, including CNS malignancies. Thus, pharmacological targeting of FGFRs may be therapeutically beneficial. A number of RTK inhibitors have been developed that show selectivity of FGFRs over other RTKs [18]. While some small-molecule inhibitors also target non-FGFR RTKs (e.g., dovitinib, levatinib, brivanib), others are selective for FGFR1–3 (e.g., PD173074, BGJ398, AZ4547, JNJ-493). To date, no small-molecule inhibitors exist with good selectivity for individual FGFR subtypes or isoforms [18].
Recently, a study identified FGFR signaling as a potential therapeutic target in pediatric glioma using a large-scale shRNA screen [94]. In this study, FGFR inhibitors (AZ4547, dovatinib, PD173074, ponatinib) were more effective in reducing the growth of pediatric glioma cells in vitro than the first-line chemotherapeutic agent Temozolomide.
The selective FGFR inhibitors AZ4547 and BGJ398 have been tested in clinical phase I/II (NCT028224133) and phase II trials (NCT01975701), respectively. AZ4547 was trialed in patients with recurrent IDH wild-type gliomas with FGFR1–TACC1 or FGFR3–TACC3 fusions, but this trial was suspended after analysis of the data from the first 12 patients. A trial of BGJ398 in malignant glioma patients with FGFR1–TACC1 or FGFR3–TACC3 fusion, and/or activating mutation in FGFR1, -2, or -3, was completed, but so far, no results have been published.
A phase I/II trial of the irreversible FGFR inhibitor TAS-120 (NCT02052778) is currently recruiting patients with advanced solid tumors, including brain tumors. As with the AZ4547 and BGJ398 trials, the focus of this trial is on patients with FGFR gene fusions or activating mutations.
Due to the prevalence of FGFRs on many CNS cells, and the importance of FGFR signaling for CNS cell survival, as well as the apparent intratumoral and intertumoral heterogeneity of FGFR expression in GBM, it will be interesting to see whether FGFR inhibitors are successful as monotherapy. Based on the evidence implicating FGFR1 as a GSC regulator, it is further tempting to speculate whether the targeted inhibition of FGFR1 in combination with conventional chemo/radiotherapy could prevent or delay recurrence in GBM.
7. Conclusions
Gene expression profiling and whole-genome sequencing data indicate that all four FGFRs are expressed to varying degrees in GBM, underlining the heterogeneity of this disease. Several studies have documented that high-grade gliomas show an increased expression of FGFR1, and decreased expression of FGFR2 [13,39,43,72,83], but in almost all cases, FGFR expression was detected at a global level, and limited or no effort was made to further identify FGFR splice isoforms. A recent report investigating gene expression at the single-cell level [92] found that FGFR3 was expressed the second-highest in invasive GBM cells. This illustrates that much more research is needed to unravel the functions of individual FGFRs and their splice isoforms in brain cancers in general, and GBM in particular.
To what extent signaling through individual FGFRs contributes to disease progression, and whether individual FGFRs and/or isoforms activate specific pathways linked to different pathobiological aspects of these cancers (e.g., invasion, tumor initiation, therapy resistance) remains largely unknown. Yet, the fact that FGFR subtypes are differentially expressed on cellular subpopulations within the same tumor suggests that individual FGFRs may have divergent functions in GBM [72,92].
The strongest evidence by far indicates that FGFR1 is an important contributor to poor outcome in GBM, and FGFR1 signaling is linked to cancer stemness, invasion, and radioresistance [70,72,85]. However, recent evidence from TCGA datasets highlights that FGFR1–4 are expressed to varying degrees and in different combinations in patient samples [72]. This calls for a more detailed analysis of FGFR distribution across GBM patients and within individual tumors. It is conceivable that different combinations of FGFR subtypes and splice isoforms mediate and/or modulate different aspects of FGF signaling in GBM cells. Only a comprehensive analysis of the cell-surface expression of FGFRs at the single-cell level will dissect the intratumoral heterogeneity of these receptors, and thus provide the foundation for new, targeted approaches to blocking FGFR signaling for glioma therapy.
Author Contributions
Conceptualization, A.J.-P. and F.A.S.; writing—original draft preparation, A.J.-P. and F.A.S.; writing—review and editing, A.J.-P. and F.A.S.; visualization, A.J.-P.; supervision, F.A.S.; project administration, F.A.S.; funding acquisition, F.A.S.
Funding
This research was funded by MRC, grant number MR/S007709/1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Trowell O.A., Willmer E.N. Studies on the growth of tissues in vitro VI. The effects of some tissue extracts on the growth of periosteal fibroblasts. J. Exp. Biol. 1939;16:60–70. [Google Scholar]
- 2.Lee P.L., Johnson D.E., Cousens L.S., Fried V.A., Williams L.T. Purification and complementary DNA cloning of a receptor for basic fibroblast growth factor. Science. 1989;245:57–60. doi: 10.1126/science.2544996. [DOI] [PubMed] [Google Scholar]
- 3.Beenken A., Mohammadi M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug. Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillemot F., Zimmer C. From cradle to grave: The multiple roles of fibroblast growth factors in neural development. Neuron. 2011;71:574–588. doi: 10.1016/j.neuron.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Miyake A., Hattori Y., Ohta M., Itoh N. Rat oligodendrocytes and astrocytes preferentially express fibroblast growth factor receptor-2 and -3 mRNAs. J. Neurosci. Res. 1996;45:534–541. doi: 10.1002/(SICI)1097-4547(19960901)45:5<534::AID-JNR3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez A.M., Berry M., Maher P.A., Logan A., Baird A. A comprehensive analysis of the distribution of FGF-2 and FGFR1 in the rat brain. Brain Res. 1995;701:201–226. doi: 10.1016/0006-8993(95)01002-X. [DOI] [PubMed] [Google Scholar]
- 7.Frinchi M., Bonomo A., Trovato-Salinaro A., Condorelli D.F., Fuxe K., Spampinato M.G., Mudo G. Fibroblast growth factor-2 and its receptor expression in proliferating precursor cells of the subventricular zone in the adult rat brain. Neurosci. Lett. 2008;447:20–25. doi: 10.1016/j.neulet.2008.09.059. [DOI] [PubMed] [Google Scholar]
- 8.Haugsten E.M., Wiedlocha A., Olsnes S., Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol. Cancer Res. 2010;8:1439–1452. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- 9.Greulich H., Pollock P.M. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol. Med. 2011;17:283–292. doi: 10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiong K.H., Mah L.Y., Leong C.O. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis. 2013;18:1447–1468. doi: 10.1007/s10495-013-0886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa R., Carneiro B.A., Taxter T., Tavora F.A., Kalyan A., Pai S.A., Chae Y.K., Giles F.J. FGFR3-TACC3 fusion in solid tumors: Mini review. Oncotarget. 2016;7:55924–55938. doi: 10.18632/oncotarget.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasorella A., Sanson M., Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017;19:475–483. doi: 10.1093/neuonc/now240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison R.S., Yamaguchi F., Saya H., Bruner J.M., Yahanda A.M., Donehower L.A., Berger M. Basic fibroblast growth factor and fibroblast growth factor receptor I are implicated in the growth of human astrocytomas. J. Neuro Oncol. 1994;18:207–216. doi: 10.1007/BF01328955. [DOI] [PubMed] [Google Scholar]
- 14.Yamada S.M., Yamaguchi F., Brown R., Berger M.S., Morrison R.S. Suppression of glioblastoma cell growth following antisense oligonucleotide-mediated inhibition of fibroblast growth factor receptor expression. Glia. 1999;28:66–76. doi: 10.1002/(SICI)1098-1136(199910)28:1<66::AID-GLIA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Dienstmann R., Rodon J., Prat A., Perez-Garcia J., Adamo B., Felip E., Cortes J., Iafrate A.J., Nuciforo P., Tabernero J. Genomic aberrations in the FGFR pathway: Opportunities for targeted therapies in solid tumors. Ann. Oncol. 2014;25:552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S., et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L., et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dieci M.V., Arnedos M., Andre F., Soria J.C. Fibroblast growth factor receptor inhibitors as a cancer treatment: From a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–279. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 19.Wiedemann M., Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg F., Zhuang L., Beyeler M., Kalin R.E., Mullis P.E., Brandli A.W., Trueb B. The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in Xenopus embryos. J. Biol. Chem. 2010;285:2193–2202. doi: 10.1074/jbc.M109.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmer N.J., Ilag L.L., Mulloy B., Pellegrini L., Robinson C.V., Blundell T.L. Towards a resolution of the stoichiometry of the fibroblast growth factor (FGF)-FGF receptor-heparin complex. J. Mol. Biol. 2004;339:821–834. doi: 10.1016/j.jmb.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad I., Iwata T., Leung H.Y. Mechanisms of FGFR-mediated carcinogenesis. Biochim. Biophys. Acta. 2012;1823:850–860. doi: 10.1016/j.bbamcr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Ornitz D.M., Marie P.J. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463–1486. doi: 10.1101/gad.266551.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammadi M., Olsen S.K., Ibrahimi O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Olsen S.K., Ibrahimi O.A., Raucci A., Zhang F., Eliseenkova A.V., Yayon A., Basilico C., Linhardt R.J., Schlessinger J., Mohammadi M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. USA. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiselyov V.V., Kochoyan A., Poulsen F.M., Bock E., Berezin V. Elucidation of the mechanism of the regulatory function of the Ig1 module of the fibroblast growth factor receptor 1. Protein Sci. 2006;15:2318–2322. doi: 10.1110/ps.062206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalinina J., Dutta K., Ilghari D., Beenken A., Goetz R., Eliseenkova A.V., Cowburn D., Mohammadi M. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure. 2012;20:77–88. doi: 10.1016/j.str.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Kan M., Yan G., Xu J., McKeehan W.L. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 1995;270:10231–10235. doi: 10.1074/jbc.270.17.10231. [DOI] [PubMed] [Google Scholar]
- 29.Yeh B.K., Igarashi M., Eliseenkova A.V., Plotnikov A.N., Sher I., Ron D., Aaronson S.A., Mohammadi M. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl. Acad. Sci. USA. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapraeger A.C., Krufka A., Olwin B.B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 31.Yayon A., Klagsbrun M., Esko J.D., Leder P., Ornitz D.M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-W. [DOI] [PubMed] [Google Scholar]
- 32.Gong S.G. Isoforms of receptors of fibroblast growth factors. J. Cell Physiol. 2014;229:1887–1895. doi: 10.1002/jcp.24649. [DOI] [PubMed] [Google Scholar]
- 33.Schlessinger J., Plotnikov A.N., Ibrahimi O.A., Eliseenkova A.V., Yeh B.K., Yayon A., Linhardt R.J., Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell. 2000;6:743–750. doi: 10.1016/S1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 34.Miki T., Bottaro D.P., Fleming T.P., Smith C.L., Burgess W.H., Chan A.M., Aaronson S.A. Determination of ligand-binding specificity by alternative splicing: Two distinct growth factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chellaiah A.T., McEwen D.G., Werner S., Xu J., Ornitz D.M. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J. Biol. Chem. 1994;269:11620–11627. [PubMed] [Google Scholar]
- 36.Kurosu H., Ogawa Y., Miyoshi M., Yamamoto M., Nandi A., Rosenblatt K.P., Baum M.G., Schiavi S., Hu M.C., Moe O.W., et al. Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 38.Adams A.C., Cheng C.C., Coskun T., Kharitonenkov A. FGF21 requires betaklotho to act in vivo. PLoS ONE. 2012;7:e49977. doi: 10.1371/journal.pone.0049977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi F., Saya H., Bruner J.M., Morrison R.S. Differential expression of two fibroblast growth factor-receptor genes is associated with malignant progression in human astrocytomas. Proc. Natl. Acad. Sci. USA. 1994;91:484–488. doi: 10.1073/pnas.91.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomlinson D.C., Knowles M.A. Altered splicing of FGFR1 is associated with high tumor grade and stage and leads to increased sensitivity to FGF1 in bladder cancer. Am. J. Pathol. 2010;177:2379–2386. doi: 10.2353/ajpath.2010.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Holzmann K., Grunt T., Heinzle C., Sampl S., Steinhoff H., Reichmann N., Kleiter M., Hauck M., Marian B. Alternative Splicing of Fibroblast Growth Factor Receptor IgIII Loops in Cancer. J. Nucleic Acids. 2012;2012:950508. doi: 10.1155/2012/950508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohashi R., Matsuda Y., Ishiwata T., Naito Z. Downregulation of fibroblast growth factor receptor 2 and its isoforms correlates with a high proliferation rate and poor prognosis in high-grade glioma. Oncol. Rep. 2014;32:1163–1169. doi: 10.3892/or.2014.3283. [DOI] [PubMed] [Google Scholar]
- 44.Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi A., Ishii H., Morita I., Oota I., Takeda H. mRNA expression of fibroblast growth factors and hepatocyte growth factor in rat plantaris muscle following denervation and compensatory overload. Pflugers Arch. 2004;448:539–546. doi: 10.1007/s00424-004-1282-5. [DOI] [PubMed] [Google Scholar]
- 46.Burgar H.R., Burns H.D., Elsden J.L., Lalioti M.D., Heath J.K. Association of the signaling adaptor FRS2 with fibroblast growth factor receptor 1 (Fgfr1) is mediated by alternative splicing of the juxtamembrane domain. J. Biol. Chem. 2002;277:4018–4023. doi: 10.1074/jbc.M107785200. [DOI] [PubMed] [Google Scholar]
- 47.Sarabipour S., Hristova K. FGFR3 unliganded dimer stabilization by the juxtamembrane domain. J. Mol. Biol. 2015;427:1705–1714. doi: 10.1016/j.jmb.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Gorry M.C., Post J.C., Ehrlich G.D. Genomic organization of the human fibroblast growth factor receptor 2 (FGFR2) gene and comparative analysis of the human FGFR gene family. Gene. 1999;230:69–79. doi: 10.1016/S0378-1119(99)00047-5. [DOI] [PubMed] [Google Scholar]
- 49.Lew E.D., Furdui C.M., Anderson K.S., Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Touat M., Ileana E., Postel-Vinay S., Andre F., Soria J.C. Targeting FGFR Signaling in Cancer. Clin. Cancer Res. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 51.Ori A., Wilkinson M.C., Fernig D.G. The heparanome and regulation of cell function: Structures, functions and challenges. Front. Biosci. 2008;13:4309–4338. doi: 10.2741/3007. [DOI] [PubMed] [Google Scholar]
- 52.Opalinski L., Sokolowska-Wedzina A., Szczepara M., Zakrzewska M., Otlewski J. Antibody-induced dimerization of FGFR1 promotes receptor endocytosis independently of its kinase activity. Sci. Rep. 2017;7:7121. doi: 10.1038/s41598-017-07479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auciello G., Cunningham D.L., Tatar T., Heath J.K., Rappoport J.Z. Regulation of fibroblast growth factor receptor signalling and trafficking by Src and Eps8. J. Cell Sci. 2013;126:613–624. doi: 10.1242/jcs.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandilands E., Akbarzadeh S., Vecchione A., McEwan D.G., Frame M.C., Heath J.K. Src kinase modulates the activation, transport and signalling dynamics of fibroblast growth factor receptors. EMBO Rep. 2007;8:1162–1169. doi: 10.1038/sj.embor.7401097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kundu S., Xiong A., Spyrou A., Wicher G., Marinescu V.D., Edqvist P.D., Zhang L., Essand M., Dimberg A., Smits A., et al. Heparanase promotes glioma progression and is inversely correlated with patient survival. Mol. Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-16-0223. [DOI] [PubMed] [Google Scholar]
- 56.Latko M., Czyrek A., Porebska N., Kucinska M., Otlewski J., Zakrzewska M., Opalinski L. Cross-talk between fibroblast growth factor receptors and other cell surface proteins. Cells. 2019;8:455. doi: 10.3390/cells8050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Liberto V., Mudo G., Belluardo N. Crosstalk between receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCR) in the brain: Focus on heteroreceptor complexes and related functional neurotrophic effects. Neuropharmacology. 2019;152:67–77. doi: 10.1016/j.neuropharm.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 58.Asimaki O., Leondaritis G., Lois G., Sakellaridis N., Mangoura D. Cannabinoid 1 receptor-dependent transactivation of fibroblast growth factor receptor 1 emanates from lipid rafts and amplifies extracellular signal-regulated kinase 1/2 activation in embryonic cortical neurons. J. Neurochem. 2011;116:866–873. doi: 10.1111/j.1471-4159.2010.07030.x. [DOI] [PubMed] [Google Scholar]
- 59.Borroto-Escuela D.O., Carlsson J., Ambrogini P., Narvaez M., Wydra K., Tarakanov A.O., Li X., Millon C., Ferraro L., Cuppini R., et al. Understanding the role of GPCR heteroreceptor complexes in modulating the brain networks in health and disease. Front. Cell Neurosci. 2017;11:37. doi: 10.3389/fncel.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belcheva M.M., Haas P.D., Tan Y., Heaton V.M., Coscia C.J. The fibroblast growth factor receptor is at the site of convergence between mu-opioid receptor and growth factor signaling pathways in rat C6 glioma cells. J. Pharmacol. Exp. Ther. 2002;303:909–918. doi: 10.1124/jpet.102.038554. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez-Heras E., Howell F.V., Williams G., Doherty P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J. Biol. Chem. 2006;281:35208–35216. doi: 10.1074/jbc.M608655200. [DOI] [PubMed] [Google Scholar]
- 62.Williams E.J., Furness J., Walsh F.S., Doherty P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron. 1994;13:583–594. doi: 10.1016/0896-6273(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 63.Condic M.L., Letourneau P.C. Ligand-induced changes in integrin expression regulate neuronal adhesion and neurite outgrowth. Nature. 1997;389:852–856. doi: 10.1038/39878. [DOI] [PubMed] [Google Scholar]
- 64.Boscher C., Mege R.M. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal. 2008;20:1061–1072. doi: 10.1016/j.cellsig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 65.Meiri K.F., Saffell J.L., Walsh F.S., Doherty P. Neurite outgrowth stimulated by neural cell adhesion molecules requires growth-associated protein-43 (GAP-43) function and is associated with GAP-43 phosphorylation in growth cones. J. Neurosci. 1998;18:10429–10437. doi: 10.1523/JNEUROSCI.18-24-10429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doherty P., Cohen J., Walsh F.S. Neurite outgrowth in response to transfected N-CAM changes during development and is modulated by polysialic acid. Neuron. 1990;5:209–219. doi: 10.1016/0896-6273(90)90310-C. [DOI] [PubMed] [Google Scholar]
- 67.Bianconi D., Unseld M., Prager G.W. Integrins in the spotlight of cancer. Int. J. Mol. Sci. 2016;17:2037. doi: 10.3390/ijms17122037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lathia J.D., Gallagher J., Heddleston J.M., Wang J., Eyler C.E., Macswords J., Wu Q., Vasanji A., McLendon R.E., Hjelmeland A.B., et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas T.L., Sciuto M.R., Brunetto L., Valvo C., Signore M., Fiori M.E., di Martino S., Giannetti S., Morgante L., Boe A., et al. Integrin alpha7 is a functional marker and potential therapeutic target in glioblastoma. Cell Stem Cell. 2017;21:35–50. doi: 10.1016/j.stem.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 70.Kowalski-Chauvel A., Gouaze-Andersson V., Baricault L., Martin E., Delmas C., Toulas C., Cohen-Jonathan-Moyal E., Seva C. Alpha6-integrin regulates FGFR1 expression through the ZEB1/YAP1 transcription complex in glioblastoma stem cells resulting in enhanced proliferation and stemness. Cancers. 2019;11:406. doi: 10.3390/cancers11030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen T., Mege R.M. N-cadherin and fibroblast growth factor receptors crosstalk in the control of developmental and cancer cell migrations. Eur. J. Cell Biol. 2016;95:415–426. doi: 10.1016/j.ejcb.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 72.Hale J.S., Jimenez-Pascual A., Kordowski A., Pugh J., Rao S., Silver D.J., Alban T., Watson D.B., Chen R., McIntyre T.M., et al. ADAMDEC1 maintains a novel growth factor signaling loop in cancer stem cells. bioRxiv. 2019 doi: 10.1101/531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Francavilla C., Loeffler S., Piccini D., Kren A., Christofori G., Cavallaro U. Neural cell adhesion molecule regulates the cellular response to fibroblast growth factor. J. Cell Sci. 2007;120:4388–4394. doi: 10.1242/jcs.010744. [DOI] [PubMed] [Google Scholar]
- 74.Amoureux M.C., Coulibaly B., Chinot O., Loundou A., Metellus P., Rougon G., Figarella-Branger D. Polysialic acid neural cell adhesion molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer. 2010;10:91. doi: 10.1186/1471-2407-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ligon K.L., Huillard E., Mehta S., Kesari S., Liu H., Alberta J.A., Bachoo R.M., Kane M., Louis D.N., Depinho R.A., et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bribian A., Barallobre M.J., Soussi-Yanicostas N., de Castro F. Anosmin-1 modulates the FGF-2-dependent migration of oligodendrocyte precursors in the developing optic nerve. Mol. Cell Neurosci. 2006;33:2–14. doi: 10.1016/j.mcn.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 77.Garcia-Gonzalez D., Clemente D., Coelho M., Esteban P.F., Soussi-Yanicostas N., de Castro F. Dynamic roles of FGF-2 and Anosmin-1 in the migration of neuronal precursors from the subventricular zone during pre- and postnatal development. Exp. Neurol. 2010;222:285–295. doi: 10.1016/j.expneurol.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 78.Murcia-Belmonte V., Esteban P.F., Garcia-Gonzalez D., De Castro F. Biochemical dissection of Anosmin-1 interaction with FGFR1 and components of the extracellular matrix. J. Neurochem. 2010;115:1256–1265. doi: 10.1111/j.1471-4159.2010.07024.x. [DOI] [PubMed] [Google Scholar]
- 79.Mohanan V., Temburni M.K., Kappes J.C., Galileo D.S. L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor. Clin. Exp. Metastasis. 2013;30:507–520. doi: 10.1007/s10585-012-9555-4. [DOI] [PubMed] [Google Scholar]
- 80.Chen P.Y., Simons M., Friesel R. FRS2 via fibroblast growth factor receptor 1 is required for platelet-derived growth factor receptor beta-mediated regulation of vascular smooth muscle marker gene expression. J. Biol. Chem. 2009;284:15980–15992. doi: 10.1074/jbc.M809399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yokote H., Fujita K., Jing X., Sawada T., Liang S., Yao L., Yan X., Zhang Y., Schlessinger J., Sakaguchi K. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc. Natl. Acad. Sci. USA. 2005;102:18866–18871. doi: 10.1073/pnas.0509741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukai J., Yokote H., Yamanaka R., Arao T., Nishio K., Itakura T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer Ther. 2008;7:2768–2778. doi: 10.1158/1535-7163.MCT-07-2263. [DOI] [PubMed] [Google Scholar]
- 83.Morrison R.S., Yamaguchi F., Bruner J.M., Tang M., McKeehan W., Berger M.S. Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 1994;54:2794–2799. [PubMed] [Google Scholar]
- 84.Loilome W., Joshi A.D., ap Rhys C.M., Piccirillo S., Vescovi A.L., Gallia G.L., Riggins G.J. Glioblastoma cell growth is suppressed by disruption of fibroblast growth factor pathway signaling. J. Neuro Oncol. 2009;94:359–366. doi: 10.1007/s11060-009-9885-5. [DOI] [PubMed] [Google Scholar]
- 85.Gouaze-Andersson V., Delmas C., Taurand M., Martinez-Gala J., Evrard S., Mazoyer S., Toulas C., Cohen-Jonathan-Moyal E. FGFR1 induces glioblastoma radioresistance through the PLCgamma/Hif1alpha pathway. Cancer Res. 2016;76:3036–3044. doi: 10.1158/0008-5472.CAN-15-2058. [DOI] [PubMed] [Google Scholar]
- 86.Miraux S., Lemiere S., Pineau R., Pluderi M., Canioni P., Franconi J.M., Thiaudiere E., Bello L., Bikfalvi A., Auguste P. Inhibition of FGF receptor activity in glioma implanted into the mouse brain using the tetracyclin-regulated expression system. Angiogenesis. 2004;7:105–113. doi: 10.1007/s10456-004-1037-0. [DOI] [PubMed] [Google Scholar]
- 87.Daido S., Takao S., Tamiya T., Ono Y., Terada K., Ito S., Ouchida M., Date I., Ohmoto T., Shimizu K. Loss of heterozygosity on chromosome 10q associated with malignancy and prognosis in astrocytic tumors, and discovery of novel loss regions. Oncol. Rep. 2004;12:789–795. doi: 10.3892/or.12.4.789. [DOI] [PubMed] [Google Scholar]
- 88.Di Stefano A.L., Fucci A., Frattini V., Labussiere M., Mokhtari K., Zoppoli P., Marie Y., Bruno A., Boisselier B., Giry M., et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin. Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frattini V., Pagnotta S.M., Fan J.J., Russo M.V., Lee S.B., Garofano L., Zhang J., Shi P., Lewis G., Zhang J., et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018;553:222–227. doi: 10.1038/nature25171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoang-Minh L.B., Siebzehnrubl F.A., Yang C., Suzuki-Hatano S., Dajac K., Loche T., Andrews N., Schmoll Massari M., Patel J., Amin K., et al. Infiltrative and drug-resistant slow-cycling cells support metabolic heterogeneity in glioblastoma. EMBO J. 2018;37:e98772. doi: 10.15252/embj.201798772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z., Zhang C., Sun L., Liang J., Liu X., Li G., Yao K., Zhang W., Jiang T. FGFR3, as a receptor tyrosine kinase, is associated with differentiated biological functions and improved survival of glioma patients. Oncotarget. 2016;7:84587–84593. doi: 10.18632/oncotarget.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Darmanis S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamada S.M., Yamada S., Hayashi Y., Takahashi H., Teramoto A., Matsumoto K. Fibroblast growth factor receptor (FGFR) 4 correlated with the malignancy of human astrocytomas. Neurol. Res. 2002;24:244–248. doi: 10.1179/016164102101199864. [DOI] [PubMed] [Google Scholar]
- 94.Schramm K., Iskar M., Statz B., Jager N., Haag D., Slabicki M., Pfister S.M., Zapatka M., Gronych J., Jones D.T.W., et al. DECIPHER pooled shRNA library screen identifies PP2A and FGFR signaling as potential therapeutic targets for DIPGs. Neuro Oncol. 2019 doi: 10.1093/neuonc/noz057. [DOI] [PMC free article] [PubMed] [Google Scholar]