Abstract
The aim of this study was to investigate genetic diversity of Helicobacter pylori virulence markers to predict clinical outcome as well as to determine an antibiotic susceptibility of H. pylori strains in Poland. Gastric biopsies from 132 patients with gastrointestinal disorders were tested for presence of H. pylori with the use of rapid urease test, microbial culture, and polymerase chain reaction (PCR) detection. The genetic diversity of 62 H. pylori positive samples was evaluated by detection of cagA and PCR-typing of vacA and iceA virulence-associated genes. Most common H. pylori genotypes were cagA(+)vacAs1m2 (27.4%) and cagA(−)vacAs2m2 (24.2%). In logistic regression analysis, we recognized the subsequent significant associations: gastritis with ureC, i.e., H. pylori infection (p = 0.006), BMI index (p = 0.032); and negatively with iceA1 (p = 0.049) and peptic ulcer with cagA (p = 0.018). Thirty-five H. pylori strains were cultured and tested by E-test method showing that 49% of strains were resistant to at least one of the tested antibiotics. This is the first study that reports the high incidence and diversity of allelic combination of virulence genes in gastroduodenitis patients in Poland. Genotyping of H. pylori strains confirmed the involvement of cagA gene and vacAs1m1 genotype in development and severity of gastric disorder.
Keywords: pylorus infection, cagA, vacA, iceA, antibiotic patterns
1. Introduction
Helicobacter pylori is an etiological factor of the most frequent and persistent bacterial infection worldwide, which affects nearly half of the world’s population. H. pylori infections are more common in developing countries—up to 90% of population, whereas in developed countries—below 40% [1]. In northern Europe, about 30% of adults are infected, whereas in south and east Europe the prevalence of H. pylori is often higher than 50%. The highest prevalence was reported in Portugal, similarly to Poland, where 84% of adult population is infected with this bacteria, which is a serious epidemiological problem [2,3]. H. pylori is associated with gastric ulcer, peptic ulcer disease, gastric cancer, and mucosa-associated lymphoma, but about 70% of infected population are carriers who stay asymptomatic [4].
The clinical outcome of H. pylori infection was supposed to be linked to certain strains differing in virulence factors presence or subtypes [5]. The presence of the cytotoxin-associated gene A (cagA) and the most active (s1m1) form of the vacuolating toxin (vacA) are the most important features of the bacterium in relation with higher risk of gastric adenocarcinoma and its pre-malignant lesions [6,7,8,9]. H. pylori is genetically heterogeneous, suggesting a lack of its clonality. It results in every H. pylori-positive subject carrying a distinct strain. This is possibly an adaptation of H. pylori to the gastric conditions of its host, as well as to the distinct patterns of the host-mediated immune response to H. pylori infection [10].
Treatment failure of H. pylori infection is caused mainly by progressive antibiotic resistance among H. pylori strains and numerous studies have shown that the prevalence of H. pylori antibiotic resistance varies significantly between countries, and even between regions within the same country. Local surveillance of antibiotic resistance is warranted to guide clinicians in their choice of therapy [11]. In regions with low resistance rates to clarithromycin (less than 20%), a standard first line-therapy which contains clarithromycin is recommended. In regions with high resistance to clarithromycin (>20%), first-line therapy with bismuth salts is recommended and, if it is not available, sequential therapy (Proton Pump Inhibitor (PPI), amoxicillin for 5 days and PPI, clarithromycin, metronidazole, or tinidazole for the next 5 days) [3,12].
Numerous diagnostic assays for H. pylori detection are available: bacterial culture, a rapid urease test (RUT), a urea breath test, histology, polymerase chain reaction (PCR), serology, and stool antigen test [13]. All these techniques have their limitations. Here we have tried to compare three diagnostic methods—microbiological culture, rapid urease test (RUT), and detection of ureC gene by PCR in biopsies from patients with gastroduodenal disease. Moreover, the pathogenicity of H. pylori strains was evaluated by detection of cytotoxin associated gene A (cagA), and genotyping of vacuolating cytotoxin gene A (vacA) and iceA gene and correlated them with the clinical outcomes. Antibiotic resistance was assessed by H. pylori culture and antibiotic susceptibility testing to monitor effectiveness of recommended treatments.
2. Material and Methods
2.1. Patients
Gastric biopsies were collected from 132 patients with indication of endoscopy (78 women and 54 men, the mean age 57.3 years, SD 15.6, median 59 years, range 20–87 years) who attended the Department and Clinic of Gastroenterology with Endoscopic Unit of Medical University of Lublin, Poland in 2016. Gastric biopsies were evaluated according to updated Sydney System. Chronic gastritis was defined as increased number of lymphocytes and plasma cells in the lamina propria of gastric mucosa. The degree of inflammation was referred to as mild, moderate and severe. Activity of inflammation was determined on the basis of neutrophilic infiltrates of the lamina propria, pits, or surface epithelium. Mild activity was diagnosed when less than one third of pits and surface was infiltrated, moderate—one-third to two-thirds, more than two-thirds defined severe activity of gastritis. The presence of atrophy, intestinal metaplasia, and intraepithelial neoplasia were determined. Attention was paid to the presence of lymphoplasia and erosions. In 23 male and in 21 female patients, moderate chronic gastritis with moderate activity was diagnosed, 2 remaining women had severe chronic gastritis with moderate activity. In two men, intestinal metaplasia was diagnosed and in none of the patients atrophic gastritis was confirmed. The participants with unintentional weight loss within the previous 6 months, upper abdominal mass, dysphagia, previous gastric resection, bleeding tendency and pregnancy were excluded from the study. Two biopsies from the distal corpus and two biopsies from the antrum were obtained during the panendoscopy. The endoscopic examination findings were: gastritis/duodenitis, peptic ulcer (gastric or duodenal ulcer), GERD (gastroesophageal reflux disease), and normal (Table 1). From each patient, informed consent was obtained. The Ethical Committee of the Medical University of Lublin approved the study protocol (no. KE-0254/174/2014).
Table 1.
Demographic and clinical features of studied population.
| Clinical Diagnosis | No. of Patients (Mean Age in Years ± SD) | ||
|---|---|---|---|
| Male (n = 54) |
Female (n = 78) |
Total (n = 132) |
|
| Gastritis/duodenitis | 23 (57.0 ± 15.3) | 23 (57.5 ± 16.6) | 46 (57.2 ± 15.6) |
| Peptic ulcer (gastric/duodenal) | 4 (55.0 ± 12.6) | 5 (66.4 ± 10.6) | 9 (61.3 ± 12.3) |
| Gastroesophageal reflux disease | 27 (58.1 ± 15.5) | 44 (56.2 ± 16.4) | 71 (56.9 ± 16.0) |
| Normal | 1 (31.0) | 5 (57.4 ± 17.6) | 6 (53.0 ± 19.1) |
2.2. Culture, Identification, and DNA Extraction
Samples were tested for the presence of urease with the use of RUT—Rapid Urease Test (CLO test Kimberly-Clark) and placed in tubes with 0.9% NaCl solution and transported to the Department of Pharmaceutical Microbiology with Laboratory for Microbiological Diagnostics, Medical University of Lublin, Poland. Biopsies were smashed with the use of two microscope slides and spread onto Schaedler agar (BioMerieux, Marcy-l’Étoile, France) with 5% sheep blood plates and Schaedler agar with 5% sheep blood plates additionally supplemented with DENT (Oxoid), incubated for 3–7 days at 35 °C in microaerophilic (5–10% CO2, 80–90% N2, 5–10% O2, Generbag microaer, BioMerieux) conditions. H. pylori isolates were identified by colony morphology, Gram-staining and positive catalase (3% H2O2), urease and oxidase (BBL DrySlide Oxidase—Becton Dickinson, Franklin Lakes, NJ, USA) tests.
DNA of biopsy sample and bacterial isolates were extracted using QIAGEN QIAamp DNA Mini Kit (Qiagen, USA) according to the manufacturer’s instructions. The identification in biopsy samples and isolates were further confirmed as H. pylori using ureC gene amplification with specific primers [14]. Extracted DNA was frozen to −70 °C until its use.
2.3. Amplification Experiments and Gene Detection
Determination of the presence cagA and genotypes of the genes vacA s, vacA m, iceA was performed by PCR amplification of the target genes from genomic DNA, using previously described oligonucleotides and protocols [15,16]. PCR reactions were performed with the use of the REDTaq ReadyMix PCR Reaction Mix (Sigma-Aldrich, St. Louis, MO, USA), followed by electrophoresis in 1.5% agarose gel (Sigma-Aldrich, St. Louis, MO, USA).
2.4. Antibiotic Susceptibility Testing
The strains obtained during the 72-h cultivation were then suspended in the brain heart infusion broth (BHI, Becton Dickinson, Germany). Cell concentration was determined using a densitometer (BioMeriux). Bacterial suspensions with a density of three according to the McFarland scale, i.e., 108 cells (CFU)/1 mL were used for susceptibility testing. In subsequent steps, the susceptibility of the strains to the antibiotics amoxicillin (AC), clarithromycin (CH), metronidazole (MZ), tetracycline (TC), levofloxacin (LE), and rifampicin (RI) was determined by the E-test method using E-test strips (AB Biodisc, Solna). The strips were placed on Mueller–Hinton agar supplemented with 5% horse blood and 20 mg/L β-NAD. The incubation was performed in microaerophilic conditions for 3 days in 35 °C. Resistance breakpoints of H. pylori were interpreted according to EUCAST (AC—0.125 mg/L; CL—0.5 mg/L; TC—1 mg/L; MZ—8 mg/L; RI—1 mg/L; LE—1 mg/L).
2.5. Statistical Analysis
Data processing and analysis were performed using STATISTICA 13 (StatSoft. Inc., Tulsa, OK, USA). Shapiro–Wilk test was applied to test normal distribution of continuous variables. The Student t-test or Mann–Whitney U-test or Kruskal–Wallis ANOVA analysis for independent variables were used as intergroup comparisons component. The distribution of discrete variables in groups were compared with Pearson’s Chi-square test or the Fisher’s exact test. Odds ratio (OR) and their 95% confidence intervals (CI) were calculated. Logistic regression models were fitted to identify risk factors associated with different clinical diagnoses (GAST, GERD, PUD) using H. pylori genotypes and demographic data (age, gender, BMI, place of residence, smoking) as independent factors. Statistical significance was set at p < 0.05. The following indexes were calculated for RUT samples and microbial culture in relation to PCR samples: sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
3. Results
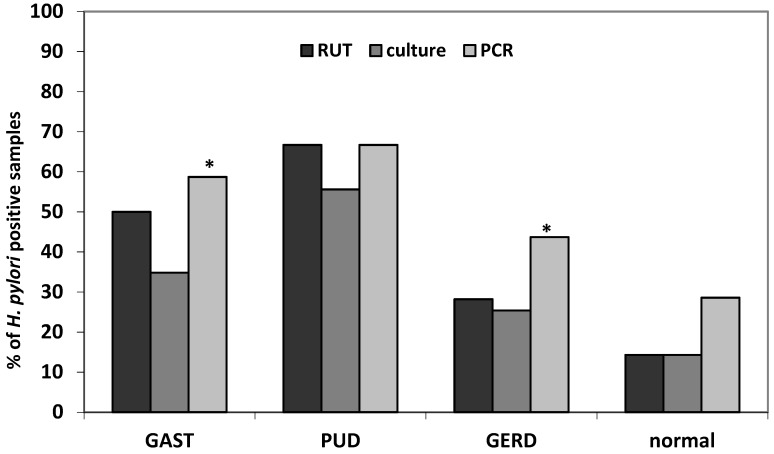
Out of 132 patients, 35 (26.5%) microbial cultures, 45 (34.1%) RUT and 62 (47.0%) PCR tests were positive for H. pylori. Significantly higher detection rates for PCR method comparing to microbial culture (RR 3.6, 95%CI 2.6–4.95, p < 0.0001) and RUT (RR 11.6, 95%CI 3.8–34.7, p < 0.0001) were observed (Figure 1). Sensitivity of microbial culture in comparison to PCR identification was 56.5% and specificity 1.0% (PPV 100.0% and NPV 72.2%). RUT test sensitivity was 67.7% and specificity—95.7% (PPV 93.3% and NPV 77.0%). However, the same analysis carried out in different clinical outcomes showed significant difference in GAST and GERD groups only (Figure 1).
Figure 1.
H. pylori identification using different methods regarding clinical diagnosis (GAST, gastritis/duodenitis; PUD, peptic ulcer disease (gastric/duodenal); GERD, gastroesophageal reflux disease); * p < 0.05 (PCR vs. culture).
Table 1 presents demographic and clinical features of studied population. Male gender (57.4% vs. 39.7%; RR 1.4, 95%CI 1.0–2.0, p = 0.046) and residence in rural areas (66.0% vs. 36.9%; RR 1.5, 95%CI 1.2–2.0, p = 0.0019) were significantly associated with H. pylori infection.
Genotype distribution of H. pylori positive samples according to PCR analysis was presented in Table 2. It was shown that 56.5% of tested H. pylori were cagA positive. Regarding iceA gene: distribution of iceA1 and iceA2 were comparable (32.3% and 35.5%, respectively), and in 4.8% of samples mixed alleles iceA1 + iceA2 were detected. The vacA s1m2 genotype was the most common in H. pylori positive patients, with 33.9%, followed by s2m2 with 29.0% and s1m1 with 19.4%. The co-infection of s1m1 with s1m2 and s2m1 with s2m2 was observed in 8% of H. pylori positive patients. All but two of the vacA s1 alleles were s1a subtype (two strains had s1b subtype). None of vacA m alleles were detected in 4.8% of samples (s1m0).
Table 2.
Distribution of H. pylori vacA genes/alleles according to cagA and iceA status.
| VacA Alleles 1 | CagA Status (%) | IceA Genotype (%) | Total (%) | ||||
|---|---|---|---|---|---|---|---|
| CagA(+) | CagA(−) | IceA1 | IceA2 | IceA1+2 | Negative | ||
| s1m1 | 11 (17.7) | 1 (1.6) | 4 (6.5) | 5 (8.1) | 0 | 3 (4.8) | 12 (19.4) |
| s1bm2 | 1 (1.6) | 0 | 0 | 0 | 1 (1.6) | 0 | 1 (1.6) |
| s1m1/s1m2 | 1 (1.6) | 2 (3.2) | 0 | 2 (3.2) | 1 (1.6) | 0 | 3 (4.8) |
| s1m2 | 16 (25.8) | 5 (8.1) | 10 (16.1) | 7 (11.3) | 0 | 4 (6.5) | 21 (33.9) |
| s2m2 | 3 (4.8) | 15 (24.2) | 5 (8.1) | 5 (8.1) | 0 | 8 (12.9) | 18 (29.0) |
| s2m1/s2m2 | 0 | 2 (3.2) | 0 | 1 (1.6) | 0 | 1 (1.6) | 2 (3.2) |
| s2m1 | 0 | 1 (1.6) | 0 | 1 (1.6) | 0 | 0 | 1 (1.6) |
| negative | 1 (1.6) | 0 | 0 | 0 | 0 | 1 (1.6) | 1 (1.6) |
| Total | 35 (56.5) | 27 (43.5) | 20 (32.3) | 22 (35.5) | 3 (4.8) | 17 (27.4) | 62 (100) |
1 untyped m-region was found in three samples (s1m0).
The most common H. pylori genotype infecting studied patients were cagA(+) vacA s1m2 (27.4%), cagA(−) vacA s2m2 (24.2%) and cagA(+) vacA s1m1 (19.4%). The prevalence of cagA gene as well as vacA and iceA genotypes varied in clinical outcome, yet without statistical significance (Table 3).
Table 3.
Genotypes of H. pylori strains and clinical outcome.
| Genotype | GAST 1 (%) | PUD 2 (%) | GERD 3 (%) | p-Value |
|---|---|---|---|---|
| n = 27 | n = 6 | n = 31 | ||
| Vac1 genotypes 4 | ||||
| s1m1 | 7 (25.9) | 3 (50) | 4 (12.9) | 0.11 |
| s1m1/s1m2 | 2 (7.4) | 0 | 1 (3.2) | 0.64 |
| s1m2 | 9 (33.3) | 3 (50) | 11 (35.5) | 0.74 |
| s2m2 | 7 (25.9) | 0 | 11 (35.5) | 0.2 |
| s2m1/s2m2 | 2 (7.4) | 0 | 1 (3.2) | 0.64 |
| s2m1 | 0 | 0 | 1 (3.2) | - |
| negative | 1 (3.7) | 0 | 0 | - |
| cagA status | ||||
| positive | 16 (59.3) | 5 (83.3) | 16 (51.6) | 0.35 |
| negative | 11 (40.7) | 1 (16.7) | 15 (48.4) | |
| iceA alleles | ||||
| iceA1 | 6 (22.2) | 3 (50) | 14 (45.2) | 0.14 |
| iceA2 | 10 (37.0) | 3 (50) | 8 (35.8) | 0.42 |
| iceA1+2 | 1 (3.7) | 0 | 2 (6.5) | 0.75 |
| negative | 10 (37.0) | 0 | 7 (22.6) | 0.14 |
| vacA s1m1/cagA+ | 7 (25.9) | 3 (50.0) | 4 (12.9) | 0.11 |
| vacAs1m2/cagA+ | 6 (22.2) | 2 (33.3) | 9 (29.0) | 0.78 |
| vacAs2m2/cagA+ | 1 (3.7) | 0 | 2 (6.5) | 0.75 |
| vacAs1m1/cagA− | 1 (3.7) | 0 | 1 (3.2) | 0.89 |
| vacAs1m2/cagA− | 4 (14.8) | 1 (16.7) | 2 (6.5) | 0.53 |
| vacAs2m2/cagA− | 6 (22.2) | 0 | 10 (32.3) | 0.23 |
1 GAST—gastritis/duodenitis; 2 PUD—peptic ulcer disease (gastric/duodenal); 3 GERD—gastroesophageal reflux disease; 4 untyped m-region was found in three samples (s1m0).
In univariate analysis, it was shown that vacAs1m1 was significantly more frequent in patients with gastritis and peptic ulcer (Table A1). However, in logistic regression analysis including epidemiological factors as potential confounders, we recognized the subsequent significant associations: gastritis with ureC presence, meaning H. pylori infection (OR 3.3, 95%CI 1.4–7.9, p = 0.006), BMI index (OR 1.13, 95%CI 1.0–1.3, p = 0.032), and negatively with iceA1 presence (OR 0.3, 95%CI 0.1–1.0, p = 0.049); peptic ulcer with cagA presence (OR 7.7, 95%CI 1.4–41.6, p = 0.018) whereas GERD with iceA1 (OR 4.5, 95%CI 1.4–15.2, p = 0.014) and negatively with cagA (OR 0.32, 95%CI 0.1–0.9, p = 0.025).
Bacterial cultures were successfully isolated in 35 patients. MIC values of examined H. pylori strains showed that all of them were susceptible to amoxicillin and tetracycline (Table 4). Among all strains, 51% were susceptible to all and 49% were resistant to at least one of the tested antibiotics. Resistance to clarithromycin, metronidazole, levofloxacin, and rifampicin was found in 5 (14.3%), 11 (31.4%), 4 (11.4%), and 9 (25.7%), respectively.
Table 4.
Susceptibility and MIC values for 35 isolated H. pylori strains.
| Antibiotics | Susceptibility (%) | MIC Range | MIC50 | MIC90 |
|---|---|---|---|---|
| Amoxicillin | 35 (100) | <0.016 | <0.016 | <0.016 |
| Clarithromycin | 30 (85.7) | <0.016–>256 | <0.016 | 16 |
| Metronidazole | 24 (68.6) | <0.016–>256 | 0.064 | >256 |
| Tetracycline | 35 (100) | <0.016–0.19 | <0.016 | 0.094 |
| Levofloxacin | 31 (88.6) | <0.002–>32 | 0.047 | 8 |
| Rifampicin | 26 (74.3) | <0.002–>256 | 0.5 | 2 |
MIC, Minimal Inhibitory Concentration; MIC50, MIC90, lowest concentration of antibiotic at which 50 and 90% of the isolates were inhibited.
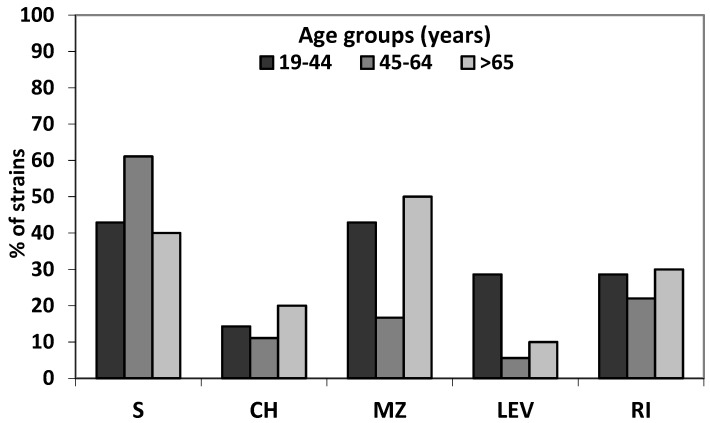
Even though there was a slight difference in amount of resistant strains isolated from patients from different age group, showing their higher frequency in the oldest group, statistical analysis revealed no significant differences (p = 0.13) in distribution of resistance patterns between age groups (Figure 2).
Figure 2.
Resistance to tested antibiotics of 35 isolates H. pylori strains in different age groups (S—sensitive to all tested antibiotics; CH—clarithromycin; MZ—metronidazole; RI—rifampicin; LEV—levofloxacin).
The multidrug resistant strains (for two or more antibiotics) were observed in eight strains (22.9%). They were resistant to metronidazole and clarithromycin (50%) and/or levofloxacin (50%) and/or rifampicin (50%) (Table 5). Most of metronidazole resistant strains were found with high MIC value.
Table 5.
Characteristics of multidrug resistant H. pylori strains.
| Age | Gender | Diagnosis | Genotype | Resistance Pattern | MIC (mg/L) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AC 4 | CH 5 | MZ 6 | LE 7 | RI 8 | TC 9 | |||||
| 53 | M | GERD 1 | cagA(+)s1m1 iceA2 | CH+MZ+LE | <0.016 | 12 | 24 | >32 | 0.125 | 0.19 |
| 64 | F | GAST 2 | cagA(+)s1m1 iceA2 | CH+MZ | <0.016 | >256 | >256 | 0.047 | 0.75 | <0.016 |
| 70 | F | GERD | cagA(−)s2m2 iceA2 | CH+MZ+LE+RI | <0.016 | 8 | 64 | 8 | 2 | <0.016 |
| 71 | F | GERD | cagA(+)s1m2 iceA1 | MZ+RI | <0.016 | <0.016 | >256 | 0.023 | >256 | <0.016 |
| 44 | M | PUD 3 | cagA(+)s1m2 iceA1 | MZ+RI | <0.016 | <0.016 | >256 | 0.125 | 2 | <0.016 |
| 34 | F | GERD | cagA(+)s2m2 iceA1 | MZ+LE | <0.016 | <0.016 | >256 | >32 | <0.002 | <0.016 |
| 29 | F | GERD | cagA(+)s1m2 iceA2 | CH+MZ+LE | <0.016 | >256 | >256 | >32 | 0.5 | 0.094 |
| 73 | F | GAST | cagA(−)s2m2 iceA2 | MZ+RI | <0.016 | <0.016 | >256 | 0.047 | 2 | 0.19 |
1 GAST—gastritis/duodenitis; 2 PUD—peptic ulcer (gastric/duodenal); 3 GERD—gastroesophageal reflux disease; 4 AC—amoxicillin; 5 CH—clarithromycin; 6 MZ—metronidazole; 7 LE—levofloxacin; 8 RI—rifampicin; 9 TC—tetracycline.
4. Discussion
There are several diagnostic assays for H. pylori infection, including bacterial culture, a rapid urease test (RUT), a urea breath test (UBT), histology, PCR, serology, and stool antigen test [10,11]. All these techniques have their limitations. The sensitivity of isolation of the bacterium has been reported to vary greatly among laboratories because of their fastidious nature [13]. Even the experienced laboratories recover the organism from only 50% to 70% of infected biopsies [17,18]. In this study, the low positivity rate of the culture (26.5%) may be due to a low number of bacterial cells, presence of non-culturable coccoid forms, contamination by other bacteria suppressing the growth of H. pylori or antibiotic therapy. Moreover, to avoid false negative results, it is recommended for patients not to consume the proton pump inhibitors (PPIs) two weeks before endoscopy, because these drugs indirectly interfere with H. pylori distribution in the stomach [19]. In our study, PCR was the most sensitive and specific method for detection of H. pylori. As the bacterial DNA was recovered from 56.5% of biopsies we consider this method as a reference. Advantages of PCR method are that it detects H. pylori in both forms i.e., spiral or coccoid forms that cannot be detected by other conventional methods [20], is able to discriminate the re-infection or recrudescence by genotyping of infective strains, enables to target pathogenic genes and assess the virulence potential of H. pylori in particular patient and does not require strict transport conditions. Therefore, PCR based methods are best for the specimens collected by invasive methods if the tests are carried out without contaminations. Nested PCR was proposed as a gold standard of H. pylori detection [13].
H. pylori infected patients develop superficial gastritis but the course of disease is partly due to environmental conditions and genetics of the host, but also due to the presence of particular virulence factors presented in the infecting H. pylori strains [21]. In this study, we focused on research of population and epidemiology of H. pylori, which provide us an information about genotypes linking them with clinical diagnosis. To our knowledge, this study presents the largest characteristic to date of H. pylori isolates collected from patients from the south east Poland. Distribution and relation between H. pylori genotypes and clinical outcomes, if derived from the different geographic regions may differ widely, then it is important to determine the most prevalent genotypes in a specific region [22].
An important application of standard PCR is the detection of specific pathogenic factors of H. pylori. There are two main pathogenic segments: the cagPAI and the polymorphism of the vacA gene. Other genes coding molecules associated with induction of inflammation (dupA, iceA) can also be detected by PCR. It was reported that cagPAI do cause more severe peptic ulceration, extra digestive diseases, and is associated with development of precancerous lesions and gastric adenocarcinoma [19]. In our study, in 56% of H. pylori positive samples cagA gene were found that was comparable to the other Polish studies [23,24], and logistic regression revealed relationship between cagA gene and clinical outcome. It was a risk factor positively associated with peptic ulcer diagnosis but negative association existed with GERD. Colonization with cagA-positive H. pylori strains was already shown to be inversely associated with reflux esophagitis and Barrett’s esophagus [25].
H. pylori also produces a vacuolating cytotoxin, VacA, which has been associated with the more severe diseases, e.g., peptic ulcer disease and gastric adenocarcinoma [26,27]. The gene encoding this cytotoxin is present in all strains but exhibits a mosaicism in the terminal (s) and median (m) regions. There are several alleles corresponding to various amounts of toxin produced: s1m1 corresponds to the highest production, followed by s1m2, while strains with the s2m2 allele do not produce any toxin. The vacA genotypes (allelic variations) are significantly different in each country and previous studies have confirmed important geographic differences in these virulence factors [27,28,29]. Among different genotypes, infection with H. pylori strains containing vacA s1m1 type was strongly associated with increased risk of gastric cancer or PUD [8,30,31]. In our study, most frequent allelic combination of vacA gene were s1am2—38.7% and s2m2—32.3%, that was similar to previous Polish studies [23,24,32]; however, statistically significant associations between vacAs1m1 type and GAST and PUD patients were observed (p = 0.042 and p = 0.038, respectively). Our findings also shown that among cagA positive strains, the combinations of vacAs1m1/cagA+ was the predominant pattern in GAST and PUD patients. When both cagA and vacA detection is performed, a strong association exists between the presence of cagA and vacA s1, corresponding to strains with the highest production of cytotoxin [29,33] as well as more severe disease [34,35,36]. In our study, traditionally, less aggressive vacAs2m2cagA(−) were most common in GERD patients (32%).
Our laboratory findings suggest that gastric mucosa is colonized by mixed virulent type of H. pylori strains. In 8% of H. pylori positive samples mixed genotypes of vacA gene were detected, i.e., at least two different alleles of m region. Mixed vacA genotypes were also observed in other studies suggesting high probability of colonization by a few different H. pylori strains [28,29,33].
The role and correlation of H. pylori iceA (a gene induced by a contact with the gastric epithelium) in clinical outcomes is unclear. Our results revealed that 32.3% of H. pylori were harboring iceA1, 35.5% were iceA2, whereas 4.8% were mixed (iceA1/iceA2) which is consistent with other data from Poland but in younger population [24] and from Eastern Europe [37]. In other studies from European countries, it was shown that iceA1 allele was two times more frequent than iceA2 [15,38]. The iceA1 allele, encoding a CATG-specific restriction endonuclease, is regulated by the contact of H. pylori with the human gastric cells, has been suggested to be related to PUD [15,16]. However, like the other authors, we doubt these findings [39,40]. In our study, iceA1 was negatively associated with gastritis whereas it was positively associated with GERD. Further research of H. pylori strains with different iceA gene alleles could lead to clarifying its role in pathogenesis of human gastrodudenal diseases.
In our study, 35 H. pylori strains were tested; 51% of them were susceptible to all, and 49% were resistant to at least one of the tested antibiotics. Multiresistant strains accounted for as many as 23%, and included those resistant to two (metronidazole and clarithromycin or metronidazole and rifampicin or metronidazole and levofloxacin) or three (metronidazole, clarithromycin, and levofloxacin) antibiotics. This study showed lower primary antibiotic resistance rate than those reported in other areas of Poland or Europe [41,42]. In Eastern and Central European countries, the prevalence of H. pylori strains resistant to metronidazole is higher than in other developed countries, reaching almost 50%, and resistance to clarithromycin is as high as 30% and is still increasing, contributing to the failure of first-line therapy in approximately 70% of patients [32,41,42]. Such a high resistance to clarithromycin observed in Poland results mainly from excessive consumption of antibiotics, especially from the group of macrolides, and their widespread use in respiratory tract infections. Moreover, rates of resistance of H. pylori strains to levofloxacin have been very low in Poland [43], but the introduction of levofloxacin to eradication therapy of H. pylori infection quickly led to the emergence of resistant strains. Most of multi-resistant strains had genotype cagA(+) (75%) and vacAs1 (62.5%).
A major limitation of our work was relatively low number of patients with specific clinical manifestations. We had only nine patients with peptic ulcer and none with gastric cancer, which can only be explained by limited number of studied patients, considering the published reports regarding its incidence in Poland [44]. We had also a very low number of H. pylori culture findings that might be related to absence of microorganisms in the gastric biopsy specimens, loss of viability during transport, and antibiotic or IPP intake.
Another limitation was that we studied only a few virulence genes although other genes involved in adherence (babA, sabA) or in pathogenesis (oipA, dupA) can be targeted to assess the virulence potential of H. pylori. To improve the significance of our study, especially the knowledge of populations from Eastern Europe, the importance of more recently disclosed vacA and cagA genotypes—such as vacA i-region/d-region and cagA EPIYA-region—would be worthy of investigation.
5. Conclusions
This is the first study that reports the high prevalence and diversity of allelic combination of cagA, vacA, and icaA genes in gastroduodenitis patients in southeast Poland. In our study, most frequent allelic combination of vacA gene were s1m2 and s2m2 and correlation between vacAs1m1 type and development of GAST and PUD in patients was shown. Our findings also noted a lower primary antibiotic resistance rate than those reported in other areas of Poland. However, new antimicrobial resistance studies should be conducted periodically and regionally in our country to provide information that may help to monitor effective eradication programs.
Acknowledgments
The paper was developed using the equipment purchased within the agreement no. POPW.01.03.00-06-010/09-00 Operational Program Development of Eastern Poland 2007–2013, Priority Axis I, Modern Economy, Operations 1.3. Innovations Promotion.
Appendix A
Table A1.
Univariate analysis of factors associated with clinical diagnosis
| Factor | GAST | PUD | GERD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAST 2 (n = 46) | Others (n = 86) | OR (95%CI) | p Value | PUD 3 (n = 9) | Others (n = 123) | OR (95%CI) | p-Value | GERD 4 (n = 71) | Others (n = 61) | OR (95%CI) | p-Value | |
| Age (mean ± SD) | 57.2 ± 15.8 | 57.4 ± 15.7 | 1.0 (0.97–1.0) | 0.81 | 61.3 ± 12.3 | 57.1 ± 15.8 | 1.0 (0.97–1.1) | 0.43 | 56.9 ± 16.0 | 57.9 ± 15.3 | 1.0 (0.97–1.0) | 0.72 |
| Male gender | 23 (50.0) | 31 (36.1) | 1.9 (0.9–3.9) | 0.88 | 4 (44.4) | 50 (40.7) | 1.2 (0.3–4.5) | 0.82 | 27 (38.0) | 27 (44.3) | 0.8 (0.4–1.6) | 0.47 |
| BMI 1 (mean ± SD) | 27.0 ± 3.8 | 25.8 ± 3.4 | 1.1 (1.0–1.2) | 0.06 | 27.2 ± 3.8 | 26.1 ± 3.6 | 1.1 (0.9–1.3) | 0.42 | 25.9 ± 3.5 | 26.6 ± 3.8 | 0.9 (0.8–1.0) | 0.22 |
| rural residence | 20 (42.5) | 27 (31.8) | 1.7 (0.8–3.5) | 0.18 | 3 (33.3) | 44 (35.8) | 1.1 (0.2–4.7) | 0.92 | 24 (33.8) | 23 (38.3) | 0.8 (0.4–1.7) | 0.59 |
| Smoking | 12 (26.1) | 28 (32.9) | 0.7 (0.3–1.6) | 0.42 | 3 (33.3) | 37 (30.1) | 1.4 (0.3–6.1) | 0.66 | 20 (28.2) | 20 (33.3) | 0.8 (0.4–1.7) | 0.52 |
| ureC | 27 (58.7) | 35 (40.7) | 1.9 (0.9–4.0) | 0.075 | 6 (66.7) | 56 (45.5) | 2.4 (0.6–10.0) | 0.23 | 31 (43.7) | 31 (50.8) | 0.8 (0.4–1.5) | 0.41 |
| cagA | 16 (34.8) | 19 (22.1) | 2.0 (0.9–4.4) | 0.093 | 5 (55.6) | 30 (24.4) | 3.9 (1.0–15.3) | 0.054 | 16 (22.5) | 19 (31.2) | 0.6 (0.3–1.4) | 0.27 |
| vacA s1m2 | 10 (21.7) | 13 (15.1) | 2.0 (0.7–5.2) | 0.17 | 3 (33.3) | 20 (16.3) | 3.4 (0.6–18.1) | 0.15 | 11 (15.5) | 12 (19.7) | 0.7 (0.3–1.8) | 0.48 |
| vacA s1m1 | 8 (17.4) | 6 (7.0) | 3.4 (1.0–11.0) | 0.042 | 3 (33.3) | 11 (8.9) | 6.2 (1.1–34.6) | 0.038 | 5 (7.0) | 9 (14.8) | 0.4 (0.1–1.4) | 0.17 |
| vacA s2m2 | 7 (15.2) | 13 (15.2) | 1.4 (0.5–3.9) | 0.56 | 0 (0) | 20 (16.3) | - | - | 12 (16.9) | 8 (13.1) | 1.2 (0.4–3.2) | 0.77 |
| iceA1 | 7 (15.2) | 16 (18.6) | 0.8 (0.3–2.1) | 0.63 | 3 (33.3) | 20 (16.3) | 2.6 (0.6–11.1) | 0.21 | 16 (22.5) | 7 (11.5) | 2.2 (0.9–5.9) | 0.1 |
| iceA2 | 11 (23.9) | 14 (16.3) | 1.6 (0.7–3.9) | 0.29 | 3 (33.3) | 22 (17.9) | 2.3 (0.5–9.8) | 0.26 | 10 (14.1) | 15 (24.6) | 0.5 (0.2–1.2) | 0.13 |
1 BMI—body mass index; 2 GAST—gastritis/duodenitis; 3 PUD—peptic ulcer disease (gastric/duodenal); 4 GERD—gastroesophageal reflux disease.
Author Contributions
Conceptualization, I.K.-G., A.M., and H.C.-L.; Methodology, I.K.-G. and S.A.; Formal analysis, I.K.-G.; Investigation, I.K.-G., R.S., and S.A.; Resources, H.C.-L., A.G., and P.M.; Writing—original draft preparation, I.K.-G., H.C.-L., R.S., and A.M.; Writing—review and editing, I.K.-G, H.C.-L., A.M., R.S., S.A., A.G., and P.M.; Supervision, H.C.-L. and A.M.; Funding acquisition, A.M. and H.C.-L.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Atherton J.C. The pathogenesis of Helicobacter pylori-induced gastro-duodenal disease. Annu. Rev. Pathol. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 2.Eusebi L.H., Zagari R.M., Bazzoli F. Epidemiology of Helicobacter pylori infection. Helicobacter. 2014;19:1–5. doi: 10.1111/hel.12165. [DOI] [PubMed] [Google Scholar]
- 3.Malfertheiner P., Megraud F., O’Morain C.A., Gisbert J.P., Kuipers E.J., Axon A.T., Bazzoli F., Gasbarrini A., Atherton J., Graham D.Y., et al. Management of Helicobacter pylori infection—The Maastricht V/Florence consensus report. Gut. 2017;66:646–664. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 4.Makola D., Petura D.A., Crowe S.E. Helicobacter pylori infection and related gastrointestinal diseases. J. Clin. Gastroenterol. 2007;41:548–558. doi: 10.1097/MCG.0b013e318030e3c3. [DOI] [PubMed] [Google Scholar]
- 5.Wen S., Moss S.F. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1–8. doi: 10.1016/j.canlet.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covacci A., Censini S., Bugnoli M., Petracca R., Burroni D., Macchia G., Massone A., Papini E., Xiang Z., Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cover T.L., Blaser M.J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 8.Atherton J.C., Cao P., Peek R.M., Tummuru M.K., Blaser M.J., Cover T.L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 9.Yamaoka Y., Graham D.Y. Helicobacter pylori virulence and cancer pathogenesis. Future Oncol. 2014;10:1487–1500. doi: 10.2217/fon.14.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lage A.P., Godfroid E., Fauconnier A., Burette A., Butzler J.P., Bollen A., Glupczynski Y. Diagnosis of Helicobacter pylori infection by PCR: Comparison with other invasive techniques and detection of cagA gene in gastric biopsy specimens. J. Clin. Microbiol. 1995;33:2752–2756. doi: 10.1128/jcm.33.10.2752-2756.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith S.M., O’Morain C., McNamara D. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World J. Gastroenterol. 2014;20:9912–9921. doi: 10.3748/wjg.v20.i29.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartnik W., Celińska-Cedro D., Dzieniszewski J., Laszewicz W., Mach T., Przytulski K., Skrzydlo-Radomanska B. Guidelines from the polish society of gastroenterology for the diagnosis and treatment of Helicobacter pylori infection. Gastroenterol. Klin. 2014;2:41–49. [Google Scholar]
- 13.Patel S.K., Pratap C.B., Jain A.K., Gulati A.K., Nath G. Diagnosis of Helicobacter pylori: What should be the gold standard? World J. Gastroenterol. 2014;20:12847–12859. doi: 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa M.G.C., Vazquez R.G., Mendez I.M., Vargas C.R., Cerezo S.G. Detection of the glmM gene in Helicobacter pylori isolates with novel primer by PCR. J. Clin. Microbiol. 2011;49:1650–1652. doi: 10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Doorn L., Figueiredo C., Sanna R., Plaisier A., Schneeberger P., De Boer W., Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/S0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 16.Peek R.M., Jr., Thompson S.A., Donahue J.P., Tham T.K., Atherton J.C., Blaser M.J., Miller G.G. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 17.Grove D.I., Koutsouridis G., Cummins A.G. Comparison of culture, histopatology and urease testing for diagnosis of Helicobacter pylori gastritis and susceptibility to amoxicillin, clarithromycin, metronidazole and tetracycline. Pathology. 1998;30:183–187. doi: 10.1080/00313029800169206. [DOI] [PubMed] [Google Scholar]
- 18.Loffeld R.J., Stobbering E., Flendrig J.A., Arends J.W. Helicobacter pylori in gastric biopsy specimens. Comparison of culture, modified giemsa stain, and immunohistochemistry. A retrospective study. J. Pathol. 1991;165:69–73. doi: 10.1002/path.1711650111. [DOI] [PubMed] [Google Scholar]
- 19.Mégraud F., Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 2007;20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duś I., Dobosz T., Manzin A., Loi G., Serra C., Radwan-Oczko M. Role of PCR in Helicobacter pylori diagnostics and research—New approaches for study of coccoid and spiral forms of bacteria. Postepy Hig. Med. Dosw. (Online) 2013;67:261–268. doi: 10.5604/17322693.1044005. [DOI] [PubMed] [Google Scholar]
- 21.Correa P., Piazuelo M.B. The gastric precancerous cascade. J. Dig. Dis. 2012;13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Módena J.L.P., Sales A.I.L., Acrani G.O., Russo R., Ribeiro M.A.V., Fukuhara Y., Da Silveira W.D., De Oliveira R.B., Brocchi M. Association between Helicobacter pylori genotypes and gastric disorders in relation to the cag pathogenicity island. Diagn. Microbiol. Infect. Dis. 2007;59:7–16. doi: 10.1016/j.diagmicrobio.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Zalewska-Ziob M., Wiczkowski A., Strzelczyk J.K., Adamek B., Gawron K., Trapp G., Sieron A., Gadowska-Cicha A. The prevalence of Helicobacter pylori vacA alleles in patients with chronic gastritis. Adv. Clin. Exp. Med. 2007;16:29–33. [Google Scholar]
- 24.Biernat M.M., Gościniak G., Iwańczak B. Prevalence of Helicobacter pylori cagA, vacA, iceA, babA2 genotypes in Polish children and adolescents with gastroduodenal disease. Postepy Hig. Med. Dośw. 2014;68:1015–1021. doi: 10.5604/17322693.1118211. [DOI] [PubMed] [Google Scholar]
- 25.Loffeld R.J., Werdmuller B.F., Kuster J.G., Perez-Perez G.I., Blaser M.J., Kuipers E.J. Colonization with cagA-positive Helicobacter pylori strains inversely associated with reflux esophagitis and Barrett’s esophagus. Digestion. 2000;62:95–99. doi: 10.1159/000007801. [DOI] [PubMed] [Google Scholar]
- 26.Figura N., Guglielmetti P., Rossolini A., Barberi A., Cusi G., A Musmanno R., Russi M., Quaranta S. Cytotoxin production by Campylobacter pylori strains isolated from patients with peptic ulcers and from patients with chronic gastritis only. J. Clin. Microbiol. 1989;27:225–226. doi: 10.1128/jcm.27.1.225-226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miehlke S., Kirsch C., Agha-Amiri K., Günther T., Lehn N., Malfertheiner P., Stolte M., Ehninger G., Bayerdörffer E. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer. 2000;87:322–327. doi: 10.1002/1097-0215(20000801)87:3<322::AID-IJC3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Latifi-Navid S., Mohammadi S., Maleki P., Zahri S., Yazdanbod A., Siavoshi F., Massarrat S. Helicobacter pylori vacA d1/-i1 genotypes and geographic differentiation between high and low incidence areas of gastric cancer in Iran. Arch. Iran. Med. 2013;16:330–337. [PubMed] [Google Scholar]
- 29.Van Doorn L., Figueiredo C., Mégraud F., Peña S., Midolo P., Queiroz D.M.D.M., Carneiro F., VanderBorght B., Pegado M.D.G.F., Sanna R., et al. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/S0016-5085(99)70065-X. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Reyes M., Tamayo E., Rojas-Rengifo D., Fischer W., Carrasco-García E., Alonso M., Lizasoain J., Bujanda L., Cosme Á., Montes M. Helicobacter pylori and primary antimicrobial resistance in Northern Spain. Eur. J. Clin. Investig. 2019:13150. doi: 10.1111/eci.13150. [DOI] [PubMed] [Google Scholar]
- 31.Idowu A., Mzukwa A., Harrison U., Palamides P., Haas R., Mbao M., Mamdoo R., Bolon J., Jolaiya T., Smith S., et al. Detection of Helicobacter pylori and its virulence genes (cagA, dupA, and vacA) among patients with gastroduodenal diseases in Chris Hani Baragwanath Academic Hospital, South Africa. BMC Gastroenterol. 2019;19:73. doi: 10.1186/s12876-019-0986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwańczak B.W., Laszewicz F., Iwańczak K., Dzierzanowska-Fangrat K., Rozynek M., Dzierzanowska D., Gosciniak G., Dlugosz J., Task force of The Polish Society of Gastroenterology Genotypic and clinical differences of seropositive Helicobacter pylori children and adults in the Polish population. J. Physiol. Pharmacol. 2014;65:801–807. [PubMed] [Google Scholar]
- 33.Audibert C., Janvier B., Grignon B., Salaüna L., Burucoa C., Lecron J.-C., Fauchere J.-L. Correlation between IL-8 induction, cagA status and vacA genotypes in 153 French Helicobacter pylori isolates. Res. Microbiol. 2000;151:191–200. doi: 10.1016/S0923-2508(00)00139-X. [DOI] [PubMed] [Google Scholar]
- 34.Molina-Castro S., Garita-Combronero J., Malespin-Bendana W., Une C., Ramirez V. Virulence factor genotyping of Helicobacter pylori isolated from Costa Rican dyspeptic patients. Microb. Pathogen. 2019;128:276–280. doi: 10.1016/j.micpath.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Matsunari O., Miftahussurur M., Shiota S., Suzuki R., Vilaichone R.-K., Uchida T., Ratanachu-Ek T., Tshering L., Mahachai V., Yamaoka Y. Rare Helicobacter pylori virulence genotypes in Bhutan. Sci. Rep. 2016;6:22584. doi: 10.1038/srep22584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akeel M., Shehata A., Elhafey A., Elmakki E., Aboshouk T., Ageely H., Mahfouz M. Helicobacter pylori vacA, cagA and iceA genotypes in dyspeptic patients from southwestern region, Saudi Arabia: Distribution and association with clinical outcomes and histopathological changes. BMC Gastroenterol. 2019;19:16. doi: 10.1186/s12876-019-0934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyanova L., Yordanov D., Gergova G., Markowska R., Mitov I. Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristics. Antonie van Leeuwenhoek. 2010;98:343–350. doi: 10.1007/s10482-010-9448-y. [DOI] [PubMed] [Google Scholar]
- 38.Homan M., Luzar B., Kocjan B.J., Orel R., Močilnik T., Shrestha M., Kveder M., Poljak M. Prevalence and clinical relevance of cagA, vacA, and iceA genotypes of Helicobacter pylori isolated from Slovenian children. J. Pediatr. Gastroenterol. Nutr. 2009;49:289–296. doi: 10.1097/MPG.0b013e31818f09f2. [DOI] [PubMed] [Google Scholar]
- 39.Lin H.-J., Perng C.-L., Lo W.-C., Wu C.-W., Tseng G.-Y., Li A.F.-Y., Sun I.-C., Ou Y.-H., Lin C.-L.P.H.-J. Helicobacter pylori cagA, iceA and vacA genotypes in patients with gastric cancer in Taiwan. World J. Gastroenterol. 2004;10:2493–2497. doi: 10.3748/wjg.v10.i17.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almeida N., Donato M.M., Romaozinho J.M., Luxo C., Cardoso O., Cipriano M.A., Marinho C., Fernandes A., Sofia C. Correlation of Helicobacter pylori genotypes with gastric histopathology in the central region of a south-european country. Dig. Dis. Sci. 2015;60:74–85. doi: 10.1007/s10620-014-3319-8. [DOI] [PubMed] [Google Scholar]
- 41.Mégraud F. Time to change approaches to Helicobacter pylori management. Lancet Gastroenterol. Hepatol. 2017;2:692–693. doi: 10.1016/S2468-1253(17)30245-5. [DOI] [PubMed] [Google Scholar]
- 42.Bińkowska A., Biernat M.M., Łaczmański Ł., Gościniak G. Molecular patterns of resistance among Helicobacter pylori strains in south-western Poland. Front. Microbiol. 2018;9:3154. doi: 10.3389/fmicb.2018.03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karczewska E., Klesiewicz K., Wojtas-Bonior I., Skiba I., Sito E., Czajecki K., Zwolińska-Wcisło M., Budak A. Levofloxacin resistance of Helicobacter pylori strains isolated from patients in Southern Poland, between 2006-2012. Acta Pol. Pharm. 2014;71:477–483. [PubMed] [Google Scholar]
- 44.GLOBOCAN 2018. [(accessed on 6 July 2019)]; Available online: https://gco.iarc.fr.