Abstract
Human immunodeficiency virus type-1 and hepatitis C virus (HIV/HCV) coinfection occurs when a patient is simultaneously infected with both human immunodeficiency virus type-1 (HIV-1) and hepatitis C virus (HCV), which is common today in certain populations. However, the treatment of coinfection is a challenge because of the special considerations needed to ensure hepatic safety and avoid drug–drug interactions. Multitarget inhibitors with less toxicity may provide a promising therapeutic strategy for HIV/HCV coinfection. However, the identification of one molecule that acts on multiple targets simultaneously by experimental evaluation is costly and time-consuming. In silico target prediction tools provide more opportunities for the development of multitarget inhibitors. In this study, by combining Naïve Bayes (NB) and support vector machine (SVM) algorithms with two types of molecular fingerprints, MACCS and extended connectivity fingerprints 6 (ECFP6), 60 classification models were constructed to predict compounds that were active against 11 HIV-1 targets and four HCV targets based on a multiple quantitative structure–activity relationships (multiple QSAR) method. Five-fold cross-validation and test set validation were performed to measure the performance of the 60 classification models. Our results show that the 60 multiple QSAR models appeared to have high classification accuracy in terms of the area under the ROC curve (AUC) values, which ranged from 0.83 to 1 with a mean value of 0.97 for the HIV-1 models and from 0.84 to 1 with a mean value of 0.96 for the HCV models. Furthermore, the 60 models were used to comprehensively predict the potential targets of an additional 46 compounds, including 27 approved HIV-1 drugs, 10 approved HCV drugs and nine selected compounds known to be active against one or more targets of HIV-1 or HCV. Finally, 20 hits, including seven approved HIV-1 drugs, four approved HCV drugs, and nine other compounds, were predicted to be HIV/HCV coinfection multitarget inhibitors. The reported bioactivity data confirmed that seven out of nine compounds actually interacted with HIV-1 and HCV targets simultaneously with diverse binding affinities. The remaining predicted hits and chemical-protein interaction pairs with the potential ability to suppress HIV/HCV coinfection are worthy of further experimental investigation. This investigation shows that the multiple QSAR method is useful in predicting chemical-protein interactions for the discovery of multitarget inhibitors and provides a unique strategy for the treatment of HIV/HCV coinfection.
Keywords: HIV/HCV coinfection, machine learning, multiple QSAR method, multitarget inhibitors, polypharmacology, drug discovery
1. Introduction
Human immunodeficiency virus type-1 (HIV-1) is the causative factor of acquired immunodeficiency syndrome (AIDS), a pandemic disease [1]. In addition, hepatitis C virus (HCV) infection causes acute and chronic liver disease, including cirrhosis and hepatocellular carcinoma [2]. Unfortunately, since HIV-1 and HCV have similar routes of transmission, the risk of HIV/HCV coinfection is very high [3]. According to the World Health Organization (WHO), approximately 2.3 million of the estimated 36.7 million people living with HIV globally had been infected with HCV as of 2015. It is necessary for these people to be diagnosed and provided with effective and reasonable treatment for both HIV and hepatitis as a priority [4]. However, in the treatment of HIV/HCV coinfection, special considerations must be made for potential drug–drug interactions and toxicities. The standard treatment for HIV-1 is highly active antiretroviral therapy (HAART), which improves the quality of life of AIDS patients by reducing viral load [5]. HAART utilizes a cocktail of drugs to inhibit multiple viral proteins involved in the viral life cycle, including reverse transcriptase, integrase, and protease [6]. Although it is effective, HAART requires the sequential administration of single-target drugs that may result in drug–drug interactions, poor treatment adherence and the emergence of drug resistance. The current therapy for HIV/HCV coinfection includes regular injections of pegylated α-interferon (PEG-IFN) plus daily ribavirin (RBV) in combination with HAART [7]. As with the HARRT treatment regimen, additive drug toxicity, high cost and the long-term therapy cycle should be taken into account during HIV/HCV coinfection treatment. To avoid these undesirable factors, multitarget therapy using a single drug capable of simultaneously inhibiting two or more viral targets has been proposed to reduce the complexity of treatment [5].
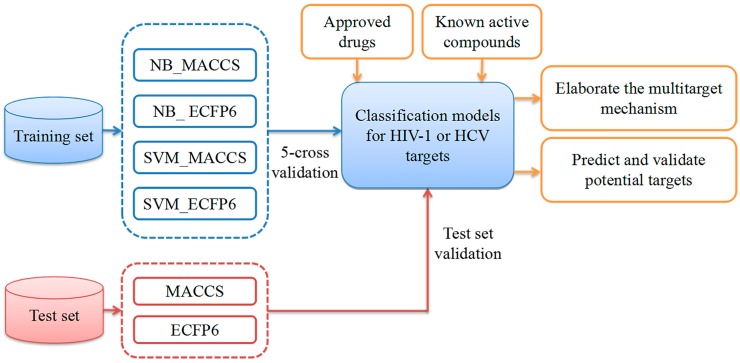
As a result of the development of computer-aided drug design (CADD) technology, several in silico methods have been exploited to predict chemical-protein interaction (CPI) pairs between drugs and target proteins [8,9,10,11,12,13,14]. These methods can be approximately classified into two categories: ligand-based [15,16,17] and structure-based [18,19]. Conventional quantitative structure-activity relationships (QSAR) methods generally use a group of analogs against one target to elucidate the relationship between chemical structure and biological activity. In 2009, Vina et al. developed a multitarget quantitative structure–activity relationships (mt-QSAR) classification model to predict the probability of drug binding for more than 60 different receptors based on molecular connectivity and receptor sequences [20]. In 2014, Fang et al. generated 100 mt-QSAR models for 25 key targets for the treatment of Alzheimer’s disease (AD) to predict the chemical–protein interactions and discover highly potent multitarget-directed ligands (MTDLs) [21]. In their research, the goal of mt-QSAR was to solve multiple binary classification problems instead of solving multilabel classification problems. In 2017, Antanasijević et al. proposed a regression multitasking quantitative structure–activity relationships (rmtk-QSAR) model based on the mt-QSAR methodology for the simultaneous prediction of the anticonvulsant activity and neurotoxicity of succinimides [22]. Hence, the mt-QSAR method has played a key role in the rational design of drugs for different biological targets. In this study, a multiple QSAR method intended for constructing multiple binary classification models was applied to predict the chemical–protein interactions (CPIs) for 11 key targets related to HIV-1 and four targets related to HCV. The workflow is depicted in Figure 1. First, by combining two machine learning algorithms (Naïve Bayes (NB) [23,24] and the support vector machine (SVM) [25]) and two molecular fingerprinting methods (MACCS [26] and extended connectivity fingerprints 6 (ECFP6) [27]), we developed multiple QSAR models for identifying inhibitors against 11 HIV-1-related and four HCV-related targets. Second, five-fold cross-validation and test set validation were used to evaluate the performance of all models. Third, to verify the application of this strategy, the multiple QSAR models that were developed were employed to predict the CPI for 27 approved HIV-1 drugs and 10 approved HCV drugs, as well as nine compounds known to be active against one or more targets of HIV-1 or HCV. The predicted results showed 20 hits to be potential HIV/HCV coinfection multitarget inhibitors, which were further confirmed by the assessment of bioactivity, resulting in seven compounds that actually interacted with HIV-1 and HCV targets simultaneously with diverse binding affinities. Our study indicates that machine learning-based multiple QSAR approaches could potentially be applied to the discovery of HIV/HCV co-inhibitors and other multitarget drugs.
Figure 1.
Flowchart showing the process of generating multiple quantitative structure–activity relationships (multiple QSAR) models for human immunodeficiency virus type-1 (HIV-1)/hepatitis C virus (HCV) coinfection (the blue, red and orange objects represent the phases of construction, validation and application of multiple QSAR models, respectively).
2. Results
2.1. Experimental Dataset Analysis
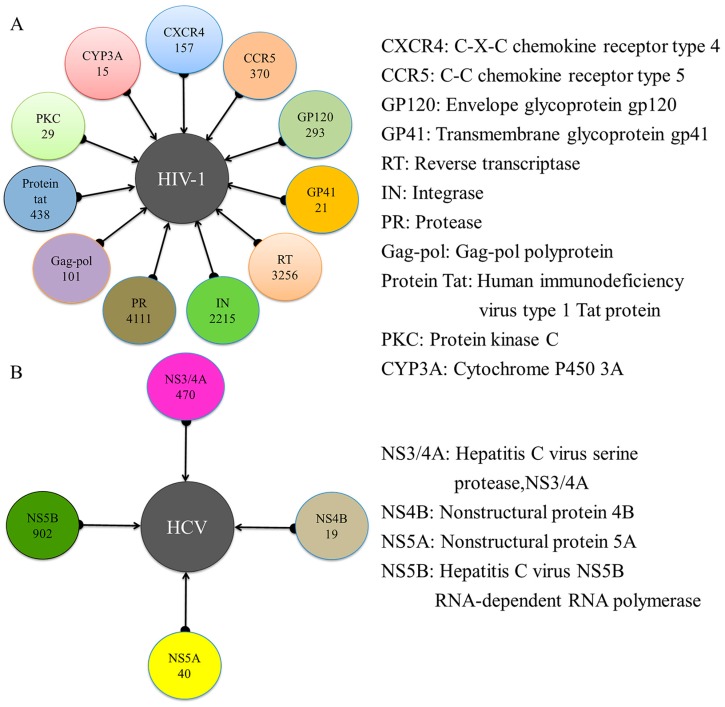
In this study, a total of 11 targets related to HIV-1 and four targets related to HCV were obtained from the Therapeutic Target Database (TTD) [28] and ChEMBL Database (version 23, https://www.ebi.ac.uk/chembl/). The numbers of known inhibitors retrieved from the ChEMBL Database [29] that act on HIV-1 and HCV targets were 11,006 and 1431, respectively. The number of active compounds for each target is shown in Figure 2. A total of 11 datasets were generated for the HIV-1 system, including 157 inhibitors and 471 decoys for C-X-C chemokine receptor type 4 (CXCR4), 370 inhibitors and 1110 decoys for C-C chemokine receptor type 5 (CCR5), 293 inhibitors and 879 decoys for envelope glycoprotein gp120 (GP120), 21 inhibitors and 63 decoys for transmembrane glycoprotein gp41 (GP41), 3256 inhibitors and 9768 decoys for reverse transcriptase (RT), 2215 inhibitors and 6645 decoys for integrase (IN), 4111 inhibitors and 12,333 decoys for protease (PR), 101 inhibitors and 303 decoys for Gag-pol, 438 inhibitors and 1314 decoys for the protein Tat, 29 inhibitors and 87 decoys for protein kinase C (PKC), and 15 inhibitors and 45 decoys for cytochrome P450 3A (CYP3A). There were four datasets generated for the HCV system, including 470 inhibitors and 1410 decoys for hepatitis C virus serine protease (NS3/4A), 19 inhibitors and 57 decoys for nonstructural protein 4B (NS4B), 40 inhibitors and 120 decoys for nonstructural protein 5A (NS5A), and 902 inhibitors and 2706 decoys for hepatitis C Virus NS5B (NS5B). Each of the 15 datasets described above was divided into a training set and a test set.
Figure 2.
Overview of the targets related to HIV-1 (A) and HCV (B) and the total number of active compounds identified for each target.
The ideal training set used to create predictive classification models that could be used to identify potent molecules from a massive database of structurally diverse compounds is diverse and large. Before modeling implementation, the Tanimoto similarity index, which is a generally used measure for evaluating molecular similarity among numerous compounds, was used to evaluate the fingerprint-based similarity using the ECFP4 fingerprint [30,31]. In general, more dissimilar compounds tend to have a smaller Tanimoto similarity index. The Tanimoto similarity indexes for a set of compounds in the training and test sets for each of 15 datasets were computed, and the results are shown in Table 1. For HIV-1, the Tanimoto similarity indexes of the compounds in the training and test sets from the 11 datasets ranged from 0.118 to 0.202 and 0.117 to 0.215, respectively. For HCV, the training sets and test sets from the four datasets had similar Tanimoto similarity indexes that ranged from 0.141 to 0.192. Table 1 shows that the compounds in the training and test sets from the 15 datasets were involved in diverse chemotypes. The dissimilarity and diversity of the training/test sets play a critical role in the performance of the classifiers.
Table 1.
Detailed statistical analysis of the 15 datasets.
| Object | Target | Training Set | Test Set | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Inhibitors | Decoys | Total | Tanimoto Similarity Index | Inhibitors | Decoys | Total | Tanimoto Similarity Index | ||
| HIV-1 | CXCR4 | 113 | 339 | 452 | 0.152 | 44 | 132 | 176 | 0.158 |
| CCR5 | 255 | 766 | 1021 | 0.161 | 115 | 344 | 459 | 0.160 | |
| GP120 | 208 | 624 | 832 | 0.142 | 85 | 255 | 340 | 0.147 | |
| GP41 | 14 | 42 | 56 | 0.172 | 7 | 21 | 28 | 0.189 | |
| RT | 2279 | 6837 | 9116 | 0.124 | 977 | 2931 | 3908 | 0.124 | |
| IN | 1617 | 4850 | 6467 | 0.134 | 598 | 1795 | 2393 | 0.135 | |
| PR | 2837 | 8509 | 11,346 | 0.141 | 1274 | 3824 | 5098 | 0.141 | |
| Gag-pol | 69 | 205 | 274 | 0.147 | 32 | 98 | 130 | 0.146 | |
| Protein tat | 302 | 906 | 1208 | 0.118 | 136 | 408 | 544 | 0.117 | |
| PKC | 20 | 58 | 78 | 0.162 | 9 | 29 | 38 | 0.169 | |
| CYP3A | 10 | 30 | 40 | 0.202 | 5 | 15 | 20 | 0.215 | |
| HCV | NS5B | 649 | 1948 | 2597 | 0.141 | 253 | 758 | 1011 | 0.141 |
| NS4B | 13 | 37 | 50 | 0.178 | 6 | 20 | 26 | 0.192 | |
| NS3/4A | 334 | 1000 | 1334 | 0.145 | 136 | 410 | 546 | 0.145 | |
| NS5A | 27 | 80 | 107 | 0.182 | 13 | 40 | 53 | 0.182 | |
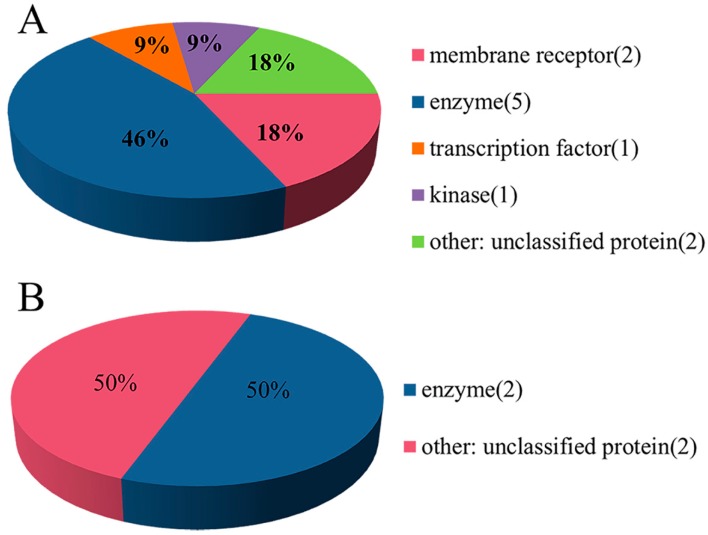
In addition, according to the protein target classification rules in the ChEMBL database, 11 targets related to HIV-1 and four targets related to HCV were spatially distributed into five and two categories, respectively. As shown in Figure 3A, the HIV-1 target space (n = 11) consisted of five categories, including membrane receptor (n = 2), enzyme (n = 5), transcription factor (n = 1), kinase (n = 1) and unclassified protein (n = 2). Similarly, the HCV target space (n = 4) was divided into two categories, including enzyme (n = 2) and unclassified protein (n = 2) in Figure 3B. Thus, the two datasets for HIV-1 and HCV had diverse target coverage.
Figure 3.
Target classification of the HIV-1 (A) and HCV (B) data sets.
2.2. Performance Evaluation and the Comparison of the Models
Machine learning algorithms have been successfully applied to drug discovery [32]. Naïve Bayes (NB) is one of the most popular classifiers and uses the hypothesis of attribution conditional independence [23]. The support vector machine (SVM) was developed based on Vapnik’s structural risk minimization (SRM) principle [33] and shows excellent classification performance with a reduced risk of overfitting. The construction of all the classification models in this study was primarily based on the use of two classifiers (NB and SVM) and two types of fingerprints (MACCS and ECFP6). Afterwards, internal five-fold cross-validation and external test set validation were conducted.
The performance of the multiple QSAR models as determined by five-fold cross-validation is shown in Table 2. Among the 44 models of HIV-1, 86.36% models (38 out of 44) had an area under the ROC curve (AUC) value exceeding 0.9, and 79.55% of the models (35 out of 44) had a non-error rate (NER) value exceeding 0.8. In addition, the AUC values ranged from 0.834 to 1, with a median of 0.983, and the NER value ranged from 0.737 to 1, with a median of 0.946. The sixteen models of HCV had AUC and NER values exceeding 0.9 and 0.8, respectively. In addition, the AUC value ranged from 0.924 to 1, with a median of 1, and the NER value ranged from 0.801 to 1, with a median of 0.990. Thus, these results suggest that the multiple QSAR models for both HIV-1 and HCV had a remarkable capability for identifying inhibitors with low false positive rates. The detailed performance data for the training sets is given in Table S1 in the Supporting Data. Similarly, for the models of HIV-1, 75% of the models (33 out of 44) had sensitivity (SE) values exceeding 0.8, with a median of 0.918, and 93.18% of the models (41 out of 44) had specificity (SP) values exceeding 0.8, with a median of 0.997. For the models of HCV, 87.5% of the models (14 out of 16) had SE values exceeding 0.8, with a median of 0.988, and 93.8% of the models (15 out of 16) had SP values exceeding 0.8, with a median of 1.
Table 2.
Performance results for the 60 multiple QSAR models determined by five-fold cross-validation.
| Object | Target | MACCS | ECFP6 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NB | SVM | NB | SVM | ||||||
| AUC | NER | AUC | NER | AUC | NER | AUC | NER | ||
| HIV-1 | CXCR4 | 0.992 | 0.962 | 0.998 | 0.978 | 0.994 | 0.969 | 0.997 | 0.973 |
| CCR5 | 0.977 | 0.878 | 0.997 | 0.990 | 0.986 | 0.936 | 0.996 | 0.976 | |
| GP120 | 0.845 | 0.799 | 0.926 | 0.851 | 0.880 | 0.775 | 0.923 | 0.737 | |
| GP41 | 0.941 | 0.964 | 0.958 | 0.964 | 0.963 | 0.964 | 0.934 | 0.786 | |
| RT | 0.838 | 0.763 | 0.994 | 0.960 | 0.898 | 0.797 | 0.994 | 0.955 | |
| IN | 0.930 | 0.863 | 0.996 | 0.976 | 0.949 | 0.878 | 0.994 | 0.960 | |
| PR | 0.980 | 0.908 | 0.999 | 0.987 | 0.982 | 0.925 | 0.997 | 0.987 | |
| Gag-pol | 0.966 | 0.942 | 0.984 | 0.949 | 0.967 | 0.925 | 0.980 | 0.935 | |
| Protein tat | 0.834 | 0.745 | 0.909 | 0.792 | 0.870 | 0.805 | 0.901 | 0.739 | |
| PKC | 0.991 | 0.991 | 1 | 0.975 | 0.994 | 0.925 | 0.988 | 0.925 | |
| CYP3A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.950 | |
| HCV | NS3/4A | 0.990 | 0.853 | 1 | 0.993 | 0.994 | 0.953 | 1 | 0.987 |
| NS4B | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.885 | |
| NS5A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| NS5B | 0.924 | 0.863 | 1 | 0.987 | 0.946 | 0.801 | 1 | 0.984 | |
To assess the generality of the multiple QSAR models in terms of distinguishing inhibitors from decoys, external test sets were used to perform a more rigorous validation of the training models. The assessment results for the test sets are listed in Table 3. For the 44 models of HIV-1, the AUC value ranged from 0.833 to 1, with a median value of 0.997, and the NER value ranged from 0.745 to 1, with a median value of 0.954. For the 16 models of HCV, the AUC value ranged from 0.842 to 1, with a median value of 0.994, and the NER value ranged from 0.788 to 1, with a median value of 0.959. The detailed performance data obtained for the test set validation is presented in Table S2 in the Supporting Data. The median values of SE and SP for the 11 datasets obtained for HIV-1 are 0.938 and 0.999, respectively. The median values of SE and SP for the four datasets obtained for HCV are 0.963 and 1, respectively. Moreover, the prediction accuracy (Q) is an important index for evaluating the predictive capability of models. Among the models of HIV-1 and HCV, 93.18% and 93.75% of the models, respectively, had an accuracy higher than 0.8. According to the results of the test sets, the comparable performance of the multiple QSAR models for the training and test sets indicated that the models showed no obvious overfitting. Thus, the 60 multiple QSAR models had a substantial capability to distinguish inhibitors from decoys with comparable yields and low false positive rates.
Table 3.
The test results for the performance of the 60 multiple QSAR models determined by test set validation.
| Object | Target | MACCS | ECFP6 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NB | SVM | NB | SVM | ||||||
| AUC | NER | AUC | NER | AUC | NER | AUC | NER | ||
| HIV-1 | CXCR4 | 1 | 0.989 | 1 | 0.989 | 1 | 0.989 | 1 | 0.989 |
| CCR5 | 0.981 | 0.891 | 0.999 | 0.990 | 0.995 | 0.926 | 1 | 0.978 | |
| GP120 | 0.881 | 0.784 | 0.940 | 0.861 | 0.883 | 0.775 | 0.910 | 0.759 | |
| GP41 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| RT | 0.837 | 0.745 | 0.995 | 0.964 | 0.899 | 0.799 | 0.994 | 0.963 | |
| IN | 0.934 | 0.856 | 0.997 | 0.979 | 0.947 | 0.876 | 0.996 | 0.972 | |
| PR | 0.975 | 0.897 | 1 | 0.991 | 0.982 | 0.932 | 0.999 | 0.987 | |
| Gag-pol | 0.977 | 0.938 | 0.988 | 0.969 | 0.975 | 0.923 | 1 | 0.969 | |
| Protein tat | 0.833 | 0.771 | 0.920 | 0.810 | 0.864 | 0.803 | 0.926 | 0.749 | |
| PKC | 1 | 1 | 1 | 0.889 | 1 | 0.944 | 1 | 0.889 | |
| CYP3A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0.900 | |
| HCV | NS3/4A | 0.986 | 0.836 | 1 | 0.996 | 0.987 | 0.946 | 1 | 0.971 |
| NS4B | 0.842 | 0.833 | 0.925 | 0.833 | 0.867 | 0.833 | 0.908 | 0.833 | |
| NS5A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| NS5B | 0.907 | 0.837 | 1 | 0.984 | 0.944 | 0.788 | 1 | 0.988 | |
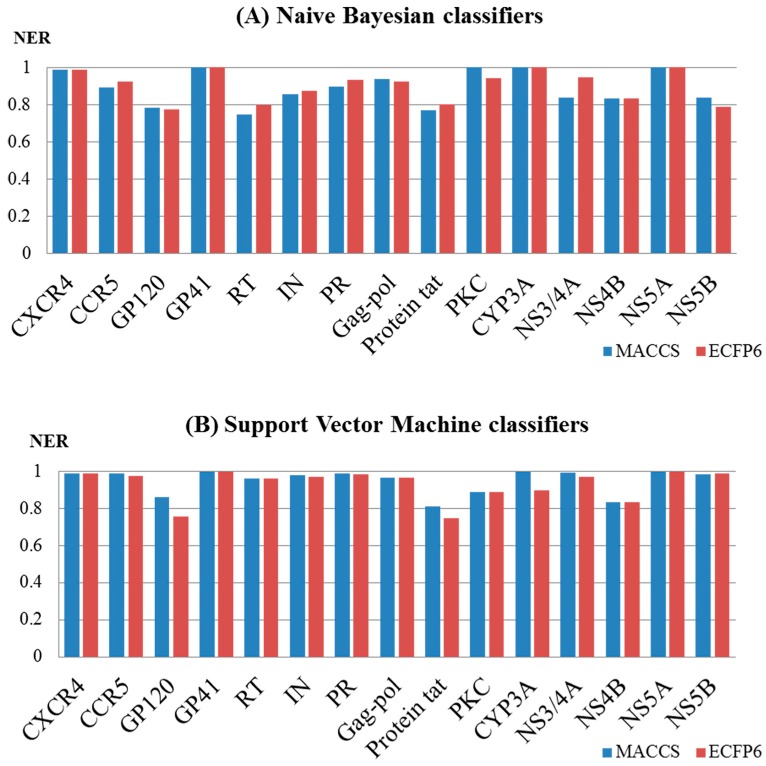
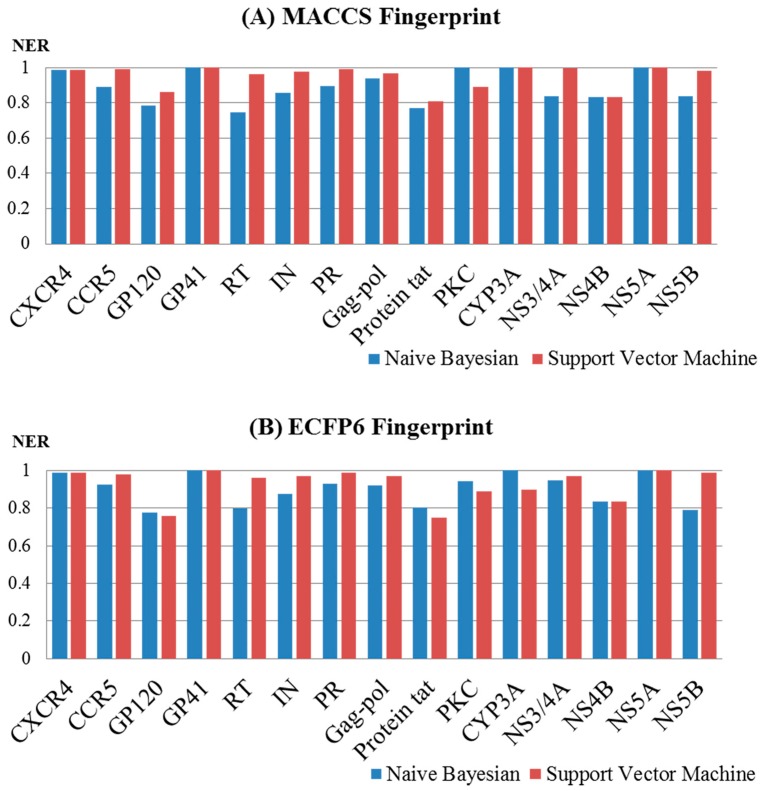
To investigate the influence of different types of molecular fingerprints (MACCS and ECFP6) on the performance of the Naïve Bayes and support vector machine models, the corresponding NER values for the test sets were compared. As shown in Figure 4A,B, the NER values obtained with the MACCS and ECFP6 fingerprints based on a single machine learning method (support vector machine or Naïve Bayes) were approximately equivalent. For the models built by the support vector machine, the NER values for MACCS ranged from 0.810 to 1 with a median of 0.984, and those for ECFP6 ranged from 0.749 to 1 with a median of 0.971. For the models built according to Naïve Bayes, the NER values for MACCS ranged from 0.745 to 1 with a median of 0.891, and those for ECFP6 ranged from 0.775 to 1 with a median of 0.926. It should be noted that different fingerprints obtained different structural representations of a chemical structure, and multiple QSAR models based on different fingerprints could have an impact on the performance in terms of yield.
Figure 4.
Performance of 60 multiple QSAR models built based on Naïve Bayes (A) and the support vector machine (B) using different fingerprints determined by test set validation.
Moreover, the performance of the different algorithms (NB and SVM) was also evaluated. For the MACCS fingerprint (Figure 5A), the performance of the SVM models was generally superior to that of the NB models. For example, for the models of HIV-1, the NER values for the SVM models ranged from 0.81 to 1 with a median of 0.979, and this was superior to that of the NB models, which ranged from 0.745 to 1 with a median of 0.897. For the models of HCV, the NER values for the SVM models ranged from 0.833 to 1 with a median of 0.990, and this was superior to that of the NB models, which ranged from 0.833 to 1 with a median value of 0.837. For the ECFP6 fingerprint (Figure 5B), the SVM models broadly outperformed the NB models. For the models of HIV-1, the NER values for the SVM models ranged from 0.749 to 1 with a median of 0.969, and this was superior to that of the NB models, which ranged from 0.775 to 1 with a median of 0.926. For the HCV models, the NER values for the SVM models ranged from 0.833 to 1 with a median of 0.980, and these were different from those obtained for the NB models, which ranged from 0.788 to 1 with a median value of 0.890.
Figure 5.
Performance of the 60 multiple QSAR models built based on MACCS (A) and ECFP6 (B) using different algorithms determined by test set validation.
In this study, on the basis of two machine learning algorithms (support vector machine and Naïve Bayes) and two fingerprints (MACC and ECFP6), four classifiers (NB_MACCS, NB_ECFP6, SVM_MACCS, SVM_ECFP6) were built for each target. As described above, for the models built by the same machine learning algorithm, the performances of the models that used MACCS were approximately equivalent to those that used ECFP6. When using the same fingerprint, the performance of the models built by the support vector machine was better than those of models built by Naïve Bayes. However, it is theoretically difficult to determine which fingerprint or algorithm is better because each algorithm or fingerprint has its own advantages and disadvantages. For example, the models built by different fingerprints or algorithms may make opposite predictions for the same molecule. Therefore, combining the results of a single classifier to predict CPIs is intelligent. One compound was predicted as positive when at least two of the four single classifiers predicted it to be active, and the corresponding CPIs were defined as potential interactions.
2.3. Case 1: Prediction and Analysis of the Polypharmacology of Known HIV-1 and HCV Drugs
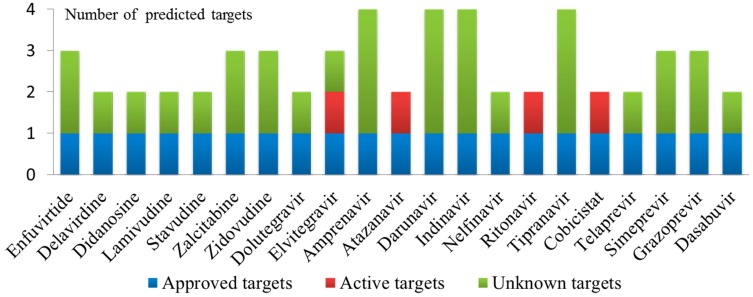
To explore the multiple bioactivities of known drugs, 60 multiple QSAR models based on 15 key targets were used to predict potential targets for 27 approved HIV-1 drugs and 10 approved HCV drugs. If the drug was predicted to be active by at least two of the four classifiers for each target, then the relevant CPI was defined as a potential interaction. The detailed prediction results of the polypharmacology of the known drugs are presented in Table S3 in the Supporting Data. The results show that 17 approved HIV-1 drugs and four approved HCV drugs were predicted to interact with more than one target, of which 7 approved HIV-1 drugs and four approved HCV drugs were predicted to interact with HIV-1 and HCV targets simultaneously. To verify the prediction results, the predicted targets for each drug were validated using the PubChem BioAssay database [34] and the ChEMBL database, and the corresponding binding values are presented in Table 4. Figure 6 shows a histogram of the number of predicted targets and established targets for 17 approved HIV-1 drugs and four approved HCV drugs. For example, elvitegravir, which was predicted to be active against HIV-1 RT, HIV-1 IN and HCV NS5B, could inhibit HIV-1 RT (active target) and IN (approved target) with IC50 values of 91 μM and 28 nM, respectively. Tipranavir, which was predicted to be active against HIV-1 GP120, HIV-1 RT, HIV-1 PR and HCV NS5B, could inhibit HIV-1 PR (approved target) with an IC50 value of 0.03 μM. Telaprevir, which was predicted to be active against HIV-1 PR and HCV NS3/4A, could inhibit HCV NS3/4A (approved target) with an IC50 value of 0.2 μM.
Table 4.
Known interactions among the CPIs predicted by the multiple QSAR models for approved drugs.
| Drug Name | Predicted Target | Known Activity | Reference |
|---|---|---|---|
| Enfuvirtide | GP41 | IC50 = 90.2 nM | [35] |
| Delavirdine | RT | IC50 = 0.17 μM | [36] |
| Didanosine | RT | IC50 = 4.6 μM | [42] |
| Lamivudine | RT | IC50 = 0.60 μM | [42] |
| Stavudine | RT | IC50 = 1.9 μM | [42] |
| Zalcitabine | RT | IC50 = 0.13 μM | [42] |
| Zidovudine | RT | IC50 = 0.13 μM | [42] |
| Dolutegravir | RT | Inactive | [50] |
| Dolutegravir | IN | IC50 = 2.7 nM | [43] |
| Elvitegravir | RT | IC50 = 91 μM | [44] |
| Elvitegravir | IN | IC50 = 28 nM | [44] |
| Amprenavir | PR | IC50 = 0.15 μM | [45] |
| Atazanavir | RT | IC50 = 26 nM | [46] |
| Atazanavir | PR | IC50 = 3.5 nM | [47] |
| Darunavir | PR | IC50 = 3.0 nM | [47] |
| Indinavir | RT | Inconclusive | [51] |
| Indinavir | PR | IC50 < 0.01 μM | [51] |
| Nelfinavir | PR | IC50 = 0.53 nM | [48] |
| Ritonavir | PR | IC50 = 0.6 nM | [49] |
| Ritonavir | CYP3A | IC50 = 0.11 μM | [49] |
| Tipranavir | PR | IC50 = 0.03 μM | [37] |
| Cobicistat | PR | IC50 = 0.15 μM | [49] |
| Cobicistat | CYP3A | IC50 > 30 μM | [49] |
| Telaprevir | NS3/4A | IC50 = 0.2 μM | [38] |
| Simeprevir | NS3/4A | IC50 = 10 nM | [39] |
| Grazoprevir | NS3/4A | IC50 = 0.07 nM | [40] |
| Dasabuvir | NS5B | IC50 = 0.6 μM | [41] |
Figure 6.
Number of predicted targets and known targets of 17 approved HIV-1 drugs and four approved HCV drugs (Approved targets: predicted targets are approved drug targets for drugs that are their inhibitor; Active targets: predicted targets for drugs with validated bioactivity; Unknown targets: predicted targets for drugs without validated bioactivity).
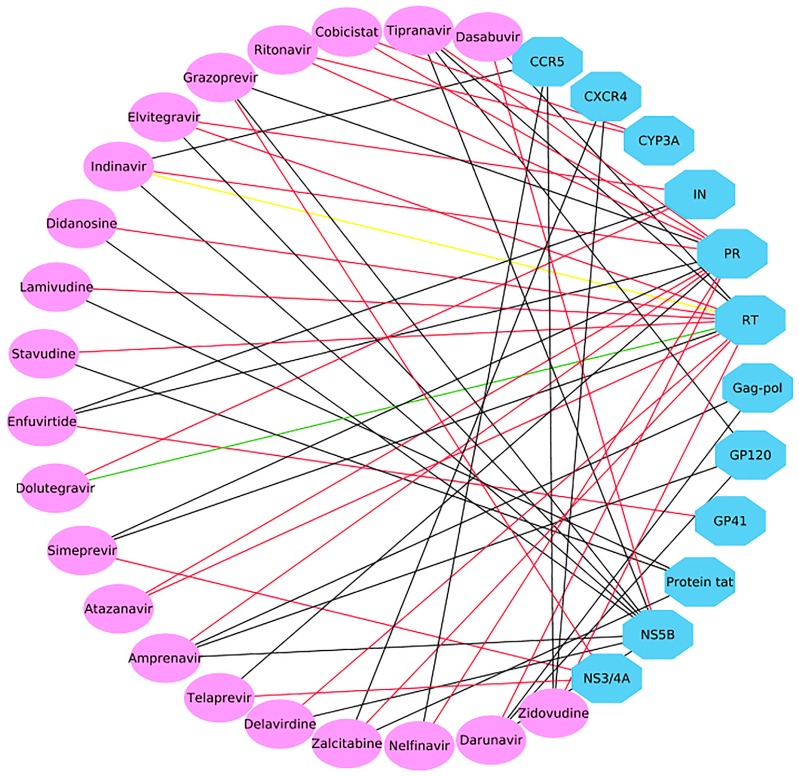
In addition, 56 CPIs were determined, as shown in Table S4 in the Supporting Data. Among them, 25 out of 56 predicted CPIs have been verified in the literature [35,36,37,38,39,40,41,42,43,44,45,46,47,48,49]; two out of 56 CPIs were predicted to be active but were validated as “inconclusive” and “inactive” [50,51]. A success rate of 44.6% (25/56) and a failure rate of 1.8% (1/56) illustrate the reliability of the multiple QSAR method. A polypharmacological interaction network of 21 drugs and predicted targets (56 CPIs) was constructed with Cytoscape 3.6 [52] (Figure 7). Figure 7 shows that each drug was predicted to have potential activity against two or three targets, which is in line with concept that “one drug can hit multiple targets”. Subsequently, the predicted CPI network identified many potential drug-target interactions that have yet to be explored, and these unverified CPIs are worthy of further experimental validation. The predicted targets may serve as new targets for approved drugs and provide promise as possible treatments for HIV/HCV coinfection.
Figure 7.
Polypharmacological analysis of 17 approved HIV-1 drugs and four approved HCV drugs based on the 60 multiple QSAR models generated in this study. Ellipses and octagons represent drug nodes and protein nodes, respectively. A red line indicates that the chemical–protein interaction (CPI) was experimentally verified as active. A black line indicates that the interaction was not proven to be worthy of further experimental validation. A green or yellow line indicates that the interaction was experimentally validated as inactive or inconclusive, respectively.
2.4. Case 2: Target prediction and Analysis of Known Inhibitors
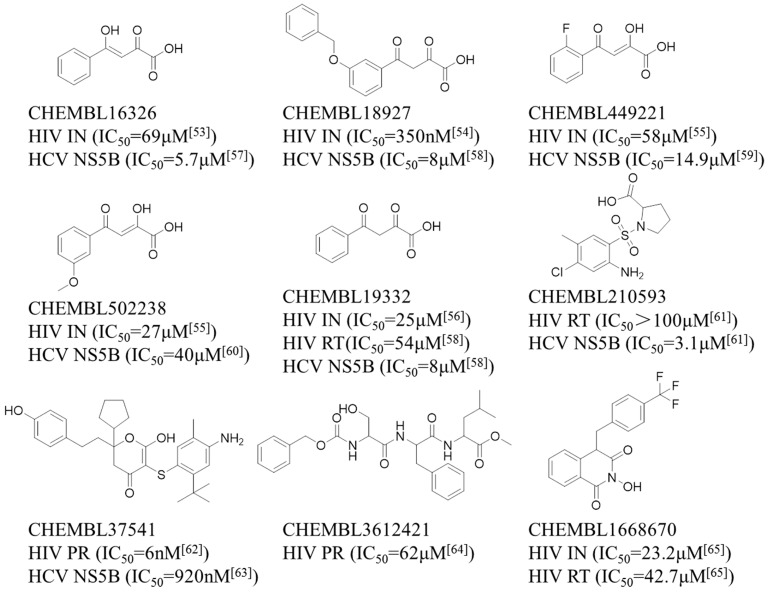
A set of nine compounds (Figure 8) with known activity targeting at least one of the four targets (HIV-1 PR, RT, IN and HCV NS5B) that was not involved in the generation of the multiple QSAR models was retrieved from the ChEMBL database. To evaluate whether the new models could correctly differentiate between these active compounds, the classifiers built in this study were used to predict chemical interactions with HIV-1 PR, RT and IN and HCV NS5B. According to the above rules, a potential interaction is defined as a CPI when at least two out of four single classifiers predict a compound to be positive. The specific predicted results are given in Table S5 in the Supporting Data, and the comparison of the predicted and experimental results is shown in Table 5. Moreover, molecular docking provided a more detailed inspection of the binding mode for each compound. The scoring values of the top scoring poses are available in Table S6 in the Supporting Data.
Figure 8.
The structure and bioactivity data of the nine selected compounds.
Table 5.
Predicted and experimental activity of the nine selected compounds against HIV-1 protease (PR), reverse transcriptase (RT), integrase (IN) and HCV NS5B.
| CHEMBL ID | PR/Pred 1 | PR/Exp 2 | IN/Pred 3 | IN/Exp 4 | RT/Pred 5 | RT/Exp 6 | NS5B/Pred 7 | NS5B/Exp 8 |
|---|---|---|---|---|---|---|---|---|
| CHEMBL16326 | false | — 9 | true | active | false | — | true | active |
| CHEMBL18927 | false | — | true | active | false | — | true | active |
| CHEMBL449221 | false | — | true | active | false | — | true | active |
| CHEMBL502238 | false | — | true | active | false | — | true | active |
| CHEMBL19332 | false | — | true | active | false | active | true | active |
| CHEMBL210593 | false | — | false | — | true | active | true | active |
| CHEMBL37541 | true | active | false | — | false | — | true | active |
| CHEMBL3612421 | true | active | false | — | false | — | true | — |
| CHEMBL1668670 | false | — | true | active | true | active | true | — |
1 “PR/Pred” shows the predicted result of a compound against PR. 2 “PR/Exp” shows the experimental result of a compound against PR.3 “IN/Pred” shows the predicted result of a compound against IN. 4 “IN/Exp” shows the experimental result of a compound against IN. 5 “RT/Pred” shows the predicted result of a compound against RT. 6 “RT/Exp” shows the experimental result of a compound against RT. 7 “NS5B/Pred” shows the predicted result of a compound against NS5B. 8 “NS5B/Exp” shows the experimental result of a compound against NS5B. 9 “—” indicates that the predicted result has not been verified by experiments.
Table 5 shows that all nine compounds were predicted to act against two or three targets, and seven compounds were reported to simultaneously interact with HIV-1 and HCV targets with different binding affinities. New predictions of the inhibitory activity of CHEMBL3612421 and CHEMBL1668670 against NS5B have yet to be reported experimentally. However, CHEMBL19332, a known active HIV-1 RT inhibitor, was incorrectly predicted to be inactive toward HIV-1 RT. Thus, a total of 19 CPI pairs were predicted to be positive, of which 17 CPI pairs were experimentally confirmed to be active [53,54,55,56,57,58,59,60,61,62,63,64,65]. Therefore, the prediction success rate of 89.5% (17/19) indicates the reliability of the multiple QSAR models. It is worth mentioning that the remaining two CPI pairs associated with two compounds deserve further experimental investigation, and the predicted targets might be new targets for known inhibitors. Moreover, one CPI pair shows false negatives in the prediction result.
The results in Table 5 show that five compounds (CHEMBL16326, CHEMBL18927, CHEMBL449221, CHEMBL502238 and CHEMBL19332) were predicted to be potentially active against HIV-1 IN and HCV NS5B, and all of these were also experimentally confirmed to be HIV-1 IN and HCV NS5B inhibitors in the literature [53,54,55,56,57,58,59,60]. In particular, five compounds with analogous structures have similar protein-ligand interactions with the active regions of HIV-1 IN and HCV NS5B. As shown in Figure 9, the hydrophobic groups in those five compounds, such as the benzene ring in CHEMBL16326 and the monofluorobenzene ring in CHEMBL449221, are located in hydrophobic pockets formed by Leu102, Ala128, Trp132, Thr174 and Met178 of HIV-1 IN (see Figure 9A) and Val52, Leu159 and Ile160 of HCV NS5B (see Figure 9B). The diketo acid moieties of these five compounds form hydrogen bonds with Glu170 and His171 in the active site of HIV-1 IN (see Figure 9A) and with Phe224 and Asp225 and via Mn2+ ions (metal chelation) in the catalytic activity site of HCV NS5B (see Figure 9B). Thus, HIV-1 IN and HCV NS5B can be affected by diketo acid inhibitors, which disturb enzymatic function during the process of virus replication.
Figure 9.
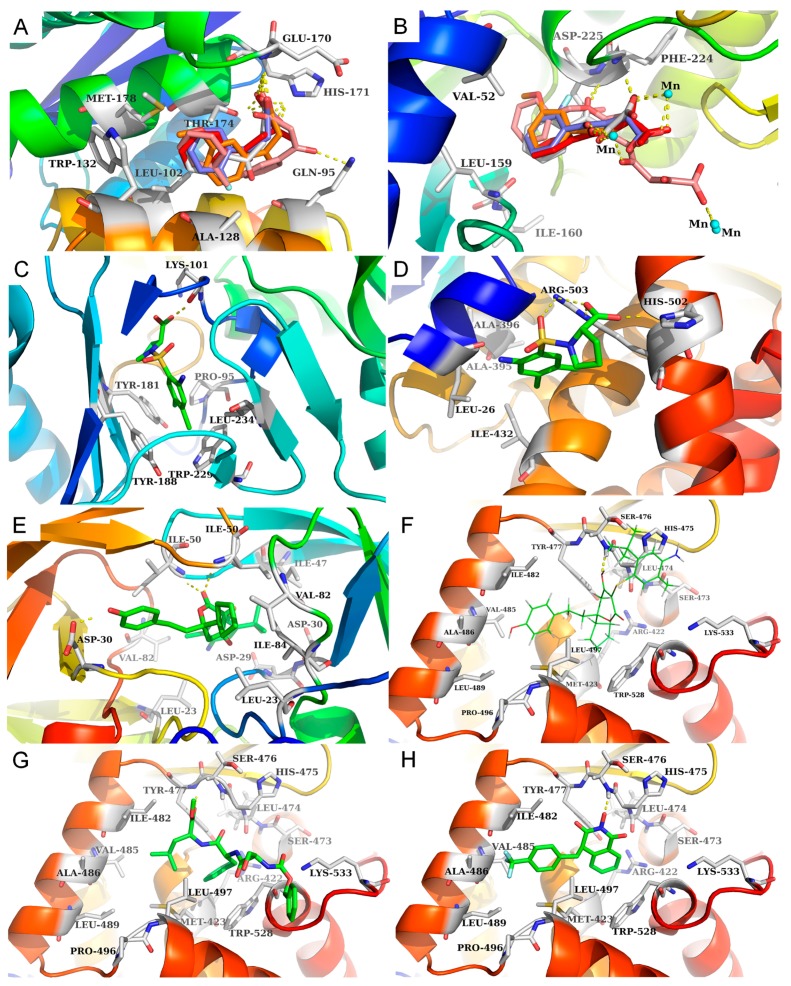
The binding modes of nine compounds in their respective binding sites. (A,B) show the binding modes of five compounds with HIV-1 IN (PDB: 4NYF) and HCV NS5B (PDB: 1GX6), respectively, including CHEMBL16326 (red), CHEMBL18927 (pink), CHEMBL449221 (purple), CHEMBL502238 (orange) and CHEMBL19332 (gray). (C,D) show the binding modes of CHEMBL210593 with HIV-1 RT (PDB: 1TL3) and HCV NS5B (PDB: 1OS5). (E,F) show the binding modes of CHEMBL37541 with HIV-1 PR (PDB: 4MC1) and HCV NS5B (PDB: 1OS5). (G,H) show the binding modes of two compounds (CHEMBL3612421 and CHEMBL1668670) with HCV NS5B (PDB 1OS5). (The cocrystallized molecules are shown as lines, whereas the molecules modeled into the receptors are shown as sticks).
CHEMBL210593 was predicted to be a HIV-1 RT and HCV NS5B inhibitor with an IC50 > 100 μM and 3.1 μM, respectivelyn [61]. Figure 9 shows that the 3-chloro-p-methylaniline in CHEMBL210593 occupies a hydrophobic pocket formed by Tyr181, Tyr188 and Trp229 in HIV-1 RT (see Figure 9C) and Leu26, Ala395, Ala396 and Ile432 in HCV NS5B (see Figure 9D). The carboxyl group of CHEMBL210593 forms hydrogen bonds with the main chain carbonyl oxygen of Lys101 in HIV-1 RT (see Figure 9C) and the side chains of Arg503 and His502 in HCV NS5B (see Figure 9D). Therefore, proline sulfonamide inhibitors may affect HIV-1 RT and HCV NS5B and influence their enzymatic function.
CHEMBL37541 and CHEMBL3612421 were also predicted to be active against HIV-1 PR and HCV NS5B. CHEMBL37541 is a known HIV-1 PR and HCV NS5B inhibitor with IC50 values of 6 nM and 920 nM, respectively [62,63]. CHEMBL3612421 is a specific HIV-1 PR inhibitor with an IC50 of 62 μM [64], but inhibitory activity against NS5B by CHEMBL3612421 has not yet been reported. In CHEMBL37541, the dihydropyrone moiety forms hydrogen bonds directly with Ile50 in HIV-1 PR (see Figure 9E) and Ser476 in HCV NS5B (see Figure 9F). The 4-ethylphenol and cyclopentane groups fit into hydrophobic pockets consisting of the residues Leu23, Leu23′, Val82, Val82′, Ile84 and Ile84′ in HIV-1 PR (see Figure 9E) and the residues Met423, Ile482, Val485, Ala486, Leu489, Leu497 and Trp528 in HCV NS5B (see Figure 9F). A newly predicted CPI pair involving CHEMBL3612421 and HCV NS5B is presented in Figure 9G. CHEMBL3612421 forms a hydrogen bond with Tyr477 in the thumb II region of HCV NS5B, and its benzene ring and dimethyl group are located in a hydrophobic region consisting of the residues Met423, Ile482, Val485, Ala486, Leu489, Leu497 and Trp528. Similarly, dihydropyrone inhibitors can affect HIV-1 PR and HCV NS5B and influence enzymatic function.
CHEMBL1668670 was also predicted to be an HIV-1 RT, IN and HCV NS5B inhibitor, and it has demonstrated bioactivity only against HIV-1 RT (IC50 = 42.7 μM) and IN (IC50 = 23.2 μM) [65]. There have been no reports on the inhibitory activity of CHEMBL1668670 against NS5B. A newly predicted CPI pair involving CHEMBL1668670 and NS5B is shown in Figure 9H. CHEMBL1668670 forms a hydrogen bond with Ser476 in the thumb II region of HCV NS5B, and its trifluorobenzene ring interacts with a hydrophobic region consisting of the residues Met423, Ile482, Val485, Ala486, Leu489 and Leu497.
Thus, the results show that it is possible to design specific inhibitors targeting HIV-1 targets and HCV targets simultaneously. In addition, an individual compound was determined to have coinhibitory ability against both virus targets if it was able to satisfy the requirements for binding the HIV/HCV target active sites in terms of the molecular shape, size and physical–chemical properties. In short, multiple QSAR models are expected to contribute to the design and identification of different types of co-inhibitors with excellent affinities that selectively and simultaneously target multiple HIV-1 and HCV targets for the treatment of HIV/HCV coinfection.
3. Discussion
In this study, based on the multiple QSAR method, 44 binary classifiers for 11 targets related to HIV-1 and 16 binary classifiers for four targets related to HCV were established to predict the CPIs. In addition, the predictive reliability of the models was confirmed by five-fold cross-validation and test set validation.
To illustrate the application of the models, two different cases were investigated to systematically predict the multiple bioactivities of 27 approved HIV-1 drugs and 10 approved HCV drugs (case 1) and nine compounds that were known to be active against at least one target of HIV-1 and HCV (case 2). For case 1, 21 approved drugs were predicted to be active against more than one biological target. The predictions were confirmed on the basis of reported bioactivity data with a success rate of 44.6% and a failure rate of 1.8%. For case 2, nine active compounds against at least one target (HIV-1 PR, RT, IN or HCV NS5B) were predicted to be multitarget inhibitors by multiple QSAR models with a success rate of 89.5%. The prediction results demonstrate that the machine learning models had high specific and low false negative rates in this study. Two possible explanations for the occurrence of false negatives are as follows. First, the molecular fingerprint was unable to sufficiently represent the molecular structural information. Second, the inherent limitations of machine learning and the composition of the training set had a significant impact on the accuracy of the models. In short, the predictive results of the above two cases indicate that multiple QSAR models showed high performance in the prediction of multitarget inhibitors with activity against both HIV and HCV.
4. Conclusions
In the past, “one gene, one drug, one disease” was the main paradigm of drug discovery. However, with the introduction of the concept of polypharmacology, multitarget therapy using a single drug that simultaneously inhibits two or more targets may provide a new strategy. Here, multiple QSAR models were used to identify compounds acting on multiple targets for the treatment of HIV/HCV coinfection.
As mentioned above, the multiple QSAR method achieved satisfactory prediction accuracy, and the machine learning models were applicable to potential target identification in this study. The limited chemical diversity of the training set makes the QSAR models suitable for predicting the knowledge or information that served as the basis on which the models were developed in the chemical space. Thus, it is unreasonable to predict a whole chemical space using a single QSAR model. A prediction based on QSAR is significant only if the specific compound being predicted falls within the HIV/HCV coinfection basis of the model. In addition, QSAR modeling neglects the interactions between receptors and ligands, unlike molecular docking. Therefore, it is necessary to choose a reasonable single method or combination of methods according to the specific situation.
In summary, the computational methods utilized in this study could effectively detect multitargeted co-inhibitors for the suppression of HIV/HCV coinfection and have potential applications for the identification of new targets, the virtual screening of multitarget drugs and drug repurposing for the treatment of other diseases. In addition, we hope that our research will be helpful in the clinical treatment of HIV/HCV coinfection.
5. Materials and Methods
5.1. Dataset Preparation
The Therapeutic Target Database (TTD) [28] and ChEMBL Database (version 23, https://www.ebi.ac.uk/chembl/) were used to determine the clinical targets of anti-HIV and anti-HCV therapy. We used keywords with ‘HIV-1′, ‘Human Immunodeficiency Virus type-1′, ‘HCV’ and ‘Hepatitis C Virus’ as the retrieval conditions, and the search results were further filtered to identify drug candidates that had at minimum entered phase I clinical trials.
In this study, the known small molecule inhibitors of the HIV-1 or HCV targets were downloaded from the ChEMBL Database. The datasets were processed according to the following requirements: (1) the compound, with an IC50, EC50 or Ki ≤ 20 μM, was retained as positive (expressed as ‘+1′); (2) salts were converted into the homologous acid or base and water molecules in hydrates were eliminated; (3) duplicate molecules were deleted. DecoyFinder software [66,67] was applied to prepare a set of decoy compounds (three decoys for each known inhibitor) [21] by using information from the ZINC database (version 12, http://zinc.docking.org/), and the decoy compounds were denoted as negative (expressed as ‘−1’) for each target. Specifically, the decoy compounds were selected based on the following constraints: (i) the MW was within 25 Da of that of the corresponding positive compound; (ii) the number of RB, HBDs, HBAs were ±1, ±1 and ±2 of those of the positive compound, respectively; (iii) the LogP was ±1 of that of the positive compound; (iv) the Tanimoto coefficient of a potential decoy vs that of the active compound and that of a potential decoy vs a previously selected decoy were, by default, 0.75 and 0.9, respectively [66]. Finally, the dataset including inhibitors and decoys for each target was divided into a training set (70%) and a test set (30%) using the default values of the parameters of the partitioning node in KNIME (version 3.4.2, https://www.knime.com) [68]. The above data processing procedures were performed in KNIME, an open-source data analysis and cheminformatic software.
5.2. Molecular Representation
Each compound in the above data sets was represented using two different types of binary fingerprint descriptors (MACCS and ECFP6) as chemical descriptors calculated in KNIME. MACCS is a common 2D molecular fingerprint descriptor composed of 166 structural keys. Most of the important chemical features are covered despite the small length [69]. Binary fingerprints (FPs) completely characterize all of the structural fragments in the molecule in a binary form [70]. It is different from classical descriptors, as fingerprints encode molecular structural information as a range of binary bits and can only be deciphered used as a whole. Additionally, extended-connectivity fingerprints (ECFPs), also known as “circular fingerprints”, are state-of-the-art topological fingerprints [27]. In this study, the ECFPs were generated with the default parameters of 1024 bits, 2 bits per pattern and a radius of 3 bonds, which was equivalent to ECFP6, using Fingerprint node [71,72] in KNIME. To date, MACCS and ECFPs have been widely used in drug discovery on the basis of their characteristics of rapid calculation and their representation of many different molecular features and stereochemical information.
5.3. Multiple QSAR Models Generation
In this study, all classification models were built by combining two machine learning algorithms (Naïve Bayes and support vector machine) and two molecular fingerprints (MACCS and ECFP6). Therefore, four classifiers (NB_MACCS, NB_ECFP6, SVM_MACCS and SVM_ECFP6) were generated for each target. The number of multiple QSAR models generated for the 11 HIV-1 targets and four HCV targets were 44 and 16, respectively.
5.3.1. Naïve Bayes (NB)
The Naïve Bayes classification models were built using KNIME (version 3.4.2). Bayesian classification is widely used to solve classification problems based on Bayesian theory, such as the discrimination between active and inactive compounds, by building a classifier. Naïve Bayes is based on the occurrence frequency of various descriptors found in two or more groups of molecules and is used to distinguish optimally between these datasets. The details of the Bayesian theorem are given in Equations (1)–(3):
| (1) |
| (2) |
| (3) |
In general, x represents molecular descriptors such as molecular weight, logP or molecular fingerprint, y represents whether a compound will exhibit a desired biological activity such as enzyme inhibition, P(yi) is the prior probability of yi being true without any knowledge of x, and P(x|yi) is the conditional probability of yi being true given x. P(x1,x2, …, xm) represents the marginal probabilities of the molecular descriptors that will appear in the training set. Because of the assumption of attribution independence, Equation (2) is simplified to Equation (3), which is also known as the Naïve Bayes formula.
5.3.2. Support Vector Machine (SVM)
Based on the principle of structural risk minimization (SRM) [73], SVM has been developed as a useful classification technique. The main idea is to convert input sample space into a high-dimensional space, in which a hyperplane with a maximal margin is generated to describe a nonlinear classification boundary. Importantly, the classification is achieved through a kernel function. There are four basic kernels: linear, polynomial, radial basis function (RBF) and sigmoid [30]. In most cases, RBF is preferred [74]. In this study, the RBF kernel function was used. The penalty parameter C and kernel parameter γ that were used in the SVM were optimized through five-fold cross-validation in SVM, and the final optimal values of C and γ that provided the best accuracy were selected for generating the final classifiers [75].
5.4. Performance Evaluation of the Multiple QSAR Models
All models were evaluated by internal five-fold cross-validation and external test set validation. In five-fold cross-validation, the training set was divided into five subsets equally. Four subsets were used to train the model, and the remaining subset was used as an internal validation set.
The major evaluation indexes included true positives (TP), true negatives (TN), false positives (FP), false negatives (FN), sensitivity (SE), specificity (SP), the overall prediction accuracy (Q) and the receiver operating characteristic curve (ROC). Among these, ROC curves are often used to estimate the merits of a binary classifier, in which the false positive rate (FPR) and true positive rate (TPR) are used as the abscissa and the ordinate, respectively. Generally, the AUC value ranges from 0 to 1, and the higher the value, the better the classification capability of the model is.
| (4) |
In addition, the average value of the sensitivity for all classes, also called the non-error rate (NER), was calculated according to Equation (4). The value of NER ranges from 0 to 1, and the value of NER is 1, indicating a perfect classification [76].
5.5. Molecular Docking
The docking algorithm Glide [77] was used for molecular docking and the analysis of the binding modes between the small molecules and target proteins. The crystal structures of HIV-1 IN (PDB: 4NYF) [78], HIV-1 RT (PDB: 1TL3) [79], HIV-1 PR (PDB: 4MC1) [80] and HCV NS5B (PDB: 1OS5 and 1GX6) [81,82] were prepared and used to build the receptor grid. The grid was centered on the ligand in the co-crystallized complex or the key residues of the proteins, and then the Glide SP protocol was utilized for docking. For grid generation and docking, all of the parameters used were the default values.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/14/3572/s1.
Author Contributions
Conceptualization, Y.W. and J.L.; Data curation, W.L. and T.D.; Formal analysis, W.L. and T.D.; Investigation, W.L. and T.D.; Methodology, W.L. and T.D.; Project administration, Y.W.; Supervision, Z.H. and J.L.; Writing—original draft, Y.W. and W.L.; Writing—review & editing, Z.H. and J.L.
Funding
This study was supported by the National Key R&D Program of China (No. 2017YFC1104400).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wang S., Milne G.W.A., Yan X., Posey I.J., Nicklaus M.C., Graham L., Rice W.G. Discovery of novel, non-peptide HIV-1 protease inhibitors by pharmacophore searching. J. Med. Chem. 1996;39:2047–2054. doi: 10.1021/jm950874+. [DOI] [PubMed] [Google Scholar]
- 2.Therese P.J., Manvar D., Kondepudi S., Battu M.B., Sriram D., Basu A., Yogeeswari P., Kaushik-Basu N. Multiple e-pharmacophore modeling, 3D-QSAR, and high-throughput virtual screening of hepatitis C virus NS5B polymerase inhibitors. J. Chem. Inf. Model. 2014;54:539–552. doi: 10.1021/ci400644r. [DOI] [PubMed] [Google Scholar]
- 3.Laguno M., Murillas J., Blanco J.L., Martínez E., Miquel R., Sánchez-Tapias J.M., Bargallo X., García-Criado A., de Lazzari E., Larrousse M., et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for treatment of HIV/HCV co-infected patients. AIDS. 2004;18:F27–F36. doi: 10.1097/00002030-200409030-00003. [DOI] [PubMed] [Google Scholar]
- 4.WHO Global Hepatitis Report. [(accessed on 24 March 2019)];2017 Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
- 5.Wang Z., Bennett E.M., Wilson D.J., Salomon C., Vince R. Rationally designed dual inhibitors of HIV reverse transcriptase and integrase. J. Med. Chem. 2007;50:3416–3419. doi: 10.1021/jm070512p. [DOI] [PubMed] [Google Scholar]
- 6.Cox B.D., Prosser A.R., Sun Y., Li Z., Lee S., Huang M.B., Bond V.C., Snyder J.P., Krystal M., Wilson L.J., et al. Pyrazolo-Piperidines Exhibit Dual Inhibition of CCR5/CXCR4 HIV Entry and Reverse Transcriptase. ACS Med. Chem. Lett. 2015;6:753–757. doi: 10.1021/acsmedchemlett.5b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Operskalski E.A., Kovacs A. HIV/HCV co-infection: Pathogenesis, clinical complications, treatment, and new therapeutic technologies. Curr. HIV/AIDS Rep. 2011;8:12–22. doi: 10.1007/s11904-010-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajorath J. Computational analysis of ligand relationships within target families. Curr. Opin. Chem. Biol. 2008;12:352–358. doi: 10.1016/j.cbpa.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 9.Rognan D. Chemogenomic approaches to rational drug design. Br. J. Pharmacol. 2007;152:38–52. doi: 10.1038/sj.bjp.0707307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L., Wang K.J., Wang L.S., Jegga A.G., Qin S.Y., He G., Chen J., Xiao Y., He L. Chemical-protein interactome and its application in off-target identification. Interdiscip. Sci. Comput. Life Sci. 2012;3:22–30. doi: 10.1007/s12539-011-0051-8. [DOI] [PubMed] [Google Scholar]
- 11.Cheng F., Zhou Y., Li J., Li W., Liu G., Tang Y. Prediction of chemical-protein interactions: multitarget-QSAR versus computational chemogenomic methods. Mol. Biosyst. 2012;8:2373–2384. doi: 10.1039/c2mb25110h. [DOI] [PubMed] [Google Scholar]
- 12.Sawada R., Iwata H., Mizutani S., Yamanishi Y. Target-Based Drug Repositioning Using Large-Scale Chemical-Protein Interactome Data. J. Chem. Inf. Model. 2015;55:2717–2730. doi: 10.1021/acs.jcim.5b00330. [DOI] [PubMed] [Google Scholar]
- 13.Luo H., Chen J., Shi L., Mikailov M., Zhu H., Wang K., He L., Yang L. DRAR-CPI: A server for identifying drug repositioning potential and adverse drug reactions via the chemical-protein interactome. Nucleic Acids Res. 2011;39:492–498. doi: 10.1093/nar/gkr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins J.L., Bender A., Davies J.W. In silico target fishing: Predicting biological targets from chemical structure. Drug Discov. Today Technol. 2006;3:413–421. doi: 10.1016/j.ddtec.2006.12.008. [DOI] [Google Scholar]
- 15.Wang X., Shen Y., Wang S., Li S., Zhang W., Liu X., Lai L., Pei J., Li H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keiser M.J., Roth B.L., Armbruster B.N., Ernsberger P., Irwin J.J., Shoichet B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 17.Bauer R.A., Bourne P.E., Formella A., Frömmel C., Gille C., Goede A., Guerler A., Hoppe A., Knapp E.-W., Pöschel T., et al. Superimpose: A 3D structural superposition server. Nucleic Acids Res. 2008;36:W47–W54. doi: 10.1093/nar/gkn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Gao Z., Kang L., Zhang H., Yang K., Yu K., Luo X., Zhu W., Chen K., Shen J., et al. TarFisDock: A web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34:W219–W224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y.Z., Zhi D.G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins. 2001;43:217–226. doi: 10.1002/1097-0134(20010501)43:2<217::AID-PROT1032>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Viña D., Uriarte E., Orallo F., González-Díaz H. Alignment-free prediction of a drug-target complex network based on parameters of drug connectivity and protein sequence of receptors. Mol. Pharm. 2009;6:825–835. doi: 10.1021/mp800102c. [DOI] [PubMed] [Google Scholar]
- 21.Fang J., Li Y., Liu R., Pang X., Li C., Yang R., He Y., Lian W., Liu A.-L., Du G.-H. Discovery of multitarget-directed ligands against Alzheimer’s disease through systematic prediction of chemical-protein interactions. J. Chem. Inf. Model. 2015;55:149–164. doi: 10.1021/ci500574n. [DOI] [PubMed] [Google Scholar]
- 22.Antanasijević D., Antanasijević J., Trišović N., Ušćumlić G., Pocajt V. From Classification to Regression Multitasking QSAR Modeling Using a Novel Modular Neural Network: Simultaneous Prediction of Anticonvulsant Activity and Neurotoxicity of Succinimides. Mol. Pharm. 2017;14:4476–4484. doi: 10.1021/acs.molpharmaceut.7b00582. [DOI] [PubMed] [Google Scholar]
- 23.Porwal A., Carranza E.J.M., Hale M. Bayesian network classifiers for mineral potential mapping. Comput. Geosci. 2006;32:1–16. doi: 10.1016/j.cageo.2005.03.018. [DOI] [Google Scholar]
- 24.Xia X., Maliski E.G., Gallant P., Rogers D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004;47:4463–4470. doi: 10.1021/jm0303195. [DOI] [PubMed] [Google Scholar]
- 25.Jorissen R.N., Gilson M.K. Virtual Screening of Molecular Databases Using a Support Vector Machine. J. Chem. Inf. Model. 2005;45:549–561. doi: 10.1021/ci049641u. [DOI] [PubMed] [Google Scholar]
- 26.Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 27.Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 28.Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., Fu T., Zhang X., Cui X., Tu G., et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46:D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaulton A., Hersey A., Nowotka M.L., Patricia Bento A., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrian-Uhalte E., et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajusz D., Rácz A., Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 2015;7:20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maggiora G.M., Shanmugasundaram V. Molecular Similarity Measures. Chemoinformatics. 2004;275:1–50. doi: 10.1385/1-59259-802-1:001. [DOI] [PubMed] [Google Scholar]
- 32.Lavecchia A. Machine-learning approaches in drug discovery: Methods and applications. Drug Discov. Today. 2015;20:318–331. doi: 10.1016/j.drudis.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Vapnik V., Sterin A. On structural risk minimization or overall risk in a problem of pattern recognition. Autom. Remote Control. 1977;10:1495e1503. [Google Scholar]
- 34.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Zhou Z., Han L., Karapetyan K., Dracheva S., Shoemaker B.A., et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chong H., Qiu Z., Su Y., He Y. The N-Terminal T-T Motif of a Third-Generation HIV-1 Fusion Inhibitor Is Not Required for Binding Affinity and Antiviral Activity. J. Med. Chem. 2015;58:6378–6388. doi: 10.1021/acs.jmedchem.5b00109. [DOI] [PubMed] [Google Scholar]
- 36.Nugent R.A., Schlachter S.T., Murphy M.J., Cleek G.J., Poel T.J., Wishka D.G., Graber D.R., Yagi Y., Keiser B.J., Olmsted R.A., et al. Pyrimidine thioethers: A novel class of HIV-1 reverse transcriptase inhibitors with activity against BHAP-resistant HIV. J. Med. Chem. 1998;41:3793–3803. doi: 10.1021/jm9800806. [DOI] [PubMed] [Google Scholar]
- 37.Thaisrivongs S., Romero D.L., Tommasi R. a, Janakiraman M.N., Strohbach J.W., Turner S.R., Biles C., Morge R.R., Johnson P.D., Aristoff P. a, et al. Structure-based design of HIV protease inhibitors: 5,6-dihydro-4-hydroxy-2-pyrones as effective, nonpeptidic inhibitors. J. Med. Chem. 1996;39:4630–4642. doi: 10.1021/jm960228q. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., Müh U., Hanzelka B.L., Bartels D.J., Wei Y., Rao B.G., Brennan D.L., Tigges A.M., Swenson L., Kwong A.D., et al. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: Sensitivity to telaprevir (VX-950) and interferon alpha. J. Biol. Chem. 2007;282:22619–22628. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 39.Ding C.Z., Zhang Y.-K., Li X., Liu Y., Zhang S., Zhou Y., Plattner J.J., Baker S.J., Liu L., Duan M., et al. Synthesis and biological evaluations of P4-benzoxaborole-substituted macrocyclic inhibitors of HCV NS3 protease. Bioorg. Med. Chem. Lett. 2010;20:7317–7322. doi: 10.1016/j.bmcl.2010.10.071. [DOI] [PubMed] [Google Scholar]
- 40.Neelamkavil S.F., Agrawal S., Bara T., Bennett C., Bhat S., Biswas D., Brockunier L., Buist N., Burnette D., Cartwright M., et al. Discovery of MK-8831, A Novel Spiro-Proline Macrocycle as a Pan-Genotypic HCV-NS3/4a Protease Inhibitor. ACS Med. Chem. Lett. 2016;7:111–116. doi: 10.1021/acsmedchemlett.5b00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meguellati A., Ahmed-Belkacem A., Nurisso A., Yi W., Brillet R., Berqouch N., Chavoutier L., Fortuné A., Pawlotsky J.-M., Boumendjel A., et al. New pseudodimeric aurones as palm pocket inhibitors of Hepatitis C virus RNA-dependent RNA polymerase. Eur. J. Med. Chem. 2016;115:217–229. doi: 10.1016/j.ejmech.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Sluis-Cremer N., Koontz D., Bassit L., Hernandez-Santiago B.I., Detorio M., Rapp K.L., Amblard F., Bondada L., Grier J., Coats S.J., et al. Anti-human immunodeficiency virus activity, cross-resistance, cytotoxicity, and intracellular pharmacology of the 3′-azido-2′,3′- dideoxypurine nucleosides. Antimicrob. Agents Chemother. 2009;53:3715–3719. doi: 10.1128/AAC.00392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi M., Yoshinaga T., Seki T., Wakasa-Morimoto C., Brown K.W., Ferris R., Foster S.A., Hazen R.J., Miki S., Suyama-Kagitani A., et al. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 2011;55:813–821. doi: 10.1128/AAC.01209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costi R., Métifiot M., Chung S., Cuzzucoli Crucitti G., Maddali K., Pescatori L., Messore A., Madia V.N., Pupo G., Scipione L., et al. Basic quinolinonyl diketo acid derivatives as inhibitors of HIV integrase and their activity against RNase H function of reverse transcriptase. J. Med. Chem. 2014;57:3223–3234. doi: 10.1021/jm5001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherrill R.G., Furfine E.S., Hazen R.J., Miller J.F., Reynolds D.J., Sammond D.M., Spaltenstein A., Wheelan P., Wright L.L. Synthesis and antiviral activities of novel N-alkoxy-arylsulfonamide-based HIV protease inhibitors. Bioorganic Med. Chem. Lett. 2005;15:3560–3564. doi: 10.1016/j.bmcl.2005.05.101. [DOI] [PubMed] [Google Scholar]
- 46.Bold G., Fässler A., Capraro H.G., Cozens R., Klimkait T., Lazdins J., Mestan J., Poncioni B., Rösel J., Stover D., et al. New aza-dipeptide analogues as potent and orally absorbed HIV-1 protease inhibitors: Candidates for clinical development. J. Med. Chem. 1998;41:3387–3401. doi: 10.1021/jm970873c. [DOI] [PubMed] [Google Scholar]
- 47.Eissenstat M., Guerassina T., Gulnik S., Afonina E., Silva A.M., Ludtke D., Yokoe H., Yu B., Erickson J. Enamino-oxindole HIV protease inhibitors. Bioorg. Med. Chem. Lett. 2012;22:5078–5083. doi: 10.1016/j.bmcl.2012.05.120. [DOI] [PubMed] [Google Scholar]
- 48.Murphy P.V., O’Brien J.L., Gorey-Feret L.J., Smith A.B. Structure-based design and synthesis of HIV-1 protease inhibitors employing β-d-mannopyranoside scaffolds. Bioorg. Med. Chem. Lett. 2002;12:1763–1766. doi: 10.1016/S0960-894X(02)00220-2. [DOI] [PubMed] [Google Scholar]
- 49.Liu H., Xu L., Hui H., Vivian R., Callebaut C., Murray B.P., Hong A., Lee M.S., Tsai L.K., Chau J.K., et al. Structure-activity relationships of diamine inhibitors of cytochrome P450 (CYP) 3A as novel pharmacoenhancers, part I: Core region. Bioorganic Med. Chem. Lett. 2014;24:989–994. doi: 10.1016/j.bmcl.2013.12.058. [DOI] [PubMed] [Google Scholar]
- 50.Tamamura H., Omagari A., Hiramatsu K., Gotoh K., Kanamoto T., Xu Y., Kodama E., Matsuoka M., Hattori T., Yamamoto N., et al. Development of specific CXCR4 inhibitors possessing high selectivity indexes as well as complete stability in serum based on an anti-HIV peptide T140. Bioorg. Med. Chem. Lett. 2001;11:1897–1902. doi: 10.1016/S0960-894X(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 51.Dong M., Zhang J., Peng X., Lu H., Yun L., Jiang S., Dai Q. Tricyclononene carboxamide derivatives as novel anti-HIV-1 agents. Eur. J. Med. Chem. 2010;45:4096–4103. doi: 10.1016/j.ejmech.2010.05.070. [DOI] [PubMed] [Google Scholar]
- 52.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sechi M., Bacchi A., Carcelli M., Compari C., Duce E., Fisicaro E., Rogolino D., Gates P., Derudas M., Al-Mawsawi L.Q., et al. From ligand to complexes: Inhibition of human immunodeficiency virus type 1 integrase by beta-diketo acid metal complexes. J. Med. Chem. 2006;49:4248–4260. doi: 10.1021/jm060193m. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Pais G.C.G., Svarovskaia E.S., Marchand C., Johnson A.A., Karki R.G., Nicklaus M.C., Pathak V.K., Pommier Y., Burke T.R. Azido-containing aryl beta-diketo acid HIV-1 integrase inhibitors. Bioorg. Med. Chem. Lett. 2003;13:1215–1219. doi: 10.1016/S0960-894X(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 55.Zeng L.-F., Jiang X.-H., Sanchez T., Zhang H.-S., Dayam R., Neamati N., Long Y.-Q. Novel dimeric aryldiketo containing inhibitors of HIV-1 integrase: Effects of the phenyl substituent and the linker orientation. Bioorg. Med. Chem. 2008;16:7777–7787. doi: 10.1016/j.bmc.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X., Marchand C., Pommier Y., Burke T.R. Design and synthesis of photoactivatable aryl diketo acid-containing HIV-1 integrase inhibitors as potential affinity probes. Bioorg. Med. Chem. Lett. 2004;14:1205–1207. doi: 10.1016/j.bmcl.2003.12.064. [DOI] [PubMed] [Google Scholar]
- 57.Stansfield I., Avolio S., Colarusso S., Gennari N., Narjes F., Pacini B., Ponzi S., Harper S. Active site inhibitors of HCV NS5B polymerase. The development and pharmacophore of 2-thienyl-5,6-dihydroxypyrimidine-4-carboxylic acid. Bioorg. Med. Chem. Lett. 2004;14:5085–5088. doi: 10.1016/j.bmcl.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 58.Summa V., Petrocchi A., Pace P., Matassa V.G., De Francesco R., Altamura S., Tomei L., Koch U., Neuner P. Discovery of alpha,gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 2004;47:14–17. doi: 10.1021/jm0342109. [DOI] [PubMed] [Google Scholar]
- 59.Bhatt A., Gurukumar K.R., Basu A., Patel M.R., Kaushik-Basu N., Talele T.T. Synthesis and SAR optimization of diketo acid pharmacophore for HCV NS5B polymerase inhibition. Eur. J. Med. Chem. 2011;46:5138–5145. doi: 10.1016/j.ejmech.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Santo R., Fermeglia M., Ferrone M., Paneni M.S., Costi R., Artico M., Roux A., Gabriele M., Tardif K.D., Siddiqui A., et al. Simple but highly effective three-dimensional chemical-feature-based pharmacophore model for diketo acid derivatives as hepatitis C virus RNA-dependent RNA polymerase inhibitors. J. Med. Chem. 2005;48:6304–6314. doi: 10.1021/jm0504454. [DOI] [PubMed] [Google Scholar]
- 61.Gopalsamy A., Chopra R., Lim K., Ciszewski G., Shi M., Curran K.J., Sukits S.F., Svenson K., Bard J., Ellingboe J.W., et al. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J. Med. Chem. 2006;49:3052–3055. doi: 10.1021/jm060168g. [DOI] [PubMed] [Google Scholar]
- 62.Vara Prasad J.V.N., Boyer F.E., Domagala J.M., Ellsworth E.L., Gajda C., Hagen S.E., Markoski L.J., Tait B.D., Lunney E.A., Tummino P.J., et al. Nonpeptidic HIV protease inhibitors: 6-alkyl-5, 6-dihydropyran-2-ones possessing achiral 3-(4-amino/carboxamide-2-t-butyl, 5-methylphenyl thio) moiety: Antiviral activities and pharmacokinetic properties. Bioorg. Med. Chem. Lett. 1999;9:1481–1486. doi: 10.1016/S0960-894X(99)00237-1. [DOI] [PubMed] [Google Scholar]
- 63.Hao W., Herlihy K.J., Zhang N.J., Fuhrman S.A., Doan C., Patick A.K., Duggal R. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob. Agents Chemother. 2007;51:95–102. doi: 10.1128/AAC.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei Y., Li J., Chen Z., Wang F., Huang W., Hong Z., Lin J. Multistage virtual screening and identification of novel HIV-1 protease inhibitors by integrating SVM, shape, pharmacophore and docking methods. Eur. J. Med. Chem. 2015;101:409–418. doi: 10.1016/j.ejmech.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 65.Billamboz M., Bailly F., Lion C., Calmels C., Andréola M.L., Witvrouw M., Christ F., Debyser Z., De Luca L., Chimirri A., et al. 2-Hydroxyisoquinoline-1,3(2H,4H)-diones as inhibitors of HIV-1 integrase and reverse transcriptase RNase H domain: Influence of the alkylation of position 4. Eur. J. Med. Chem. 2011;46:535–546. doi: 10.1016/j.ejmech.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 66.Cereto-Massagué A., Guasch L., Valls C., Mulero M., Pujadas G., Garcia-Vallvé S. DecoyFinder: An easy-to-use python GUI application for building target-specific decoy sets. Bioinformatics. 2012;28:1661–1662. doi: 10.1093/bioinformatics/bts249. [DOI] [PubMed] [Google Scholar]
- 67.Adrià C.M., Garcia-Vallvé S., Pujadas G. DecoyFinder, a tool for finding decoy molecules. J. Cheminform. 2012;4:P2. doi: 10.1186/1758-2946-4-S1-P2. [DOI] [Google Scholar]
- 68.Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Thiel K., Wiswedel B. KNIME—The Konstanz information miner: Version 2 and Beyond. ACM SIGKDD Explor. Newsl. 2009;11:26–31. doi: 10.1145/1656274.1656280. [DOI] [Google Scholar]
- 69.Muegge I., Mukherjee P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug Discov. 2016;11:137–148. doi: 10.1517/17460441.2016.1117070. [DOI] [PubMed] [Google Scholar]
- 70.Grisoni F., Consonni V., Todeschini R. Impact of Molecular Descriptors on Computational Models. Methods Mol. Biol. 2018;1825:171–209. doi: 10.1007/978-1-4939-8639-2_5. [DOI] [PubMed] [Google Scholar]
- 71.Steinbeck C., Han Y., Kuhn S., Horlacher O., Luttmann E., Willighagen E. The Chemistry Development Kit (CDK): An open-source Java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinbeck C., Hoppe C., Kuhn S., Floris M., Guha R., Willighagen E. Recent Developments of the Chemistry Development Kit (CDK)—An Open-Source Java Library for Chemo- and Bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., Yang L. Improving SVM through a Risk Decision Rule Running on MATLAB. J. Softw. 2012;7:2252–2257. doi: 10.4304/jsw.7.10.2252-2257. [DOI] [Google Scholar]
- 74.Yang C., Odvody G.N., Fernandez C.J., Landivar J.A., Minzenmayer R.R., Nichols R.L. Evaluating unsupervised and supervised image classification methods for mapping cotton root rot. Precis. Agric. 2015;16:201–215. doi: 10.1007/s11119-014-9370-9. [DOI] [Google Scholar]
- 75.Bouzalmat A., Kharroubi J., Zarghili A. Face Recognition Using SVM Based on LDA. Int. J. Comput. Sci. Issues. 2013;10:171–179. [Google Scholar]
- 76.Ballabio D., Grisoni F., Todeschini R. Multivariate comparison of classification performance measures. Chemom. Intell. Lab. Syst. 2018;174:33–44. doi: 10.1016/j.chemolab.2017.12.004. [DOI] [Google Scholar]
- 77.Glide . Schrödinger, Version 6.7. LLC; New York, NY, USA: 2015. [Google Scholar]
- 78.Fader L.D., Malenfant E., Parisien M., Carson R., Bilodeau F., Landry S., Pesant M., Brochu C., Morin S., Chabot C., et al. Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1. ACS Med. Chem. Lett. 2014;5:422–427. doi: 10.1021/ml500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Freeman G.A., Andrews Iii C.W., Hopkins A.L., Lowell G.S., Schaller L.T., Cowan J.R., Gonzales S.S., Koszalka G.W., Hazen R.J., Boone L.R., et al. Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2. J. Med. Chem. 2004;47:5923–5936. doi: 10.1021/jm040072r. [DOI] [PubMed] [Google Scholar]
- 80.Ganguly A.K., Alluri S.S., Wang C.-H., Antropow A., White A., Caroccia D., Biswas D., Kang E., Zhang L.-K., Carroll S.S., et al. Structural optimization of cyclic sulfonamide based novel HIV-1 protease inhibitors to picomolar affinities guided by X-ray crystallographic analysis. Tetrahedron. 2014;70:2894–2904. doi: 10.1016/j.tet.2014.03.038. [DOI] [Google Scholar]
- 81.Love R.A., Parge H.E., Yu X., Hickey M.J., Diehl W., Gao J., Wriggers H., Ekker A., Wang L., Thomson J.A., et al. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 2003;77:7575–7581. doi: 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bressanelli S., Tomei L., Rey F.A., De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.