Abstract
As the core components of abscisic acid (ABA) signal pathway, Clade A PP2C (PP2C-A) phosphatases in ABA-dependent stress responses have been well studied in Arabidopsis. However, the roles and natural variations of maize PP2C-A in stress responses remain largely unknown. In this study, we investigated the expression patterns of ZmPP2C-As treated with multiple stresses and generated transgenic Arabidopsis plants overexpressing most of the ZmPP2C-A genes. The results showed that the expression of most ZmPP2C-As were dramatically induced by multiple stresses (drought, salt, and ABA), indicating that these genes may have important roles in response to these stresses. Compared with wild-type plants, ZmPP2C-A1, ZmPP2C-A2, and ZmPP2C-A6 overexpression plants had higher germination rates after ABA and NaCl treatments. ZmPP2C-A2 and ZmPP2C-A6 negatively regulated drought responses as the plants overexpressing these genes had lower survival rates, higher leaf water loss rates, and lower proline accumulation compared to wild type plants. The natural variations of ZmPP2C-As associated with drought tolerance were also analyzed and favorable alleles were detected. We widely studied the roles of ZmPP2C-A genes in stress responses and the natural variations detected in these genes have the potential to be used as molecular markers in genetic improvement of maize drought tolerance.
Keywords: Zea mays, phosphatase, ZmPP2C-A, abiotic stresses, natural variation
1. Introduction
Reversible protein phosphorylation is one of the most universal post-translational modifications, which is involved in many signal pathways. Protein kinases (PKs) and protein phosphatases (PPs) work antagonistically in this process. Both families were originally discovered in animal systems, and researchers in recent years have revealed the significant structure and function difference of the two gene families in plant [1]. In Arabidopsis, more than 1000 kinase genes have been annotated, while only 112 phosphatase gene have been identified [2]. Based on the different substrates of phosphatases, plant PPs are classified into serine/threonine (Ser/Thr) phosphatases and tyrosine phosphatases. Ser/Thr phosphatases are further categorized into two subfamilies, PPs consisting of PP1, PP2A, PP2B, PP4, PP5, PP6, and PP7, and PPs containing PP2C and other metal ion-dependent phosphatases [3]. According to the result of phylogenetic analysis, the PP2C family was further divided into 11 subclades (A-K) [4]. A series of studies on PP2C-A genes uncovered that PP2C-As participate in the core regulatory network in ABA responses as negative regulators [2,3,5,6].
Osmotic stress caused by drought leads to the accumulation of ABA in plant. ABA is the key regulator of plant drought resistance [7]. The core signaling pathway of ABA containing three family genes, pyrabactin resistance1/PYR1-like/regulatory components of ABA receptors (PYR1/PYL/RCAR) receptor, PP2C phosphatases, and sucrose non-fermenting-1 related protein kinase 2 (SnRK2) kinases. PP2Cs interact with SnRK2s to inhibit the activities of SnRK2s under normal conditions, while the accumulation of ABA caused by drought stress leads to the interaction between PYR/PYL/RCAR receptors and PP2C phosphatases, thus, SnRK2 kinases are released to phosphorylate downstream ABA-responsive elements-binding protein/ABA-responsive elements-binding factor (AREB/ABF) proteins [5,8,9,10]. Apart from ABA signaling pathways, mitogen-activated protein kinase (MAPK) signaling pathways are also important parts in response to abiotic stress, and crosstalk between them has been reposted. Dephosphorylation of MAPKs catalyzed by PP2Cs results in the inactivation of MAPKs [11]. In addition, autophagy plays roles in abiotic stress responses including oxidative stress, drought, and salt [12]. Two PP2Cs were reported to promote macroautophagy by dephosphorylating Atg1 complex [13]. Besides, tremendous transcription factors (like MYB, WRKY, NAC, and so on) play vital roles in regulating plant response to abiotic stress [14,15].
There are nine members in clade A of PP2C in Arabidopsis, namely ABI1, ABI2, HAB1, HAB2, AHG1, AHG3/AtPP2CA, HAI1, HAI2, and HAI3 [7]. ABI1 and ABI2 were firstly identified by screening ABA insensitive mutants [16]. HAB1 was also a member reported early. HAB1 and HAB2 are both homology to ABI1. The protein sequence of HAB1 is 55% and 54% identical, respectively, to those of ABI1 and ABI2. Nevertheless, expression of HAB1 is induced by ABA treatment [17]. Over-expression of HAB1 in Arabidopsis results in tolerance to ABA, both in seeds and vegetative tissues, while the mutant of HAB1 shows the contrary phenotype to ABA. Both results demonstrate the negative role of HAB1 in ABA signal transduction [18]. Among the ABA hypersensitive mutants, ahg3 displays the strongest phenotype during germination, but no significant phenotypes were observed in adult plants, implying the different functional patterns of PP2C-A individuals [19]. Similar to ahg3, ahg1 was hypersensitive to ABA, NaCl, KCl, mannitol, glucose, and sucrose in seedling stage, but showed no difference to wild-type at adult stage. Moreover, ahg1 ahg3 double mutant showed stronger phenotypes to its single parental mutant, suggesting that AHG1 and AHG3 work together to regulate the ABA signal pathway [20]. All nine PP2C-A genes were upregulated after the treatment of ABA, among which three genes, named HAI1, HAI2, and HAI3, were strongly induced in the vegetative phase. No expression of HAI1 was detected in seeds, and the interaction between HAI1 and SnRK2 was clarified by bimolecular fluorescence complementation (BiFC) experiment, revealing HAI1 function in the vegetative stage [21]. Compared to other members, mutants of HAI genes showed the most modest phenotype to ABA treatment. Single hai mutant had no difference to wild-type in ABA sensitivity, while the double and triple mutants were insensitive to ABA in germination stage, but sensitive to ABA in post-germination stage [22]. All these studies from Arabidopsis indicated pleiotropic roles of clade A PP2C phosphatases in plant development and stress responses. How clade A PP2C phosphatases in maize (ZmPP2C-As) regulate stress responses and agronomic traits remain largely known.
Few progresses have been made in maize PP2C-A research in recent years. ZmPP2C-A10 was identified through a candidate gene association analysis and was proven to be associated with drought tolerance. Similar to the mechanism in Arabidopsis, ZmPP2C-A10 is involved in ABA signaling pathway by interacting with some ZmPYL and ZmSnRK2 [23]. In research about maize protein phosphatase gene family, 16 ZmPP2C-A gene were identified [24]. So far, a total of 13 ZmPYLs and four ZmSnRK2s, together with 16 PP2C-As, in maize were identified, and their patterns of gene expression, subcellular localization and interaction network were investigated. All interaction combinations between ZmPYLs and ZmPP2Cs, and ZmPP2Cs and ZmSnRKs were detected by yeast two-hybrid assays, resulting in the discovery of a comprehensive core ABA signaling network in maize [25].
Many natural variations in drought tolerance in crops were discovered through association analysis in recent years [26]. With a 82-bp miniature inverted-repeat transposable element (MITE) inserted in the promoter, the expression of ZmNAC111 was repressed via RNA-directed DNA methylation and H3K9 dimethylation, contributing to the reduced drought tolerance in maize [27]. The expression of ZmVPP1 is regulated by a 366-bp insertion in the promoter containing three MYB cis elements, resulting in the improved drought tolerance in maize inbred lines with tolerant allele [28]. In the 5′UTR region of ZmPP2C-A10, a deletion of endoplasmic reticulum stress response element (ERSE) related to endoplasmic reticulum stress-induced gene expression, resulted in more tolerance of maize plant to drought stress [23]. These studies imply the feasibility of association studies in the discovery of natural variations concerned with complex agronomic traits in maize.
The main purpose of this study was to investigate the function of ZmPP2C-As in response to multiple abiotic stresses. Expression patterns of ZmPP2C-As with multiple stress treatments had been detected. Overexpressing most of ZmPP2C-As in Arabidopsis led to the discovery of their broad roles in stress responses. In addition, candidate gene association analysis revealed the natural variations of ZmPP2C-As associated with drought tolerance and the development of other agronomic traits, which may be used as molecular makers in maize genetic improvement.
2. Results
2.1. Expression Analyses of ZmPP2C-A Genes under ABA Treatment
In a previous research, 13 putative maize genes were identified belong to ZmPP2C clade A gene family, namely ZmPP2C-A1 to ZmPP2C-A13, and their expression levels in drought response were recovered [23]. In order to know how these genes respond to other abiotic stresses, we studied the gene expression patterns of ZmPP2C-A under ABA treatments excluding ZmPP2C-A8 and ZmPP2C-A9 due to their low expression levels in our experiments.
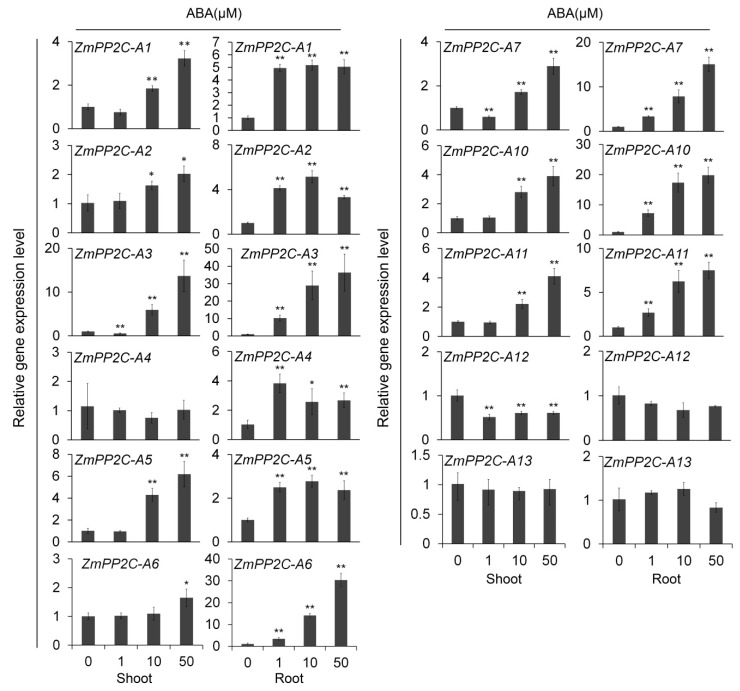
The results showed that most ZmPP2C-A genes were induced either in shoot or in root, or in both, by external ABA application except ZmPP2C-A13 (Figure 1). After being treated with different concentrations of ABA, ZmPP2C-A3’s expression levels in shoot and root varied significantly in a concentration-dependent manner (Figure 1). Similar expression patterns were observed for ZmPP2C-A1(shoot), ZmPP2C-A2 (shoot), ZmPP2C-A5 (shoot), ZmPP2C-A6 (root), ZmPP2C-A7 (root), ZmPP2C-A10 (root), and ZmPP2C-A11(shoot and root) (Figure 1), indicating significant roles of these genes in ABA response in shoot or root or both organs. These results revealed the different expression patterns among different genes and organs under ABA treatment and suggested the functional segmentation of ZmPP2C-As in ABA pathways (Figure 1).
Figure 1.
Relative expression of ZmPP2C-A genes to ABA treatment. The relative expression levels of ZmPP2C-A genes were analyzed by quantitative reverse transcription-PCR (qRT-PCR) in maize seedlings after ABA treatment. ZmActin5 gene was used as an internal control. Seedlings at three-leaf stage were subjected to various concentrations of ABA (0, 1, 10, 50 μM) solution. Both leaf and root tissues were collected three hours after the treatments. Data represent the mean ± standard deviation (SD) of three replicates. Asterisks indicate the significance of T-test, * p < 0.05, ** p < 0.01.
2.2. Expression Patterns of ZmPP2C-A Genes in Response to Salt Treatment
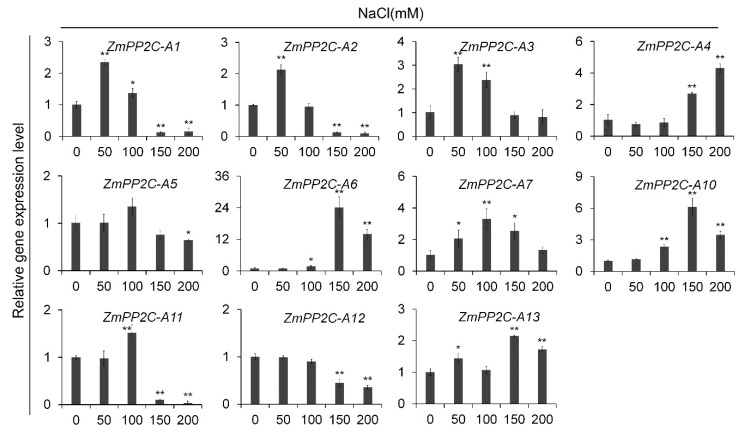
We next investigated the expression patterns of ZmPP2C-A genes under sodium chloride (NaCl) treatment. Interestedly, we observed that the expression patterns of ZmPP2C-A genes varied significantly in response to salt treatments. As shown in Figure 2, ZmPP2C-A1, ZmPP2C-A2, and ZmPP2C-A11 were induced by NaCl at lower concentrations (≤100 mM), while inhibited by NaCl at higher concentrations (≥150 mM). ZmPP2C-A3 was only induced by NaCl at lower concentrations; nevertheless ZmPP2C-A5 was suppressed by NaCl at the highest concentration. ZmPP2C-A7 was induced by NaCl from 50 mM to 150 mM, with a peak occurring at 100 mM; ZmPP2C-A4, ZmPP2C-A6, and ZmPP2C-A10 were more strongly induced by NaCl at higher concentrations; ZmPP2C-A12 and ZmPP2C-A13 showed opposite expression patterns in response to NaCl stresses, namely ZmPP2C-A12 was inhibited while ZmPP2C-A13 was induced by NaCl at higher concentrations. In addition, more than twenty-fold changes were observed in ZmPP2C-A6 under treatment with 150 mM NaCl, which was outstanding among ZmPP2C-A genes (Figure 2). These expression patterns suggested different roles of ZmPP2C-A genes in regulation of maize salt stress responses.
Figure 2.
Relative expression of ZmPP2C-A genes to salt treatment. The relative expression levels of ZmPP2C-A genes were analyzed by qRT-PCR in maize seedlings after salt treatment. ZmActin5 gene was used as an internal control. Seedlings at three-leaf stage were subjected with various concentrations of NaCl (0, 50, 100, 150, 200 mM) solution, and samples were harvested three hours after the treatments. Data represent the mean ± SD of three replicates. Asterisks indicate the significance of T-test, * p < 0.05, ** p < 0.01.
2.3. Roles of ZmPP2C-A Genes in Regulation of ABA and Salt Stress Responses
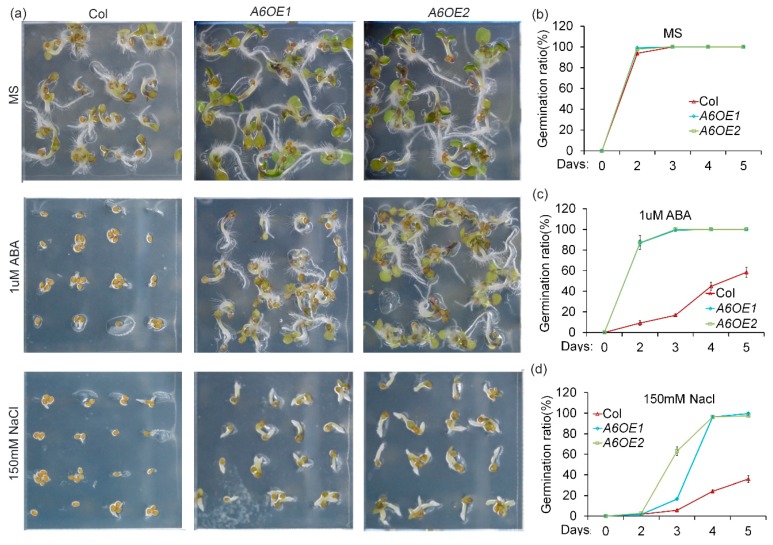
To determine the biological roles of ZmPP2C-A genes in abiotic stress responses, we overexpressed ZmPP2C-As in Arabidopsis, and multiple stresses experiments were conducted on the transgenic plants. In Arabidopsis and other plants, ABA has been widely known as a negative regulator of seed germination [29,30]. Salt is a natural stress that inhibits seed germination [31]. To know how ZmPP2C-As regulate ABA response, seeds of Col and ZmPP2C-A6 overexpression lines were sown on a Murashige and Skoog (MS) medium, supplemented with or without 1 μM ABA. After 48 h of cultivation, germination rates of both Col and ZmPP2C-A6 transgenic seeds were almost 100% on MS plates without ABA (Figure 3a,b). Although germination rates of both materials were decreased in MS plates with 1 μM ABA, germination rates of Col seeds were reduced more as compared to those of ZmPP2C-A6 overexpression lines (Figure 3a,c). These data suggested that ZmPP2C-A6 is a positive regulator of ABA responses. On MS plates supplied with 150 mM sodium chloride, ZmPP2C-A6 overexpression lines germinated faster than Col, although there were no different germination rates between Col and ZmPP2C-A6 overexpression transgenic seeds on MS plates without salt (Figure 3a,d), suggesting a positive role of ZmPP2C-A6 in plant salt tolerance. The similar germination tendencies were also observed for transgenic Arabidopsis plants overexpressing ZmPP2C-A1 (Figure S1) or ZmPP2C-A2 (Figure S2) under ABA and salt stresses. Taken together, these data implied that ZmPP2C-A1, ZmPP2C-A2, as well as ZmPP2C-A6, played positive roles in ABA and salt stress responses.
Figure 3.
Overexpression of ZmPP2C-A6 in Arabidopsis increased tolerance to ABA and salt stresses. Germination phenotypes (a) (Bar = 0.5 cm) and statistical analyses (b–d) of Col and two ZmPP2C-A6 overexpressing lines (A6OE1 and A6OE2) sown on MS medium (b) or MS medium supplemented with 1 μM ABA (c) or 150 mM sodium chloride (d). Data represent the mean ± SD of three replicates in (b–d).
2.4. ZmPP2C-A Genes Act as Negative Regulators in Plant Drought Responses
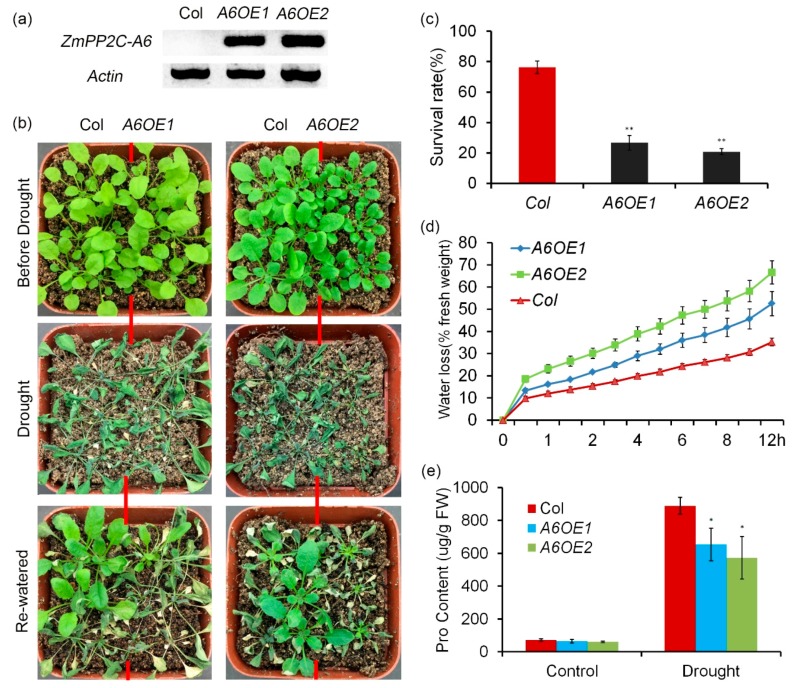
Previously we reported that ZmPP2C-A10 attenuated plant drought tolerance [23]. In order to know the roles of other ZmPP2C-A genes in regulation of plant drought tolerance, we treated the ZmPP2C-A transgenic Arabidopsis plants with drought stresses. Twenty-day-old seedlings of Col and ZmPP2C-A6 overexpression lines were stressed with severe drought stress. After being re-watered, survival rates were counted. We observed that more than 70% seedlings of Col survived, while only ~20% seedlings of ZmPP2C-A6 transgenic lines survived (Figure 4a–c), implying that ZmPP2C-A6 negatively regulates drought resistance. Water loss of excised leaves from Col and ZmPP2C-A6 transgenic lines were measured every 30 min. The results showed that ZmPP2C-A6 transgenic lines lost water faster than Col (Figure 4d). In addition, the proline contents of the two materials were detected with or without drought stresses. Under well-watered conditions (without drought stress), no obvious difference was observed between the two materials (Figure 4e). But under severe drought stress, the levels of osmolyte proline in Col was significantly higher than those in ZmPP2C-A6 overexpression lines (Figure 4e). Therefore, compared to Col, ZmPP2C-A6 overexpression lines were more sensitive to drought stress, which resulted from faster water loss and less accumulation of osmolyte proline. We conducted similar drought stress and water loss assay to ZmPP2C-A2 transgenic lines, and similar phenotypes were observed to these transgenic lines in response to drought stresses (Figure S3). All these data suggested that both ZmPP2C-A2 and ZmPP2C-A6 played negative roles in plant drought stress responses.
Figure 4.
Overexpression of ZmPP2C-A6 in Arabidopsis decreased tolerance to drought. (a) RT-PCR analysis of ZmPP2C-A6 overexpression lines. (b) Drought tolerance assay of Col and two ZmPP2C-A6 overexpression lines. Twenty-day-old plants were drought-stressed for ten days and then re-watered for three days. (c) Survival rates of Col and ZmPP2C-A6 transgenic plants after drought stress. Asterisks indicate the significance of T-test, ** p < 0.01 (d) Water loss assay indicates that leaves from ZmPP2C-A6 transgenic plants more easily lost water after detached. (e) Proline contents of well-watered and drought-stressed Col and ZmPP2C-A6 transgenic plants. Asterisks indicate the significance of T-test, * p < 0.05 Data represent the mean ± SD of three replicates in (c–e).
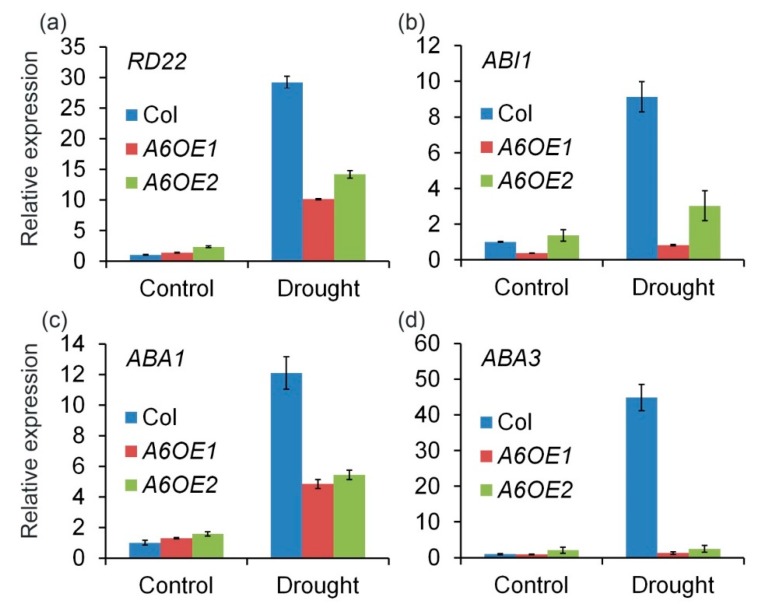
To further confirm the role of ZmPP2C-A6 in drought stresses, the expression levels of several marker genes, including RD22, ABI1, ABA1, and ABA3 which were known to respond to drought and ABA, were analyzed. Expression levels of these marker genes were measured in Col and ZmPP2C-A6 transgenic lines, with or without drought stresses. We observed that the expression levels of these marker genes were significantly and dramatically inhibited in ZmPP2C-A6 transgenic lines as compared to Col plants after drought stresses (Figure 5). These results suggest that the accumulation of ABA or ABA signaling were attenuated in ZmPP2C-A6 overexpression lines, which resulted in the sensitivity of these lines to drought stresses.
Figure 5.
ZmPP2C-A6 altered expression of stress-responsive genes in transgenic Arabidopsis. (a) The RD22 were involved in responding to drought and ABA treatment; (b) The ABI1 is the ABA signaling regulators gene which was also had a great effect on the ABA biosynthesis in the plants; (c,d) The ABA1 and ABA3 genes are the main function of ABA biosynthesis. Expression of stress-responsive genes in ZmPP2C-A6 transgenic plants and Col. Overexpression of ZmPP2C-A6 reduced the expression of drought responsible marker genes. The expression of all marker genes under normal and drought conditions was examined by qRT-PCR. Three-week-old plants were either watered or drought-stressed for 8 days.
2.5. Identification of Natural Variation in ZmPP2C-A Genes Associated with Stress Responses
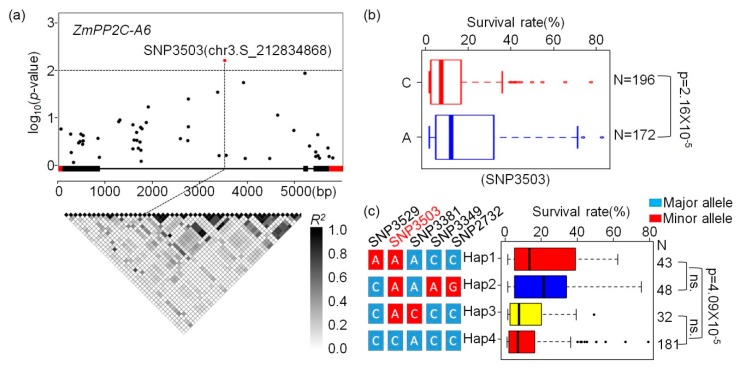
Previously, an association mapping panel containing 527 maize inbred lines with various genetic backgrounds were collected [32], and 368 inbred lines of which were re-sequenced through RNA-sequencing [33]. In addition, 1.25 M high quality single nuclear polymorphisms (SNPs) were called out from this association panel by integrating reduced genome sequencing (GBS), high-density array technologies (600 K), and RNA-sequencing [34]. Survival rates of this association panel after salt and drought stresses were reported by us and other researchers recently [35,36]. In order to mine the natural variations of ZmPP2C-A1, 2, 6 associated with drought and salt tolerance, SNPs located within ZmPP2C-A genes were collected, and association analyses were performed using these SNPs and the survival rates of 368 maize inbred lines stressed with drought or salt, respectively. Three algorithms, namely general linear model (GLM), GLM + Q, and mixed linear model (MLM), were used in these analyses. We found that there were no SNP located in these ZmPP2C-A genes that were detected with all three algorithms associated with survival rates after salt stress (Table S1). Meanwhile, there were different amounts of SNPs within ZmPP2C-A6 that were identified to be associated with survival rates after drought stresses with all three statistical models (Table S2). The lead SNP (SNP3503) was located in the intron of ZmPP2C-A6, which has the A and C alleles (Figure 6a). According to this lead SNP, the 368 inbred lines could be divided into two groups, the A and C groups, and the survival rates between these two groups were significantly different in that plants with allele A had much higher survival rates than those with C alleles (Figure 6b). Based on the lead SNP, we analyzed the haplotypes (Hap) of ZmPP2C-A6 in the association panel. A total of four Haps were detected, and Hap1 and Hap2 have obviously higher survival rates than those of Hap3 or Hap4 (Figure 6c).
Figure 6.
Association analysis of ZmPP2C-A6 natural variations with drought tolerance. (a) ZmPP2C-A6 association analysis and pairwise linkage disequilibrium (LD) among the SNP markers. The significantly associated SNPs are shown in red (p < 0.01). The gene structure is shown in the middle. Exons and introns are shown as filled boxes and dark lines, respectively. (b) Association of two alleles in SNP3503 with drought tolerance. (c) Association of haplotypes (Hap) in ZmPP2C-A6 with drought tolerance. ns. means no significant.
3. Discussion
Plants have developed complicated regulatory networks to respond to abiotic stresses, and plant hormone ABA plays pivotal roles in these processes [7,37]. The core regulatory network of ABA consists of three members, PYR/PYL receptor proteins as receptors, Clade A PP2C phosphatases as negative regulators, and SNF1-related protein kinase 2 (SnRK2) as positive regulators [5,9,10]. Similar to the signal pathway in Arabidopsis, ZmPP2C-A10 has been reported to take part in ABA signal transduction by interacting with ZmPYLs and ZmSnRK2s [23]. Furthermore, the research on gene expression, subcellular localization, and interaction network of ZmPYLs, ZmPP2Cs, and ZmSnRK2s resulted in the discovery of core ABA signaling networks in maize [17]. ZmPP2C-A10 is a negative regulator in drought resistance [23], but the biological function of the rest of the members in the ZmPP2C-A gene family remains unclear. To solve this problem, we analyzed the expression patterns of ZmPP2C-As to multiple stresses, and overexpressed ZmPP2C-As in Arabidopsis to survey their roles in abiotic stress responses.
After ABA treatment, most ZmPP2C-As were induced either in different tissues or by different concentration of ABA except ZmPP2C-A13 (Figure 1). ZmPP2C-A12 was the only ZmPP2C-A that was not induced by any concentration of sodium chloride (Figure 2), suggesting its different role from other ZmPP2C-As in response to abiotic stresses. It will be important to study how ZmPP2C-A12 functions in stress responses in future. In terms of fold changes in expression, the biggest change occurred in ZmPP2C-A3 under ABA treatment (Figure 2), and in ZmPP2C-A6 under salt treatment (Figure 2), indicating their vital roles in the corresponding stress responses. Differences in expression patterns among ZmPP2C-As suggested intricate regulatory mechanisms exist in these genes in response to abiotic stresses.
It has been reported that plant PP2C-As were involved in many abiotic stress response pathways [18,38,39]. To seek for the potential regulatory factors of ZmPP2C-As, we analyzed the 1.5 kb promoter region of all ZmPP2C-As. We found that all of these promoter regions have many kinds of potential transcription factor (TF) binding sites (Figure S4). While many TF binding sites like basic region-leucine zipper (bZIP), auxin response factor (ARF), and ethylene responsive factor (ERF) binding sites commonly exist in all ZmPP2C-A promoter regions, some of them only exist in part of them. Meanwhile, binding sites of many transcription factors involved in other pathways such as endoplasmic reticulum (ER) stress, auxin response, and ethylene response were also found in most of ZmPP2C-A promoters. These results suggested that as an upstream signal receptor element in ABA response pathway, ZmPP2C-As may be regulated by many other abiotic stress response pathways and further change the phosphorylation level of downstream targets. Our study indicates the important role of ZmPP2CAs in crosstalk between abiotic stress signaling.
The germination experiments showed that, compared to the Col, ZmPP2C-A6 overexpression lines were insensitive to ABA and salt (Figure 3). The same phenotypes were observed in ZmPP2C-A1 and ZmPP2C-A2 (Figures S2 and S3), demonstrating the functional redundancy among these ZmPP2C-A genes. As for drought stress, ZmPP2C-A6 transgenic lines were more sensitive than the Col, resulting from the faster water loss and less accumulation of proline (Figure 4). The other members of ZmPP2C-As including ZmPP2C-A2 (Figure S3) and ZmPP2C-A10 [23] seem to play the similar roles in drought resistance, which is opposite to the roles of ZmPYLs, as ZmPYLs act as positive regulators in drought resistance [40]. Mutant analyses have been conducted to all members of Arabidopsis PP2C-As. Most single mutants displayed hypersensitivity to ABA in germination stage, and mutants of AHG were sensitive to salt and osmotic stress during germination [19,20,22]. As for function in abiotic stress, proline levels in hai mutants were higher than that in Col, indicating the negative role of HAI in drought resistance [22]. Additionally, double mutants of Arabidopsis PP2C-As were generated, and showed reduced water loss and resistance to drought stress [41]. Except for mutant analysis, HAI1 overexpressing lines were insensitive to ABA treatment, and were sensitive to drought treatment [18]. The results are consistent with the phenotype of ZmPP2C-A1, ZmPP2C-A2, ZmPP2C-A6 overexpression plants in our excrement, revealing that function of these PP2C-As are conserved between Arabidopsis and maize.
Four marker genes were chosen for expression analysis in ZmPP2C-A6 transgenic plants, namely RD22, ABI1, ABA1, and ABA3. As a member of the BNM2, USP, RD22 and polygalacturonase isozyme (BURP) protein family, RD22 was reported to be induced by drought, salt stress, and endogenous ABA [42], and has been used as a marker gene for abiotic stress [43,44,45]. ABI1 is the key component in ABA signaling pathway in Arabidopsis [2,3,6], while ABA1 and ABA3 both participate in ABA biosynthesis [46]. All four marker genes were down-regulated in ZmPP2C-A6 transgenic plants compared with Col plants after drought treatment, implying that both ABA biosynthesis and signaling were inhibited in ZmPP2C-A6 transgenic plants. Overexpression of ZmPP2C-A6 may lead to the negative feedback regulation of ABA accumulation and signal transduction in Arabidopsis. Considering the ABA function in promoting drought stress tolerance, the expression levels of marker genes were consistent with the phenotypes that ZmPP2C-A6 transgenic plants were sensitive to drought stress.
To investigate the natural variation associated with drought and salt stress, candidate gene association analyses were performed between SNP within ZmPP2C-As and survival rate after salt or drought stress (Tables S1 and S2). The SNP3503 located in intron of ZmPP2C-A6 was found to associate with drought stress, but the mechanism of how this SNP regulates ZmPP2C-A6 function needs further studies. Hap1 and Hap2 were the favorable haplotypes in this locus, which may be useful in drought-resistant maize breading programs. In addition, candidate gene association analyses were also conducted between ZmPP2C-A genes and 17 maize agronomic traits, the associations were observed in 11 members (except ZmPP2C-A12, ZmPP2C-A13) of the gene family (Table S3). These findings suggest that ZmPP2C-As may participate in maize growth and development, and the potential use of these natural variations in maize genetic improvement.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Maize inbred line B73 was cultured and used for gene cloning and expression analysis. B73 seeds were sown and grown in a greenhouse under 16 h light/8 h dark photoperiod at 28 °C.
The Arabidopsis ecotype Col-0 was used as the wild-type. The pRCS2(bar)-ZmPP2C-As plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Arabidopsis Col-0 ecotype using the floral dip method [47]. Seeds of transformed Arabidopsis were selected using MS medium supplemented with 25 mg/L hygromycin. Homozygous lines of T4 generations were used for further analysis.
Arabidopsis seeds were surface-sterilized with 10% bleach and washed three to four times with sterile water. The seeds were pretreated in a refrigerator at 4 °C for 3 days, then sowed on MS medium and placed in a plant growth incubator under a 16 h light/8 h dark photoperiod at 22 °C. After growing in the incubator for a week, Arabidopsis seedlings were transferred into soil in a growth chamber with the same light periods and temperature.
4.2. ABA and NaCl Treatments
For expression analysis, B73 inbred lines were grown in aerated Hoagland’s nutrient solution. The nutrient solution was replaced every three days. Seedlings at the three-leaf stage were subjected to two different treatments: ABA (0, 1, 10, 50 μM) and NaCl (0, 50, 100, 150, 200 mM). After 3 h, all samples collected were immediately frozen in liquid nitrogen and stored in a refrigerator at −80 °C until RNA extraction.
For the germination rate assay, full and uniform Arabidopsis seeds of wild-type and independent transgenic ZmPP2C-A-overexpression lines were evenly spotted in MS medium supplemented with 1 μM ABA or 150 mM NaCl. The normal MS medium was used as a control.
4.3. Germination Rate Analyses
For germination test, plants of wild-type and different transgenic lines were grown under the same growth chamber, and Arabidopsis seeds were collected at the same time. Seeds were put on plates containing MS medium with ABA or NaCl solution. Seed germination was determined based on the appearance of an embryonic axis protrusion, as observed under a microscope, and the germination rate was counted from the 2nd day to the end of the 5th to determine the percentage of stress-tolerant germinated seeds.
4.4. Drought Tolerance, Proline, and Water Loss Assays
After transplanting the seedlings into the soil, the wild-type and homozygous transgenic plants continued to grow under normal watering conditions in the growth chamber for 20 days. Watering was then halted. Samples were taken for marker gene and proline assays after 8 days. When plants began to exhibit lethal effects of dehydration after a further 2 weeks, watering was then resumed and the plants were allowed to grow for a subsequent 3 days, and the survival rate was investigated.
Proline content was measured using the colorimetric determination based on proline’s reaction with ninhydrin [48]. To assay proline, 50 mg of rosette leaves or cauline leaves were shredded into 10 mL centrifuge tubes using scissors. Each sample was incubated with 5 mL of 3% sulfosalicylic acid solution and boiled for 10–30 min to obtain the extract solution. After cooling to room temperature, 2 mL of supernatant were pipetted into a new 10 mL centrifuge tube and mixed with 2 mL acetic acid and 2 mL ninhydrin at 100 °C for 30 min. After mixture had cooled down, the proline was extracted by 4 mL methylbenzene and assayed by UV-vis spectrophotometry (UV-1800 spectrophotometer, Shanghai, China) at 520 nm.
For the water loss assays, the Arabidopsis rosette leaves with normal growth of about 20 days were detached on the filter paper to lose water, and the nodes were weighed at different times to calculate the water loss rate of the leaves.
4.5. Constructs
For Arabidopsis transformation, the ZmPP2C-A genes were cloned from cDNA of maize line B73 individually, then digested with EcoR I and Xho I. The digested product was ligated to the binary vector pJET1.2 vector, and inserted into the pSAT6 vector to produce pSAT6-ZmPP2C-A. The expression cassettes using 2X35S promoters to drive expression of the ZmPP2C-A cDNAs were released from the pSAT6 vectors by digestion with PI-Psp I and inserted into the pRCS2-Bar-OCS binary vector and then transformed into Arabidopsis ecotype Columbia-0 [49].
4.6. RNA Purification and Expression Analysis
Samples were collected after ABA and salt treatments. Total RNA was extracted using Trizol reagent (TransGen, Beijing, China) from more than three seedlings for each treatment. RNA was treated with DNase I (Thermo Scientific, Waltham, MA, USA) for purification, and single-stranded cDNA was then synthesized using M-MLV reverse transcriptase (Promega, Madison, WI, USA). The maize and Arabidopsis actin genes were used as the internal control.
4.7. Association Analysis
Candidate-gene association mapping was carried out to identify the causal variants of ZmPP2C-A in a set of 368 diverse maize lines. All phenotypic data used in the study were obtained in a previous study. Among 1.25 million high-quality SNP markers with Minor Allele Frequency (MAF) less than 0.05, 149 SNPs were found in the regions of ZmPP2C-A1, ZmPP2C-A2, and ZmPP2C-A6 genes. Association analysis was performed using a mixed model (MLM), considering population structure and relative kinship, in TASSEL 5.0 (Available online: https://tassel.bitbucket.io/) [50].
4.8. Statistical Analyses
All of the experiments in this study were repeated three times, and the values presented are mean ± SD. Asterisks above the columns in figures indicate the significance of T-test, * p < 0.05, ** p < 0.01. Statistical analyses were performed using Excel 2013 (Microsoft, Redmond, WA, USA). Figures were plotted by using Adobe Photoshop CC 2018 (Adobe Systems, San Jose, CA, USA).
Acknowledgments
We thank Lingli Song and Yanli Xiang for reading and suggestions of this article.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/14/3573/s1.
Author Contributions
Conceptualization, Z.H. and M.D.; methodology, Z.H.; software, X.S.; validation, Z.H., J.W., and M.D.; formal analysis, Z.H.; investigation, Z.H. and J.W.; resources, J.W.; data curation, Z.H. and X.S.; writing—original draft preparation, Z.H., J.W., and M.D.; writing—review and editing, M.D.; visualization, X.S.; supervision, M.D.; project administration, M.D.; funding acquisition, M.D.
Funding
This research was funded by National Key Research and Development Program of China (2016YFD0100600), Thousand Talents Plan of China and the Fundamental Research Funds for the Central Universities of China (2662015PY170).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Luan S. Protein phosphatases in plants. Annu. Rev. Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 2.Schweighofer A., Hirt H., Meskiene I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Singh A., Pandey A., Srivastava A.K., Tran L.S.P., Pandey G.K. Plant protein phosphatases 2C: From genomic diversity to functional multiplicity and importance in stress management. Crit. Rev. Biotechnol. 2016;36:1023–1035. doi: 10.3109/07388551.2015.1083941. [DOI] [PubMed] [Google Scholar]
- 4.Singh A., Giri J., Kapoor S., Tyagi A.K., Pandey G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs S., Grill E., Meskiene I., Schweighofer A. Type 2C protein phosphatases in plants. FEBS J. 2013;280:681–693. doi: 10.1111/j.1742-4658.2012.08670.x. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K., Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013;32:959–970. doi: 10.1007/s00299-013-1418-1. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Guzman M., Betz K., Dupeux F., Márquez J.A., Rodriguez P.L., Santiago J., Rodriguez L., Antoni R. Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci. 2010;182:3–11. doi: 10.1016/j.plantsci.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 10.Miyakawa T., Tanokura M. Regulatory mechanism of abscisic acid signaling. Biophysics. 2011;7:123–128. doi: 10.2142/biophysics.7.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smékalová V., Doskočilová A., Komis G., Šamaj J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 2014;32:2–11. doi: 10.1016/j.biotechadv.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Han S., Yu B., Wang Y., Liu Y. Role of plant autophagy in stress response. Protein Cell. 2011;2:784–791. doi: 10.1007/s13238-011-1104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memisoglu G., Eapen V.V., Yang Y., Klionsky D.J., Haber J.E. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc. Natl. Acad. Sci. USA. 2019;116:1613–1620. doi: 10.1073/pnas.1817078116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan S.A., Li M.Z., Wang S.M., Yin H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018;19:1634. doi: 10.3390/ijms19061634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali M.A., Azeem F., Nawaz M.A., Acet T., Abbas A., Imran Q.M., Shah K.H., Rehman H.M., Chung G., Yang S.H., et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018;226:12–21. doi: 10.1016/j.jplph.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Koornneef M., Reuling G., Karssen C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;61:377–383. doi: 10.1111/j.1399-3054.1984.tb06343.x. [DOI] [Google Scholar]
- 17.Rodriguez P.L., Leube M.P., Grill E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 1998;38:879–883. doi: 10.1023/A:1006012218704. [DOI] [PubMed] [Google Scholar]
- 18.Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M.P., Nicolas C., Lorenzo O., Rodriguez P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313X.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida T. ABA-Hypersensitive Germination3 Encodes a Protein Phosphatase 2C (AtPP2CA) That Strongly Regulates Abscisic Acid Signaling during Germination among Arabidopsis Protein Phosphatase 2Cs. Plant Physiol. 2005;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., Hirayama T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamada K., Ito T., Katagiri T., Nakashima K., Kobayashi M., Ishiyama K., Nakasone S., Kanamori N., Yamaguchi-Shinozaki K., Yoshida T., et al. Three SnRK2 Protein Kinases are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 22.Bhaskara G.B., Nguyen T.T., Verslues P.E. Unique Drought Resistance Functions of the Highly ABA-Induced Clade A Protein Phosphatase 2Cs. Plant Physiol. 2012;160:379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang Y., Sun X., Gao S., Qin F., Dai M. Deletion of an Endoplasmic Reticulum Stress Response Element in a ZmPP2C-A Gene Facilitates Drought Tolerance of Maize Seedlings. Mol. Plant. 2017;10:456–469. doi: 10.1016/j.molp.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Wei K., Pan S. Maize protein phosphatase gene family: Identification and molecular characterization. BMC Genom. 2014;15:1–20. doi: 10.1186/1471-2164-15-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y.G., Fu F.L., Yu H.Q., Hu T., Zhang Y.Y., Tao Y., Zhu J.K., Zhao Y., Li W.C. Interaction network of core ABA signaling components in maize. Plant Mol. Biol. 2018;96:245–263. doi: 10.1007/s11103-017-0692-7. [DOI] [PubMed] [Google Scholar]
- 26.Wang H., Qin F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017;8:1–12. doi: 10.3389/fpls.2017.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao H., Wang H., Liu S., Li Z., Yang X., Yan J., Li J., Tran L.S.P., Qin F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015;6:1–13. doi: 10.1038/ncomms9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Wang H., Liu S., Ferjani A., Li J., Yan J., Yang X., Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016;48:1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- 29.Gubler F., Millar A.A., Jacobsen J.V. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Finkelstein R., Reeves W., Ariizumi T., Steber C. Molecular Aspects of Seed Dormancy. Annu. Rev. Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 31.Foolad M.R., Lin G.Y. Genetic potential for salt tolerance during germination in Lycopersicon species. HortScience. 1997;32:296–300. doi: 10.21273/HORTSCI.32.2.296. [DOI] [Google Scholar]
- 32.Yang X., Gao S., Xu S., Zhang Z., Prasanna B.M., Li L., Li J., Yan J. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 2011;28:511–526. doi: 10.1007/s11032-010-9500-7. [DOI] [Google Scholar]
- 33.Fu J., Cheng Y., Linghu J., Yang X., Kang L., Zhang Z., Zhang J., He C., Du X., Peng Z., et al. RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 2013;4:1–12. doi: 10.1038/ncomms3832. [DOI] [PubMed] [Google Scholar]
- 34.Liu H., Luo X., Niu L., Xiao Y., Chen L., Liu J., Wang X., Jin M., Li W., Zhang Q., et al. Distant eQTLs and Non-coding Sequences Play Critical Roles in Regulating Gene Expression and Quantitative Trait Variation in Maize. Mol. Plant. 2017;10:414–426. doi: 10.1016/j.molp.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Liu S., Wang X., Wang H., Xin H., Yang X., Yan J., Li J., Tran L.S.P., Shinozaki K., Yamaguchi-Shinozaki K., et al. Genome-Wide Analysis of ZmDREB Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of Zea mays L. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo X., Wang B., Gao S., Zhang F., Terzaghi W., Dai M. Genome-wide association study dissects the genetic bases of salt tolerance in maize seedlings. J. Integr. Plant Biol. 2019;61:658–674. doi: 10.1111/jipb.12797. [DOI] [PubMed] [Google Scholar]
- 37.Qin F., Shinozaki K., Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- 38.Lu T., Zhang G., Wang Y., He S., Sun L., Hao F. Genome-wide characterization and expression analysis of PP2CA family members in response to ABA and osmotic stress in Gossypium. PeerJ. 2019;7:e7105. doi: 10.7717/peerj.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F., Wei Q., Shi J., Jin X., He Y., Zhang Y., Luo Q., Wang Y., Chang J., Yang G., et al. Brachypodium distachyon BdPP2CA6 Interacts with BdPYLs and BdSnRK2 and Positively Regulates Salt Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2017;8:1–15. doi: 10.3389/fpls.2017.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Z., Zhong J., Sun X., Wang B., Terzaghi W., Dai M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 2018;9:1–12. doi: 10.3389/fpls.2018.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.-H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. Triple Loss of Function of Protein Phosphatases Type 2C Leads to Partial Constitutive Response to Endogenous Abscisic Acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi-Shinozaki K., Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. MGG Mol. Gen. Genet. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- 43.Park M.Y., Chung M.S., Koh H.S., Lee D.J., Ahn S.J., Kim C.S. Isolation and functional characterization of the Arabidopsis salt-tolerance 32 (AtSAT32) gene associated with salt tolerance and ABA signaling. Physiol. Plant. 2009;135:426–435. doi: 10.1111/j.1399-3054.2008.01202.x. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez J., Chua N. Arabidopsis PLC1 Is required for secondary responses to abscisic acid signals. Plant Cell. 2001;13:1143–1154. doi: 10.1105/tpc.13.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song H., Zhao R., Fan P., Wang X., Chen X., Li Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta. 2009;229:955–964. doi: 10.1007/s00425-008-0886-y. [DOI] [PubMed] [Google Scholar]
- 46.Dong T., Park Y., Hwang I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015;58:29–48. doi: 10.1042/bse0580029. [DOI] [PubMed] [Google Scholar]
- 47.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 48.Bates L.S., Waldren R.P., Teare I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 49.Dai M., Zhang C., Kania U., Chen F., Xue Q., Mccray T., Li G., Qin G., Wakeley M., Terzaghi W., et al. A PP6-Type Phosphatase Holoenzyme Directly Regulates PIN Phosphorylation and Auxin Efflux in Arabidopsis. Plant Cell. 2012;24:2497–2514. doi: 10.1105/tpc.112.098905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.