Abstract
Extracellular vesicles (EVs) secreted in biological fluids contain several transcripts of the cell of origin, which may modify the functions and phenotype of proximal and distant cells. Cancer-derived EVs may promote a favorable microenvironment for cancer growth and invasion by acting on stroma and endothelial cells and may favor metastasis formation. The transcripts contained in cancer EVs may be exploited as biomarkers. Protein and extracellular RNA (exRNA) profiling in patient bio-fluids, such as blood and urine, was performed to identify molecular features with potential diagnostic and prognostic values. EVs are concentrated in saliva, and salivary EVs are particularly enriched in exRNAs. Several studies were focused on salivary EVs for the detection of biomarkers either of non-oral or oral cancers. The present paper provides an overview of the available studies on the diagnostic potential of exRNA profiling in salivary EVs.
Keywords: miRNA, non coding RNA, exosomes, microvesicles, cancer, saliva
1. Introduction
The aim of liquid biopsy is to identify biomarkers with diagnostic, predictive and prognostic values in bio-fluids, to avoid more invasive approaches. Researchers focused on different types of biomarkers, including proteins, circulating DNA fragments and cells, and extracellular RNAs (exRNAs). ExRNAs are more sensitive and specific biomarkers than proteins and better reflect the cell dynamic than DNA does [1]. However, several limitations in the use of exRNA as biomarkers still remain, related to their heterogeneity, the incomplete definition of their multiple targets and functions, and their stability in different biological fluids [2].
Nowadays, the recently developed techniques of sequencing allow for an accurate evaluation of RNA expression, which reflects cellular genetic and functional states. Different types of RNA biomarkers have been considered in cancer. Differential mRNA expression profiles may reflect the positive and negative regulation of tumor-associated genes in several cancers and may provide suitable biomarkers for monitoring the clinical outcome of patients [3,4,5]. Non-coding RNAs, such as microRNAs (miRNAs), piwi-interacting RNA (piRNA), small nucleolar RNA (snoRNA), circular RNA (circRNA) and long non-coding RNAs (lncRNAs), have also been investigated as potential biomarkers in cancer [1]. Moreover, the detection of chimeric RNAs may allow for the identification of chromosomal aberrations [6,7]. The stability of different exRNAs in the biological fluids depends on protection from exonucleases, provided either by RNA binding proteins, such as those of the Argonaute family, and high- and low-density lipoproteins, and by encapsulation in membrane vesicles [8,9,10].
Membrane vesicles released by cells in the extracellular space have recently emerged as a good evolutionarily preserved mechanism of inter-cellular communication. The vesicles are able to share genetic information among cells by delivering proteins, bio-active lipids and nucleic acids protected from degrading enzymes [11,12,13]. These vesicles, termed extracellular vesicles (EVs), are abundant in all biological fluids and can be exploited for searching biomarkers since they retain the molecular signature of the cell of origin.
One challenge of liquid biopsy is the choice of the bio-fluid that better reflects the occurrence of cancer. Most studies have focused on blood, but several other bio-fluids are now gaining attention, including saliva. Saliva is enriched in EVs and may represent a bio-fluid suitable for searching for markers of oral and systemic diseases.
2. EVs as Carriers of exRNA
EVs are a heterogeneous population, which includes membrane vesicles of different sizes and biogenesis. The three main categories of EVs include exosomes, ectosomes, and apoptotic bodies [14,15,16,17]. Exosomes are nano-sized vesicles (35–100 nm), which originate from the multivesicular bodies and are secreted by a process of exocytosis. This process requires the inward budding of multivesicular bodies-membrane, followed by fusion with plasma membrane and release in the extracellular space. The endosomal sorting complex required for transport (ESCRT) machinery, and several components of the Ras GTPases (RAB) family [18,19] and of the tetraspanin family [18], are involved in such processes. Vesicles generated by the budding of surface plasma membrane with the inclusion of cytoplasmic constituents have been termed microvesicles. This term is misleading as these vesicles include a large population of vesicles within the nano-range (60–250 nm), such as those released from healthy cells. It has been therefore suggested that one should name these vesicles ectosomes or shedding vesicles [14]. Shedding vesicles also include larger vesicles that may reach 1000 nm, and some of them may derive from cells in a pre-apoptotic phase. Microvesicle formation is related to the modification of plasma membrane curvature due to changes in lipid and protein interactions involving the arrestin domain-containing protein-1 (ARRDC1) and the late endosomal protein tumor susceptibility gene 101 (TSG101). The cytoskeleton rearrangements controlled by the signaling cascade of Ras-related GTPase ADP-ribosylation factor 6 (ARF6) promote vesiculation and release [20]. The apoptotic bodies released by cells undergoing programmed death are vesicles with a diameter of 1000–5000 nm and may contain nuclear fragments and intact chromosomes [21].
Most of the studies on the use of EVs as potential biomarkers have been performed on exosomes and microvesicles, as both types of vesicles may encapsulate fragments of genomic and mitochondrial DNA origin [22,23,24,25], and different classes of RNA, such as mRNA, miRNA, lncRNA, mitochondrial RNA, transfer RNA, and ribosomal RNA [26,27,28,29]. Furthermore, nano-sized vesicles may be released by the same cell by exocytosis or by surface membrane budding, and it may be difficult to discriminate vesicles discharged by non-apoptotic cells on the basis of mechanisms of origin. In fact, some molecules constituent of the endosomal sorting complex required for transport (ESCRT) and some ancillary proteins such as TSG101, Alix and Vacuolar protein sorting-associated protein 4 (VPS4) implicated in the formation of exosomes, are also reported in the literature to also be shared by shedding vesicles [30]. The discharge of exosomes may involve some constituents of the RAB family of GTPase proteins implicated in the MVBs/plasma membrane interaction [31,32]. Furthermore, the biogenesis of shedding vesicles may depend on a reorganization of the proteins of the cytoskeleton myosin and actin under the control of the ARF6 signaling [30]. Some tetraspanins and some ESCRT proteins are often reported as common exosome and shedding vesicle markers and cannot represent a peculiarity principle [33]. However, CD9, CD63 and CD81 tetraspanins are reported to be enhanced in exosomes [18], while annexin A1 is considered a marker for microvesicles [34]. Due to the heterogeneity of EVs produced by different cell types and present in the biological fluids, the protocols used for EV purification assume a critical relevance. For this reason, the public available databases [35,36,37] take into account the procedures used for the purification of EVs when describing the lipid, protein and nucleic acid composition. Of interest, the comparative lipidomic, proteomic and genomic analyses between the cells of origin and their released EVs highlight the presence of qualitative and quantitative differences, in both basal and stimulated condition. These data suggest that the EV cargo is actively modulated [38,39,40]. The EV mediated transfer of their cargo into recipient cells can induce epigenetic and functional changes into the recipient cells [41]. Some studies indicate that genetic materials encapsulated in EV include mitochondrial [24,42] and genomic [22] DNAs. Other studies performed on exosomal sub-fractions of EVs suggest that the DNA release is not related to the small vesicle release but to the autophagy- and multivesicular-endosome-dependent mechanism [34]. Several RNA species were found to be associated with EVs. EVs contain intact mRNA that can be translated into proteins in the recipient cells [27,28], but also many fragments of 200 nucleotides [43] that may have a biological role as scavengers and/or values as biomarkers. The exRNA enriched in EVs include miRNAs, ribosomal RNAs, tRNA fragments piRNA, snoRNA, Y-RNA, circRNA and lncRNAs [44,45,46,47,48].
Little is known about the process of nucleic acid compartimentalization into EVs [39,49,50,51,52]. Some proteins involved in EV biogenesis are potential candidates for RNA encapsulation in EVs. For instance, it has been shown that in EVs purified by differential ultracentrifugation from liver stem cells, Alix coprecipitate with Argonate 2 (Ago2) protein and miRNAs. The significant reduction of EV-associated miRNAs in Alix knock-down cells suggests that Alix can have a role as a component of ESCRT in the export of the Ago2-miRNA complex [53]. Through a high-resolution density gradient fractionation coupled with an immunoaffinity capture of exosomes, Argonaute proteins were detected in the non-vesicular compartment [34], which may contain components of the multivesicular body membranes. In breast cancer-derived EVs, miRNAs associated with Ago2 were shown to induce an alteration in the transcriptome of the recipient cells [54]. By regulating the Ago2 secretion [55], GTPase KRas (KRAS) has been involved in the miRNA compartmentalization into EVs released by colorectal cancer cells [56]. Moreover, miRNA packing into EVs depends on the interaction with the heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) [57] and with the RNA-binding protein Y-box protein I (YBX1) [58].
3. EVs in Cancer Biology
EVs released by cancer cells may act both locally, contributing to create a favorable microenvironment for cancer growth, and at distance, promoting the metastatic niche formation. Several studies have shown that cancer EVs contribute to the induction of chemo-resistance [59,60,61,62,63], to the remodeling of extracellular matrix [64,65], to tumor vascularization [66] and to epithelial-mesenchymal transition with a consequent enhanced migration/invasion and metastasis formation [67,68,69]. EVs also participate as active players in the bi-directional crosstalk between cancer cells and cells present in the microenvironment, such as fibroblasts [70,71], which may secrete EVs conferring chemo-resistance [72,73,74] and invasiveness to cancer cells [70,71].
Several mechanisms of action involving the EV-mediated transfer of proteins and exRNA have been described and exploited as diagnostic markers. In particular, the miRNA-mediated effects have been extensively studied. Several miRNAs present in EVs released from breast cancer (miR-100, miR-222, miR30a and miR-17), lung cancer (miR-100-5p), and ovarian cancer (miR-21) or released from stromal cells (miR-21 and miR-146a) were shown to confer chemo-resistance [60,61,62,73,74] (Table 1). Cancer EVs may contribute to new blood vessel formation by transferring to recipient fibroblasts and endothelial cells pro-angiogenic miRNAs such as miR-155, miR-210 and miR-494, which are under the regulation of the hypoxia-inducible factor (HIF) 1α [75,76,77,78,79] (Table 2). Moreover, several studies on EVs released by cancer cells indicate that they promote the development of a pre-metastatic niche by transferring either proteins or oncogenic miRNAs [80,81,82,83]. For instance, some miRNAs (miR-125b, miR-130b and miR-155) present in prostate cancer and released by EVs have been shown to confer a protumorigenic phenotype to adipose-derived mesenchymal stem cells [84].
Table 1.
The role of EVs in chemo-resistance and immune-modulation. Several proteins and exRNAs have been described to be involved in tumor chemo-resistance and immune-modulation.
| Biological Effect | Mechanism of Action | Cell Source | Target | References |
|---|---|---|---|---|
| Resistance to chemotherapy | Transfer of MDR-1/P-gp | Docetaxel-resistant prostate cancer | Docetaxel-sensitive prostate cancer | [59] |
| Transfer of miR-100, miR-222, miR-30a and miR-17 | Adriamycin and docetaxel-resistant breast cancer | Adriamycin and docetaxel-sensitive breast cancer | [60] | |
| Transfer of miR-21 | Platinum-resistant ovarian cancer | Platinum-sensitive ovarian cancer | [61] | |
| Transfer of miR-100-5p, miR-21 and miR-133b | Cisplatin-resistant lung cancer | Cisplatin-sensitive lung cancer | [62,63] | |
| Transfer of miR-21, which downregulates APAF1 | Stroma | Ovarian cancer | [74] | |
| Transfer of miR-146a with Snail mRNA | Cancer-Associated Fibroblasts | Pancreatic cancer | [73] | |
| Activation of the antiviral/ NOTCH3 signaling pathway | Stroma | Breast cancer | [72] | |
| Tumor immune-escape | Release of pro-inflammatory cytokines by macrophages, possibly mediated by miR-21 and miR-29a | Breast and lung cancer, melanoma | Tumor cells, fibroblasts, endothelial cells, and immune cells | [85,86,87] |
| Inhibition of dendritic cell maturation and functions, by delivering specific miRNAs (e.g., miR-203, miR-212-3p) | Renal carcinoma, pancreatic cancer, melanoma | Dendritic and T cells | [87,88,89] | |
| MDSCs activation, which leads to TGF-β-mediated suppression of T cell activity | Melanoma and colorectal carcinoma | CD14+ monocytes | [90,91] | |
| Suppression of the T-cell activity mediated by PDL-1, TGF-β, Fas ligand and TRAIL | Melanoma, colorectal, gastric and prostate cancer, head and neck squamous cell carcinoma | CD8+T cells | [92,93,94,95,96] | |
| Inhibition of NK cell cytotoxic activity, possibly mediated by MIC A ligand of NKG2D receptor | Mammary carcinoma, melanoma, cervical, head and neck, liver cancer | NK cells | [97,98,99] | |
| Enhancement of immune response | Activation of a tumor antigen-specific immune response in humans | Melanoma and non-small cell lung cancer patients-derived dendritic cells | systemic administration | [100,101] |
EVs: extracellular vesicles, MDR: multidrug resistance protein, APAF1: apoptotic protease-activating factor 1, MDSCs: myeloid-derived suppressor cells, TGF-β: trasforming growth factor-β, PDL-1: programmed death-ligand 1, TRAIL: tumor necrosis factor-related apoptosis-inducing ligand, NK: natural killer, MIC: MHC class I–related chain, NKG2D: NKG2-D type II integral membrane protein.
Table 2.
Role of EVs as biomarkers of tumor progression. Cancer-derived EV content has been proposed as a tumor biomarker and has been related to several processes involved in tumor aggressiveness.
| Biological Effect | Mechanism of Action | Cell Source | Target | References |
|---|---|---|---|---|
| Tumor biomarkers | Transfer of miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205 and miR-214 | Ovarian cancer | Serum | [102] |
| Transfer of miR-17-3p, miR-21, miR-29a, miR-106a, miR-146 miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212 and miR-214 | Lung cancer | Serum | [85,103] | |
| Transfer of miR-18a, miR-221 and miR-224 | Hepatocellular carcinoma | Serum | [104] | |
| Pro-angiogenic effect | Transfer of proangiogenic miRNAs, mostly regulated by HIF-1α (miR-155-5p, miR-210 and miR-494) | Melanoma, hepatocellular, lung and renal adenocarcinoma | CAFs and endothelial cells | [75,77,78,79,81] |
| Decrease cell-to-cell adhesion | Reduction of E-cadherin, let-7i and β-catenin expression, and increase of Snail1-2, Twist1-2, Sip1, vimentin, ZEB2 and N-cadherin expression, activation of MAPK pathway | Breast and bladder cancer, melanoma | Mammary and urothelial cells epithelial cells, primary melanocytes | [67,68,69] |
| Increase in cell migration/invasion | Lipids and proteins (e.g., CD81)-dependent stimulation of the cancer cell motility via Wnt signaling | Cancer Associated Fibroblasts | Melanoma, breast and prostate cancer | [70,71] |
| Development of premetastatic niche | Delivery of TYRP2, VLA4, HSP70, an HSP90 isoform and the MET oncoprotein | Melanoma | Bone marrow progenitor cell | [83] |
| Exosomal expression of tumor-specific integrin patterns | Osteosarcoma, rhabdomyosarcoma, Wilms tumor, skin and uveal melanoma, breast, colorectal, pancreatic and gastric cancer | Brain, lung and liver epithelium | [82] | |
| Delivery of MIF | Pancreatic ductal adenocarcinoma | Kupffer cell | [80] | |
| Delivery of specific oncogenic miRNAs, e.g., miR-125b, miR-130b and miR-155, which induce a neoplastic reprogramming of recipient cells | Prostate, renal cancer | Adipose-derived stem cells, lung epithelium | [81,84] |
HIF-1a: hypoxia inducible factor 1α, HSP90: heat shock protein 90, MET: hepatocyte growth factor receptor., TYRP2: tyrosinase-related protein-2, VLA4: very late antigen 4, HSP70: heat shock protein 70, MIF: macrophage migration inhibitory factor.
A contribution in favoring the tumor immune-escape of EVs released by cancer cells has been also suggested [105,106] (Table 1). The mechanisms involved the activation of tumor-associated macrophages [85,86,87], suppressor myeloid cells [90,91] and the inhibition of NK cell activity [97,98,99]. By expressing PDL1 [92,93], the transforming growth factor (TGF) beta [94], the tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) and the Fas ligand [95,96], cancer EVs exhibit an immunosuppressive activity on T cells. Moreover, cancer EVs inhibit the maturation of dendritic cells through a mechanism involving the expression of HLA-G [88] and specific miRNAs, such as miR-203 and miR-212-3p [89]. Despite tumor EVs have been mainly implicated in the tumor immune escape, they can also be exploited to cross-present tumor antigens to the antigen presenting cells eliciting an antigen-specific cytotoxic lymphocyte anti-tumoral response [100,101]. However, clinical trials based on this assumption have provided conflicting results [107,108,109].
At present, most of the studies looking for exRNAs as cancer biomarkers have been performed on whole blood, urine and cerebrospinal fluids. Recently, several studies have explored the detection of exRNAs associated with the EVs. Despite the fact that the quantitative and stoichiometric analyses revealed that many miRNAs are present in less than one single copy per single exosome [110], several studies indicate a potential utility as cancer biomarkers [102,103]. For instance, in hepatocellular carcinoma, the expression by serum EVs of miR-18a, miR-221and miR-224 has been suggested as potential diagnostic biomarkers [104]. Fabbri et al. demonstrated that EV-associated miR-21 and miR-29a bound to a Toll-like receptor family favoring an inflammatory pro-metastatic response in lung [85]. On the other hand, the expression of miR-21 in serum EVs in patients with breast cancer correlates with a favorable outcome [111]. By comparing the miRNA signature of ovarian cancer EVs with that of EVs from normal subjects, Taylor and colleagues suggested a potential utility to screening asymptomatic patients [102]. A significant similarity of EV-associated miRNAs was observed with tumor-derived miRNAs in lung adenocarcinoma [103]. Moreover, the miRNA patterns of patients were clearly distinct from those of normal controls, suggesting that circulating EV-associated miRNAs might be useful as a non-invasive screening test [103].
4. Salivary EVs as Biomarkers
EVs are particularly enriched in saliva, which in respect to blood does not undergo coagulation. This is an important issue because many studies have been performed on serum. Coagulation induces a consistent release of EVs from platelets, thus modifying the composition of circulating EVs [112]. Salivary EVs should derive in part from salivary glands and in part from circulation: indeed, about a 30% similarity of salivary and plasma proteome has been described by a few studies [113,114,115]. In particular, using liquid chromatography and mass spectrometry, 19,474 unique peptides have been isolated from whole saliva in a multicenter study [113]. Protein annotation was assessed by matching the identified peptides with a recently published dataset of the human plasma proteome [116], and 1939 different proteins were identified as commonly expressed in blood and saliva. However, a puzzling aspect is the expression of neuronal markers in salivary EVs with significant changes in the miRNA pattern and in the proteomic profile after a head concussion [117] and in neurological diseases [118,119]. Moreover, the EV composition may be affected by the presence in saliva of viruses, including the human papillomavirus (HPV) [120,121,122,123,124] and the neurotropic human herpesviruses (e.g., HHV-6), which are detectable in the saliva of infected subjects [125].
A critical aspect in the use of salivary EVs as biomarkers is the purification technique that is used (Table 3). In fact, results may vary depending on the purified subpopulations and the presence of contaminants, such as bacterial flora. Therefore, accurate mouth washing, careful standardization on saliva collection and sample filtration are recommended to abate the bacterial load.
Table 3.
Biomarkers detected in salivary EVs. Salivary EVs can be purified using different EV isolation techniques and can be exploited as biomarkers because they contain disease-related proteins and exRNA.
| Disease | Isolation Method | EV Biomarkers | Type of Biomarker | References |
|---|---|---|---|---|
| Brain injury and neurological disorders | Differential ultracentrifugation | CDC2, CSNK1A1, and CTSD | mRNA | [117] |
| XYCQ EV Enrichment KIT | α-synuclein | protein | [119] | |
| Oral squamous cell carcinoma | Differential ultracentrifugation | CD63 | protein | [126,127] |
| Differential ultracentrifugation | PPIA | protein | [128] | |
| Charge-based precipitation | miR-412-3p, miR-512-3p, miR-27a-3p, miR-494-3p, miR-302b-3p, miR-517b-3p | miRNA | [129] | |
| Lung cancer | Affinity chromatography column combined with filter system (ACCF) | Annexin A1, A2, A3, A5, A6, A11; NPRL2; CEACAM1; MUC1; PROM1; HIST1H4A; TNFAIP3 | protein | [130] |
| Affinity chromatography column combined with filter system (ACCF) | BPIFA1, CRNN, MUC5B, IQGAP | protein | [131] | |
| Head and neck carcinoma | Differential ultracentrifugation | miR-486-5p, miR-486-3p, miR-10b-5p, miR-122 | miRNA | [132] |
| Pancreatic cancer | Total Exosome Isolation Reagent (Invitrogen) | miR-1246, miR-4644 | miRNA | [133] |
| Differential ultracentrifugation | Apbb1ip, Aspn, BCO31781, Daf2, Foxp1, Gng2, Incenp | mRNA | [134] |
CDC2: Cyclin-dependent kinase A-1, CSNK1A1: Casein Kinase 1 Alpha 1, CTSD: Cathepsin D, PPIA: Peptidyl-prolyl cis-trans isomerase A, NPRL2: GATOR complex protein NPRL2, CEACAM1: Carcinoembryonic antigen-related cell adhesion molecule 1, MUC1: Mucin 1, PROM1: Prominin 1, HIST1H4A: Histone H4, TNFAIP3: Tumor necrosis factor alpha-induced protein 3, BPIFA1: BPI fold-containing family A member 1, CRNN: Cornulin, MUC5B: Mucin 5b, IQGAP: Ras GTPase-activating-like protein IQGAP1.
Differential ultracentrifugation or density gradient ultracentrifugation are considered the gold standard for the purification of EV subpopulations. These techniques have been further implemented with the combined use of the immune-affinity capture of exosomes [34]. To improve the separation of vesicles from non-vesicular components a floating technique has been proposed, based on gradient fractioning centrifugation, with samples applied to the bottom of tubes [135]. However, the standardization of these techniques may be difficult, as the results are influenced not only by the centrifugal radius of the rotor and g force type, but also by the viscosity of the starting solution. In addition, due to mechanical damage, membrane debris are generated, as seen by electron microscopy. Moreover, the difficult detection of proteins and RNAs has been described [136,137,138,139]. To avoid shear stress due to ultracentrifugation, size exclusion chromatography has been employed with the aim to separate small vesicles from protein contaminants [140,141,142]. Immuno-affinity purification allows for the recovery of sub fractions of EVs based on the expression of surface markers [139,143,144,145], and several kits are commercially available. Microfiltration has also been used with membranes with appropriate pore sizes to remove cell debris and apoptotic bodies [143]. However, this technique is limited by EV adhesion to membranes and pore clogging. In addition, to isolate small biological samples, all these techniques may have a low efficient recovery of EVs. Another approach for isolating EVs from biological liquids is based on polymeric precipitation [146,147,148,149,150,151]. This approach allows for a rapid precipitation of EVs, but it is limited by the co-precipitation of proteins of a non-vesicular origin such as lipoproteins [136,152,153]. Recently, a new technique based on electric field-induced release and measurement has been successfully applied to liquid biopsy in saliva [154]. Using this technique, the mutation of epidermal growth factor receptor (EGFR) in patients with lung cancers was detected and matched with biopsy genotyping [154,155]. Moreover, the electric field-induced release has been combined with the magnetic beads immune-capturing of exosomes [156,157], resulting in a highly sensitive and specific method of exRNA extraction and analysis. Compared to polymeric precipitation and differential centrifugation, this approach is less time consuming, requires smaller sample volumes and does not involve sample lysis that may reduce exRNA yield. However, for each EV extraction, the capture probe that is attached to the magnetic beads allows for the isolation of only those EVs containing the exosome-specific surface marker used for capturing EVs [156]. In fact, EVs are a heterogeneous population of vesicles, and individual EV analyses show that not all EVs co-express the same tetraspanin. Therefore, this technique may not include the whole pattern of EV-associated exRNA.
By quantitative nano-structural and single molecule force spectroscopy, Sharma et al. [126] performed a bio-molecular analysis of exosomes present in the saliva from patients with oral cancer. They demonstrated that exosomes were augmented in number and size, displayed a dissimilar morphology and showed an increased expression of CD63. Similarly, Zlotogorski-Hurvitz et al. [127] described a bigger salivary exosome concentration and size in patients with oral cancers in comparison with healthy subjects, a higher expression of CD63 and a decreased expression of CD9 and CD81. Few other studies performed a proteomic analysis of salivary exosomes in search of potential biomarkers of oral [128] and lung carcinomas [130]. A higher expression of the CD63 molecule was observed in EVs from the saliva of patients with oral cancers in respect to normal subjects [126]. Sun et al. performed a comparative proteomic analysis of salivary EVs in normal subjects and lung cancer patients [131]. In this study, several proteins were found to be dysregulated, and four of them were present in both salivary microvesicles and exosomes, suggesting their potential use for the detection of lung cancer.
It has been reported that in saliva, the bulk of miRNAs is packaged in exosomes [13]. In fact, miRNAs are easily detectable in EVs present in saliva [158,159]. Several studies focused on the possibility of exRNA isolation from saliva and oral samples [160,161,162] and in particular on salivary EV associated miRNAs in patients with oral cancer [160,161,162,163,164].
Langevin et al. performed a comprehensive miRNA sequence analysis of EVs derived from the saliva of patients with head and neck carcinomas and identified a distinct pattern of secretion and, in particular, miRNAs secreted only by cancer cells [132]. Some miRNAs, such as miR-486-5p, miR-486-3p and miR-10b-5p, were specifically overexpressed in the EVs of a subset of head and neck carcinomas. Machida and colleagues showed that miR-1246 and miR-4644 present in salivary EVs are potential biomarkers of cancers of the pancreato-biliary tract [133]. Taken together, these analyses may provide the bases for the development of new tumor biomarkers (Table 3).
A transcriptomic signature specific for pancreatic [165] and ovarian cancers [166] and proteomic signature modifications in lung cancer [167] have been described in whole saliva. Zhang et al. [165] demonstrated that the combination of KRAS, metyl CpG binding domain protein 3 like 2 (MBD3L2), acrosomal vesicle 1 (ACRV1), and dolichyl-phosphate mannosyltransferase subunit 1 (DPM1) mRNAs in saliva may differentiate patients with pancreatic carcinomas from patients with chronic pancreatitis and healthy subjects with a high sensitivity and specificity. Moreover, a transcriptomic analysis of salivary EVs by next generation sequencing showed the presence of many coding and non-coding RNAs, such as mRNAs for several proteins, miRNAs, snoRNAs, piRNAs, and lncRNAs [48,168]. Palanisamy et al. [169] found, in exosomes isolated from saliva, 509 mRNA transcripts, which once incorporated in keratinocytes were able to modify the protein expression in these cells. Moreover, exosomes from adenocarcinoma of the pancreatic ducts were able to modify the biology of exosomes derived from the salivary gland and induce changes in the salivary biomarker profiles [134]. Similarly, they showed an interaction between exosomes derived from the human metastatic mammary gland epithelial adenocarcinoma cell line MDA-MB-231 cells and exosomes derived from the human submandibular gland (HSG) cells. This interaction induced an activation of the HSG cell transcriptional machinery with an increase of total cellular RNA and transcriptomic and proteomic changes [170]. Salivary EVs derived from patients with pancreatic carcinoma were shown to inhibit NK cell activation, thus favoring tumor immune escape [171].
We analyzed the zeta potential of salivary EVs and, based on their negative charge, we developed a charge-based precipitation protocol. This technique allows for the efficient recovery of exRNA from a salivary EV population with a very homogeneous size and shape [159] (Figure 1). In a recent work [129], we used the charge-based precipitation method to isolate EVs from the saliva of patients with oral squamous cell carcinoma (OSCC) to investigate the presence of exRNAs suitable as biomarkers. Our aim was to assess whether this quick, simple and efficient technique could be useful for detecting exRNA in the salivary EVs of patients with OSCC. The diagnosis of OSCC is based on oral examination and histological analysis. However, the identification of salivary biomarkers may have potential prognostic and therapeutic values. To exclude misleading results due to a different exposition to risk factors, at the time of patients’ recruitment, subjects included in our study were checked for their habits regarding smoking and alcohol consumption. In fact, smoke and alcohol consumption has been described as potentially affecting the composition of EV-associated exRNA [172,173]. Therefore, patients and controls were matched to obtain a similar distribution of risk factors among the two groups to reduce this bias. Moreover, 5 out 21 patients were positive to the Human Papilloma Virus (HPV). To avoid the detection of exRNA of viral origin, we screened EVs for the presence of about 800 miRNAs with human-specific primers. Although HPV infection may alter the EV release and cargo, we did not observe any significant change in the size and concentration of EVs from HPV-positive patients compared to negative patients. A differential expression of the EV miRNA signature in OSCC cells infected or not by HPV has been previously shown [124]. In this study, the authors observed that HPV infected cells released EVs enriched with 14 miRNAs, whereas non-infected cells overexpressed 19 miRNAs. The cohorts of patients we studied were too small to draw any conclusions. However, we did not observe the differential expression of miRNAs, which has been previously described for EVs released by in vitro infected OSCC cells.
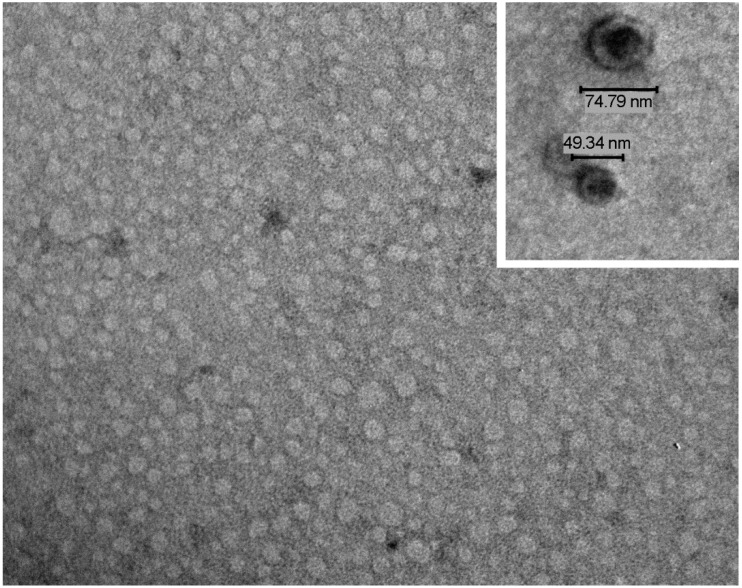
Figure 1.
Salivary EVs characterization. A representative transmission electron microscopy image of EVs isolated by a charge-based precipitation method, showing a carpet of vesicles in the nano-range. In the inset, the bars indicate the size of the extracellular vesicles (EVs). The preparation was stained with NanoVan (JEOL Jem-1010 electron microscope, original magnification ×75,000; inset ×150,000).
By comparing the miRNA expression of cancer patients and matched controls, we observed an up-regulation of miR-412-3p, miR-512-3p, miR-27a-3p and miR-494-3p in patients with oral squamous cell carcinoma. MiR-512-3p and miR-412-3p were also potentially sensitive and specific biomarkers, as indicated by the high AUC values (0.847 and 0.871 respectively, with p values < 0.02) and a maximum Youden’s Index. Interestingly, we also observed an exclusive expression of miR-302b-3p and miR-517b-3p in cancer EVs. Moreover, we performed a bio-informatic analysis to better understand whether the tumor-enriched miRNAs could be functionally related to the tumor. We observed that eight tumor-related pathways were potentially targeted by these miRNAs. In particular, miR-512-3p and miR-27a-3p may target 7 and 20 genes, respectively, of the ErbB signaling pathway, which is known to promote cell proliferation and survival in cancer [174] and is activated in oral carcinomas [175,176,177]. MiR-512-3p, miR-27a-3p, and miR-302b-3p could potentially target proteoglycan genes and CD44 involved in c-Fos-mediated cell invasion and migration [178], ERK1/2 phosphorylation [179] and the phenotype of oral cancer stem cells [180]. Moreover, miR-512-3p, miR-412-3p, miR-27a-3p, and miR-302b-3p reduced the expression of TGFβR2, frequently reduced in cancer and stroma cells in patients with oral squamous carcinomas [181]. Increased levels of the oncogenic miR-27a-3p has also been detected in EVs obtained from the plasma of OSCC patients [182]. In this study, a comparable miRNA signature was observed between plasma EVs and EVs released by OSCC cells in vitro.
Recent studies have shown that EVs also contain lncRNAs [183]. The expression of lncRNAs has not been investigated in salivary EVs. However, salivary lncRNAs may represent a potential marker for OSSC [184]. In fact, a subset of lncRNAs was correlated with high metastatic OSCC. In particular, the lncRNA HOTAIR was found to be highly expressed in the saliva of patients with lymph node metastasis. Therefore, besides miRNAs, the search for lncRNAs in salivary EVs could be a valuable diagnostic and prognostic tool for OSCC.
5. Conclusions
Taken together, these studies suggest that EVs derived from cancer cells may modulate the function and may induce epigenetic changes in neighboring or distant cells. These biological effects are related to the delivery of transcripts that are specific of the originator cells. Several studies have shown a prominent role of exRNAs associated with vesicles. Since EVs may retain the molecular signature of the cell of origin, it has been suggested that they are a potential diagnostic exploitation. The salivary EV composition may reflect the presence of local or systemic diseases and has been investigated as a potential biomarker for both oral and non-oral cancers. Changes in the molecular composition of the EVs of non-oral cancers may either depend on their derivation from blood (since salivary glands are vascularized) or be the consequence of phenotypic changes occurring in gland cells (as the results of the stimulation by circulating cancer EVs). However, so far, available studies are relatively few and include a low number of patients. Further studies are necessary to optimize the protocol of EV isolation from saliva in order to obtain reproducible results. Moreover, the use of the EV content as a biomarker should take into account that this may be influenced by a number of cancer-associated risk factors, such as viral infections, smoking, alcohol abuse, as well as a number of non-cancer-associated factors related to concomitant diseases. However, these limitations in the use of EVs as biomarkers are not restricted to saliva, but may influence EVs derived from any biological fluid. Since saliva is an easily obtainable non-invasive bio-fluid particularly enriched in EVs, it may represent a new approach for cancer biomarker discovery. However, to define whether salivary EVs have a real clinical diagnostic and prognostic potential would require comparative studies between EVs derived from tumor cells, blood and saliva, which are not at present available.
Author Contributions
All authors equally contributed to the conceptualization of the article. The research of the pertinent literature was performed by M.C.D., G.C.; writing—original draft preparation, G.C.; review and editing, C.G., M.C.D., and G.C.
Funding
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC).
Conflicts of Interest
G.C. is a component of Scientific Advisory Board of Unicyte AG. The authors declare no conflict of interest.
References
- 1.Xi X., Li T., Huang Y., Sun J., Zhu Y., Yang Y., Lu Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. NonCoding RNA. 2017;3:9. doi: 10.3390/ncrna3010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan H., Lu H., Wang X., Jin H. MicroRNAs as Potential Biomarkers in Cancer: Opportunities and Challenges. Biomed. Res. Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Ledesma E., Verhaak R.G.W., Treviño V. Identification of a Multi-Cancer Gene Expression Biomarker for Cancer Clinical Outcomes Using a Network-Based Algorithm. Sci. Rep. 2015;5:11966. doi: 10.1038/srep11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network Comprehensive Molecular Portraits of Human Breast Tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuzick J., Swanson G.P., Fisher G., Brothman A.R., Berney D.M., Reid J.E., Mesher D., Speights V.O., Stankiewicz E., Foster C.S., et al. Prognostic Value of an RNA Expression Signature Derived from Cell Cycle Proliferation Genes in Patients with Prostate Cancer: A Retrospective Study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asmann Y.W., Necela B.M., Kalari K.R., Hossain A., Baker T.R., Carr J.M., Davis C., Getz J.E., Hostetter G., Li X., et al. Detection of Redundant Fusion Transcripts as Biomarkers or Disease-Specific Therapeutic Targets in Breast Cancer. Cancer Res. 2012;72:1921–1928. doi: 10.1158/0008-5472.CAN-11-3142. [DOI] [PubMed] [Google Scholar]
- 7.Attard G., Clark J., Ambroisine L., Fisher G., Kovacs G., Flohr P., Berney D., Foster C.S., Fletcher A., Gerald W.L., et al. Duplication of the Fusion of TMPRSS2 to ERG Sequences Identifies Fatal Human Prostate Cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guallar D., Wang J. RNA-Binding Proteins in Pluripotency, Differentiation, and Reprogramming. Front. Biol. (Beijing) 2014;9:389–409. doi: 10.1007/s11515-014-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quesenberry P.J., Aliotta J.M. Cellular Phenotype Switching and Microvesicles. Adv. Drug Deliv. Rev. 2010;62:1141–1148. doi: 10.1016/j.addr.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Liang H., Zhang J., Zen K., Zhang C.-Y. Secreted MicroRNAs: A New Form of Intercellular Communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Gallo A., Tandon M., Alevizos I., Illei G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocucci E., Meldolesi J. Ectosomes and Exosomes: Shedding the Confusion between Extracellular Vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Bobrie A., Colombo M., Krumeich S., Raposo G., Théry C. Diverse Subpopulations of Vesicles Secreted by Different Intracellular Mechanisms Are Present in Exosome Preparations Obtained by Differential Ultracentrifugation. J. Extracell. Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratajczak M.Z., Ratajczak J. Extracellular Microvesicles as Game Changers in Better Understanding the Complexity of Cellular Interactions-From Bench to Clinical Applications. Am. J. Med. Sci. 2017;354:449–452. doi: 10.1016/j.amjms.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratajczak J., Wysoczynski M., Hayek F., Janowska-Wieczorek A., Ratajczak M.Z. Membrane-Derived Microvesicles: Important and Underappreciated Mediators of Cell-to-Cell Communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 18.Kowal J., Tkach M., Théry C. Biogenesis and Secretion of Exosomes. Curr. Opin. Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Hyenne V., Apaydin A., Rodriguez D., Spiegelhalter C., Hoff-Yoessle S., Diem M., Tak S., Lefebvre O., Schwab Y., Goetz J.G., et al. RAL-1 Controls Multivesicular Body Biogenesis and Exosome Secretion. J. Cell Biol. 2015;211:27–37. doi: 10.1083/jcb.201504136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tricarico C., Clancy J., D’Souza-Schorey C. Biology and Biogenesis of Shed Microvesicles. Small Gtpases. 2017;8:220–232. doi: 10.1080/21541248.2016.1215283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hristov M., Erl W., Linder S., Weber P.C. Apoptotic Bodies from Endothelial Cells Enhance the Number and Initiate the Differentiation of Human Endothelial Progenitor Cells in Vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 22.Balaj L., Lessard R., Dai L., Cho Y.-J., Pomeroy S.L., Breakefield X.O., Skog J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lázaro-Ibáñez E., Sanz-Garcia A., Visakorpi T., Escobedo-Lucea C., Siljander P., Ayuso-Sacido A., Yliperttula M. Different GDNA Content in the Subpopulations of Prostate Cancer Extracellular Vesicles: Apoptotic Bodies, Microvesicles, and Exosomes. Prostate. 2014;74:1379–1390. doi: 10.1002/pros.22853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guescini M., Genedani S., Stocchi V., Agnati L.F. Astrocytes and Glioblastoma Cells Release Exosomes Carrying MtDNA. J. Neural Transm. (Vienna) 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 25.Sansone P., Savini C., Kurelac I., Chang Q., Amato L.B., Strillacci A., Stepanova A., Iommarini L., Mastroleo C., Daly L., et al. Packaging and Transfer of Mitochondrial DNA via Exosomes Regulate Escape from Dormancy in Hormonal Therapy-Resistant Breast Cancer. Proc. Natl. Acad. Sci. USA. 2017;114:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P., Ratajczak M.Z. Embryonic Stem Cell-Derived Microvesicles Reprogram Hematopoietic Progenitors: Evidence for Horizontal Transfer of MRNA and Protein Delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 27.Deregibus M.C., Cantaluppi V., Calogero R., Lo Iacono M., Tetta C., Biancone L., Bruno S., Bussolati B., Camussi G. Endothelial Progenitor Cell Derived Microvesicles Activate an Angiogenic Program in Endothelial Cells by a Horizontal Transfer of MRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 28.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 29.Fatima F., Nawaz M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA. 2017;3:10. doi: 10.3390/ncrna3010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Niel G., D’Angelo G., Raposo G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 31.Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. Syndecan-Syntenin-ALIX Regulates the Biogenesis of Exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 32.D’Souza-Schorey C., Chavrier P. ARF Proteins: Roles in Membrane Traffic and Beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 33.Nabhan J.F., Hu R., Oh R.S., Cohen S.N., Lu Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of Exosome Composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borràs F.E., Breakefield X., Budnik V., et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D.-K., Kang B., Kim O.Y., Choi D.-S., Lee J., Kim S.R., Go G., Yoon Y.J., Kim J.H., Jang S.C., et al. EVpedia: An Integrated Database of High-Throughput Data for Systemic Analyses of Extracellular Vesicles. J. Extracell. Vesicles. 2013;2:20384. doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathivanan S., Simpson R.J. ExoCarta: A Compendium of Exosomal Proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C.C.Y., Eaton S.A., Young P.E., Lee M., Shuttleworth R., Humphreys D.T., Grau G.E., Combes V., Bebawy M., Gong J., et al. Glioma Microvesicles Carry Selectively Packaged Coding and Non-Coding RNAs Which Alter Gene Expression in Recipient Cells. RNA Biol. 2013;10:1333–1344. doi: 10.4161/rna.25281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villarroya-Beltri C., Baixauli F., Gutiérrez-Vázquez C., Sánchez-Madrid F., Mittelbrunn M. Sorting It out: Regulation of Exosome Loading. Semin. Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quesenberry P.J., Aliotta J., Deregibus M.C., Camussi G. Role of Extracellular RNA-Carrying Vesicles in Cell Differentiation and Reprogramming. Stem Cell Res. 2015;6:153. doi: 10.1186/s13287-015-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guescini M., Guidolin D., Vallorani L., Casadei L., Gioacchini A.M., Tibollo P., Battistelli M., Falcieri E., Battistin L., Agnati L.F., et al. C2C12 Myoblasts Release Micro-Vesicles Containing MtDNA and Proteins Involved in Signal Transduction. Exp. Cell Res. 2010;316:1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Batagov A.O., Kurochkin I.V. Exosomes Secreted by Human Cells Transport Largely MRNA Fragments That Are Enriched in the 3’-Untranslated Regions. Biol. Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng L., Sun X., Scicluna B.J., Coleman B.M., Hill A.F. Characterization and Deep Sequencing Analysis of Exosomal and Non-Exosomal MiRNA in Human Urine. Kidney Int. 2014;86:433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 45.Crescitelli R., Lässer C., Szabó T.G., Kittel A., Eldh M., Dianzani I., Buzás E.I., Lötvall J. Distinct RNA Profiles in Subpopulations of Extracellular Vesicles: Apoptotic Bodies, Microvesicles and Exosomes. J. Extracell. Vesicles. 2013;2:20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill A.F., Pegtel D.M., Lambertz U., Leonardi T., O’Driscoll L., Pluchino S., Ter-Ovanesyan D., Nolte-’t Hoen E.N.M. ISEV Position Paper: Extracellular Vesicle RNA Analysis and Bioinformatics. J. Extracell. Vesicles. 2013;2:22859. doi: 10.3402/jev.v2i0.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M., et al. Characterization of Human Plasma-Derived Exosomal RNAs by Deep Sequencing. BMC Genom. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawa Y., Taketomi Y., Murakami M., Tsujimoto M., Yanoshita R. Small RNA Transcriptomes of Two Types of Exosomes in Human Whole Saliva Determined by next Generation Sequencing. Biol. Pharm. Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 49.Abels E.R., Breakefield X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., et al. Secreted Monocytic MiR-150 Enhances Targeted Endothelial Cell Migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Collino F., Deregibus M.C., Bruno S., Sterpone L., Aghemo G., Viltono L., Tetta C., Camussi G. Microvesicles Derived from Adult Human Bone Marrow and Tissue Specific Mesenchymal Stem Cells Shuttle Selected Pattern of MiRNAs. PLoS ONE. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldie B.J., Dun M.D., Lin M., Smith N.D., Verrills N.M., Dayas C.V., Cairns M.J. Activity-Associated MiRNA Are Packaged in Map1b-Enriched Exosomes Released from Depolarized Neurons. Nucleic Acids Res. 2014;42:9195–9208. doi: 10.1093/nar/gku594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iavello A., Frech V.S.L., Gai C., Deregibus M.C., Quesenberry P.J., Camussi G. Role of Alix in MiRNA Packaging during Extracellular Vesicle Biogenesis. Int. J. Mol. Med. 2016;37:958–966. doi: 10.3892/ijmm.2016.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melo S.A., Sugimoto H., O’Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenzie A.J., Hoshino D., Hong N.H., Cha D.J., Franklin J.L., Coffey R.J., Patton J.G., Weaver A.M. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cha D.J., Franklin J.L., Dou Y., Liu Q., Higginbotham J.N., Demory Beckler M., Weaver A.M., Vickers K., Prasad N., Levy S., et al. KRAS-Dependent Sorting of MiRNA to Exosomes. eLife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated HnRNPA2B1 Controls the Sorting of MiRNAs into Exosomes through Binding to Specific Motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R. Y-Box Protein 1 Is Required to Sort MicroRNAs into Exosomes in Cells and in a Cell-Free Reaction. ELife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corcoran C., Rani S., O’Brien K., O’Neill A., Prencipe M., Sheikh R., Webb G., McDermott R., Watson W., Crown J., et al. Docetaxel-Resistance in Prostate Cancer: Evaluating Associated Phenotypic Changes and Potential for Resistance Transfer via Exosomes. PLoS ONE. 2012;7:e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen W., Liu X., Lv M., Chen L., Zhao J., Zhong S., Ji M., Hu Q., Luo Z., Wu J., et al. Exosomes from Drug-Resistant Breast Cancer Cells Transmit Chemoresistance by a Horizontal Transfer of MicroRNAs. PLoS ONE. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crow J., Atay S., Banskota S., Artale B., Schmitt S., Godwin A.K. Exosomes as Mediators of Platinum Resistance in Ovarian Cancer. Oncotarget. 2017;8:11917–11936. doi: 10.18632/oncotarget.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin X., Yu S., Zhou L., Shi M., Hu Y., Xu X., Shen B., Liu S., Yan D., Feng J. Cisplatin-Resistant Lung Cancer Cell-Derived Exosomes Increase Cisplatin Resistance of Recipient Cells in Exosomal MiR-100-5p-Dependent Manner. Int. J. Nanomed. 2017;12:3721–3733. doi: 10.2147/IJN.S131516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao X., Yu S., Li S., Wu J., Ma R., Cao H., Zhu Y., Feng J. Exosomes: Decreased Sensitivity of Lung Cancer A549 Cells to Cisplatin. PLoS ONE. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawaz M., Shah N., Zanetti B.R., Maugeri M., Silvestre R.N., Fatima F., Neder L., Valadi H. Extracellular Vesicles and Matrix Remodeling Enzymes: The Emerging Roles in Extracellular Matrix Remodeling, Progression of Diseases and Tissue Repair. Cells. 2018;7:167. doi: 10.3390/cells7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sung B.H., Ketova T., Hoshino D., Zijlstra A., Weaver A.M. Directional Cell Movement through Tissues Is Controlled by Exosome Secretion. Nat. Commun. 2015;6:7164. doi: 10.1038/ncomms8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song W., Yan D., Wei T., Liu Q., Zhou X., Liu J. Tumor-Derived Extracellular Vesicles in Angiogenesis. Biomed. Pharm. 2018;102:1203–1208. doi: 10.1016/j.biopha.2018.03.148. [DOI] [PubMed] [Google Scholar]
- 67.Galindo-Hernandez O., Serna-Marquez N., Castillo-Sanchez R., Salazar E.P. Extracellular Vesicles from MDA-MB-231 Breast Cancer Cells Stimulated with Linoleic Acid Promote an EMT-like Process in MCF10A Cells. Prostaglandins Leukot. Essent. Fat. Acids. 2014;91:299–310. doi: 10.1016/j.plefa.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Franzen C.A., Blackwell R.H., Todorovic V., Greco K.A., Foreman K.E., Flanigan R.C., Kuo P.C., Gupta G.N. Urothelial Cells Undergo Epithelial-to-Mesenchymal Transition after Exposure to Muscle Invasive Bladder Cancer Exosomes. Oncogenesis. 2015;4:e163. doi: 10.1038/oncsis.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao D., Barry S., Kmetz D., Egger M., Pan J., Rai S.N., Qu J., McMasters K.M., Hao H. Melanoma Cell-Derived Exosomes Promote Epithelial-Mesenchymal Transition in Primary Melanocytes through Paracrine/Autocrine Signaling in the Tumor Microenvironment. Cancer Lett. 2016;376:318–327. doi: 10.1016/j.canlet.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Santi A., Caselli A., Ranaldi F., Paoli P., Mugnaioni C., Michelucci E., Cirri P. Cancer Associated Fibroblasts Transfer Lipids and Proteins to Cancer Cells through Cargo Vesicles Supporting Tumor Growth. Biochim. Biophys. Acta. 2015;1853:3211–3223. doi: 10.1016/j.bbamcr.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 71.Luga V., Zhang L., Viloria-Petit A.M., Ogunjimi A.A., Inanlou M.R., Chiu E., Buchanan M., Hosein A.N., Basik M., Wrana J.L. Exosomes Mediate Stromal Mobilization of Autocrine Wnt-PCP Signaling in Breast Cancer Cell Migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 72.Boelens M.C., Wu T.J., Nabet B.Y., Xu B., Qiu Y., Yoon T., Azzam D.J., Twyman-Saint Victor C., Wiemann B.Z., Ishwaran H., et al. Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards K.E., Zeleniak A.E., Fishel M.L., Wu J., Littlepage L.E., Hill R. Cancer-Associated Fibroblast Exosomes Regulate Survival and Proliferation of Pancreatic Cancer Cells. Oncogene. 2017;36:1770–1778. doi: 10.1038/onc.2016.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Au Yeung C.L., Co N.-N., Tsuruga T., Yeung T.-L., Kwan S.-Y., Leung C.S., Li Y., Lu E.S., Kwan K., Wong K.-K., et al. Exosomal Transfer of Stroma-Derived MiR21 Confers Paclitaxel Resistance in Ovarian Cancer Cells through Targeting APAF1. Nat. Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui H., Seubert B., Stahl E., Dietz H., Reuning U., Moreno-Leon L., Ilie M., Hofman P., Nagase H., Mari B., et al. Tissue Inhibitor of Metalloproteinases-1 Induces a pro-Tumourigenic Increase of MiR-210 in Lung Adenocarcinoma Cells and Their Exosomes. Oncogene. 2015;34:3640–3650. doi: 10.1038/onc.2014.300. [DOI] [PubMed] [Google Scholar]
- 76.Dang K., Myers K.A. The Role of Hypoxia-Induced MiR-210 in Cancer Progression. Int. J. Mol. Sci. 2015;16:6353–6372. doi: 10.3390/ijms16036353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao G., Liu Y., Fang X., Liu Y., Fang L., Lin L., Liu X., Wang N. Tumor-Derived MicroRNA-494 Promotes Angiogenesis in Non-Small Cell Lung Cancer. Angiogenesis. 2015;18:373–382. doi: 10.1007/s10456-015-9474-5. [DOI] [PubMed] [Google Scholar]
- 78.Matsuura Y., Wada H., Eguchi H., Gotoh K., Kobayashi S., Kinoshita M., Kubo M., Hayashi K., Iwagami Y., Yamada D., et al. Exosomal MiR-155 Derived from Hepatocellular Carcinoma Cells Under Hypoxia Promotes Angiogenesis in Endothelial Cells. Dig. Dis. Sci. 2019;64:792–802. doi: 10.1007/s10620-018-5380-1. [DOI] [PubMed] [Google Scholar]
- 79.Zhou X., Yan T., Huang C., Xu Z., Wang L., Jiang E., Wang H., Chen Y., Liu K., Shao Z., et al. Melanoma Cell-Secreted Exosomal MiR-155-5p Induce Proangiogenic Switch of Cancer-Associated Fibroblasts via SOCS1/JAK2/STAT3 Signaling Pathway. J. Exp. Clin. Cancer Res. 2018;37:242. doi: 10.1186/s13046-018-0911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. Pancreatic Cancer Exosomes Initiate Pre-Metastatic Niche Formation in the Liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grange C., Tapparo M., Collino F., Vitillo L., Damasco C., Deregibus M.C., Tetta C., Bussolati B., Camussi G. Microvesicles Released from Human Renal Cancer Stem Cells Stimulate Angiogenesis and Formation of Lung Premetastatic Niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 82.Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells toward a Pro-Metastatic Phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abd Elmageed Z.Y., Yang Y., Thomas R., Ranjan M., Mondal D., Moroz K., Fang Z., Rezk B.M., Moparty K., Sikka S.C., et al. Neoplastic Reprogramming of Patient-Derived Adipose Stem Cells by Prostate Cancer Cell-Associated Exosomes. Stem Cells. 2014;32:983–997. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., et al. MicroRNAs Bind to Toll-like Receptors to Induce Prometastatic Inflammatory Response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chow A., Zhou W., Liu L., Fong M.Y., Champer J., Van Haute D., Chin A.R., Ren X., Gugiu B.G., Meng Z., et al. Macrophage Immunomodulation by Breast Cancer-Derived Exosomes Requires Toll-like Receptor 2-Mediated Activation of NF-ΚB. Sci. Rep. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marton A., Vizler C., Kusz E., Temesfoi V., Szathmary Z., Nagy K., Szegletes Z., Varo G., Siklos L., Katona R.L., et al. Melanoma Cell-Derived Exosomes Alter Macrophage and Dendritic Cell Functions in Vitro. Immunol. Lett. 2012;148:34–38. doi: 10.1016/j.imlet.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 88.Grange C., Tapparo M., Tritta S., Deregibus M.C., Battaglia A., Gontero P., Frea B., Camussi G. Role of HLA-G and Extracellular Vesicles in Renal Cancer Stem Cell-Induced Inhibition of Dendritic Cell Differentiation. BMC Cancer. 2015;15:1009. doi: 10.1186/s12885-015-2025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.101 Ding G., Zhou L., Qian Y., Fu M., Chen J., Chen J., Xiang J., Wu Z., Jiang G., Cao L. Pancreatic Cancer-Derived Exosomes Transfer MiRNAs to Dendritic Cells and Inhibit RFXAP Expression via MiR-212-3p. Oncotarget. 2015;6:29877–29888. doi: 10.18632/oncotarget.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chalmin F., Ladoire S., Mignot G., Vincent J., Bruchard M., Remy-Martin J.-P., Boireau W., Rouleau A., Simon B., Lanneau D., et al. Membrane-Associated Hsp72 from Tumor-Derived Exosomes Mediates STAT3-Dependent Immunosuppressive Function of Mouse and Human Myeloid-Derived Suppressor Cells. J. Clin. Investig. 2010;120:457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valenti R., Huber V., Filipazzi P., Pilla L., Sovena G., Villa A., Corbelli A., Fais S., Parmiani G., Rivoltini L. Human Tumor-Released Microvesicles Promote the Differentiation of Myeloid Cells with Transforming Growth Factor-Beta-Mediated Suppressive Activity on T Lymphocytes. Cancer Res. 2006;66:9290–9298. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 92.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theodoraki M.-N., Yerneni S.S., Hoffmann T.K., Gooding W.E., Whiteside T.L. Clinical Significance of PD-L1+ Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. 2018;24:896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yen E.-Y., Miaw S.-C., Yu J.-S., Lai I.-R. Exosomal TGF-Β1 Is Correlated with Lymphatic Metastasis of Gastric Cancers. Am. J. Cancer Res. 2017;7:2199–2208. [PMC free article] [PubMed] [Google Scholar]
- 95.Abusamra A.J., Zhong Z., Zheng X., Li M., Ichim T.E., Chin J.L., Min W.-P. Tumor Exosomes Expressing Fas Ligand Mediate CD8+ T-Cell Apoptosis. Blood Cells Mol. Dis. 2005;35:169–173. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 96.Huber V., Fais S., Iero M., Lugini L., Canese P., Squarcina P., Zaccheddu A., Colone M., Arancia G., Gentile M., et al. Human Colorectal Cancer Cells Induce T-Cell Death through Release of Proapoptotic Microvesicles: Role in Immune Escape. Gastroenterology. 2005;128:1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 97.Ashiru O., Boutet P., Fernández-Messina L., Agüera-González S., Skepper J.N., Valés-Gómez M., Reyburn H.T. Natural Killer Cell Cytotoxicity Is Suppressed by Exposure to the Human NKG2D Ligand MICA*008 That Is Shed by Tumor Cells in Exosomes. Cancer Res. 2010;70:481–489. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu C., Yu S., Zinn K., Wang J., Zhang L., Jia Y., Kappes J.C., Barnes S., Kimberly R.P., Grizzle W.E., et al. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function. J. Immunol. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 99.Ludwig S., Floros T., Theodoraki M.-N., Hong C.-S., Jackson E.K., Lang S., Whiteside T.L. Suppression of Lymphocyte Functions by Plasma Exosomes Correlates with Disease Activity in Patients with Head and Neck Cancer. Clin. Cancer Res. 2017;23:4843–4854. doi: 10.1158/1078-0432.CCR-16-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Escudier B., Dorval T., Chaput N., André F., Caby M.-P., Novault S., Flament C., Leboulaire C., Borg C., Amigorena S., et al. Vaccination of Metastatic Melanoma Patients with Autologous Dendritic Cell (DC) Derived-Exosomes: Results of Thefirst Phase I Clinical Trial. J. Transl. Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morse M.A., Garst J., Osada T., Khan S., Hobeika A., Clay T.M., Valente N., Shreeniwas R., Sutton M.A., Delcayre A., et al. A Phase I Study of Dexosome Immunotherapy in Patients with Advanced Non-Small Cell Lung Cancer. J. Transl. Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor D.D., Gercel-Taylor C. MicroRNA Signatures of Tumor-Derived Exosomes as Diagnostic Biomarkers of Ovarian Cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 103.Rabinowits G., Gerçel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 104.Sohn W., Kim J., Kang S.H., Yang S.R., Cho J.-Y., Cho H.C., Shim S.G., Paik Y.-H. Serum Exosomal MicroRNAs as Novel Biomarkers for Hepatocellular Carcinoma. Exp. Mol. Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taylor D.D., Gercel-Taylor C. Exosomes/Microvesicles: Mediators of Cancer-Associated Immunosuppressive Microenvironments. Semin. Immunopathol. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 106.Jabalee J., Towle R., Garnis C. The Role of Extracellular Vesicles in Cancer: Cargo, Function, and Therapeutic Implications. Cells. 2018;7:93. doi: 10.3390/cells7080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bu N., Wu H., Sun B., Zhang G., Zhan S., Zhang R., Zhou L. Exosome-Loaded Dendritic Cells Elicit Tumor-Specific CD8+ Cytotoxic T Cells in Patients with Glioma. J. Neurooncol. 2011;104:659–667. doi: 10.1007/s11060-011-0537-1. [DOI] [PubMed] [Google Scholar]
- 108.Graner M.W., Alzate O., Dechkovskaia A.M., Keene J.D., Sampson J.H., Mitchell D.A., Bigner D.D. Proteomic and Immunologic Analyses of Brain Tumor Exosomes. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kunigelis K.E., Graner M.W. The Dichotomy of Tumor Exosomes (TEX) in Cancer Immunity: Is It All in the ConTEXt? Vaccines (Basel) 2015;3:1019–1051. doi: 10.3390/vaccines3041019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chevillet J.R., Kang Q., Ruf I.K., Briggs H.A., Vojtech L.N., Hughes S.M., Cheng H.H., Arroyo J.D., Meredith E.K., Gallichotte E.N., et al. Quantitative and Stoichiometric Analysis of the MicroRNA Content of Exosomes. Proc. Natl. Acad. Sci. USA. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anfossi S., Giordano A., Gao H., Cohen E.N., Tin S., Wu Q., Garza R.J., Debeb B.G., Alvarez R.H., Valero V., et al. High Serum MiR-19a Levels Are Associated with Inflammatory Breast Cancer and Are Predictive of Favorable Clinical Outcome in Patients with Metastatic HER2+ Inflammatory Breast Cancer. PLoS ONE. 2014;9:e83113. doi: 10.1371/journal.pone.0083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gemmell C.H., Sefton M.V., Yeo E.L. Platelet-Derived Microparticle Formation Involves Glycoprotein IIb-IIIa. Inhibition by RGDS and a Glanzmann’s Thrombasthenia Defect. J. Biol. Chem. 1993;268:14586–14589. [PubMed] [Google Scholar]
- 113.Yan W., Apweiler R., Balgley B.M., Boontheung P., Bundy J.L., Cargile B.J., Cole S., Fang X., Gonzalez-Begne M., Griffin T.J., et al. Systematic Comparison of the Human Saliva and Plasma Proteomes. Proteom. Clin. Appl. 2009;3:116–134. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bandhakavi S., Stone M.D., Onsongo G., Van Riper S.K., Griffin T.J. A Dynamic Range Compression and Three-Dimensional Peptide Fractionation Analysis Platform Expands Proteome Coverage and the Diagnostic Potential of Whole Saliva. J. Proteome Res. 2009;8:5590–5600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao M., Yang Y., Guo Z., Shao C., Sun H., Zhang Y., Sun Y., Liu Y., Song Y., Zhang L., et al. A Comparative Proteomics Analysis of Five Body Fluids: Plasma, Urine, Cerebrospinal Fluid, Amniotic Fluid, and Saliva. Proteom. Clin. Appl. 2018;12:e1800008. doi: 10.1002/prca.201800008. [DOI] [PubMed] [Google Scholar]
- 116.Omenn G.S., States D.J., Adamski M., Blackwell T.W., Menon R., Hermjakob H., Apweiler R., Haab B.B., Simpson R.J., Eddes J.S., et al. Overview of the HUPO Plasma Proteome Project: Results from the Pilot Phase with 35 Collaborating Laboratories and Multiple Analytical Groups, Generating a Core Dataset of 3020 Proteins and a Publicly-Available Database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 117.Cheng Y., Pereira M., Raukar N., Reagan J.L., Queseneberry M., Goldberg L., Borgovan T., LaFrance W.C., Dooner M., Deregibus M., et al. Potential Biomarkers to Detect Traumatic Brain Injury by the Profiling of Salivary Extracellular Vesicles. J. Cell. Physiol. 2019;234:14377–14388. doi: 10.1002/jcp.28139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saeedi S., Israel S., Nagy C., Turecki G. The Emerging Role of Exosomes in Mental Disorders. Transl. Psychiatry. 2019;9:122. doi: 10.1038/s41398-019-0459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao Z., Wu Y., Liu G., Jiang Y., Wang X., Wang Z., Feng T. α-Synuclein in Salivary Extracellular Vesicles as a Potential Biomarker of Parkinson’s Disease. Neurosci. Lett. 2019;696:114–120. doi: 10.1016/j.neulet.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 120.Jin Y., Guan Z., Wang X., Wang Z., Zeng R., Xu L., Cao P. ALA-PDT Promotes HPV-Positive Cervical Cancer Cells Apoptosis and DCs Maturation via MiR-34a Regulated HMGB1 Exosomes Secretion. Photodiagn. Photodyn. 2018;24:27–35. doi: 10.1016/j.pdpdt.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 121.Harden M.E., Munger K. Human Papillomavirus 16 E6 and E7 Oncoprotein Expression Alters MicroRNA Expression in Extracellular Vesicles. Virology. 2017;508:63–69. doi: 10.1016/j.virol.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Honegger A., Leitz J., Bulkescher J., Hoppe-Seyler K., Hoppe-Seyler F. Silencing of Human Papillomavirus (HPV) E6/E7 Oncogene Expression Affects Both the Contents and the Amounts of Extracellular Microvesicles Released from HPV-Positive Cancer Cells. Int. J. Cancer. 2013;133:1631–1642. doi: 10.1002/ijc.28164. [DOI] [PubMed] [Google Scholar]
- 123.Chiantore M.V., Mangino G., Iuliano M., Zangrillo M.S., De Lillis I., Vaccari G., Accardi R., Tommasino M., Columba Cabezas S., Federico M., et al. Human Papillomavirus E6 and E7 Oncoproteins Affect the Expression of Cancer-Related MicroRNAs: Additional Evidence in HPV-Induced Tumorigenesis. J. Cancer Res. Clin. Oncol. 2016;142:1751–1763. doi: 10.1007/s00432-016-2189-1. [DOI] [PubMed] [Google Scholar]
- 124.Peacock B., Rigby A., Bradford J., Pink R., Hunter K., Lambert D., Hunt S. Extracellular Vesicle MicroRNA Cargo Is Correlated with HPV Status in Oropharyngeal Carcinoma. J. Oral Pathol. Med. 2018;47:954–963. doi: 10.1111/jop.12781. [DOI] [PubMed] [Google Scholar]
- 125.Hukin J., Farrell K., MacWilliam L.M., Colbourne M., Waida E., Tan R., Mroz L., Thomas E. Case-Control Study of Primary Human Herpesvirus 6 Infection in Children with Febrile Seizures. Pediatrics. 1998;101:E3. doi: 10.1542/peds.101.2.e3. [DOI] [PubMed] [Google Scholar]
- 126.Sharma S., Gillespie B.M., Palanisamy V., Gimzewski J.K. Quantitative Nanostructural and Single-Molecule Force Spectroscopy Biomolecular Analysis of Human-Saliva-Derived Exosomes. Langmuir. 2011;27:14394–14400. doi: 10.1021/la2038763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zlotogorski-Hurvitz A., Dayan D., Chaushu G., Salo T., Vered M. Morphological and Molecular Features of Oral Fluid-Derived Exosomes: Oral Cancer Patients versus Healthy Individuals. J. Cancer Res. Clin. Oncol. 2016;142:101–110. doi: 10.1007/s00432-015-2005-3. [DOI] [PubMed] [Google Scholar]
- 128.Winck F.V., Prado Ribeiro A.C., Ramos Domingues R., Ling L.Y., Riaño-Pachón D.M., Rivera C., Brandão T.B., Gouvea A.F., Santos-Silva A.R., Coletta R.D., et al. Insights into Immune Responses in Oral Cancer through Proteomic Analysis of Saliva and Salivary Extracellular Vesicles. Sci. Rep. 2015;5:16305. doi: 10.1038/srep16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gai C., Camussi F., Broccoletti R., Gambino A., Cabras M., Molinaro L., Carossa S., Camussi G., Arduino P.G. Salivary Extracellular Vesicle-Associated MiRNAs as Potential Biomarkers in Oral Squamous Cell Carcinoma. BMC Cancer. 2018;18:439. doi: 10.1186/s12885-018-4364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun Y., Xia Z., Shang Z., Sun K., Niu X., Qian L., Fan L.-Y., Cao C.-X., Xiao H. Facile Preparation of Salivary Extracellular Vesicles for Cancer Proteom. Sci. Rep. 2016;6:24669. doi: 10.1038/srep24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun Y., Huo C., Qiao Z., Shang Z., Uzzaman A., Liu S., Jiang X., Fan L.-Y., Ji L., Guan X., et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J. Proteome Res. 2018;17:1101–1107. doi: 10.1021/acs.jproteome.7b00770. [DOI] [PubMed] [Google Scholar]
- 132.Langevin S., Kuhnell D., Parry T., Biesiada J., Huang S., Wise-Draper T., Casper K., Zhang X., Medvedovic M., Kasper S. Comprehensive MicroRNA-Sequencing of Exosomes Derived from Head and Neck Carcinoma Cells in Vitro Reveals Common Secretion Profiles and Potential Utility as Salivary Biomarkers. Oncotarget. 2017;8:82459–82474. doi: 10.18632/oncotarget.19614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Machida T., Tomofuji T., Maruyama T., Yoneda T., Ekuni D., Azuma T., Miyai H., Mizuno H., Kato H., Tsutsumi K., et al. MiR-1246 and MiR-4644 in Salivary Exosome as Potential Biomarkers for Pancreatobiliary Tract Cancer. Oncol. Rep. 2016;36:2375–2381. doi: 10.3892/or.2016.5021. [DOI] [PubMed] [Google Scholar]
- 134.Lau C., Kim Y., Chia D., Spielmann N., Eibl G., Elashoff D., Wei F., Lin Y.-L., Moro A., Grogan T., et al. Role of Pancreatic Cancer-Derived Exosomes in Salivary Biomarker Development. J. Biol. Chem. 2013;288:26888–26897. doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Momen-Heravi F., Balaj L., Alian S., Trachtenberg A.J., Hochberg F.H., Skog J., Kuo W.P. Impact of Biofluid Viscosity on Size and Sedimentation Efficiency of the Isolated Microvesicles. Front. Physiol. 2012;3:162. doi: 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jeppesen D.K., Hvam M.L., Primdahl-Bengtson B., Boysen A.T., Whitehead B., Dyrskjøt L., Orntoft T.F., Howard K.A., Ostenfeld M.S. Comparative Analysis of Discrete Exosome Fractions Obtained by Differential Centrifugation. J. Extracell. Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cvjetkovic A., Lötvall J., Lässer C. The Influence of Rotor Type and Centrifugation Time on the Yield and Purity of Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:23111. doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line LIM1863-Derived Exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 140.Müller G. Novel Tools for the Study of Cell Type-Specific Exosomes and Microvesicles. J. Bioanal. Biomed. 2012;4:46–60. doi: 10.4172/1948-593X.1000063. [DOI] [Google Scholar]
- 141.Taylor D.D., Lyons K.S., Gerçel-Taylor C. Shed Membrane Fragment-Associated Markers for Endometrial and Ovarian Cancers. Gynecol. Oncol. 2002;84:443–448. doi: 10.1006/gyno.2001.6551. [DOI] [PubMed] [Google Scholar]
- 142.Böing A.N., van der Pol E., Grootemaat A.E., Coumans F.A.W., Sturk A., Nieuwland R. Single-Step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J. Extracell. Vesicles. 2014;3:23430. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Taylor D.D., Shah S. Methods of Isolating Extracellular Vesicles Impact Down-Stream Analyses of Their Cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 144.Caby M.-P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like Vesicles Are Present in Human Blood Plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 145.Zarovni N., Corrado A., Guazzi P., Zocco D., Lari E., Radano G., Muhhina J., Fondelli C., Gavrilova J., Chiesi A. Integrated Isolation and Quantitative Analysis of Exosome Shuttled Proteins and Nucleic Acids Using Immunocapture Approaches. Methods. 2015;87:46–58. doi: 10.1016/j.ymeth.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 146.Szatanek R., Baran J., Siedlar M., Baj-Krzyworzeka M. Isolation of Extracellular Vesicles: Determining the Correct Approach (Review) Int. J. Mol. Med. 2015;36:11–17. doi: 10.3892/ijmm.2015.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Alvarez M.L., Khosroheidari M., Kanchi Ravi R., DiStefano J.K. Comparison of Protein, MicroRNA, and MRNA Yields Using Different Methods of Urinary Exosome Isolation for the Discovery of Kidney Disease Biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 148.Alvarez M.L. Isolation of Urinary Exosomes for RNA Biomarker Discovery Using a Simple, Fast, and Highly Scalable Method. Methods Mol. Biol. 2014;1182:145–170. doi: 10.1007/978-1-4939-1062-5_13. [DOI] [PubMed] [Google Scholar]
- 149.Rekker K., Saare M., Roost A.M., Kubo A.-L., Zarovni N., Chiesi A., Salumets A., Peters M. Comparison of Serum Exosome Isolation Methods for MicroRNA Profiling. Clin. Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 150.Zlotogorski-Hurvitz A., Dayan D., Chaushu G., Korvala J., Salo T., Sormunen R., Vered M. Human Saliva-Derived Exosomes: Comparing Methods of Isolation. J. Histochem. Cytochem. 2015;63:181–189. doi: 10.1369/0022155414564219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kanchi Ravi R., Khosroheidari M., DiStefano J.K. A Modified Precipitation Method to Isolate Urinary Exosomes. J. Vis. Exp. 2015;95:e51158. doi: 10.3791/51158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Witwer K.W., Buzás E.I., Bemis L.T., Bora A., Lässer C., Lötvall J., Nolte-’t Hoen E.N., Piper M.G., Sivaraman S., Skog J., et al. Standardization of Sample Collection, Isolation and Analysis Methods in Extracellular Vesicle Research. J. Extracell. Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yuana Y., Levels J., Grootemaat A., Sturk A., Nieuwland R. Co-Isolation of Extracellular Vesicles and High-Density Lipoproteins Using Density Gradient Ultracentrifugation. J. Extracell. Vesicles. 2014;3:23262. doi: 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wei F., Lin C.-C., Joon A., Feng Z., Troche G., Lira M.E., Chia D., Mao M., Ho C.-L., Su W.-C., et al. Noninvasive Saliva-Based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014;190:1117–1126. doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pu D., Liang H., Wei F., Akin D., Feng Z., Yan Q., Li Y., Zhen Y., Xu L., Dong G., et al. Evaluation of a Novel Saliva-Based Epidermal Growth Factor Receptor Mutation Detection for Lung Cancer: A Pilot Study. Thorac. Cancer. 2016;7:428–436. doi: 10.1111/1759-7714.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei F., Yang J., Wong D.T.W. Detection of Exosomal Biomarker by Electric Field-Induced Release and Measurement (EFIRM) Biosens. Bioelectron. 2013;44:115–121. doi: 10.1016/j.bios.2012.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheng J., Nonaka T., Wong D.T.W. Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials (Basel) 2019;12:654. doi: 10.3390/ma12040654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Michael A., Bajracharya S.D., Yuen P.S.T., Zhou H., Star R.A., Illei G.G., Alevizos I. Exosomes from Human Saliva as a Source of MicroRNA Biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Deregibus M.C., Figliolini F., D’Antico S., Manzini P.M., Pasquino C., De Lena M., Tetta C., Brizzi M.F., Camussi G. Charge-Based Precipitation of Extracellular Vesicles. Int. J. Mol. Med. 2016;38:1359–1366. doi: 10.3892/ijmm.2016.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Y., St John M.A.R., Zhou X., Kim Y., Sinha U., Jordan R.C.K., Eisele D., Abemayor E., Elashoff D., Park N.-H., et al. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin. Cancer Res. 2004;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 161.Zhou Y., Kolokythas A., Schwartz J.L., Epstein J.B., Adami G.R. MicroRNA from Brush Biopsy to Characterize Oral Squamous Cell Carcinoma Epithelium. Cancer Med. 2017;6:67–78. doi: 10.1002/cam4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yap T., Vella L.J., Seers C., Nastri A., Reynolds E., Cirillo N., McCullough M. Oral Swirl Samples—A Robust Source of MicroRNA Protected by Extracellular Vesicles. Oral Dis. 2017;23:312–317. doi: 10.1111/odi.12603. [DOI] [PubMed] [Google Scholar]
- 163.Mestdagh P., Van Vlierberghe P., De Weer A., Muth D., Westermann F., Speleman F., Vandesompele J. A Novel and Universal Method for MicroRNA RT-QPCR Data Normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kaczor-Urbanowicz K.E., Martin Carreras-Presas C., Aro K., Tu M., Garcia-Godoy F., Wong D.T. Saliva Diagnostics-Current Views and Directions. Exp. Biol. Med. (Maywood) 2017;242:459–472. doi: 10.1177/1535370216681550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang L., Farrell J.J., Zhou H., Elashoff D., Akin D., Park N.-H., Chia D., Wong D.T. Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology. 2010;138:e1–e7. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee Y.-H., Kim J.H., Zhou H., Kim B.W., Wong D.T. Salivary Transcriptomic Biomarkers for Detection of Ovarian Cancer: For Serous Papillary Adenocarcinoma. J. Mol. Med. 2012;90:427–434. doi: 10.1007/s00109-011-0829-0. [DOI] [PubMed] [Google Scholar]
- 167.Xiao H., Zhang L., Zhou H., Lee J.M., Garon E.B., Wong D.T.W. Proteomic Analysis of Human Saliva from Lung Cancer Patients Using Two-Dimensional Difference Gel Electrophoresis and Mass Spectrometry. Mol. Cell Proteom. 2012;11:M111.012112. doi: 10.1074/mcp.M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ogawa Y., Tsujimoto M., Yanoshita R. Next-Generation Sequencing of Protein-Coding and Long Non-Protein-Coding RNAs in Two Types of Exosomes Derived from Human Whole Saliva. Biol. Pharm. Bull. 2016;39:1496–1507. doi: 10.1248/bpb.b16-00297. [DOI] [PubMed] [Google Scholar]
- 169.Palanisamy V., Sharma S., Deshpande A., Zhou H., Gimzewski J., Wong D.T. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS ONE. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lau C.S., Wong D.T.W. Breast Cancer Exosome-like Microvesicles and Salivary Gland Cells Interplay Alters Salivary Gland Cell-Derived Exosome-like Microvesicles in Vitro. PLoS ONE. 2012;7:e33037. doi: 10.1371/journal.pone.0033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Katsiougiannis S., Chia D., Kim Y., Singh R.P., Wong D.T.W. Saliva Exosomes from Pancreatic Tumor-Bearing Mice Modulate NK Cell Phenotype and Antitumor Cytotoxicity. FASEB J. 2017;31:998–1010. doi: 10.1096/fj.201600984R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Serban K.A., Rezania S., Petrusca D.N., Poirier C., Cao D., Justice M.J., Patel M., Tsvetkova I., Kamocki K., Mikosz A., et al. Structural and Functional Characterization of Endothelial Microparticles Released by Cigarette Smoke. Sci. Rep. 2016;6:31596. doi: 10.1038/srep31596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kodidela S., Wang Y., Patters B.J., Gong Y., Sinha N., Ranjit S., Gerth K., Haque S., Cory T., McArthur C., et al. Proteomic Profiling of Exosomes Derived from Plasma of HIV-Infected Alcohol Drinkers and Cigarette Smokers. J. Neuroimmune Pharmacol. 2019:1–19. doi: 10.1007/s11481-019-09853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Appert-Collin A., Hubert P., Crémel G., Bennasroune A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front. Pharm. 2015;6:283. doi: 10.3389/fphar.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li X., Sun R., Geng X., Wang S., Zen D., Pei J., Yang J., Fan Y., Jiang H., Yang P., et al. A Comprehensive Analysis of Candidate Gene Signatures in Oral Squamous Cell Carcinoma. Neoplasma. 2017;64:167–174. doi: 10.4149/neo_2017_201. [DOI] [PubMed] [Google Scholar]
- 176.Ohnishi Y., Yasui H., Kakudo K., Nozaki M. Lapatinib-Resistant Cancer Cells Possessing Epithelial Cancer Stem Cell Properties Develop Sensitivity during Sphere Formation by Activation of the ErbB/AKT/Cyclin D2 Pathway. Oncol. Rep. 2016;36:3058–3064. doi: 10.3892/or.2016.5073. [DOI] [PubMed] [Google Scholar]
- 177.Li S.-X., Yang Y.-Q., Jin L.-J., Cai Z.-G., Sun Z. Detection of Survivin, Carcinoembryonic Antigen and ErbB2 Level in Oral Squamous Cell Carcinoma Patients. Cancer Biomark. 2016;17:377–382. doi: 10.3233/CBM-160651. [DOI] [PubMed] [Google Scholar]
- 178.Dong C., Ye D.-X., Zhang W.-B., Pan H.-Y., Zhang Z.-Y., Zhang L. Overexpression of C-Fos Promotes Cell Invasion and Migration via CD44 Pathway in Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. 2015;44:353–360. doi: 10.1111/jop.12296. [DOI] [PubMed] [Google Scholar]
- 179.Judd N.P., Winkler A.E., Murillo-Sauca O., Brotman J.J., Law J.H., Lewis J.S., Dunn G.P., Bui J.D., Sunwoo J.B., Uppaluri R. ERK1/2 Regulation of CD44 Modulates Oral Cancer Aggressiveness. Cancer Res. 2012;72:365–374. doi: 10.1158/0008-5472.CAN-11-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ghuwalewala S., Ghatak D., Das P., Dey S., Sarkar S., Alam N., Panda C.K., Roychoudhury S. CD44(High)CD24(Low) Molecular Signature Determines the Cancer Stem Cell and EMT Phenotype in Oral Squamous Cell Carcinoma. Stem Cell Res. 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 181.Meng W., Xia Q., Wu L., Chen S., He X., Zhang L., Gao Q., Zhou H. Downregulation of TGF-Beta Receptor Types II and III in Oral Squamous Cell Carcinoma and Oral Carcinoma-Associated Fibroblasts. BMC Cancer. 2011;11:88. doi: 10.1186/1471-2407-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Momen-Heravi F., Bala S. Extracellular vesicles in oral squamous carcinoma carry oncogenic miRNA profile and reprogram monocytes via NF-κB pathway. Oncotarget. 2018;9:34838–34854. doi: 10.18632/oncotarget.26208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Arantes L.M.R.B., De Carvalho A.C., Melendez M.E., Lopes Carvalho A. Serum, Plasma and Saliva Biomarkers for Head and Neck Cancer. Expert Rev. Mol. Diagn. 2018;18:85–112. doi: 10.1080/14737159.2017.1404906. [DOI] [PubMed] [Google Scholar]
- 184.Tang H., Wu Z., Zhang J., Su B. Salivary LncRNA as a Potential Marker for Oral Squamous Cell Carcinoma Diagnosis. Mol. Med. Rep. 2013;7:761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]