ABSTRACT
Salamanders have been hailed as champions of regeneration, exhibiting a remarkable ability to regrow tissues, organs and even whole body parts, e.g. their limbs. As such, salamanders have provided key insights into the mechanisms by which cells, tissues and organs sense and regenerate missing or damaged parts. In this Primer, we cover the evolutionary context in which salamanders emerged. We outline the varieties of mechanisms deployed during salamander regeneration, and discuss how these mechanisms are currently being explored and how they have advanced our understanding of animal regeneration. We also present arguments about why it is important to study closely related species in regeneration research.
KEY WORDS: CNS, Axolotl, Genome, Limb, Model organism, Newt
Summary: This Primer provides an overview of salamanders as a model system for studying regeneration, outlining the mechanisms deployed during salamander regeneration and highlighting how the study of salamanders has increased our understanding of regeneration more broadly.
Introduction
During the Devonian period, around 400 million years ago, the first tetrapods began exploring land. During this period, they radiated into several branches, including two types of amphibians: frogs (anurans – without tail) and salamanders (urodeles – tail showing). While all amphibians exhibit regenerative capacities, some of these are more regenerative than others. Indeed, compared with their tailless peers, salamanders excel in regenerating damaged or lost body parts throughout their entire life. In fact, among tetrapods, salamanders exhibit the widest range of regenerative capacity, with an impressive ability to regrow tissues, organs and entire body parts (Tanaka, 2003; Yun, 2015).
The histories of both regeneration and developmental biology are rooted in the question of reproduction: how is a new organism, or part of an organism, formed? During the Enlightenment, two opposing views attempted to explain how animals develop: preformation and epigenesis. Was animal development a matter of growth from a preformed miniature version (a germ) or a matter of forces that assembled simpler units to gradually generate more- complex organisms (Dinsmore, 1995)? Within the context of this debate, Lazzaro Spallanzani (1729-1799) investigated salamander regeneration. By amputating salamander tails and limbs, Spallanzani built on three lines of inquiry. The first was based on the ancient observation that lizards can regrow their tails, a phenomenon that gained renewed interest in the late 17th century. Second, the earlier works of French naturalist René-Antoine Ferchault de Réaumur (1683-1757) had demonstrated reproducible regeneration of crustacean appendages (crayfish claws). The third line of inquiry was a rush of experiments that systematically bisected Hydra, sliced worms and severed snail heads to explore the prevalence of regeneration among animals. The father of this tradition was Abraham Trembley (1710-1784) who, by means of (in retrospect) a faulty hypothesis, cut Hydra into two to see whether they would regrow as plants or die as animals. However, the value of this experiment – arguably the foundation of experimental biology – was not in classifying Hydra as flora or fauna; rather it posed a formidable challenge to both preformation and epigenesis, as either theory of generation had to be reconciled with regeneration. Taking the challenge to the extreme, Spallanzani investigated regeneration in more-complex animals and departed from experiments on simpler invertebrates to actual tetrapods that resembled human anatomy (Dinsmore, 1996). Here, the challenge was acute: how is a new limb regenerated in a vertebrate with an anatomy similar to our own? The questions provoked by salamander regeneration have since been refined to address the regeneration-specific mechanisms involved in sensing which cells, tissues, organs or entire appendages are missing in a mature body, and in triggering the appropriate regenerative response to recreate the original structure.
In this Primer, we provide an overview of salamanders as a model for the study of regeneration. We discuss the life cycle (Fig. 1), the genomic and experimental accessibility of different species, as well as their regenerative capabilities. We also present arguments for why it is important to study several types of salamander in regeneration research, including closely related species. Finally, we outline the variety of mechanisms deployed during salamander regeneration, highlight how these mechanisms are currently being investigated and how their study is informing us more broadly about regenerative mechanisms and capabilities.
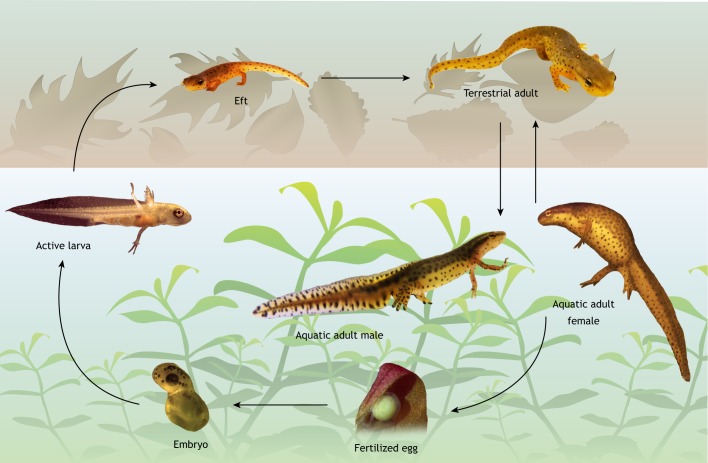
Fig. 1.
Salamanders display complex life cycles in both terrestrial and aquatic habitats. A typical salamander life cycle (exemplified here by that of Notophthalmus viridescens) involves both terrestrial and aquatic stages. Adult newts alternate facultative aquatic/terrestrial lifestyles, but they mate and lay fertilized eggs in the water. These eggs then develop into embryos that hatch as aquatic larvae. The larvae are ferocious zooplankton hunters that undergo metamorphosis prior to leaving the aquatic milieu and becoming terrestrial juveniles (termed efts), which seasonally return to water to breed after they reach sexual maturity. Many salamander species are entirely land living without an aquatic larval stage.
Salamander species: variations on a theme
The regenerative capabilities of salamanders
All salamanders demonstrate the potential to regenerate complex structures: they can regrow, among other parts, entire limbs, a tail, ocular tissues, substantial parts of their central nervous system and heart (Joven and Simon, 2018; Tanaka, 2016). Importantly, by studying various species, differences among salamanders – with regard to both their regenerative potential and their regenerative mechanisms – have been discovered.
Model systems for regeneration.
This article is part of a series entitled ‘Model systems for regeneration’. This series of articles aims to highlight key model systems and species that are currently being used to study tissue and organ regeneration. Each article provides background information about the phylogenetic position of the species, its life-cycle and habitat, the different organs and tissues that regenerate, and the experimental tools and techniques that are available for studying these organisms in a regenerative context. Importantly, these articles also give examples of how the study of these models has increased our understanding of regenerative mechanisms more broadly, and how some of the open questions in the field of regeneration may be answered using these organisms. To see the full collection as it grows, please visit: https://dev.biologists.org/collection/regeneration_models
The salamander species used most often in regeneration research are the axolotl (Ambystoma mexicanum) and three species of newts (Notophthalmus viridescens, Eastern red-spotted newt; Cynops phyrrogaster, Japanese fire-belly newt; and Pleurodeles waltl, Iberian ribbed newt). These animals have similar, although not completely overlapping, natural regeneration capacities (Fig. 2). For example, newts regenerate more body parts than axolotls. This is exemplified by regeneration of the eye lens throughout the entire life-span of newts. Lens regeneration in axolotls occurs during the first 2 weeks after hatching but is lost thereafter (Sousounis et al., 2014). In Cynops, however, lens regeneration does not decline with age nor with the number of lens removal/regeneration cycles (Eguchi et al., 2011). Another distinctive feature of newt regeneration is the prominence of injury-evoked reversal of the terminally differentiated state. For example, lens regeneration in newts is dependent on iris pigmented epithelial cells that dedifferentiate and proliferate, and the subsequent transdifferentiation of a subset of these cells into a new lens (Eguchi et al., 2011; Sousounis et al., 2014). A similarly radical manifestation of in vivo reprogramming as a response to injury also occurs during newt limb regeneration when postmitotic, multinucleated muscle cells break up into mononucleate progeny, which subsequently re-enter the cell cycle and contribute to the new appendage. In contrast, the axolotl does not exhibit muscle dedifferentiation during limb regeneration; instead, new muscle fibres seem to be entirely derived from the activation of a resident Pax7-expressing stem cell population (Fei et al., 2017; Sandoval-Guzmán et al., 2014; Wang and Simon, 2016). Whether these differences between newts and axolotl in lens regeneration capacity and in myogenic dedifferentiation reflect a higher degree of cell plasticity in newts remains to be determined. Similarly, the extent to which stem cells contribute to newt regeneration in general is not clearly understood. Importantly, the reprogramming of cells derived from mature tissues towards an undifferentiated state does play a role in axolotl regeneration. Recent work profiling connective tissue during limb regeneration has demonstrated that these heterogeneous cells transit to an embryonic-like state that is more homogenous in the blastema (the cell mass that gives rise to the new limb), before redifferentiating to build the new limb (Gerber et al., 2018) (discussed below).
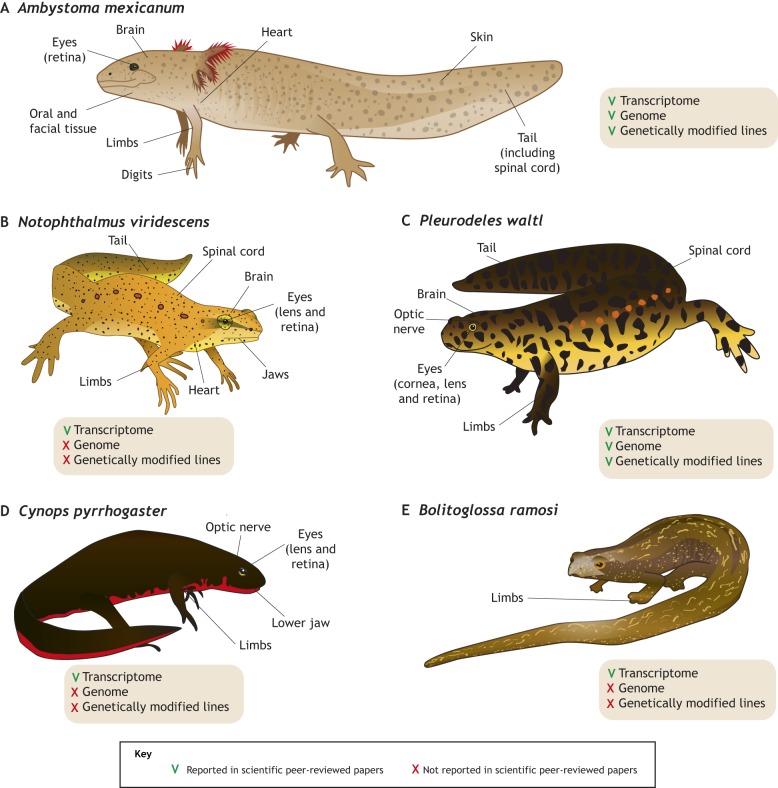
Fig. 2.
Salamander species in regeneration research. Both established and emerging species are shown, highlighting regenerative organs/tissues and major resources available for each species. (A) The Mexican axolotl, Ambystoma mexicanum, is a paedomorphic salamander that retains fully aquatic features throughout its entire life cycle. The axolotl is easy to breed under laboratory conditions, and is the most commonly used salamander model organism in regeneration research, mostly owing to the availability of several genetically modified lines (Tanaka, 2016). (B) The Eastern red spotted newt, Notophthalmus viridescens, has contributed significantly to our understanding of multiple regeneration processes with reference transcriptomes available. Genetically modified lines are difficult to establish due to its long generation time and complex life cycle (Abdullayev et al., 2013; Looso et al., 2013). (C) The Iberian ribbed newt, Pleurodeles waltl, is a highly regenerative, emerging model species. It can be maintained in a fully aquatic habitat throughout its entire life cycle and has a similar generation time to the axolotl. Transcriptomes and genome assemblies are now available, as well as genetically modified lines (Elewa et al., 2017; Hayashi and Takeuchi, 2015; Hayashi et al., 2013; Joven et al., 2015, 2018). (D) The Japanese fire-bellied newt, Cynops pyrrhogaster, has been used to study eye, limb, jaw and brain regeneration. A transcriptome focusing on lens and neural retina regeneration has been assembled (Casco-Robles et al., 2016; Kurosaka et al., 2008; Nakamura et al., 2014). (E) The plethodontid Bolitoglossa ramosi is a fully terrestrial, direct developer (no larval stage) for which a limb regeneration transcriptome has been reported (Arenas Gomez et al., 2017, 2018).
Life cycle and experimental accessibility
There are also considerable differences in the life cycles of salamanders (Fig. 1). The life cycle of newts recapitulates the evolutionary conquest of land. Fertilized eggs are laid in water, where embryos develop and hatch, starting their life as aquatic larvae. They usually become terrestrial after metamorphosis, and return to water as adults to produce the next generation (Fig. 1). Among other salamanders, there are several variations in this process, with some species exhibiting viviparous (Salamandra salamandra), fully aquatic (Ambystoma mexicanum) or fully terrestrial (Plethodontidae) life cycles (Bonett and Blair, 2017; Griffiths, 1995).
Notophthalmus and Cynops have very complex life cycles, with both aquatic and terrestrial phases, which makes them cumbersome to breed under laboratory conditions. Even if possible, their long generation time (more than 2 years) restricts efficient production of genetically modified lines. On the other hand, the axolotl is a fully aquatic paedomorphic animal, meaning that it retains larval features, such as external gills, throughout its entire life span. Axolotls are therefore easy to maintain in laboratory conditions and to breed in captivity, as they provide offspring in a season-independent manner (Khattak et al., 2014). The axolotl has therefore emerged as the prime salamander model for regeneration studies. This has also been aided by the feasibility of germline transgenesis in axolotls, which has enabled germline mutagenesis and Cre-loxP reporter-mediated lineage tracking (Bryant et al., 2017; Fei et al., 2018, 2017; Flowers et al., 2017; Leigh et al., 2018; Nowoshilow et al., 2018). In addition, although the axolotl genome is gigantic (32 Gb, discussed below), it is now assembled and annotated with impressive contiguity (Nowoshilow et al., 2018; Smith et al., 2019). While these technological advantages (transgenesis and genomic resources) have been a major driving force for focusing on the axolotl, a conceptual consideration is whether it is truly possible to examine the ‘adult mode’ of regeneration in axolotls, given their paedomorphic nature. Indeed, it may be possible that larval animals are more prone to reactivate developmental programs than post-metamorphic adults. In fact, recent single cell RNAseq (scRNA-seq) profiling of the axolotl regenerating limb demonstrated that the majority of the blastema reverts to a limb bud-like transcriptional profile (Gerber et al., 2018). However, reservations about how adult the axolotl is should be treated with care. Rather than classifying animals as either ‘adult’ or ‘growing’, one should instead determine the actual constraints for each experimental paradigm in relation to the question under investigation. For example, even if the axolotl is paedomorphic, its limbs have all the structural elements found in a fully metamorphosed salamander. Conversely, just because newts undergo metamorphosis, it does not necessarily mean that they would lose all embryonic features as adults.
Recently, substantial efforts have been made to establish a newt model species that is amenable to genetic manipulations on par with the axolotl. The Iberian ribbed newt (Pleurodeles waltl) fulfils all necessary criteria: these animals are easy to breed in the laboratory because they do not require a terrestrial habitat after metamorphosis, they have a generation time similar to the axolotl (of 9-12 months) and they possess the same regeneration spectrum as other newts (Chevallier et al., 2004; Joven et al., 2015; Tassava et al., 1993; Urata et al., 2018). Moreover, a reference transcriptome and preliminary genome assembly are now available for Pleurodeles, as well as several genetically modified lines, enabling functional studies and reporter lineage tracking (Elewa et al., 2017; Hayashi and Takeuchi, 2016; Joven et al., 2018). Together, the axolotl and Pleurodeles offer two accessible systems in which genome editing can be performed to interrogate the roles of specific genes, to initiate cell-type specific lineage tracing, and to construct genome assemblies that enable gene expression and chromatin landscape studies.
Salamander genomes
Salamander genomes are vast, ranging between 14 and 120 Gb (Brockes, 2015), and their sheer size has delayed their characterization. Why salamander genomes became gigantic is a matter of discussion but recent sequencing data have shed some light on this matter (Sun and Mueller, 2014; Elewa et al., 2017; Nowoshilow et al., 2018). These data have revealed that a disproportionate expansion of repetitive sequences – predominantly long terminal repeat (LTR) retrotransposons – contributes significantly to salamander genome gigantism. Repeated elements are often located in introns whose median size in the axolotl is on average an order of magnitude longer than introns in the human genome. In addition, intergenic regions in the axolotl genome are an order of magnitude longer than those in other vertebrates. An exception from this rule is the HoxA gene cluster: despite the general increase in intron length, the sizes of introns in the axolotl HoxA locus are very similar to those in other vertebrates (Nowoshilow et al., 2018).
In the Pleurodeles genome, Gypsy LTR retrotransposons are the most frequent repetitive elements followed by the Harbinger transposons, which together account for about two-thirds of the genome's repetitive content. Harbinger elements are rare in vertebrate genomes, and their expansion in the Pleurodeles genome is unique. In addition, Harbinger elements are expressed after injury and during limb regeneration (Elewa et al., 2017). It will be interesting to determine whether this expansion and post-injury expression is a general feature of salamanders, restricted to newts or possibly only to Pleurodeles. Another intriguing feature of Pleurodeles is the presence of over 100 copies of the microRNA gene mir-427. This gene is also found in multiple copies in the genomes of Xenopus (Tang and Maxwell, 2008) and zebrafish (Chen et al., 2005), where it is known as mir-430. In Xenopus and zebrafish, miR-427 functions during the maternal to zygotic transition, mediating the degradation of inherited maternal mRNAs to clear out parental epigenetic instructions (Lund et al., 2009; Giraldez et al., 2006). While miR-427 expression and function was thought to be limited to early embryogenesis, its expression in adult newts after injury suggests that a similar mRNA clearance event might occur during newt regeneration, perhaps as a component of cellular reprogramming. Indeed, the mammalian counterpart of miR-427 (miR-302) has been used to reprogram fibroblasts into induced pluripotent cells (Anokye-Danso et al., 2011).
Analyses of salamander genomes have also provided clues about the genes that function during regenerative processes. As noted in the Introduction, development and regeneration are two tightly interlinked processes. For example, genes responsible for patterning and morphogenesis are re-activated during limb regeneration, although their precise regulation is not a complete recapitulation of embryonic development (Stocum, 2017). Although major signalling components of the Wnt and Hedgehog signalling pathways are present in the axolotl, a surprising finding was that Pax3 is absent in the axolotl genome (Nowoshilow et al., 2018). Loss-of-function experiments in axolotls, using TALEN and CRISPR/Cas9 genome editing, indicate that the paralogue Pax7 takes on the role that Pax3 performs in other vertebrates, as Pax7 axolotl mutants have major developmental abnormalities and lack limb muscle (Nowoshilow et al., 2018). In sharp contrast, the Pleurodeles genome harbours both Pax3 and Pax7. Loss of Pax3 in Pleurodeles leads to severe abnormalities, including skeletal muscle agenesis, again as occurs in other vertebrates (Elewa et al., 2017). Despite Pax7 being absolutely essential for successful skeletal muscle regeneration in mammals (Kuang et al., 2006), Pax7 loss of function in Pleurodeles does not cause any major regeneration phenotype. This finding might indicate that, in the absence of Pax7, skeletal muscle regeneration is fuelled by dedifferentiation of myofibres in Pleurodeles (Elewa et al., 2017).
In summary, salamanders have finally entered the post-genomic era, following the sequencing of two salamander genomes and with a growing toolbox for cell type-specific molecular interrogation. The technological developments made over the past decade have made both axolotl and newts amenable to the molecular interrogation of regeneration mechanisms. These advances now allow for systematic cross-species comparisons among salamanders, as well as between salamanders and less regenerative tetrapods.
Insights from studying regeneration in salamanders
Mechanisms of limb regeneration
The speed of salamander limb regeneration varies among species and developmental stage, but is nevertheless impressive. The best staging, based on meticulous histological series, is available for Notophthalmus, which completes adult limb regeneration within less than 2 months (Iten and Bryant, 1973) (Fig. 3). During this event, peripheral nerves retract after amputation and then regrow into the blastema, secreting factors necessary for regeneration to progress (Kumar et al., 2007). Another key step that follows amputation is the formation of a wound epidermis covering the injured site (Tassava et al., 1993). Immune cells also populate the area and become activated, and systemic depletion of macrophages during an early, sensitive period of the regeneration event results in wound closure but permanent failure of limb regeneration (Godwin et al., 2013). Underneath the wound epidermis, stump cells begin to populate the blastema. By the second week after amputation, the blastema has grown noticeably, and by the third week the initial stages of an elbow bend and a flattening hand palette can be discerned. Thus, 1 month after amputation, a salamander limb can regenerate its complex features; it then spends an additional month growing back to its original size (Iten and Bryant, 1973).
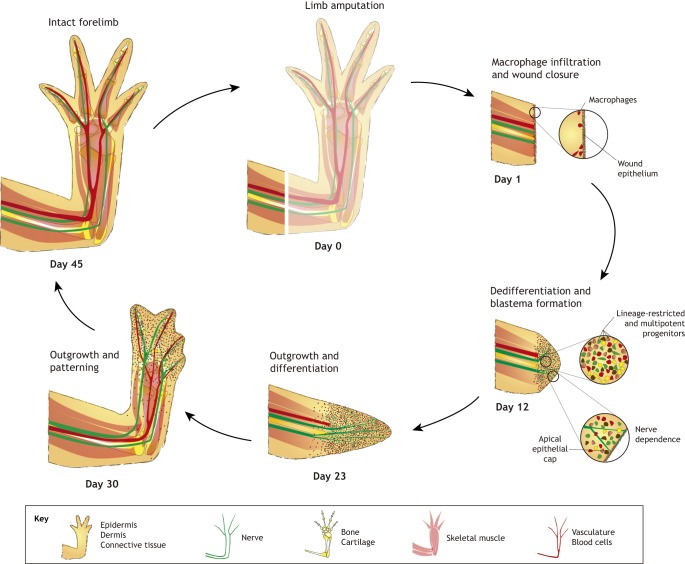
Fig. 3.
Key processes during limb regeneration. The salamander limb contains all typical structural elements of tetrapods. Upon amputation, salamander limb regeneration starts by scar-free wound healing and wound closure. Infiltrating macrophages are essential for this event, probably for clearing debris, although other signalling mechanisms cannot be excluded. Cells in the mature limb then undergo reprogramming/dedifferentiation to form a blastema. The degree of reprogramming varies between cell types and species. Nerve-derived factors are required for subsequent blastema cell proliferation and outgrowth. The cells also retain positional memory during the regeneration event, allowing them to undergo the appropriate patterning to re-from an intact limb. The time course of regeneration indicated in this figure is based on staging in adult Notophthalmus viridescens.
Notably, a small or limited wound to a salamander limb does not induce outgrowth. Instead, complete amputation, or a wound that covers the entire circumference, is necessary for limb regrowth. This means that the salamander body can compute the severity of trauma and distinguish between a minor injury and amputation. Additionally, severed nerves at the site of amputation are necessary for blastema cell proliferation, as denervation prevents blastema growth and consequently limb regeneration (Farkas and Monaghan, 2017). In the event of an amputation, only the missing part of the limb will regrow, which is the region distal to the wound site (Stocum, 2017). In other words, an amputation through the upper arm will lead to regeneration of a limb from elbow to hand, while an amputation through the lower arm will not regenerate a second upper elbow but only the more-distal structures (the wrist and hand). Grafting experiments have demonstrated that the proximo-distal positional identity, and the resulting fate of the regenerated tissue, reside in the blastema (Tanaka, 2016). Thus, four key features of salamander regeneration are: (1) distinction between minor injury and amputation; (2) immune cell infiltration; (3) nerve dependence; and (4) positional memory.
Molecular studies have identified links between nerve dependence and positional memory, notably between positional cues along both the proximo-distal and anterior-posterior axis. For example, it has been shown that limbs can regrow in the absence of nerves upon forced expression of the gene anterior gradient (AG). AG is a ligand of the cell surface receptor, Prod1, the overexpression of which confers blastemal cells a proximal identity (Kumar et al., 2007). The crucial role of nerves in limb regeneration, and their link to positional cues, was also demonstrated in the accessory limb model. In this experimental paradigm, a lateral wound to the anterior side of a limb can form a blastema if the peripheral nerves are deviated to the wound site (Endo et al., 2015). However, this accessory blastema eventually regresses unless a piece of skin from the posterior side of the limb is grafted to the anterior wound site. This juxtaposition of anterior and posterior cells then allows limb outgrowth from the accessory blastema. This may reflect the juxtaposition that occurs upon amputation: as the blastema grows, the flat transection of an amputation site becomes a dome-shaped protrusion, at the tip of which cells from distant regions (e.g. posterior and anterior) become neighbours. Studies have also revealed that posteriorly localized Hedgehog signalling supports anterior expression of FGF8, and that sustained FGF signalling is a key factor for persistent blastema cell proliferation (Nacu et al., 2016; Satoh et al., 2016; Singh et al., 2012). How and by which cells FGF and Hedgehog signalling are translated into positional values during limb regeneration remains uncertain (Bryant and Gardiner, 2018). It is possible that the relative levels of gene expression in neighbouring cells contribute to positional values. This model would suggest that, in a normal limb, the disparity of values in adjacent cells is minimal, forming gradients along the dorsal/ventral, proximo-distal and anterior/posterior axes. After amputation, however, when anterior and posterior cells are juxtaposed, a disparity between cells will arise and could stimulate proliferation to populate the gap with cells that reinstate the positional gradient. Such gap filling is termed intercalation and has been proposed as an integral part of regeneration in several species (Brockes and Kumar, 2008).
Finally, recent scRNA-seq studies have increased the resolution by which we can study limb regeneration and have offered insights into immune cell participation, wound epidermis signalling and the extent to which cells revert to an embryonic-like state (Gerber et al., 2018; Leigh et al., 2018). Using unbiased profiling and clustering of over 25,000 cells, Leigh and colleagues described the variety of immune cells that localize to the wound site and infiltrate the developing blastema. Their data elucidate different classes of innate and adaptive immunity cells including CD4+ regulatory T cells (TRegs), which have been implicated in muscle regeneration (Burzyn et al., 2013), and spinal cord, heart and retina regeneration (Hui et al., 2017) in zebrafish. Leigh et al. also described the heterogeneity of wound epidermis cells during axolotl limb homeostasis and regeneration, and identified markers for ionocytes, Langerhans cells, apical, intermediate and basal epidermis and small secretory cells. Of note, they revealed that a small population of putative Leydig cells appears during wound healing and that, during this stage, the intermediate epidermis and small secretory cells express anterior gradient protein 2 a (agr2a; a homolog of newt AG). After the completion of wound healing, the basal epidermis also expresses agr2a. Their pseudotime analysis also identified a trajectory for wound epidermis differentiation in which basal epidermal cells provide a reservoir of progenitor cells that connect the basal epidermis to outer small secretory cells via the layer of intermediate epidermis (Leigh et al., 2018). These findings support a view whereby the wound epidermis extends its homeostatic function to respond to injury without dedifferentiating and reverting to an embryonic state. In contrast, by selectively profiling lineage-traced connective tissue, Gerber and colleagues showed that connective tissue heterogeneity is temporarily lost as cells turn into a homogenous population that resembles embryonic limb-bud cells. Earlier studies identified connective tissue, which gives rise to cartilage, bone, tendons, periskeleton and dermal and interstitial fibroblasts, as the major contributor to the blastema during limb regeneration (Muneoka et al., 1986). Moreover, connective tissue cells have been identified as the cells that retain positional memory, supporting the model that they are major driving forces for limb regeneration (Bryant and Gardiner, 2018). The mechanisms by which the cells that make up connective tissue retain the memory of their cellular identity and their location along developmental axes as they dedifferentiate and respond to wound cues is a puzzle waiting to be solved.
Regeneration of neural tissues: regrowing, integrating and restoring function
Adult salamanders can regenerate various damaged neural tissues, including retinae, brain regions and the spinal cord, both in terms of structure and function (Joven and Simon, 2018; Lust and Tanaka, 2019). Damage to the central nervous system (CNS) usually affects both neuronal and non-neuronal cell types and, depending on the extent of damage, may lead to behavioural abnormalities. In order to restore function, both missing cells and damaged connections need to be regenerated. Salamanders perform remarkably well in these tasks, and do so using a variety of processes, as outlined below.
A common injury model used for regeneration studies is extirpation of the neural retina. In this model, the retinal pigmented epithelium (RPE) is detached from the photoreceptor cell layer. As a response, RPE cells start proliferating and give rise to all cell types of the neural retina (Fig. 4) (Grigoryan and Markitantova, 2016). In the case of lentectomy (lens removal, Fig. 4), pigment cells are the principal source of a new lens: in this case, also referred to as Wolffian regeneration, fully differentiated pigmented epithelial cells dedifferentiate and proliferate, but only the cells that originated from the dorsal iris transdifferentiate into a new lens (Eguchi et al., 2011). Although lens regeneration is not affected by repeated removal or by ageing in newts, regenerative capacity is lost in the axolotl 2 weeks after hatching (Eguchi et al., 2011; Henry and Hamilton, 2018; Sousounis et al., 2014). Taking advantage of this age-dependent regeneration, Sousounis and colleagues used microarrays to identify genes that are differentially expressed before and after this crucial transition, revealing a correlation between the ontogeny of immunity and the onset of differentiation with loss of regenerative ability (Sousounis et al., 2014). However, it is important to remember that immune cells infiltrate the axolotl regenerating limb and that macrophages are necessary for blastema formation (Godwin et al., 2013). If indeed the ontogeny of immunity results in loss of lens regenerative capacity, an important question is how can immune cells be refractive to lens regeneration but essential for limb regeneration in the same animal species?
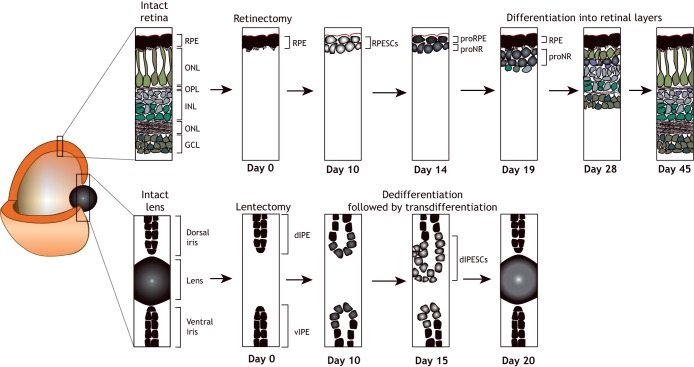
Fig. 4.
Regeneration of ocular tissues. (Top) Following retinectomy (detachment of the RPE from the photoreceptor cell layer), a new pigmented cell layer appears first. This is followed by the formation of neuro-retinal cell types in an order that recapitulates development: ganglion cells form first, followed by amacrine cells, horizontal cells and Müller glia. (Bottom) The lens can regenerate following lentectomy (lens removal), via the dedifferentiation and subsequent transdifferentiation of pigmented epithelial cells of the dorsal iris. Newts retain lens regeneration ability throughout adulthood, unlike axolotls, in which the ability to regenerate the lens is lost 2 weeks after hatching. dIPE, dorsal iris pigmented epithelium; dIPESCs, dorsal iris pigmented epithelium cells; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; OPL, outer plexiform layer; proNR, inner rudimentary layer; proRE, retinal pigmented epithelium rudimentary layer; RPE, retinal pigmented epithelium; RPESCs, retinal pigmented epithelium stem cells; vIPE, ventral iris pigmented epithelium.
Salamanders are also able to regenerate their spinal cord following injury (Diaz Quiroz and Echeverri, 2013; Tazaki et al., 2017; Joven and Simon, 2018). As such, and in contrast to mammals, spinal cord trauma in salamanders leads to only a transient loss of locomotion (Butler and Ward, 1967; Chevallier et al., 2004; Davis et al., 1990). After spinal cord transection in salamanders, a process of wound healing at each side of the injury leads to restoration of the central canal, followed by the production of new neurons and axons. The damaged axons then regrow through permissive channels formed by the extensions of ependymoglial cells (which are the counterparts of radial glial cells in mammals) that line the central canal, allowing for rewiring of the damaged circuitry (reviewed by Joven and Simon, 2018). Tail amputation in salamanders also results in the formation of a new spinal cord, and this experimental paradigm has been instrumental for the discovery of key processes and molecules implicated during spinal cord regeneration (for recent reviews, see Diaz Quiroz and Echeverri, 2013; Tazaki et al., 2017).
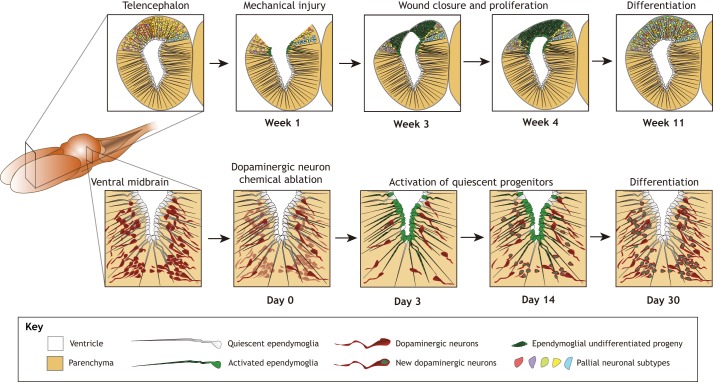
The most complex part of the CNS – the brain – can also regenerate in salamanders. The overall cytoarchitecture of the brain, with its multiple neuronal subpopulations arranged in spatially defined domains, is shared among all vertebrates, including salamanders, even though the salamander brain has undergone secondary simplification during evolution (reviewed by Joven and Simon, 2018). Two types of injury models have demonstrated significant restorative processes in the salamander brain, both in terms of tissue regeneration as well as behavioural recovery. For example, some models have removed parts of the telencephalon or dorsal midbrain, showing that this is followed by wound closure, massive proliferation, re-appearance of neuronal diversity and the formation of new inter-neuronal connexions (Fig. 5). Although salamanders do much better than mammals, careful analyses, including axonal tracing, have identified that this process of regeneration is not always an entirely faithful recapitulation of the original structure (Amamoto et al., 2016; Fujisawa, 1981; Maden et al., 2013; Minelli et al., 1987; Okamoto et al., 2007; Urata et al., 2018). By contrast, other types of injury models have addressed how individual neuronal subpopulations regenerate following intracranial injection of toxins that eliminate specific neuronal subtypes (Fig. 5). Lineage tracing in these studies has revealed that ependymoglial cells constitute the principal source for new neurons. These experiments also revealed a crucial role for neurotransmitter signalling in ependymoglia proliferation and neurogenesis in a region-specific manner (Berg et al., 2010, 2011; Joven et al., 2018; Kirkham et al., 2014). Such chemical ablations also showed remarkable recovery of locomotor performance (Parish et al., 2007). Furthermore, based on associative learning, decision making and fear behaviour assays, these studies showed that ontogenetic encoding of stereotyped behaviours is conserved between salamanders and mammals (Joven et al., 2018). Hence, brain regeneration studies in salamanders are feasible to consider in a cross-species comparative setting, which is important for testing and translating findings in mammals. As a proof of this principle, it was possible, based on studies in newts, to enhance dopamine-mediated neurogenesis in the mouse midbrain (Hedlund et al., 2016).
Fig. 5.
Brain regeneration in salamanders. (Top) Brain regeneration following injury (e.g. unilateral forebrain extirpation) presumably occurs by activation of ependymoglial cells. Neuronal diversity is restored but projections are not a faithful replication of the original. The time frame shown here is based on studies in the axolotl. (Bottom) Studies on red-spotted newts have showed that dopaminergic and cholinergic (not shown) neurons regenerate in several brain regions after the selective ablation of individual neuronal subtypes. Injury-responsive neurogenesis is fuelled by reactivation of quiescent resident neuronal progenitor cells, the ependymoglial cells, which are the equivalent of glial cells in mammals.
Conclusions and perspectives
Model organisms such as yeast, C. elegans and Drosophila are amenable to large-scale mutagenesis screens, which give rise to phenotypes of interest that can be traced back to the gene of origin. This power of genetics, along with the art of genetic screens, has been the main engine driving discovery in developmental biology during the 20th century. Salamanders do not offer such an approach to discovering molecular mechanisms, and this perhaps contributed to their transient decline as a research model. A case in point: Thomas Hunt Morgan studied regeneration in several organisms, including salamanders (Sunderland, 2010), but his decision to fully invest his efforts into Drosophila genetics during the latter part of his career captures the direction of developmental biology during the past century. However, in lieu of mutagenesis, comparative genomics now allow us to contrast different responses to injury and to identify gene expression signatures that correlate with efficient regeneration. Correlation can then inform functional studies that can determine genetic causation. Furthermore, harnessing the power of diversity and developing an art for comparative studies will be crucial for homing in on the key components enabling regeneration.
Studies of blind Mexican cave fish inspire a standard that salamander regeneration studies should be able to reach. The cave fish species Astyanax mexicanus diverged over several million years ago into populations of fish that remained in lakes and others that invaded underground environments and became confined to caves (Gross, 2012). Stockdale and colleagues demonstrated that surface fish and a number of cave fish respond differently to cardiac injury (Stockdale et al., 2018). Using quantitative trait locus (QTL) analysis, they linked the degree of cardiac regeneration to three loci in the genome, thereby identifying candidate genes fundamental to the regulation of heart regeneration. One may argue that QTL analysis is not feasible with gigantic salamander genomes, or that all salamanders regenerate and an example as clear as Mexican cave fish is untenable. However, we have highlighted above cases that establish the variety of regenerative capacities among only a few salamanders. The wealth of salamander species in the Amazons and the Appalachian Mountains (Kozak, 2017; Vences and Wake, 2007), which include cave dwelling salamanders, is an untapped resource that could help unlock the mechanisms behind such fantastic regenerative abilities where they do exist. In addition, the continuous drop in sequencing costs, including that of long-read technologies such as PacBio and Nanopore, which are essential to assembling salamander genomes, and the versatility of CRISPR/Cas9 for genome editing mean that salamanders are no longer subject to technical challenges for quantitative molecular research. Finally, salamander researchers ought to revive the lost tradition of amphibian cloning via nuclear transfer (Gurdon, 1960) to accelerate the generation of isogenic transgenic animals as a means to more-efficient functional studies, and also to offer an additional system for studying cellular reprograming (Jullien et al., 2011). Although not all salamanders are paedomorphic, research-wise, they are all late bloomers and can flourish in a research environment capable of harnessing the power of diversity. Their heyday is here.
Acknowledgements
We apologize to the authors whose work we could not cite due to limitations in the number of references.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The author's research is funded by Cancerfonden, by the Vetenskapsrådet, by Hjärnfonden, by the Knut och Alice Wallenbergs Stiftelse and by Stiftelsen Olle Engkvist Byggmästare to A.S., and by a National Institutes of Health Ruth Kirschstein Postdoctoral Fellowship (F32GM117806 to A.E.). Deposited in PMC for release after 12 months.
References
- Abdullayev I., Kirkham M., Björklund A. K., Simon A. and Sandberg R. (2013). A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp. Cell Res. 319, 1187-1197. 10.1016/j.yexcr.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Amamoto R., Huerta V. G., Takahashi E., Dai G., Grant A. K., Fu Z. and Arlotta P. (2016). Adult axolotls can regenerate original neuronal diversity in response to brain injury. Elife 5, e13998 10.7554/eLife.13998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P. J., Epstein J. A. et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376-388. 10.1016/j.stem.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas Gomez C. M., Gomez Molina A., Zapata J. D. and Delgado J. P. (2017). Limb regeneration in a direct-developing terrestrial salamander, Bolitoglossa ramosi (Caudata: Plethodontidae): limb regeneration in plethodontid salamanders. Regeneration (Oxf) 4, 227-235. 10.1002/reg2.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas Gomez C. M., Woodcock R. M., Smith J. J., Voss R. S. and Delgado J. P. (2018). Using transcriptomics to enable a plethodontid salamander (Bolitoglossa ramosi) for limb regeneration research. BMC Genomics 19, 704 10.1186/s12864-018-5076-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. A., Kirkham M., Beljajeva A., Knapp D., Habermann B., Ryge J., Tanaka E. M. and Simon A. (2010). Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development 137, 4127-4134. 10.1242/dev.055541 [DOI] [PubMed] [Google Scholar]
- Berg D. A., Kirkham M., Wang H., Frisén J. and Simon A. (2011). Dopamine controls neurogenesis in the adult salamander midbrain in homeostasis and during regeneration of dopamine neurons. Cell Stem Cell 8, 426-433. 10.1016/j.stem.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Bonett R. M. and Blair A. L. (2017). Evidence for complex life cycle constraints on salamander body form diversification. Proc. Natl. Acad. Sci. USA 114, 9936-9941. 10.1073/pnas.1703877114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes J. P. (2015). Variation in salamanders: an essay on genomes, development, and evolution. Methods Mol. Biol. 1290, 3-15. 10.1007/978-1-4939-2495-0_1 [DOI] [PubMed] [Google Scholar]
- Brockes J. P. and Kumar A. (2008). Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 24, 525-549. 10.1146/annurev.cellbio.24.110707.175336 [DOI] [PubMed] [Google Scholar]
- Bryant S. V. and Gardiner D. M. (2018). Regeneration: sooner rather than later. Int. J. Dev. Biol. 62, 363-368. 10.1387/ijdb.170269dg [DOI] [PubMed] [Google Scholar]
- Bryant D. M., Sousounis K., Farkas J. E., Bryant S., Thao N., Guzikowski A. R., Monaghan J. R., Levin M. and Whited J. L. (2017). Repeated removal of developing limb buds permanently reduces appendage size in the highly-regenerative axolotl. Dev. Biol. 424, 1-9. 10.1016/j.ydbio.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J. L., Cerletti M., Jang Y., Sefik E., Tan T. G., Wagers A. J., Benoist C. et al. (2013). A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282-1295. 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler E. G. and Ward M. B. (1967). Reconstitution of the spinal cord after ablation in adult Triturus. Dev. Biol. 15, 464-486. 10.1016/0012-1606(67)90038-3 [DOI] [PubMed] [Google Scholar]
- Casco-Robles M. M., Islam M. R., Inami W., Tanaka H. V., Kunahong A., Yasumuro H., Hanzawa S., Casco-Robles R. M., Toyama F., Maruo F. et al. (2016). Turning the fate of reprogramming cells from retinal disorder to regeneration by Pax6 in newts. Sci. Rep. 6, 33761 10.1038/srep33761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. Y., Manninga H., Slanchev K., Chien M., Russo J. J., Ju J., Sheridan R., John B., Marks D. S., Gaidatzis D. et al. (2005). The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 19, 1288-1293. 10.1101/gad.1310605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier S., Landry M., Nagy F. and Cabelguen J.-M. (2004). Recovery of bimodal locomotion in the spinal-transected salamander, Pleurodeles waltlii. Eur. J. Neurosci. 20, 1995-2007. 10.1111/j.1460-9568.2004.03671.x [DOI] [PubMed] [Google Scholar]
- Davis B. M., Ayers J. L., Koran L., Carlson J., Anderson M. C. and Simpson S. B. Jr. (1990). Time course of salamander spinal cord regeneration and recovery of swimming: HRP retrograde pathway tracing and kinematic analysis. Exp. Neurol. 108, 198-213. 10.1016/0014-4886(90)90124-B [DOI] [PubMed] [Google Scholar]
- Diaz Quiroz J. F. and Echeverri K. (2013). Spinal cord regeneration: where fish, frogs and salamanders lead the way, can we follow? Biochem. J. 451, 353-364. 10.1042/BJ20121807 [DOI] [PubMed] [Google Scholar]
- Dinsmore C. E. (1995). Animal regeneration: from fact to concept. Bioscience 45, 484-492. 10.2307/1312792 [DOI] [Google Scholar]
- Dinsmore C. E. (1996). Urodele limb and tail regeneration in early biological thought: an essay on scientific controversy and social change. Int. J. Dev. Biol. 40, 621-627. [PubMed] [Google Scholar]
- Echeverri K., Clarke J. D. and Tanaka E. M. (2001). In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol. 236, 151-164. 10.1006/dbio.2001.0312 [DOI] [PubMed] [Google Scholar]
- Eguchi G., Eguchi Y., Nakamura K., Yadav M. C., Millán J. L. and Tsonis P. A. (2011). Regenerative capacity in newts is not altered by repeated regeneration and ageing. Nat. Commun. 2, 384 10.1038/ncomms1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elewa A., Wang H., Talavera-López C., Joven A., Brito G., Kumar A., Hameed L. S., Penrad-Mobayed M., Yao Z., Zamani N. et al. (2017). Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 8, 2286 10.1038/s41467-017-01964-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Gardiner D. M., Makanae A. and Satoh A. (2015). The accessory limb model: an alternative experimental system of limb regeneration. Methods Mol. Biol. 1290, 101-113. 10.1007/978-1-4939-2495-0_8 [DOI] [PubMed] [Google Scholar]
- Farkas J. E. and Monaghan J. R. (2017). A brief history of the study of nerve dependent regeneration. Neurogenesis 4, e1302216-e1302216 10.1080/23262133.2017.1302216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J.-F., Schuez M., Knapp D., Taniguchi Y., Drechsel D. N. and Tanaka E. M. (2017). Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proc. Natl. Acad. Sci. USA 114, 12501-12506. 10.1073/pnas.1706855114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J.-F., Lou W.-P., Knapp D., Murawala P., Gerber T., Taniguchi Y., Nowoshilow S., Khattak S. and Tanaka E. M. (2018). Application and optimization of CRISPR-Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum). Nat. Protoc. 13, 2908-2943. 10.1038/s41596-018-0071-0 [DOI] [PubMed] [Google Scholar]
- Flowers G. P., Sanor L. D. and Crews C. M. (2017). Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. Elife 6, e25726 10.7554/eLife.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H. (1981). Retinotopic analysis of fiber pathways in the regenerating retinotectal system of the adult newt cynops Pyrrhogaster. Brain Res. 206, 27-37. 10.1016/0006-8993(81)90098-6 [DOI] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S. et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J. and Schier A. F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75-79. 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Godwin J. W., Pinto A. R. and Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 110, 9415-9420. 10.1073/pnas.1300290110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. (1995). Newts and Salamanders of Europe. London: T & AD Poyser natural history. [Google Scholar]
- Grigoryan E. N. and Markitantova Y. V. (2016). Cellular and molecular preconditions for retinal pigment epithelium (RPE) natural reprogramming during retinal regeneration in urodela. Biomedicines 4, 28 10.3390/biomedicines4040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B. (2012). The complex origin of Astyanax cavefish. BMC Evol. Biol. 12, 105 10.1186/1471-2148-12-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J. B. (1960). The developmental capacity of nuclei taken from differentiating endoderm cells of Xenopus laevis. J. Embryol. Exp. Morphol. 8, 505-526. [PubMed] [Google Scholar]
- Hayashi T. and Takeuchi T. (2015). Gene manipulation for regenerative studies using the Iberian ribbed newt, Pleurodeles waltl. Methods Mol. Biol. 1290, 297-305. 10.1007/978-1-4939-2495-0_23 [DOI] [PubMed] [Google Scholar]
- Hayashi T. and Takeuchi T. (2016). Mutagenesis in newts: protocol for iberian ribbed newts. Methods Mol. Biol. 1338, 119-126. 10.1007/978-1-4939-2932-0_10 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Yokotani N., Tane S., Matsumoto A., Myouga A., Okamoto M. and Takeuchi T. (2013). Molecular genetic system for regenerative studies using newts. Dev. Growth Differ. 55, 229-236. 10.1111/dgd.12019 [DOI] [PubMed] [Google Scholar]
- Hedlund E., Belnoue L., Theofilopoulos S., Salto C., Bye C., Parish C., Deng Q., Kadkhodaei B., Ericson J., Arenas E. et al. (2016). Dopamine receptor antagonists enhance proliferation and neurogenesis of midbrain Lmx1a-expressing progenitors. Sci. Rep. 6, 26448 10.1038/srep26448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J. J. and Hamilton P. W. (2018). Diverse evolutionary origins and mechanisms of lens regeneration. Mol. Biol. Evol. 35, 1563-1575. 10.1093/molbev/msy045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S. P., Sheng D. Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D. and Kikuchi K. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659-672.e655. 10.1016/j.devcel.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Iten L. E. and Bryant S. V. (1973). Forelimb regeneration from different levels of amputation in the newt, Notophthalmus viridescens: length, rate, and stages. Wilhelm Roux Arch. Entwickl. Mech. Org. 173, 263-282. 10.1007/BF00575834 [DOI] [PubMed] [Google Scholar]
- Joven A. and Simon A. (2018). Homeostatic and regenerative neurogenesis in salamanders. Prog. Neurobiol. 170, 81-98. 10.1016/j.pneurobio.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Joven A., Kirkham M. and Simon A. (2015). Husbandry of Spanish ribbed newts (Pleurodeles waltl). Methods Mol. Biol. 1290, 47-70. 10.1007/978-1-4939-2495-0_4 [DOI] [PubMed] [Google Scholar]
- Joven A., Wang H., Pinheiro T., Hameed L. S., Belnoue L. and Simon A. (2018). Cellular basis of brain maturation and acquisition of complex behaviors in salamanders. Development 145, dev160051 10.1242/dev.160051 [DOI] [PubMed] [Google Scholar]
- Jullien J., Pasque V., Halley-Stott R. P., Miyamoto K. and Gurdon J. B. (2011). Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat. Rev. Mol. Cell Biol. 12, 453-459. 10.1038/nrm3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S., Murawala P., Andreas H., Kappert V., Schuez M., Sandoval-Guzmán T., Crawford K. and Tanaka E. M. (2014). Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat. Protoc. 9, 529-540. 10.1038/nprot.2014.040 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Hameed L. S., Berg D. A., Wang H. and Simon A. (2014). Progenitor cell dynamics in the Newt Telencephalon during homeostasis and neuronal regeneration. Stem Cell Rep. 2, 507-519. 10.1016/j.stemcr.2014.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak K. H. (2017). What drives variation in plethodontid salamander species richness over space and time? Herpetologica 73, 220-228. 10.1655/HERPETOLOGICA-D-16-00085.1 [DOI] [Google Scholar]
- Kuang S., Charge S. B., Seale P., Huh M. and Rudnicki M. A. (2006). Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 172, 103-113. 10.1083/jcb.200508001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A. and Brockes J. P. (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772-777. 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka H., Takano-Yamamoto T., Yamashiro T. and Agata K. (2008). Comparison of molecular and cellular events during lower jaw regeneration of newt (Cynops pyrrhogaster) and West African clawed frog (Xenopus tropicalis). Dev. Dyn. 237, 354-365. 10.1002/dvdy.21419 [DOI] [PubMed] [Google Scholar]
- Leigh N. D., Dunlap G. S., Johnson K., Mariano R., Oshiro R., Wong A. Y., Bryant D. M., Miller B. M., Ratner A., Chen A. et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9, 5153 10.1038/s41467-018-07604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looso M., Preussner J., Sousounis K., Bruckskotten M., Michel C. S., Lignelli E., Reinhardt R., Höffner S., Krüger M., Tsonis P. A. et al. (2013). A de novo assembly of the newt transcriptome combined with proteomic validation identifies new protein families expressed during tissue regeneration. Genome Biol. 14, R16 10.1186/gb-2013-14-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Liu M., Hartley R. S., Sheets M. D. and Dahlberg J. E. (2009). Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA (New York, N.Y.) 15, 2351-2363. 10.1261/rna.1882009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lust K. and Tanaka E. M. (2019). A comparative perspective on brain regeneration in amphibians and teleost fish. Dev. Neurobiol. 10.1002/dneu.22665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M., Manwell L. A. and Ormerod B. K. (2013). Proliferation zones in the axolotl brain and regeneration of the telencephalon. Neural Dev. 8, 1 10.1186/1749-8104-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli G., Franceschini V., Del Grande P. and Ciani F. (1987). Newly-formed neurons in the regenerating optic tectum of Triturus cristatus carnifex. Basic Appl. Histochem. 31, 43-52. [PubMed] [Google Scholar]
- Muneoka K., Fox W. F. and Bryant S. V. (1986). Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 116, 256-260. 10.1016/0012-1606(86)90062-X [DOI] [PubMed] [Google Scholar]
- Nacu E., Gromberg E., Oliveira C. R., Drechsel D. and Tanaka E. M. (2016). FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407-410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Islam M. R., Takayanagi M., Yasumuro H., Inami W., Kunahong A., Casco-Robles R. M., Toyama F. and Chiba C. (2014). A transcriptome for the study of early processes of retinal regeneration in the adult newt, Cynops pyrrhogaster. PLoS ONE 9, e109831 10.1371/journal.pone.0109831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowoshilow S., Schloissnig S., Fei J.-F., Dahl A., Pang A. W. C., Pippel M., Winkler S., Hastie A. R., Young G., Roscito J. G. et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50-55. 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Ohsawa H., Hayashi T., Owaribe K. and Tsonis P. A. (2007). Regeneration of retinotectal projections after optic tectum removal in adult newts. Mol. Vis. 13, 2112-2118. [PubMed] [Google Scholar]
- Parish C. L., Beljajeva A., Arenas E. and Simon A. (2007). Midbrain dopaminergic neurogenesis and behavioural recovery in a salamander lesion-induced regeneration model. Development 134, 2881-2887. 10.1242/dev.002329 [DOI] [PubMed] [Google Scholar]
- Sandoval-Guzmán T., Wang H., Khattak S., Schuez M., Roensch K., Nacu E., Tazaki A., Joven A., Tanaka E. M. and Simon A. (2014). Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell 14, 174-187. 10.1016/j.stem.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Satoh A., Makanae A., Nishimoto Y. and Mitogawa K. (2016). FGF and BMP derived from dorsal root ganglia regulate blastema induction in limb regeneration in Ambystoma mexicanum. Dev. Biol. 417, 114-125. 10.1016/j.ydbio.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Singh B. N., Doyle M. J., Weaver C. V., Koyano-Nakagawa N. and Garry D. J. (2012). Hedgehog and Wnt coordinate signaling in myogenic progenitors and regulate limb regeneration. Dev. Biol. 371, 23-34. 10.1016/j.ydbio.2012.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Timoshevskaya N., Timoshevskiy V. A., Keinath M. C., Hardy D. and Voss S. R. (2019). A chromosome-scale assembly of the axolotl genome. Genome Res. 29, 317-324. 10.1101/gr.241901.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K., Athippozhy A. T., Voss S. R. and Tsonis P. A. (2014). Plasticity for axolotl lens regeneration is associated with age-related changes in gene expression. Regeneration 1, 47-57. 10.1002/reg2.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale W. T., Lemieux M. E., Killen A. C., Zhao J., Hu Z., Riepsaame J., Hamilton N., Kudoh T., Riley P. R., van Aerle R. et al. (2018). Heart regeneration in the Mexican cavefish. Cell Rep. 25, 1997-2007.e1997. 10.1016/j.celrep.2018.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum D. L. (2017). Mechanisms of urodele limb regeneration. Regeneration 4, 159-200. 10.1002/reg2.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C. and Mueller R. L. (2014). Hellbender genome sequences shed light on genomic expansion at the base of crown salamanders. Genome Biol. Evol. 6, 1818-1829. 10.1093/gbe/evu143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland M. E. (2010). Regeneration: Thomas Hunt Morgan's window into development. J. Hist. Biol. 43, 325-361. 10.1007/s10739-009-9203-2 [DOI] [PubMed] [Google Scholar]
- Tanaka E. M. (2003). Regeneration: if they can do it, why can't we? Cell 113, 559-562. 10.1016/S0092-8674(03)00395-7 [DOI] [PubMed] [Google Scholar]
- Tanaka E. M. (2016). The molecular and cellular choreography of appendage regeneration. Cell 165, 1598-1608. 10.1016/j.cell.2016.05.038 [DOI] [PubMed] [Google Scholar]
- Tang G.-Q. and Maxwell E. S. (2008). Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res. 18, 104-112. 10.1101/gr.6539108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassava R. A., Castilla M., Arsanto J.-P. and Thouveny Y. (1993). The wound epithelium of regenerating limbs of Pleurodeles waltl and Notophthalmus viridescens: studies with mAbs WE3 and WE4, phalloidin, and DNase 1. J. Exp. Zool. 267, 180-187. 10.1002/jez.1402670211 [DOI] [PubMed] [Google Scholar]
- Tazaki A., Tanaka E. M. and Fei J.-F. (2017). Salamander spinal cord regeneration: The ultimate positive control in vertebrate spinal cord regeneration. Dev. Biol. 432, 63-71. 10.1016/j.ydbio.2017.09.034 [DOI] [PubMed] [Google Scholar]
- Urata Y., Yamashita W., Inoue T. and Agata K. (2018). Spatio-temporal neural stem cell behavior leads to both perfect and imperfect structural brain regeneration in adult newts. Biol. Open 7, bio033142 10.1242/bio.033142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences M. and Wake D. B. (2007). Speciation, species boundaries and phylogeography of amphibians. In Amphibian Biology (ed. Heatwole H. H. and Tyler M.), pp. 2613-2669. Chipping Norton, Australia: Surrey Beatty & Sons. [Google Scholar]
- Wang H. and Simon A. (2016). Skeletal muscle dedifferentiation during salamander limb regeneration. Curr. Opin. Genet. Dev. 40, 108-112. 10.1016/j.gde.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Yun M. H. (2015). Changes in regenerative capacity through lifespan. Int. J. Mol. Sci. 16, 25392-25432. 10.3390/ijms161025392 [DOI] [PMC free article] [PubMed] [Google Scholar]