ABSTRACT
The ability of a single genome to produce distinct and often dramatically different male and female forms is one of the wonders of animal development. In Drosophila melanogaster, most sexually dimorphic traits are controlled by sex-specific isoforms of the doublesex (dsx) transcription factor, and dsx expression is mostly limited to cells that give rise to sexually dimorphic traits. However, it is unknown how this mosaic of sexually dimorphic and monomorphic organs arises. Here, we characterize the cis-regulatory sequences that control dsx expression in the foreleg, which contains multiple types of sex-specific sensory organs. We find that separate modular enhancers are responsible for dsx expression in each sexually dimorphic organ. Expression of dsx in the sex comb is co-regulated by two enhancers with distinct spatial and temporal specificities that are separated by a genitalia-specific enhancer. The sex comb-specific enhancer from D. willistoni, a species that primitively lacks sex combs, is not active in the foreleg. Thus, the mosaic of sexually dimorphic and monomorphic organs depends on modular regulation of dsx transcription by dedicated cell type-specific enhancers.
KEY WORDS: Sexual dimorphism, Drosophila, Doublesex, Sex comb, Enhancers
Summary: The expression of doublesex in sexually dimorphic structures in Drosophila legs and genitalia is regulated by tissue-specific cis-regulatory elements, providing insight into how cells obtain their sex-specific identity.
INTRODUCTION
Most animals are mosaics of sexually dimorphic and monomorphic tissues. Despite the multitude of traits that distinguish males from females, many tissues and organs lack overt sexual dimorphism. To understand how this mosaic pattern is produced, the action of sex determination pathways needs to be understood at the cellular level.
Sexually dimorphic morphologies are specified by diverse molecular mechanisms in different animal phyla (Kopp, 2012; Matson and Zarkower, 2012). One of the best-studied mechanisms is found in the fruit fly Drosophila melanogaster, in which sex-specific development of most somatic cells is controlled by an alternative pre-mRNA splicing pathway (reviewed by Christiansen et al., 2002; Cline, 1993; Erickson and Quintero, 2007; McKeown, 1992). In Drosophila, the number of X chromosomes sets off a cell-autonomous cascade of sex-specific splicing that leads to the production of functional Transformer (Tra) protein in females, but not in males. Tra controls alternative splicing of doublesex (dsx) pre-mRNAs, such that the presence of functional Tra in females leads to a female-specific dsx isoform, and the absence of functional Tra leads to the production of a male-specific isoform (dsxF and dsxM, respectively). dsx encodes transcription factors involved in the establishment of almost all morphological traits that differ between males and females (Baker and Ridge, 1980; Hildreth, 1965). The DsxM and DsxF proteins share a common DNA-binding domain and bind to the same target sequences, but have different and sometimes opposite effects on gene expression, leading to sex-specific cell differentiation (Arbeitman et al., 2004, 2016; Burtis et al., 1991; Goldman and Arbeitman, 2007; Lebo et al., 2009; Li and Baker, 1998; Yang et al., 2008). For instance, the Yolk protein 1, bric a brac, Fmo-2 and Fad2 genes are directly regulated by Dsx and are expressed at higher levels in females compared with males. dsx mutants show intermediate expression of Yolk protein 1, bric a brac and Fmo-2 in both sexes, indicating that DsxF is likely activating, and DsxM repressing, these genes (Coschigano and Wensink, 1993; Luo and Baker, 2015; Williams et al., 2008). desat-F, on the other hand, is activated by DsxF but is not affected by DsxM (Shirangi et al., 2009). At the morphological level, loss-of-function dsx mutants develop as intersexes with a mixture of male, female and intermediate traits (Baker and Ridge, 1980; Hildreth, 1965).
The details of the tra/dsx splicing cascade were characterized in the 1980s (Boggs et al., 1987; Burtis and Baker, 1989; Butler et al., 2018; McKeown et al., 1987, 1988; Nagoshi et al., 1988), and are a textbook example of the role of alternative splicing in development. In contrast, the role of transcriptional regulation of dsx in Drosophila sexual differentiation was realized only recently. In a variety of developmental contexts, dsx is only transcribed in cells that are associated with sexually dimorphic organs; often, this is only a small minority of cells within the overall tissue (Camara et al., 2008; Rideout et al., 2010; Robinett et al., 2010; Tanaka et al., 2011). This mosaicism makes it all the more remarkable that dsx regulates the development of so many different sex-specific traits, from pigmentation and genital morphology to brain neural circuits and gene expression in the gut (Goldman and Arbeitman, 2007; Keisman et al., 2001; Luo and Baker, 2015; Sanders and Arbeitman, 2008; Williams et al., 2008). To understand how this mosaic pattern is specified, we need to elucidate the mechanisms that establish dsx transcription.
Among the best-studied sexually dimorphic organs in Drosophila are the foreleg bristles. The foreleg of D. melanogaster displays two distinct sex-specific features. First, the sex comb – a row of strongly modified mechanosensory bristles – is present only in males; in females, the homologous bristles retain the typical mechanosensory bristle morphology (Kopp, 2011; Tokunaga, 1962). Second, males have a greater number of foreleg chemosensory bristles compared with females (Mellert et al., 2012; Tokunaga, 1962). The sex comb and chemosensory bristles are found in close proximity to each other, and both are involved in mating behavior (Fan et al., 2013; Hurtado-Gonzales et al., 2015; Ng and Kopp, 2008; Spieth, 1952). The male-specific chemosensory bristles are used early in the stereotypic courtship sequence to taste the female cuticular pheromones (Fan et al., 2013; Spieth, 1952), and the sex comb assists in grasping the female prior to copulation (Hurtado-Gonzales et al., 2015; Ng and Kopp, 2008; Spieth, 1952).
dsx is required both for the distinctive sex comb morphology and for the higher number of chemosensory bristles in males (Belote and Baker, 1982; Mellert et al., 2012). Expression of dsx in the developing foreleg starts in the late 3rd instar larva (Tanaka et al., 2011). During prepupal and early pupal development, this pattern resolves into several distinct clusters of cells corresponding to chemosensory bristle precursors, sex comb bristle precursors, and surrounding epithelial cells (Mellert et al., 2012; Robinett et al., 2010; Tanaka et al., 2011). This raises a question: is dsx expression in the foreleg controlled by a different, modular enhancer for each sex-specific bristle type, or by a common leg enhancer? More generally, is dsx, despite its apparently global function as a binary sex switch, subject to modular transcriptional control, with a dedicated enhancer for every cell population that expresses dsx?
To address this question, we identified and characterized the enhancers that control dsx expression in the sex comb and the foreleg chemosensory bristles. We found that three separate enhancers are responsible for regulating dsx expression in the foreleg. One enhancer drives broad foreleg expression that encompasses both sex comb and chemosensory organ primordia, whereas the other two have mutually exclusive activities – one in the sex comb, and the other in sex-specific chemosensory bristles. The two enhancers that contribute to sex comb development are separated by a genitalia-specific enhancer that has no activity in the leg. This complex cis-regulatory architecture suggests that dsx transcription, like that of other developmentally regulated transcription factors, is controlled by modular tissue-specific enhancers that can have both overlapping and non-overlapping spatial activities.
RESULTS
Three enhancers with distinct spatial and temporal activities contribute to complex dsx expression in the foreleg
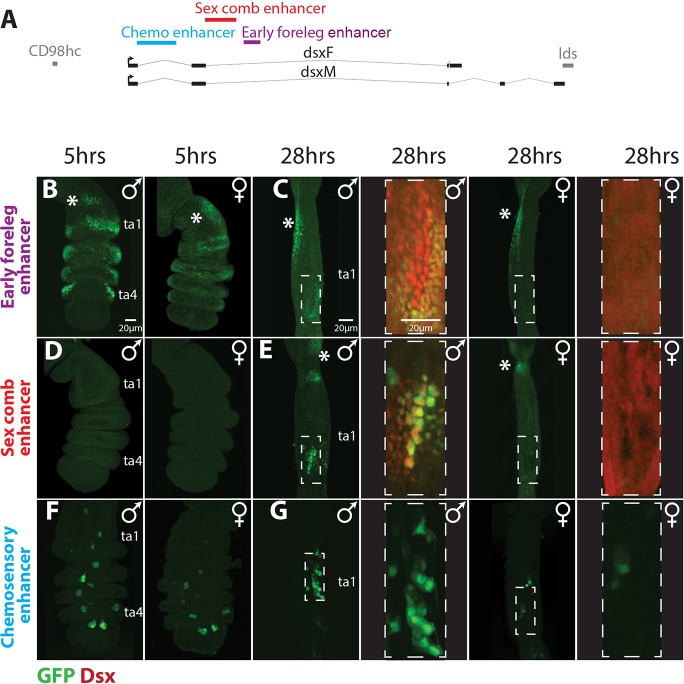
To identify the enhancers that specify the complex expression pattern of dsx during foreleg development, we generated a series of pPTGAL GAL4 reporter lines covering the nearly 50 kb of the upstream and intronic regions of the dsx locus (Fig. S1, brown boxes). In parallel, we examined an overlapping series of pBPGUw GAL4 reporters generated using a different vector and integration method (Pfeiffer et al., 2008) (Fig. S1, gray boxes). These lines were crossed to UAS-GFP.nls and screened 5 h and 24 h after puparium formation (APF). Using both sets of reporters, we identified three DNA fragments that drove expression patterns consistent with known dsx expression in the foreleg (Rideout et al., 2010; Robinett et al., 2010; Tanaka et al., 2011). We identified a chemosensory enhancer in a ∼4 kb fragment of the first dsx intron (Fig. 1F,G, Fig. S1B, blue text), a sex comb enhancer in a ∼3 kb section of the second intron (Fig. S1C, red and green text), and an early foreleg enhancer in a ∼4 kb section of the second intron downstream from the sex comb enhancer (Fig. S1C, purple and green text). In parallel, another research group had identified a genital enhancer in the overlap between the 40F03 and 42D04 reporters (Fig. S1C) (S. Yan, W. Wang, M. Arbeitman and J. Yoder, personal communication), both of which showed foreleg expression in our screen. To test whether the sex comb and early foreleg enhancers were separate, and to reduce the temporal lag between enhancer activation and reporter detection, we cloned smaller sub-fragments of the dsx second intron into a nuclear GFP expression vector. We found that both the sex comb and the early foreleg enhancers drove the same expression patterns in this assay as in the larger GAL4 reporter constructs (Fig. 1B-E).
Fig. 1.
Three modular enhancers drive doublesex expression in the foreleg. (A) A map of the doublesex locus with the positions of chemosensory (blue), sex comb (red) and foreleg (purple) dsx enhancers. Thick gray lines represent the flanking genes lds and CD98hc, thick black lines are the exons of dsx, and thin black lines are the dsx introns. (B-G) Foreleg expression patterns of the three dsx enhancers, with enhancer-driven GFP in green and magnifications of 28 h samples of the early foreleg and sex comb enhancers showing Dsx antibody staining in red. hrs, h APF. The early foreleg and sex comb enhancers constructs (B-E) are directly adjacent to GFP, whereas the chemosensory enhancer (42C06; Pfeiffer et al., 2008) construct (F,G) is a GAL4 driver crossed to a UAS-GFP.nls line. (B) At 5 h APF, the early foreleg enhancer recapitulates the Dsx expression pattern in the leg epithelium (Tanaka et al., 2011) and shows a similar expression pattern in males (left) and females (right). Both sexes show ectopic expression in the proximal first tarsal segment (ta1) (asterisks in B-E). (C) At 28 h APF, the early foreleg enhancer shows clear sexual dimorphism in the first tarsal segment. Males show strong expression in the epithelial cells surrounding the sex comb and weak to no expression in the sex comb bristle cells. Females show no expression in the distal ta1, consistent with the loss of Dsx expression by this stage (red channel shows non-specific cytoplasmic background in an over-exposed image). Both males and females show ectopic expression in the proximal ta1 (asterisks), in the same region as at 5 h APF (B), that is not seen by Dsx antibody staining (Tanaka et al., 2011). (D) The sex comb enhancer is not active in either sex at 5 h APF, except for a proximal patch of ectopic expression seen near the joint in some individuals. (E) At 28 h APF, the sex comb enhancer is active in the bristle cells of the male sex comb (large nuclei), and weakly in the epithelial cells ventral to the sex comb. Females show weak expression in the distal portion of the segment. Both sexes show ectopic expression in the joint between the tibia and ta1. (F) At 5 h APF, the chemosensory enhancer is active in small clusters of cells in ta1-ta5 in both sexes, with more GFP-positive cells in males than in females. (G) At 28 h APF, both sexes show small clusters of expression in ta1-ta5; only ta1 is shown. (No Dsx antibody staining was performed in F,G.)
The three enhancers direct distinct spatial and temporal expression of reporter genes, consistent with known dsx expression. dsx expression in the foreleg first becomes visible in wandering third instar larvae and is well developed in prepupal legs (Tanaka et al., 2011). Leg bristles are specified over several developmental periods. Most chemosensory bristles and the largest mechanosensory bristles are specified at the end of the 3rd larval instar or early in prepupal development, whereas most mechanosensory bristles (including sex comb teeth) are specified between 6 and 15 h APF (Belote and Baker, 1982; Held, 2002). At 5 h APF, Dsx is present in an anterior-ventral region of tarsal segments 1 to 4 (ta1-4), where the sex comb and sex-specific chemosensory bristles will form (Tanaka et al., 2011). At this stage, the early foreleg enhancer drives reporter expression in a pattern similar to Dsx protein (Fig. 1B). Although Dsx expression is already becoming sexually dimorphic by 5 h APF (Tanaka et al., 2011), the GFP reporter intensity at this stage was roughly monomorphic, possibly owing to the higher stability of the GFP protein (Fig. 1B). The sex comb enhancer was not active at 5 h APF (Fig. 1D), whereas the chemosensory enhancer was expressed in both sexes in small clusters of cells that match the pattern of developing chemosensory bristles (Fig. 1F).
By 28 h APF, Dsx expression is strongly dimorphic; in males, high Dsx expression is seen in sex comb bristles and surrounding epithelial cells, whereas in females, the homologous region of ta1 shows much lower Dsx expression (Tanaka et al., 2011; Fig. 1C). In the rest of the tarsus, epithelial expression of Dsx disappears by 28 h APF. At this stage in males, the early foreleg enhancer still showed weak activity in epithelial cells around the sex comb, especially on the distal-ventral side; however, it was not visibly active in sex comb bristles (Fig. 1C). The sex comb enhancer drives a complementary pattern, with strong expression in sex comb bristles and weak expression in the surrounding epithelial cells (Fig. 1E). In females, neither of these enhancers showed significant expression at 28 h APF, consistent with the sexually dimorphic expression of Dsx (Fig. 1C,E). For the chemosensory enhancer, cell clusters that reflect the pattern of chemosensory bristles continued to be observed at 28 h APF in both males and females (Fig. 1G). Thus, each of the three enhancers directs a distinct expression pattern in the foreleg.
All three enhancers contribute to sex-specific sensory organ development
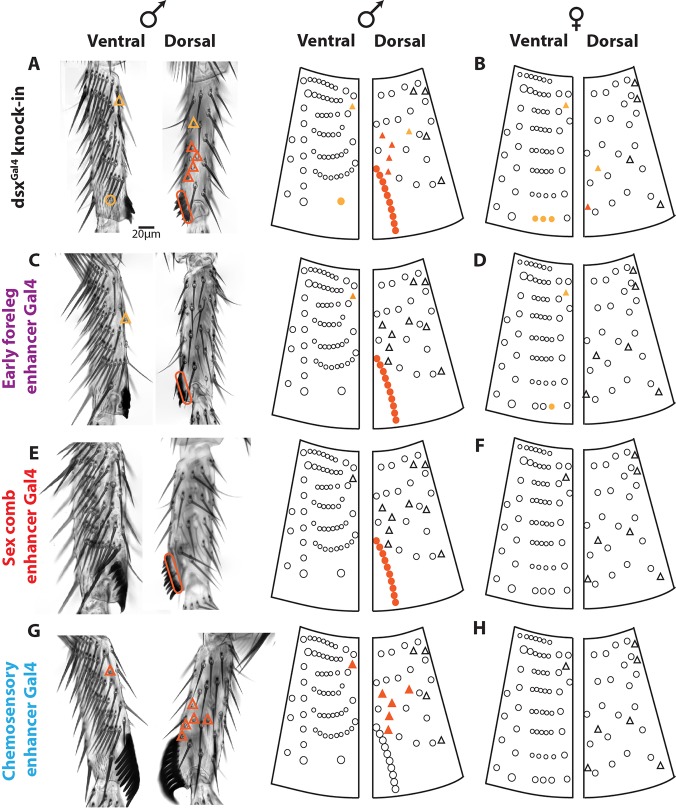
The patterns of GFP reporter expression suggest that the sex comb enhancer controls sex comb development and the chemosensory enhancer controls sex-specific chemosensory bristle development, whereas the early foreleg enhancer could potentially contribute to the development of both types of sex-specific bristles. To test these hypotheses, we used the chemosensory (42C06), foreleg (42D04) and sex comb (40F03) pBPGUw-GAL4 lines to drive a UAS-RNAi construct targeting the shaven gene (sv; also known as Pax2). Knockdown of sv in sensory organ precursors causes severe truncation of bristle shafts (Kavaler et al., 1999), allowing us to identify the bristles in which each dsx enhancer is active during prepupal and pupal development. We collected and analyzed eight male and five female samples for each of the three GAL4 lines. This approach also allowed us to test for enhancer activity at developmental stages that were not observed by visual inspection of GFP reporters. The chaetotaxy of ta1 is particularly stereotypic and has been thoroughly mapped (Tokunaga, 1962), allowing us to compare the male and female knockdown phenotypes.
First, to identify all dsx-expressing bristles, we expressed UAS-svRNAi using a dsxGAL4 knock-in line, which was generated by inserting the GAL4 coding sequence into the dsx locus and reflects the full expression pattern of dsx (Robinett et al., 2010). We found that the ta1 chemosensory bristles fall into three classes (Fig. 2). First, the dorsal-posterior chemosensory bristles are not affected by dsxGAL4/UAS-svRNAi; in the wild type, these bristles are sexually monomorphic in size and position, suggesting they do not express or require dsx for their development (Fig. 2A,B). Second, on the dorsal-anterior side, males have four chemosensory bristles on the distal half of ta1 (Fig. 2A, dark orange triangles), compared with only one in females (Fig. 2B); these bristles were always strongly affected by sv RNAi in both sexes, indicating that they express dsx (Fig. 2A,B). The third class, two chemosensory bristles (Fig. 2A,B, yellow triangles) were affected weakly and with more variability among individuals, suggesting that they may express dsx at lower levels or for a briefer period.
Fig. 2.
All three leg enhancers contribute to sex-specific bristle development. For each enhancer, a GAL4 reporter was used to drive a UAS-shaven RNAi construct, truncating the bristles where that enhancer is active. Images of male ta1 are shown next to a schematic of male and female ta1 chaetotaxy modified from Tokunaga (1962). Circles designate mechanosensory bristles; triangles designate chemosensory bristles. Dark orange symbols mark bristles that were affected in all individuals; bristles marked with yellow varied among individuals of the same genotype raised under standard conditions. (A) A GAL4 knock-in in the dsx gene, which reflects the full expression pattern of dsx (Robinett et al., 2010), affects both sex comb and sexually dimorphic chemosensory bristles in males. (B) In females, the dsxGAL4 knock-in has a variable effect on the distal TBR, which is homologous to the sex comb, and on some chemosensory bristles. (C) In males, the early foreleg enhancer affects sex comb bristles in all flies as well as a single chemosensory bristle in some individuals. (D) In females, the early foreleg enhancer affects one bristle of the distal TBR and the same chemosensory bristle as in males. (E) In males, the sex comb enhancer affects only the sex comb. (F) The sex comb enhancer does not affect any bristles in females. (G) In males, the chemosensory enhancer affects the same chemosensory bristles as the dsxGAL4 knock-in but has no effect on the sex comb. (H) The chemosensory enhancer does not affect any bristles in females.
The male sex comb was always severely affected by dsxGAL4/UAS-svRNAi, whereas the homologous female bristles were affected weakly and with more variability (Fig. 2A,B), consistent with a lower level of dsx expression in females compared with males (Tanaka et al., 2011). With the exception of the sex comb and homologous female bristles, no mechanosensory bristles were affected by dsxGAL4/UAS-svRNAi (Fig. 2A,B), confirming their sexually monomorphic nature.
Next, we used each of the three dsx leg enhancers to drive UAS-svRNAi and compared the resulting phenotypes in ta1 with that of the dsx knock-in. The early foreleg enhancer was similar to the dsx knock-in in having a strong and consistent effect on the male sex comb, but only a weak and variable effect on homologous bristles in females (Fig. 2C,D). It also affected a single chemosensory bristle in proximal ta1 in both sexes (Fig. 2C,D). As expected, the sex comb enhancer had a clear effect on sex comb development in males, but did not affect chemosensory bristles or any bristles in females (Fig. 2E,F). The chemosensory enhancer consistently affected all anterior-ventral chemosensory bristles in males, including the two proximal bristles that were only weakly affected by the dsx knock-in; it did not affect any mechanosensory bristles (Fig. 2G). Surprisingly, the chemosensory enhancer had no effect in females (Fig. 2H), even though some female chemosensory bristles express dsx (Fig. 2B), and the chemosensory enhancer drives UAS-GFP expression in ta1 in females (Fig. 1C).
In addition to their function in ta1, the foreleg and chemosensory enhancers showed expression in the more distal segments (ta2-ta4) (Fig. 1B,F). These segments also contain chemosensory bristles, some of which are sexually dimorphic (Mellert et al., 2012; Tokunaga, 1962). In ta2 and ta3, the dsx knock-in and the chemosensory enhancer affected the same subset of four chemosensory bristles, whereas the sex comb enhancer had no effect (Fig. S2). The early foreleg enhancer affected some but not all of the bristles affected by the dsx knock-in, as well as one of the chemosensory bristles that was not affected by the dsx knock-in or the chemosensory enhancer (Fig. S2). Pupal expression of the early foreleg enhancer is epithelial, not bristle specific (Fig. 1E); thus, it is difficult to say whether this phenotype reflects a true developmental function of the early foreleg enhancer in sensory organs, or the perdurance of the GAL4 protein or the RNAi effect from an earlier time in development.
Examination of adult bristle phenotypes caused by UAS-svRNAi expression suggests that the three leg enhancers of dsx make distinct contributions to sex-specific bristle development. The foreleg and sex comb enhancers function in sex comb development, whereas the chemosensory enhancer functions in the development of sexually dimorphic chemosensory organs. We also cannot rule out some contribution of the early foreleg enhancer to chemosensory bristle development.
The foreleg and sex comb enhancers are modular and flank a separate genital enhancer
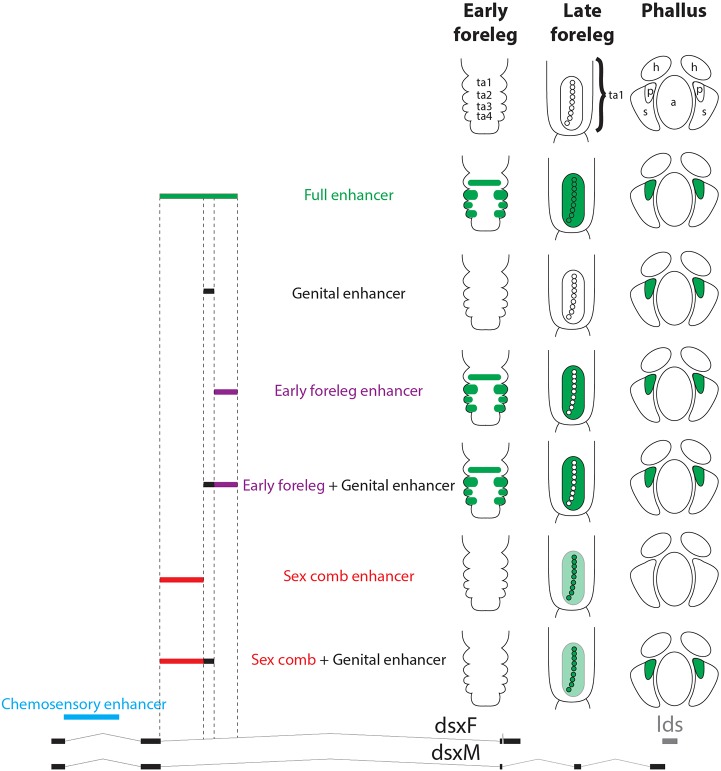
The foreleg and sex comb enhancers are located in close proximity within the second intron of dsx and are both active in the sex comb, although in different spatial patterns and at different times (Fig. 1B-E). The same broad region also contains an enhancer that is active in the third instar larval genital disc (John Yoder, University of Alabama, AL, USA personal communication). Mutations in dsx have been previously shown to disrupt clasper (also known as the surstylus) bristles (Hildreth, 1965), which have a morphology similar to sex comb teeth. Through antibody staining, we found that Dsx is expressed in several regions of the pupal male genitalia (Fig. S3). In the developing clasper, Dsx is enriched in the bristle precursor cells, similar to its enrichment in sex comb bristles. Analysis of the genital enhancer showed weak expression in the clasper as well as strong expression in the ventral postgonites (Figs S3, S4). These observations led us to inquire whether the foreleg, genital, and sex comb enhancers were fully modular and independent, or parts of a broader pleiotropic enhancer with overlapping activities in multiple tissues. To distinguish between these possibilities, we generated non-overlapping reporter constructs for each enhancer and examined each reporter both in the foreleg and in the genitalia. In addition, we generated larger constructs in which either the sex comb or the early foreleg enhancer was excluded (Fig. 3).
Fig. 3.
The foreleg and sex comb enhancers are separate and modular. To test for overlap between the foreleg, genital and sex comb enhancer activities, the region they encompass was split into six constructs: full (all three enhancers, green), genital (black), foreleg (purple), foreleg and genital (purple and black), sex comb (red), and sex comb and genital (red and black). In the drawings, green shading shows reporter expression that matches endogenous Dsx expression, with light green indicating weak expression and white a lack of expression. Representative images of each construct are shown in Fig. S3. Only the constructs that include the early foreleg enhancer drive expression in the forelegs at 5 h APF and in the epithelial cells surrounding the sex comb at 24 h. Only regions that include the sex comb enhancer drive expression in the sex comb bristles at 24 h APF. Both the genital and the early foreleg enhancer constructs are able to drive expression in the genitalia at 48 h. a, aedeagus; h, hypandrium; p, ventral postgonites; s, aedeagal sheath; ta1-ta4, tarsal segments 1-4.
We first confirmed that a 5.5 kb genomic fragment containing all three candidate enhancers (sex comb, genital and foreleg) was able to drive both early and late expression in the foreleg, as well as genital expression (Fig. 3, Fig. S4). Each separate enhancer could be seen to drive a distinct subset of that overall pattern. The late sex comb enhancer is active in the sex comb bristle cells but not in the genitalia, whereas the genital enhancer drives expression in the genitalia but has no activity in the foreleg (Fig. 3, Fig. S4). The construct encompassing the early foreleg enhancer is active in the epithelial cells surrounding the sex comb, as well as in the genitalia in a pattern that resembles the genital enhancer (Fig. S4). However, further dissection of this region shows that the foreleg and genital activities are largely separable. The sequences necessary for accurate foreleg expression are located on the 3′ side of the foreleg enhancer and do not drive genital expression, whereas the sequences that activate expression in the genitalia are located on the 5′ side and only drive ectopic expression in the foreleg (Fig. 4G,H, Fig. S3). Pairwise combinations of enhancers behave in a predictably modular fashion: a construct including the sex comb and genital enhancers drives genital and late sex comb expression, but no early epithelial expression in the foreleg, whereas the construct including the genital and early foreleg enhancers drives early foreleg and genital expression but no expression in sex comb teeth (Fig. 3, Fig. S4). The chemosensory enhancer is located in a different intron, and is therefore clearly distinct from the foreleg and sex comb enhancers (Fig. 3). In summary, we conclude that the three leg enhancers are all fully separable, modular elements, but we cannot rule out some minor overlap between the foreleg and genital enhancers.
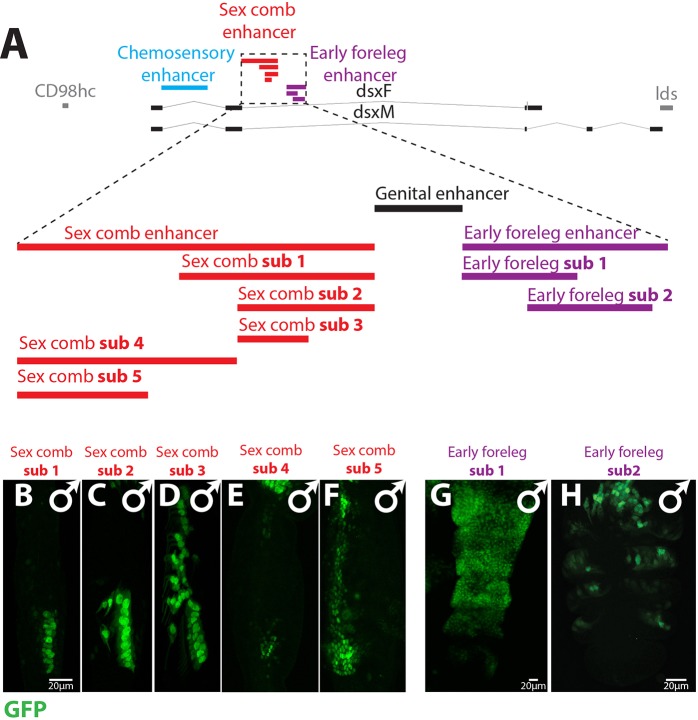
Fig. 4.
Repressor elements are necessary to limit dsx expression to sexually dimorphic cells. (A) A map of the sex comb (red), genital (black) and foreleg (purple) enhancers. (B-F) Representative images of male forelegs at 36 h APF for the sex comb sub1-sub5 constructs; only the ta1 segment is shown. (B) The sex comb sub1 enhancer drives expression in the sex comb bristle cells. (C) sex comb sub2 is active in sex comb bristles but has additional ectopic expression in the central bristle and in the second most distal TBR. (D) sex comb sub3 is expressed in all TBRs bristle cells in the ta1 segment. (E) sex comb sub4 is expressed in some of the epithelial cells that surround the sex comb, but these cells are not directly adjacent to the sex comb bristles. (F) sex comb sub5 at 24 h AFP is expressed in the epithelial cells surrounding the sex comb, but also shows ectopic expression in more proximal epithelial cells. (G,H) Foreleg expression of the foreleg sub1 and sub2 constructs at 5 h APF. The foreleg sub1 region drives broad ectopic expression throughout the ta1-ta4; foreleg sub2 shows expression in a pattern similar to the complete early foreleg enhancer (Fig. 1B).
Sequences in the foreleg and sex comb enhancers confine their activity to sexually dimorphic primordia
To identify the minimal sequences required for correct activity, we subdivided the sex comb and early foreleg enhancers into smaller reporter constructs (Fig. 4A). The ∼3 kb sex comb enhancer drives specific expression in sex comb bristles and the epithelial cells distal to the sex comb (Fig. 1E), but shorter sequences are unable to recapitulate this pattern. The 3′ ∼1.5 kb of the sex comb enhancer (sex comb enhancer sub1) drives specific expression in sex comb bristles but is not expressed in the epithelial cells (Fig. 4B). A shorter ∼1 kb 3′ fragment (sex comb enhancer sub2) retains expression in the sex comb bristles, but also causes ectopic expression in the most distal transverse bristle row (TBR) (Fig. 4C). A core sequence of ∼0.5 kb (sex comb enhancer sub3) expands expression to all ta1 TBRs (Fig. 4D). The TBRs are serially homologous to the sex comb (Kopp, 2011; Tokunaga, 1962), but are sexually monomorphic and do not express Dsx (Robinett et al., 2010; Tanaka et al., 2011).
The 5′ ∼1.5 kb of the sex comb enhancer (sex comb enhancer sub4) drives expression in some of the epithelial cells surrounding the sex comb, but this expression does not encompass all of the epithelial cells where the full sex comb enhancer is active (Fig. 4E). Reduction of this sequence to a ∼1 kb 5′ fragment (sex comb enhancer sub5) causes ectopic expression in proximal epithelial cells (Fig. 4F) that do not express Dsx (Robinett et al., 2010; Tanaka et al., 2011). Thus, sequences within the sex comb enhancer are necessary to prevent its activity in sexually monomorphic bristle and epithelial cells.
Dissection of the early foreleg enhancer identified a shorter 3′ fragment (early foreleg enhancer sub2) capable of driving expression in the correct region of the foreleg (Fig. 4H). A partially overlapping fragment (early foreleg enhancer sub1) shows broad ectopic activity throughout the tarsal segments (Fig. 4G). Thus, similar to the late sex comb enhancer, both activating and repressive sequences are needed to limit the activity of this enhancer to sexually dimorphic cell populations.
Enhancer activity reflects the evolution of sexual dimorphism
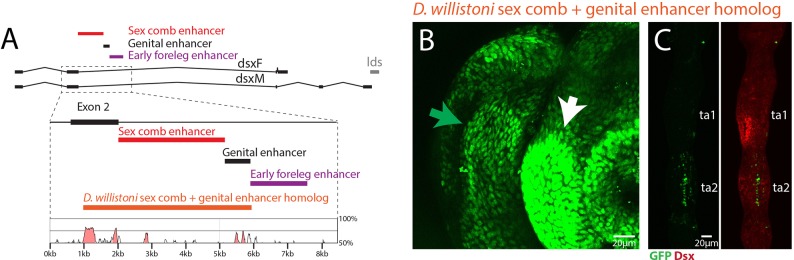
The sex comb is a new structure that evolved recently within the Drosophila genus (Kopp, 2011). D. willistoni is one of the closest relatives of D. melanogaster that primitively lacks the sex comb, and does not express Dsx in the homologous region of the pupal foreleg (Tanaka et al., 2011). We cloned the fragment of the D. willistoni dsx locus homologous to the sex comb and genital enhancers of D. melanogaster (Fig. 5A). In transgenic D. melanogaster, this fragment drives strong expression in the pupal male genitalia, in a pattern similar to the D. melanogaster genital enhancer, indicating that the genital enhancer is functionally conserved between the two species (Fig. 5B). In contrast, the D. willistoni reporter shows some ectopic expression in ta2, but does not drive any expression in the sex comb primordia in the male pupal foreleg (Fig. 5C), consistent with the lack of Dsx expression in this region in D. willistoni (Tanaka et al., 2011). This suggests that the sex comb-specific enhancer has evolved since the divergence between D. melanogaster and D. willistoni. More broadly, it suggests that changes in spatial and temporal activity of dsx enhancers contribute to the evolution of sex-specific traits, and that the modular organization of dsx enhancers facilitates the evolution of sexual dimorphism by uncoupling the evolutionary trajectories of different structures.
Fig. 5.
D. willistoni enhancers drive expression in the male genitalia but not in the sex comb. (A) Map of the sex comb (red), genital (black) and early foreleg (purple) enhancers in D. melanogaster. The D. willistoni reporter fragment (orange) encompasses part of the second exon, the genital enhancer, and the region homologous to the sex comb enhancer in between. The VISTA plot (Frazer et al., 2004) below shows regions of high sequence similarity between the two species, which were used to locate the right boundary of the genital enhancer in D. willistoni. Similarity >70% is highlighted in pink. (B) Expression of the D. willistoni reporter in in pupal male genitalia at 45 h APF. The strongest expression is seen in the aedeagal sheath (white arrow), with weaker expression in the clasper (green arrow), suggesting that the genital enhancer is functionally conserved between D. willistoni and D. melanogaster. (C) The same D. willistoni reporter in the pupal foreleg of D. melanogaster at 24 h APF (green), with Dsx counter-stained in red. The reporter shows ectopic expression in ta2, but has no activity in the sex comb region in distal ta1.
DISCUSSION
dsx expression in the foreleg is controlled by modular enhancers with both distinct and overlapping activities
Spatial and temporal restriction of dsx expression is essential for normal foreleg development. Ectopic expression of dsxM in leg bristles in either males or females converts many of these bristles into sex comb teeth, and ubiquitous expression of dsxM throughout the leg epidermis results in severe truncation of the leg (Tanaka et al., 2011). In this study, we have characterized the cis-regulatory architecture directing the precise spatiotemporal and cell type-specific expression of dsx in the foreleg. Our results help explain the origin of the developmental mosaicism that generates a mix of sex-specific and sexually monomorphic organs in Drosophila, and possibly in other insects. Given the largely cell-autonomous control of insect sexual differentiation, the essential function of dsx enhancers is to provide spatial landmarks that establish which organs will become sexually dimorphic. In principle, dsx expression in different types of foreleg bristles could be controlled either by a single, broadly acting leg enhancer, or by multiple enhancers, each dedicated to a different bristle type. For example, the proneural genes achaete and scute, which control bristle specification, have very complex cis-regulatory regions with dozens of modular enhancers responsible for different, stereotypically patterned bristles (Garcia-Bellido and de Celis, 2009). Other enhancers are pleiotropic and active in multiple tissues, reflecting a history of evolutionary co-option from the more ancient organs to the more recently evolved (Glassford et al., 2015; Nagy et al., 2018; Rice and Rebeiz, 2019). In fact, some genes contain both modular (tissue-specific) and pleiotropic (multi-tissue) enhancers (Preger-Ben Noon et al., 2018).
We observe a partial overlap in the temporal and spatial activities of the three leg enhancers (Figs 1–3). However, these are not functionally redundant ‘shadow enhancers’ of the type found in some other genes (Frankel et al., 2010; Hong et al., 2008). During prepupal development, the early foreleg enhancer is expressed in cells that give rise both to the sex comb and to the chemosensory bristles (Fig. 2, Fig. S2). Later, the early foreleg enhancer expression in the sex comb becomes complementary to the activity of the sex comb enhancer: the former is expressed only in the epithelial cells, and the latter predominantly in sex comb bristles and only weakly in the epithelial cells. At pupal stages, when the early foreleg enhancer is no longer active in bristle cells, the late-acting sex comb and chemosensory enhancers have completely non-overlapping expression patterns. Overall, the existence of these three modular enhancers suggests that dsx expression in the foreleg goes through two successive stages: relatively broad expression early in development, followed by tightly limited, cell type-specific expression later on.
These phases of dsx expression may have different developmental functions. For example, dsx knockdown in chemosensory bristles late in bristle development causes a defect in the axonal midline crossing by the bristle neurons (Mellert et al., 2012). Early dsx expression in the foreleg induces a male-specific increase in the number of bristles both in the sex comb and in the chemosensory system (Belote and Baker, 1982; Mellert et al., 2012). The broad early foreleg expression of dsx is established prior to the formation of mechanosensory bristles (Tanaka et al., 2011), suggesting that the early foreleg enhancer may control sex comb size by modulating the specification of sensory organ precursors. On the other hand, the later-acting sex comb enhancer is more likely to be involved in controlling the morphology of the individual sex comb teeth, which develop from mechanosensory bristle shafts but, in contrast to most such bristles, are thick, blunt and darkly pigmented. Indeed, knockdown of dsx late in development affects the morphology but not the number of sex comb teeth, which become masculinized in females and feminized in males (Belote and Baker, 1982). Both the foreleg and the sex comb enhancers are active in the epithelial cells around the sex comb, suggesting that both could contribute to sex comb rotation – a coordinated sequence of cell-shape changes that moves the sex comb from an initial transverse to the final longitudinal orientation (Atallah et al., 2009; Ho et al., 2018; Malagón et al., 2014; Tanaka et al., 2011). As sex comb development involves so many different cellular processes spread over multiple developmental stages, it is perhaps not surprising that it requires multiple dsx enhancers.
Not all tissues and cell types behave in a modular fashion with respect to dsx expression. In the central nervous system, only one of the pPTGAL reporters tiling the dsx region showed an overlap with endogenous dsx expression (in four neurons in the PC2 cluster) (M.A., unpublished). A separate study identified a dsx enhancer upstream of the transcription start with an activity overlapping Dsx protein expression in the brain (Zhou et al., 2014). However, most of the Dsx CNS expression pattern is not accounted for by any of the dsx reporter constructs (M.A., unpublished). This suggests that for most dsx-expressing cells in the CNS, dsx regulation may not be due to modular enhancers acting independently, but rather to more complex, long-distance sequence interactions that are not captured by ∼5 kb reporter fragments. If true, such ‘entangled’ regulatory architecture could be more constraining than the modular enhancer organization we observe in the foreleg, which could in turn lead to slower evolution of dsx expression in the brain compared with other tissues.
Cis-regulatory information limits dsx expression to sexually dimorphic cells
In contrast to some enhancers for which very short sequences are sufficient to convey correct regulatory information (Grieder et al., 1997; Manak et al., 1994; Swanson et al., 2010), both the sex comb and the early foreleg enhancers are large and cannot be reduced to a core region without perturbing their activities. Interestingly, dissecting these enhancers further leads, not to the loss of activity, but rather to its expansion. This is similar to the abdominal enhancer of the ebony gene, for which a core 0.7 kb sequence is sufficient for abdominal activity, but a much broader genomic region is necessary to prevent ectopic expression (Rebeiz et al., 2009). We find a similar pattern in our study: both the foreleg and the late sex comb enhancers of dsx have core sequences of less than 1 kb capable of driving expression in the foreleg, but require additional sequences to repress ectopic activity.
The late sex comb enhancer is particularly illuminating in this respect. The sex comb develops from the most distal TBR on ta1 (Atallah et al., 2009; Tanaka et al., 2009; Tokunaga, 1962), so the bristles that comprise it are serially homologous to the more proximal TBRs. However, whereas the distal TBR that gives rise to the sex comb undergoes dramatic male-specific modification, the remaining TBRs retain their original, sexually monomorphic morphology. Consistent with this, dsx is expressed only in the most distal TBR; ectopic expression of dsx in the more proximal TBRs converts TBR bristles into sex comb teeth (Tanaka et al., 2011). We find that the late sex comb enhancer of dsx is active only in the most distal TBR, and that loss of sequences at the 5′ end of this enhancer leads to an expansion of its activity to the proximal, sexually monomorphic TBRs. Thus, much of the regulatory information contained in the sex comb enhancer is devoted to restricting its activity to sexually dimorphic cells, from a broader population of similar cells. This may reflect a general trend for dsx expression: in each tissue or body part (such as the brain, midgut, foreleg, body wall epidermis, etc.), only a subset of cells express dsx and undergo sex-specific differentiation (Goldman and Arbeitman, 2007; Luo and Baker, 2015; Rideout et al., 2010; Robinett et al., 2010; Sanders and Arbeitman, 2008; Tanaka et al., 2011). Integrating both activating and repressive inputs in dsx enhancers may be necessary to segregate these cells from otherwise similar but sexually monomorphic cells.
Modular dsx enhancers may allow different sex-specific traits to evolve independently
Although the sex combs are present in only a subset of Drosophila species, particularly in the melanogaster and obscura species groups, most though not all Drosophila species have sex-specific chemosensory bristles on their forelegs. Thus, one could imagine a more ancestral chemosensory foreleg enhancer becoming co-opted for sex comb development in the D. melanogaster and D. obscura groups. A more extreme scenario would involve a co-option of a genital enhancer for sex-specific foreleg development. Male genitalia and legs are serially homologous and share similar developmental programs involving some of the same transcription factors and signaling pathways (Gorfinkiel et al., 1999; Keisman et al., 2001; Sánchez et al., 1997), and the male genitalia of most Drosophila species have strongly modified clasper bristles reminiscent of sex comb teeth. Indeed, we find that Dsx is expressed in the clasper during genital development. However, our results argue against a simple co-option scenario. The chemosensory bristle enhancer of dsx is clearly distinct from both enhancers that contribute to sex comb development, being located in a different intron. The situation is less clear with the early foreleg enhancer, because we cannot rule out a minor overlap with the genital enhancer. However, the late sex comb enhancer, which appears to play the dominant role in the development of sex comb teeth, is clearly separate from the genital enhancer.
The evolutionary conservation of enhancer activity appears to reflect morphological evolution. D. willistoni lacks sex combs, and does not express Dsx in pupal forelegs (Tanaka et al., 2011). We find that the fragment of the D. willistoni dsx locus corresponding to the sex comb enhancer of D. melanogaster does not drive any expression in the pupal leg. In contrast, the nearby genital enhancer has broadly similar activity in both species. Thus, the enhancer involved in the development of a structure that is sexually dimorphic in both species is functionally conserved, whereas the genomic region associated with the development of a novel sex-specific organ may have evolved its tissue-specific regulatory activity more recently. Although more extensive taxon sampling is clearly needed, these results suggest that modular dsx enhancers allow different sex-specific traits to evolve independently.
Both the sex comb and the chemosensory bristles play important roles during courtship (Hurtado-Gonzales et al., 2015; Mellert et al., 2012; Ng and Kopp, 2008) and have undergone extensive changes in number, size, morphology and spatial distribution in Drosophila evolution. The ability of all three foreleg enhancers to function and evolve independently of each other may have been a key element driving this evolutionary diversification. Analysis of these enhancers in other Drosophila species will provide important insights into the role of dsx in the evolution of sex combs and chemosensory bristles. The new GAL4 drivers that mark different types of sex-specific sensory organs may also prove valuable for dissecting the neural mechanisms of courtship behavior.
Modular transcriptional regulation of dsx may reflect an evolutionarily ancient mode of sexual development
Dsx-related transcription factors (the Dmrt gene family) play important roles in sexual differentiation in animals as different as vertebrates, insects and nematodes (Kopp, 2012; Matson and Zarkower, 2012). Outside of insects, however, Dmrt genes do not produce distinct male and female isoforms, but are regulated instead at the transcriptional level. Sex-specific splicing of dsx, and more generally the mode of sexual differentiation based on alternative splicing, is an insect innovation (Kopp, 2012; Wexler et al., 2019 preprint). In crustaceans and chelicerates, dsx homologs are transcribed exclusively or predominantly in males, particularly in male-specific organs, and are required for male but not for female sexual development (Kato et al., 2011; Li et al., 2018; Pomerantz and Hoy, 2015). In Caenorhabditis elegans, the Dmrt genes mab-3, mab-23 and dmd-3 are expressed in tightly restricted cell lineages and are essential for male-specific differentiation of these cells, but are dispensable in females (Lints and Emmons, 2002; Mason et al., 2008; Ross et al., 2005; Serrano-Saiz et al., 2017; Yi et al., 2000). In vertebrates, the Dmrt1 gene is highly expressed in the male but not in the female gonad, and is necessary to direct or maintain male-specific differentiation of the initially bipotential gonad (Matson et al., 2011).
These similarities suggest that sexual differentiation based on male-specific transcription of Dmrt genes in restricted cell populations is an evolutionarily ancient mode of establishing sexual dimorphism, and that in insects sex-specific splicing of dsx was overlaid on this ancestral mechanism (Kopp, 2012; Wexler et al., 2019 preprint). The finding that dsx transcription in Drosophila is controlled by multiple modular enhancers may reflect this more ancestral mode of sexual development. We predict that a similar modular control of dsx expression will be found in other arthropods, from basal insects and crustaceans to chelicerates.
MATERIALS AND METHODS
Fly stocks
Fly strains were obtained from the Bloomington Stock Center, including UAS-GFP.nls (Bloomington stock #4775, genotype: w[1118]; P{w[+mC]=UAS-GFP.nls}14), UAS-svRNAi P{TRiP.JF02582}attP2 (Bloomington stock #27269, genotype: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02582}attP2), and dsxGAL4 (Robinett et al., 2010) (Bloomington stock #66674, genotype: w[1118]; TI{GAL4}dsx[GAL4]/TM6B, Tb[1]).
Transgenic reporter constructs
Candidate enhancer regions were cloned into the GAL4 reporter vector pPTGAL (Sharma et al., 2002), and randomly integrated into the D. melanogaster genome by P-element transformation. Three independent lines from each of ten overlapping fragments were examined. In addition, we analyzed 15 independently generated GAL4 lines that span the same non-coding regions of dsx but have different fragment boundaries (Pfeiffer et al., 2008). These lines were constructed using the site-specific pBPGUw vector (Pfeiffer et al., 2008) and integrated into either the attP2 or attP40 genomic landing sites via the PhiC31 integrase system (Groth et al., 2004; Venken et al., 2006). Each line from both sets was crossed to a UAS-GFP.nls reporter and GFP expression was examined in the developing foreleg at 5 and at 24 h APF.
Although the high level of GFP expression driven by the GAL4/UAS system was useful for the preliminary screening of genomic fragments, subsequent analysis was performed using direct GFP reporters, which enable more accurate temporal resolution. Genomic fragments were amplified by PCR, cloned into either pCR2 or pCR8 vectors (Invitrogen), transferred into the pGreenFriend GFP destination vector (Miller et al., 2014) using either restriction/ligation or Gateway cloning, and integrated into the attP2 or attP40 genomic landing sites using the PhiC31 integrase system. Germline transformation was performed either in the Kopp lab or by BestGene (http://www.thebestgene.com/). In tests performed on a subset of constructs, we found little difference between attP2 and attP40; we subsequently used the attP40 site, which gave higher integration efficiency. In designing the sub-fragments of the sex comb and early foreleg enhancers, we used EvoPrinter (Odenwald et al., 2005) and VISTA (Frazer et al., 2004) to identify sequences within these enhancers that were conserved between D. melanogaster and D. pseudoobscura. Whenever possible, we avoided breaking up highly conserved regions.
The 4.2 kb D. willistoni genomic region homologous to the D. melanogaster sex comb and genital enhancers was identified using highly conserved sequences in the second dsx exon and at the boundary between the genital and early foreleg enhancers. This region was amplified from D. willistoni strain 14030-0811.00, cloned into the SMG4GFPR2R1 reporter vector (a gift from N. Gompel, Ludwig-Maximilians-Universität München, Munich, Germany), and inserted into the attP40 site. The boundaries of all reporter fragments and the primers used to amplify them are listed in Tables S1 and S2.
Light microscopy
Legs were dissected from adult flies and mounted in PVA-Mounting-Medium (BioQuip) or Hoyer's media to clear the samples. Images were taken under bright-field illumination using a Leica DM500B microscope with a Leica DC500 camera.
Immunohistochemistry and confocal microscopy
Samples were aged, dissected, fixed, stained and imaged as described by Tanaka et al. (2009) and Mellert et al. (2012). The primary mouse-Dsx[DBD] antibody (a gift from C. Robinett and B. Baker, Janelia Research Campus, Howard Hughes Medical Institute, USA; later obtained from the Developmental Studies Hybridoma Bank, DsxDBD, RRID:AB_2617197) was used at 1:10. The secondary goat anti-mouse Alexa Fluor 594 antibody (Invitrogen, A20185) was used at 1:200. Confocal images were taken using an Olympus FV1000 laser scanning confocal microscope.
Supplementary Material
Acknowledgements
We thank Dr Carmen Robinett, Dr Bruce Baker, and the Developmental Studies Hybridoma Bank for Dsx antibodies; Dr John Yoder and the Bloomington Stock Center for Drosophila strains; Dr Nicolas Gompel for the SMG4GFPR2R1 vector; and the FlyLight Project for pBPGUw GAL4 lines. We also thank Dr John Yoder and Dr Carmen Robinett for discussions of dsx enhancer expression, and Gabriella Wong, Dr Kohtaro Tanaka and Dr Benjamin Vincent for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: G.R.R., M.A., A.K.; Methodology: G.R.R., D.L., M.A., A.K.; Validation: G.R.R., O.B., D.L.; Formal analysis: G.R.R., O.B., A.K.; Investigation: G.R.R., O.B., D.L., K.H., A.K.; Resources: O.B., M.A.; Writing - original draft: G.R.R., A.K.; Writing - review & editing: G.R.R., M.A., A.K.; Visualization: G.R.R., O.B., A.K.; Supervision: O.B., A.K.; Project administration: O.B., A.K.; Funding acquisition: M.A., A.K.
Funding
This work was funded by National Institutes of Health (NIH) grants (R01GM105726 and R35GM122592 to A.K.), by a Collaborative Supplement to NIH grant (R01GM073039 to A.K. and M.A.), and by the National Science Foundation Graduate Research Fellowship Program and Doctoral Dissertation Improvement Grant (1501148 to G.R.R.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.178285.supplemental
References
- Arbeitman M. N., Fleming A. A., Siegal M. L., Null B. H. and Baker B. S. (2004). A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131, 2007-2021. 10.1242/dev.01077 [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., New F. N., Fear J. M., Howard T. S., Dalton J. E. and Graze R. M. (2016). Sex differences in Drosophila somatic gene expression: variation and regulation by doublesex. G3 6, 1799-1808. 10.1534/g3.116.027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah J., Liu N. H., Dennis P., Hon A., Godt D. and Larsen E. W. (2009). Cell dynamics and developmental bias in the ontogeny of a complex sexually dimorphic trait in Drosophila melanogaster. Evol. Dev. 11, 191-204. 10.1111/j.1525-142X.2009.00319.x [DOI] [PubMed] [Google Scholar]
- Baker B. S. and Ridge K. A. (1980). Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94, 383-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote J. M. and Baker B. S. (1982). Sex determination in Drosophila melanogaster: analysis of transformer-2, a sex-transforming locus. Proc. Natl. Acad. Sci. USA 79, 1568-1572. 10.1073/pnas.79.5.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs R. T., Gregor P., Idriss S., Belote J. M. and McKeown M. (1987). Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 50, 739-747. 10.1016/0092-8674(87)90332-1 [DOI] [PubMed] [Google Scholar]
- Burtis K. C. and Baker B. S. (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997-1010. 10.1016/0092-8674(89)90633-8 [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Coschigano K. T., Baker B. S. and Wensink P. C. (1991). The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10, 2577-2582. 10.1002/j.1460-2075.1991.tb07798.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler B., Pirrotta V., Irminger-Finger I. and Nöthiger R. (2018). The sex-determining gene tra of Drosophila: molecular cloning and transformation studies. EMBO J. 5, 3607-3613. 10.1002/j.1460-2075.1986.tb04689.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara N., Whitworth C. and Doren M. V. (2008). The Creation of Sexual Dimorphism in the Drosophila Soma. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Christiansen A. E., Keisman E. L., Ahmad S. M. and Baker B. S. (2002). Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 18, 510-516. 10.1016/S0168-9525(02)02769-5 [DOI] [PubMed] [Google Scholar]
- Cline T. W. (1993). The Drosophila sex determination signal: how do flies count to two? Trends Genet. 9, 385-390. 10.1016/0168-9525(93)90138-8 [DOI] [PubMed] [Google Scholar]
- Coschigano K. T. and Wensink P. C. (1993). Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 7, 42-54. 10.1101/gad.7.1.42 [DOI] [PubMed] [Google Scholar]
- Erickson J. W. and Quintero J. J. (2007). Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5, 2821-2830. 10.1371/journal.pbio.0050332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Manoli D. S., Ahmed O. M., Chen Y., Agarwal N., Kwong S., Cai A. G., Neitz J., Renslo A., Baker B. S. et al. (2013). Genetic and neural mechanisms that inhibit drosophila from mating with other species. Cell 154, 89-102. 10.1016/j.cell.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N., Davis G. K., Vargas D., Wang S., Payre F. and Stern D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490-493. 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M. and Dubchak I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, 273-279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A. and de Celis J. F. (2009). The complex tale of the achaete–scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182, 631-639. 10.1534/genetics.109.104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford W. J., Johnson W. C., Dall N. R., Smith S. J., Liu Y., Boll W., Noll M. and Rebeiz M. (2015). Co-option of an ancestral Hox-regulated network underlies a recently evolved morphological novelty article. Dev. Cell 34, 520-531. 10.1016/j.devcel.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman T. D. and Arbeitman M. N. (2007). Genomic and functional studies of drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3, e216 10.1371/journal.pgen.0030216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N., Sánchez L. and Guerrero I. (1999). Drosophila terminalia as an appendage-like structure. Mech. Dev. 86, 113-123. 10.1016/S0925-4773(99)00122-7 [DOI] [PubMed] [Google Scholar]
- Grieder N. C., Marty T., Ryoo H. D., Mann R. S. and Affolter M. (1997). Synergistic activation of a Drosophila enhancer by HOM/EXD and DPP signaling. EMBO J. 16, 7402-7410. 10.1093/emboj/16.24.7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R. and Calos M. (2004). Construction of transgenic drosophila by using the site-specific integrase from phage φC31. Genetics 166, 1775-1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held L. I. (2002). Imaginal Discs: The Genetic and Cellular Logic of Pattern Formation. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Hildreth P. E. (1965). Doublesex, recessive gene that transforms both males and females of Drosophila into intersexes. Genetics 51, 659-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E. C. Y., Malagón J. N., Ahuja A., Singh R. and Larsen E. (2018). Rotation of sex combs in Drosophila melanogaster requires precise and coordinated spatio-temporal dynamics from forces generated by epithelial cells. PLoS Comput. Biol. 14, 1-28. 10.1371/journal.pcbi.1006455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.-W., Hendrix D. A. and Levine M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Gonzales J., Gallaher W., Warner A. and Polak M. (2015). Microscale laser surgery demonstrates the grasping function of the male sex combs in drosophila melanogaster and drosophila bipectinata. Ethology 121, 45-56. 10.1111/eth.12316 [DOI] [Google Scholar]
- Kato Y., Kobayashi K., Watanabe H. and Iguchi T. (2011). Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7, 1-12. 10.1371/journal.pgen.1001345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaler J., Fu W., Duan H., Noll M. and Posakony J. W. (1999). An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development 126, 2261-2272. [DOI] [PubMed] [Google Scholar]
- Keisman E. L., Christiansen A. E. and Baker B. S. (2001). The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginai disc. Dev. Cell 1, 215-225. 10.1016/S1534-5807(01)00027-2 [DOI] [PubMed] [Google Scholar]
- Kopp A. (2011). Drosophila sex combs as a model of evolutionary innovations. Evol. Dev. 13, 504-522. 10.1111/j.1525-142X.2011.00507.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175-184. 10.1016/j.tig.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo M. S., Sanders L. E., Sun F. and Arbeitman M. N. (2009). Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics 10, 1-23. 10.1186/1471-2164-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Baker B. S. (1998). hermaphrodite and doublesex function both dependently and independently to control various aspects of sexual differentiation in Drosophila. Development 125, 2641-2651. [DOI] [PubMed] [Google Scholar]
- Li S., Li F., Yu K. and Xiang J. (2018). Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 649, 1-7. 10.1016/j.gene.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Lints R. and Emmons S. W. (2002). Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16, 2390-2402. 10.1101/gad.1012602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S. D. and Baker B. S. (2015). Constraints on the evolution of a doublesex target gene arising from doublesex's pleiotropic deployment. Proc. Natl. Acad. Sci. USA 112, E852-E861. 10.1073/pnas.1501192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagón J. N., Ahuja A., Sivapatham G., Hung J. and Lee J. (2014). Evolution of Drosophila sex comb length illustrates the inextricable interplay between selection and variation. Proc. Natl. Acad. Sci. USA 111, E4103-E4109. 10.1073/pnas.1322342111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J. R., Mathies L. D. and Scott M. P. (1994). Regulation of a decapentaplegic midgut enhancer by homeotic proteins. Development 120, 3605-3619. [DOI] [PubMed] [Google Scholar]
- Mason D. A., Rabinowitz J. S. and Portman D. S. (2008). dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135, 2373-2382. 10.1242/dev.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K. and Zarkower D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. 10.1038/nrg3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J. and Zarkower D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101-104. 10.1038/nature10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M. (1992). Sex differentiation: the role of alternative splicing. Curr. Opin. Genet. Dev. 2, 299-303. 10.1016/S0959-437X(05)80288-6 [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote J. M. and Baker B. S. (1987). A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell 48, 489-499. 10.1016/0092-8674(87)90199-1 [DOI] [PubMed] [Google Scholar]
- McKeown M., Belote J. M. and Boggs R. T. (1988). Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male drosophila. Cell 53, 887-895. 10.1016/S0092-8674(88)90369-8 [DOI] [PubMed] [Google Scholar]
- Mellert D. J., Robinett C. C. and Baker B. S. (2012). Doublesex functions early and late in gustatory sense organ development. PLoS ONE 7, e51489 10.1371/journal.pone.0051489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. W., Rebeiz M., Atanasov J. E. and Posakony J. W. (2014). Neural precursor-specific expression of multiple Drosophila genes is driven by dual enhancer modules with overlapping function. Proc. Natl. Acad. Sci. USA 111, 17194-17199. 10.1073/pnas.1415308111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M. and Baker B. S. (1988). The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 53, 229-236. 10.1016/0092-8674(88)90384-4 [DOI] [PubMed] [Google Scholar]
- Nagy O., Nuez I., Savisaar R., Peluffo A. E., Yassin A., Lang M., Stern D. L., Matute D. R., David J. R. and Courtier-Orgogozo V. (2018). Correlated evolution of two copulatory organs via a single cis-regulatory nucleotide change. Curr. Biol. 28, 1-8. 10.1016/j.cub.2018.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. S. and Kopp A. (2008). Sex combs are important for male mating success in Drosophila melanogaster. Behav. Genet. 38, 195-201. 10.1007/s10519-008-9190-7 [DOI] [PubMed] [Google Scholar]
- Odenwald W. F., Rasband W., Kuzin A. and Brody T. (2005). EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc. Natl. Acad. Sci. USA 102, 14700-14705. 10.1073/pnas.0506915102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T.-T. B., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H., Laverty T. R. et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz A. F. and Hoy M. A. (2015). Expression analysis of Drosophila doublesex, transformer-2, intersex, fruitless-like, and vitellogenin homologs in the parahaploid predator Metaseiulus occidentalis (Chelicerata: Acari: Phytoseiidae). Exp. Appl. Acarol. 65, 1-16. 10.1007/s10493-014-9855-2 [DOI] [PubMed] [Google Scholar]
- Preger-Ben Noon E., Sabarís G., Ortiz D. M., Sager J., Liebowitz A., Stern D. L. and Frankel N. (2018). Comprehensive analysis of a cis-regulatory region reveals pleiotropy in enhancer function. Cell Rep. 22, 2809-2817. 10.1016/j.celrep.2018.02.073 [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Pool J. E., Kassner V. A., Aquadro C. F. and Carroll S. B. (2009). Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663-1667. 10.1126/science.1178357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G. and Rebeiz M. (2019). How many phenotypes do regulatory mutations affect? Curr. Biol. 29, R21-R23. 10.1016/j.cub.2018.11.027 [DOI] [PubMed] [Google Scholar]
- Rideout E. J., Dornan A. J., Neville M. C., Eadie S. and Goodwin S. F. (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13, 458-466. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Vaughan A. G., Knapp J. M. and Baker B. S. (2010). Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8, e1000365 10.1371/journal.pbio.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. M., Kalis A. K., Murphy M. W. and Zarkower D. (2005). The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev. Cell 8, 881-892. 10.1016/j.devcel.2005.03.017 [DOI] [PubMed] [Google Scholar]
- Sánchez L., Casares F., Gorfinkiel N. and Guerrero I. (1997). The genital disc of Drosophila melanogaster. II. Role of the genes hedgehog, decapentaplegic and wingless. Dev. Genes Evol. 207, 229-241. 10.1007/s004270050111 [DOI] [PubMed] [Google Scholar]
- Sanders L. E. and Arbeitman M. N. (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320, 378-390. 10.1016/j.ydbio.2008.05.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Oren-Suissa M., Bayer E. A. and Hobert O. (2017). Sexually dimorphic differentiation of a C. elegans hub neuron is cell autonomously controlled by a conserved transcription factor . Curr. Biol. 27, 199-209. 10.1016/j.cub.2016.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Y., Cheung U., Larsen E. W. and Eberl D. F. (2002). PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in drosophila. Genesis 34, 115-118. 10.1002/gene.10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi T. R., Dufour H. D., Williams T. M. and Carroll S. B. (2009). Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 7, e1000168 10.1371/journal.pbio.1000168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth H. T. (1952). Mating behavior within the genus Drosophila (Diptera). Bull. AMNH 99, 395-474. [Google Scholar]
- Swanson C., Evans N. and Barolo S. (2010). Structural rules and complex regulatory circuitry constrain expression of a notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359-376. 10.1016/j.devcel.2009.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Barmina O. and Kopp A. (2009). Distinct developmental mechanisms underlie the evolutionary diversification of Drosophila sex combs. Proc. Natl. Acad. Sci. USA 106, 4764-4769. 10.1073/pnas.0807875106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Barmina O., Sanders L. E., Arbeitman M. N. and Kopp A. (2011). Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9, e1001131 10.1371/journal.pbio.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga C. (1962). Cell lineage and differentiation on the male foreleg of Drosophila melanogaster. Dev. Biol. 4, 489-516. 10.1016/0012-1606(62)90054-4 [DOI] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A. and Bellen H. J. (2006). P[acman ]: A BAC transgenic platform fragments in D. melanogaster. Science 314, 1747-1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Wexler J., Delaney E. K., Belles X., Schal C., Wada-Katsumata A., Amicucci M. and Kopp A. (2019). Hemimetabolous insects elucidate the origin of sexual development via alternative splicing bioRxiv 10.1101/587964 [DOI] [PMC free article] [PubMed]
- Williams T. M., Selegue J. E., Werner T., Gompel N., Kopp A. and Carroll S. B. (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610-623. 10.1016/j.cell.2008.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang W., Bayrer J. R. and Weiss M. A. (2008). Doublesex and the regulation of sexual dimorphism in drosophila melanogaster. J. Biol. Chem. 283, 7280-7292. 10.1074/jbc.M708742200 [DOI] [PubMed] [Google Scholar]
- Yi W., Ross J. M. and Zarkower D. (2000). mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127, 4469-4480. [DOI] [PubMed] [Google Scholar]
- Zhou C., Pan Y., Robinett C. C., Meissner G. W. and Baker B. S. (2014). Article central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83, 149-163. 10.1016/j.neuron.2014.05.038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.