ABSTRACT
Bone protrusions provide stable anchoring sites for ligaments and tendons and define the unique morphology of each long bone. Despite their importance, the mechanism by which superstructures are patterned is unknown. Here, we identify components of the genetic program that control the patterning of Sox9+/Scx+ superstructure progenitors in mouse and show that this program includes both global and regional regulatory modules. Using light-sheet fluorescence microscopy combined with genetic lineage labeling, we mapped the broad contribution of the Sox9+/Scx+ progenitors to the formation of bone superstructures. Then, by combining literature-based evidence, comparative transcriptomic analysis and genetic mouse models, we identified Gli3 as a global regulator of superstructure patterning, whereas Pbx1, Pbx2, Hoxa11 and Hoxd11 act as proximal and distal regulators, respectively. Moreover, by demonstrating a dose-dependent pattern regulation in Gli3 and Pbx1 compound mutations, we show that the global and regional regulatory modules work in a coordinated manner. Collectively, our results provide strong evidence for genetic regulation of superstructure patterning, which further supports the notion that long bone development is a modular process.
This article has an associated ‘The people behind the papers’ interview.
KEY WORDS: Cartilage, Patterning, Morphology, Superstructure, Sox9, Scleraxis, Modularity, Gli3, Pbx, Hox, Mouse
Highlighted Article: The development of protrusions, such as eminences and condyles, along long bones in mice is coordinated by a patterning mechanism involving global and regional genetic regulation.
INTRODUCTION
The vertebrate skeleton is composed of numerous bones, each displaying a unique shape and size. Yet, despite this morphological diversity, most bones are formed by a common developmental process, namely endochondral ossification (Berendsen and Olsen, 2015; Cervantes-Diaz et al., 2017; Long and Ornitz, 2013; Olsen et al., 2000). During this process, mesenchymal cells derived from the lateral plate mesoderm under the regulation of the transcription factor SRY-box 9 (SOX9) condense and differentiate into chondroprogenitors, forming the cartilaginous anlage of the future bones (Kawakami et al., 2006; Zhao et al., 1997). Next, a cascade of chondrocyte differentiation steps will give rise to growth plates on both proximal and distal ends of the anlage. Subsequently, blood vessels and bone-building cells, termed osteoblasts, invade the cartilage anlage and drive ossification of the cartilaginous template from the mid-shaft, pursuing the progression of the growth plates (Kronenberg, 2003).
Endochondral ossification has been studied extensively for more than two centuries, generating a vast amount of information on this process. Nevertheless, mechanisms that grant each bone with its distinctive shape are missing. A hallmark of the unique morphology of each long bone is the superstructures that protrude from its surface known as bone eminences, such as the greater and deltoid tuberosities, greater and lesser trochanters, etc., and condyles, such as the distal lateral and medial epicondyles of the humerus. One of the main functions of bone superstructures is to provide an attachment point for tendons and ligaments, which transmit force from the contracting muscles to the skeleton (Lessa et al., 2008; McHenry and Corruccini, 1975; Polly, 2007).
Interestingly, it was previously shown that the site where tendon attaches to bone is formed by a unique set of progenitors that co-express Sox9 and scleraxis (Scx) (Blitz et al., 2013; Sugimoto et al., 2013). Moreover, it was demonstrated that cells that contribute to the bony side of the attachment do not descend from the cells that create the bone shaft anlage, but are specified after them. Finally, it was demonstrated that these unique progenitors are specifically regulated by the TGFβ and BMP signaling pathways (Blitz et al., 2009, 2013). In addition to providing a mechanism for the development of tendon-to-bone attachment site, these findings also offer an alternative, modular model for long bone development. According to this model, one set of Sox9+ cells forms the cylindrical anlage of the future bone shaft, which will serve as the bone substructure, whereas a second pool of Sox9+/Scx+ cells will be added onto this substructure to give rise to the different superstructures.
Current literature provides ample evidence in support of the modular model of long bone development, which is based on lineage tracing, temporal initiation and cell differentiation of the Sox9+/Scx+ progenitor cells. However, this model is still missing the mechanisms that regulate the early patterning of the Sox9+/Scx+ progenitor cells, such that superstructures will form on the developing long bone at the right location, and with the correct shape and size. In this study, we examine the regulatory mechanisms underlying these aspects of modularity in skeletal development. Three-dimensional (3D) reconstruction of different long bones and their protruding superstructures allowed us to demonstrate modularity in long bone development. Comparative transcriptomic analysis and loss-of-function assays showed that GLI-Kruppel family member (Gli3) globally regulates the spatial organization of the Sox9+/Scx+ progenitors in both forelimb and hindlimb, whereas homeobox a11 (Hoxa11) and Hoxd11 regulate patterning of distal superstructures and pre-B cell leukemia homeobox 1 (Pbx1) and Pbx2 regulate proximal superstructure progenitors. Overall, we provide cellular evidence for the existence of a patterning mechanism that involves both global and regional regulation and highlight some of the genes that facilitate the patterning of superstructures along the developing long bones.
RESULTS
Sox9+/Scx+ superstructure progenitors contribute extensively to the morphology of long bone anlagen
To understand better the scope of modularity in long bone morphogenesis, we performed pulse-chase cell-lineage experiments by crossing either Sox9-CreERT2 or collagen type II alpha 1 (Col2a1)-CreERT2 transgenic reporter mice with Rosa26-tdTomato reporter mice (Madisen et al., 2010; Nakamura et al., 2006; Soeda et al., 2010). Pregnant females were administered a single dose of 0.03 mg/g tamoxifen/body weight at either embryonic day (E) 10.5 or E11.5. Previously, we showed that this time window allows the exclusive labeling of cells of the cylindrical bone substructure, leaving the superstructure cells unlabeled (described in detail by Blitz et al., 2013; Eyal et al., 2015). To map comprehensively the unlabeled superstructure cells in different skeletal elements, we have established a 3D imaging pipeline that includes clearing of the labeled limbs optically, light-sheet fluorescence microscopy and, finally, reconstruction of obtained images into a 3D object (Treweek et al., 2015; Yang et al., 2014). To visualize the superstructures better, whole-mount limbs were immunostained for COL2A1.
As seen in Fig. 1A,B (and Movies 1-3), in E14.5 Sox9-tdTomato+ or E15.5 Col2a1-tdTomato+ limbs, all superstructures of different long bones were tdTomato negative, including greater, lesser and deltoid tuberosities, olecranon, and greater, lesser and third trochanters. Interestingly, in addition to bone eminences, we observed various condyles and sesamoid bones that were also tdTomato negative, including distal medial and lateral humeral epicondyles, medial tibial condyle, patella, and medial and lateral fabella. These results demonstrate the generality of the modular process of long bone morphogenesis in the limb.
Fig. 1.
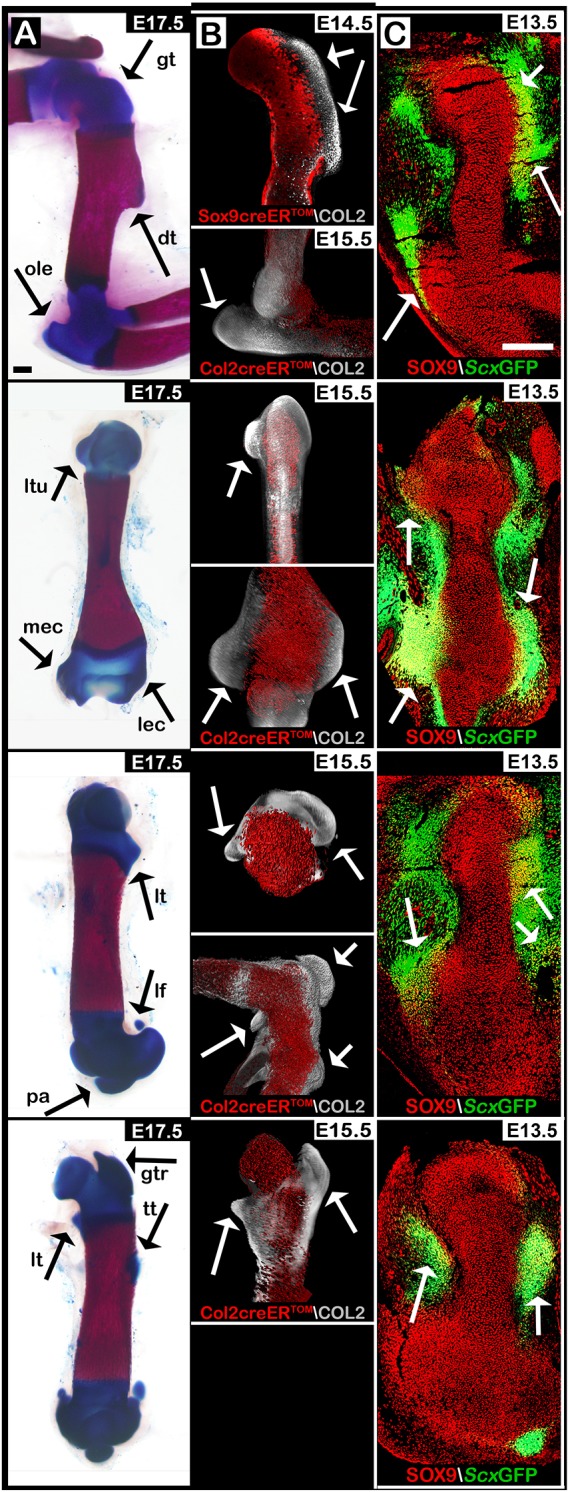
Long bone morphology is affected extensively by numerous pools of Sox9+/Scx+ progenitors. Images showing sagittal and coronal views of the superstructures that form along the forelimb and hindlimb long bones. (A) Skeletal preparations of E17.5 wild-type limbs. Superstructures are indicated by black arrows. (B) Maximum intensity projection (MIP) images of E14.5 forelimb from Sox9-CreER-tdTomato transgenic embryos and E15.5 fore- and hindlimbs from Col2a1-CreER-tdTomato transgenic embryos. Whole-mount limbs were stained against COL2A1, imaged using light-sheet microscopy and reconstructed. Whereas first-wave Sox9+ progenitors were labeled by tdTomato, the secondary wave of Sox9+/Scx+ progenitors remained unstained. tdTomato− precursors contributed exclusively to superstructure formation (white arrows). (C) Sagittal and coronal sections through E13.5 fore- and hindlimbs from ScxGFP transgenic embryos stained against SOX9. SOX9 and Scx co-expressing cells are indicated by white arrows. Sagittal sections in first and third rows; coronal sections in second and fourth rows. dt, deltoid tuberosity; gt, greater tuberosity; gtr, greater trochanter; lec, lateral epicondyle; lf, lateral fabella; lt, lesser trochanter; ltu, lesser tuberosity; mec, medial epicondyle; ole, olecranon; pa, patella; tt, third trochanter. Scale bars: 200 µm.
To demonstrate this modularity further, we examined sagittal and coronal sections of E13.5 fore- and hindlimbs from ScxGFP transgenic embryos (Pryce et al., 2007) that were stained using antibodies against SOX9. As seen in Fig. 1C, we observed an abundance of SOX9+ and Scx+ double-positive cells at anatomical locations of various future superstructures along the shafts of different bones. Taken together, these results indicate the extensive contribution of Sox9+/Scx+ superstructure progenitors to the 3D morphology of cartilaginous anlagen, highlighting the modularity of this process both temporally and from lineage analyses.
Gli3 is a global regulator of Sox9+/Scx+ progenitors patterning during superstructure development
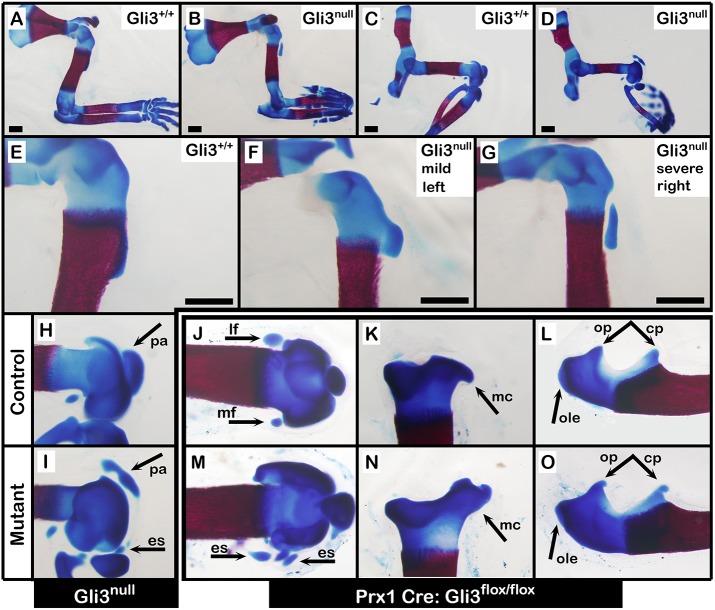
An immediate implication of the modular model is the existence of a mechanism that regulates the patterning of the Sox9+/Scx+ progenitors to position the superstructure correctly along the bone substructure. To uncover this mechanism, we first searched the literature for mutations leading to observable superstructure patterning abnormalities. We found that the skeletons of Gli3-null embryos, previously known as extra toes (Xt), exhibit a variety of superstructure abnormalities (Johnson, 1967). To characterize these defects, we crossed Gli3 heterozygous mice and examined skeletal preparations from E17.5 embryos. As seen in Fig. 2A-I, both fore- and hindlimbs of Gli3null embryos exhibited numerous dysplastic superstructures, including lesser and deltoid tuberosity, olecranon, patella, and additional ectopic sesamoids. Notably, the aberrant patterning of the deltoid tuberosity ranged in severity from mildly abnormal morphology to complete separation from the humeral shaft (compare left and right forelimbs in Fig. 2F and 2G, respectively) (Table S1). We also examined Prx1-Cre;Gli3fl/fl embryos, in which Gli3 was conditionally knocked out (cKO) in limb mesenchyme (Blaess et al., 2008; Logan et al., 2002). Similar to Gli3null mutants, limb-specific Gli3 cKO also resulted in diverse abnormally patterned superstructures, such as induction of supernumerary sesamoids around the knee, dysplastic medial tibial condyle and dysplastic ulnar olecranon and coronoid processes (Fig. 2J-O).
Fig. 2.
Gli3 regulates the patterning of superstructures globally. (A-O) Skeletal preparations of E17.5 fore- and hindlimbs from Gli3 KO and control embryos (A-I) and from limb-specific Prx1-Cre;Gli3floxed cKO and control embryos (J-O). Affected superstructures are indicated by black arrows. cp, coronoid process; es, ectopic sesamoids; lf, lateral fabella; mc, medial tibial condyle; mf, medial fabella; ole, olecranon; op, olecranon process; pa, patella. Scale bars: 500 µm.
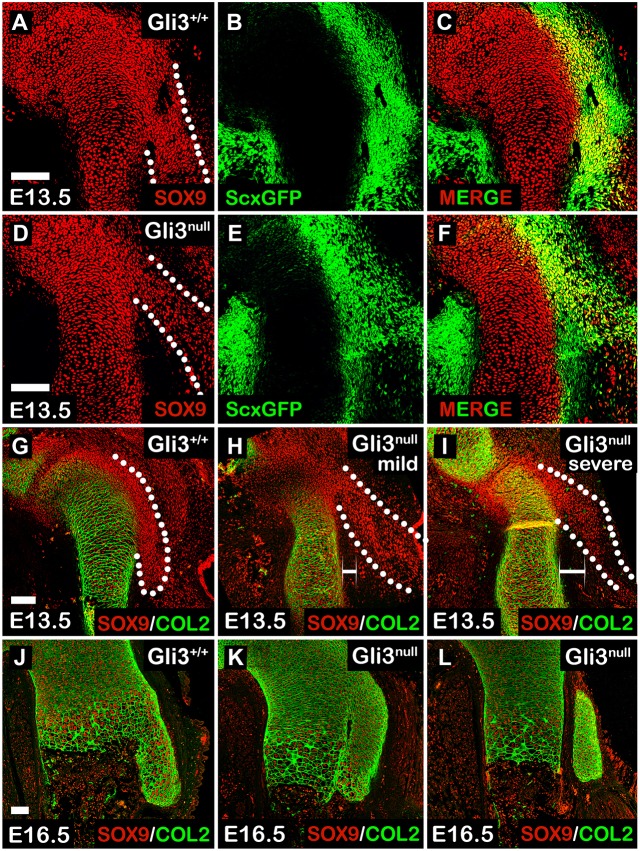
To study the possibility that Gli3 regulates the patterning of superstructure progenitors, we sought to examine the distribution of Sox9+/Scx+ cells in Gli3null limbs. For that, we crossed heterozygous Gli3 mice with Gli3+/−; ScxGFP transgenic reporter mice and stained sagittal sections of E13.5 fore- and hindlimbs against SOX9 to highlight chondrocytes. As seen in Fig. 3A-F, the spatial distribution of Sox9+/Scx+ progenitors in Gli3null limbs was abnormal. Whereas in control limbs these progenitors were patterned in juxtaposition to the humeral shaft (Fig. 3A, dotted line), in the mutant these cells were laterally spread and less condensed (Fig. 3D, dotted line). To understand how the abnormal distribution of Sox9+/Scx+ progenitors translates into superstructure abnormality, we examined the spatiotemporal differentiation of deltoid tuberosity precursors in E13.5 and E16.5 sagittal sections from control and Gli3 KO forelimbs. Sections were stained using antibodies against SOX9 and COL2A1, a marker for differentiated chondrocytes (Fig. 3G-L). As expected, at E13.5 we repeatedly observed the lateral spreading of deltoid tuberosity precursors (Fig. 3H,I, dotted lines). Moreover, we noticed that the degree of cellular lateralization varied both in pattern and in distance from the humeral shaft (Fig. 3H,I, white bars). At E16.5, the abnormal deltoid tuberosity morphologies were clearly evident and ranged from dysplastic to separate deltoid tuberosity (Fig. 3K and 3L, respectively). We concluded that Gli3 is necessary for correct spatial organization of deltoid tuberosity Sox9+/Scx+ precursors and, thus, for superstructure patterning.
Fig. 3.
Organization but not specification of Sox9+/Scx+ progenitors is regulated by Gli3. (A-F) Sagittal sections through the proximal humeri of E13.5 Gli3;ScxGFP transgenic embryos that were stained against SOX9. Although SOX9 and Scx co-expressing cells are observed in control and mutant embryos (C,F), their spatial organization is abnormal in Gli3null mutants (A,D, dotted lines). (G-L) Sagittal sections through the proximal humeri of E13.5 (G-I) and E16.5 (J-L) Gli3 KO and control embryos that were stained against SOX9 and COL2A1. At E13.5, spatial organization of deltoid tuberosity precursors is abnormal in mutants (G-I, dotted lines), displaying varying degrees of lateralization and distance from the humeral shaft (H,I, white brackets). In E16.5 mutants, aberrant deltoid tuberosity morphology ranges from aplasia (K) to detachment from the humeral shaft (L). Scale bars: 100 µm.
The finding that Gli3 regulates superstructure patterning raises the question whether Gli3 acts autonomously within the Sox9+/Scx+ progenitors. To directly examine whether Gli3 operates autonomously within the Sox9+/Scx+ progenitor population, we blocked the expression of Gli3 in Scx- or Sox9-expressing cells by crossing Scx-Cre or Sox9-Cre mice to Gli3fl/fl mice. As seen in Fig. 4, skeletal preparation of both Scx-Cre;Gli3fl/fl (Fig. 4A-B′) and Sox9-Cre;Gli3fl/fl cKOs (Fig. 4C-D′) revealed hypoplasia of the deltoid tuberosity and lateral fabella. In addition, we observed dysplasia of the patella in Sox9-Cre;Gli3fl/fl mutants (Fig. 4D′). This finding clearly indicates an autonomous role of Gli3 in superstructure patterning; yet, the milder phenotypes that we observed in the skeletons of both Scx-Cre;Gli3fl/fl and Sox9-Cre;Gli3fl/fl cKOs relative to the Gli3null mice suggest a potential additional, non-autonomous effect of Gli3 in superstructure patterning.
Fig. 4.
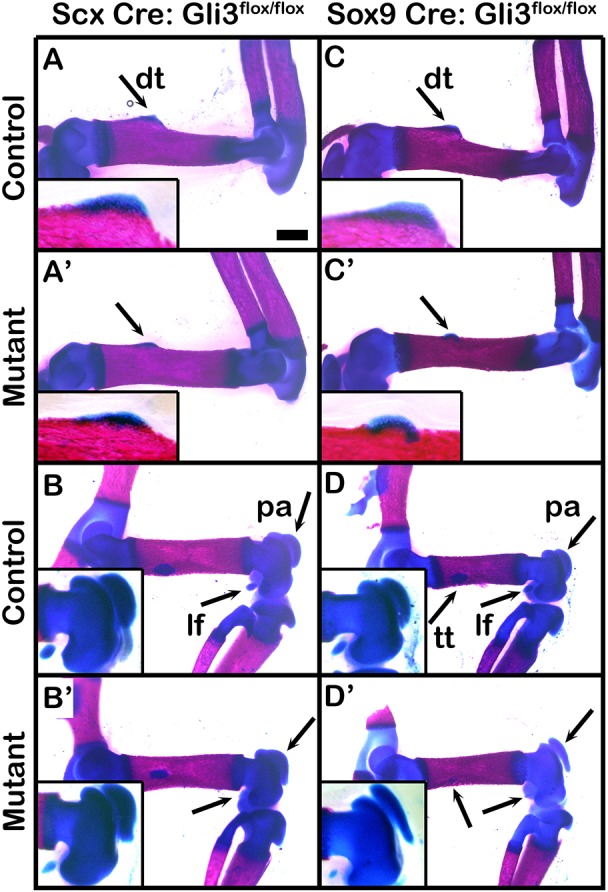
Autonomous Gli3 regulation has a mild effect on superstructure patterning. (A-D′) Skeletal preparations of E17.5 fore- and hindlimbs from tenocyte-specific Scx-Cre;Gli3floxed cKO and control embryos (A-B′) and from chondrocyte-specific Sox9-Cre;Gli3floxed cKO and control embryos (C-D′). Affected superstructures are indicated by black arrows. Insets show deltoid tuberosity or knee areas. dt, deltoid tuberosity; lf, lateral fabella; pa, patella; tt, third trochanter. Scale bar: 500 µm.
Comparative transcriptomic analysis of proximal and distal Sox9+/Scx+ progenitors predicts regional regulation of superstructure patterning
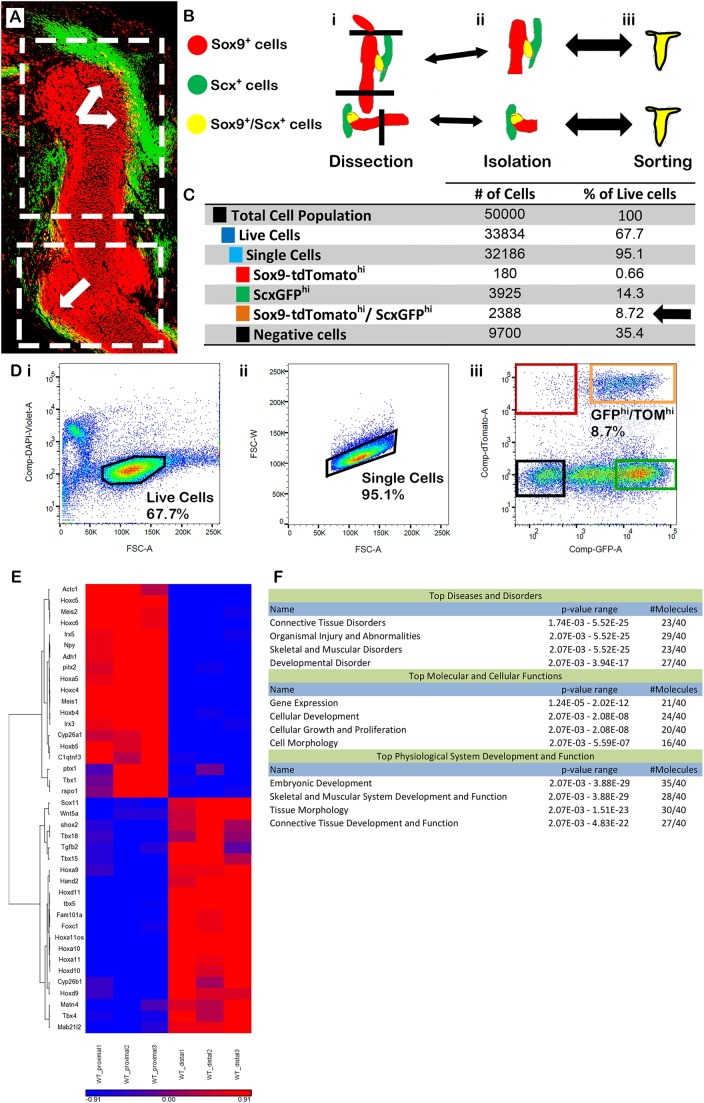
Although Gli3 had a global effect on superstructure patterning, we suspected that regional regulatory circuitry also exists, such that patterning of specific superstructures could be regulated independently. To search for genes potentially involved in this mechanism, we developed a protocol for prospective isolation of Sox9-tdTomato+/ScxGFP+ progenitors by fluorescence-activated cell sorting (FACS). Moreover, to enable distinction between global and regional patterning regulators, we designed our system to allow separate isolation of proximal and distal cell populations. To this end, we crossed Sox9-CreERT2;Rosa26-tdTomato reporter mice with ScxGFP transgenic reporter mice. Pregnant females were administered a single dose of 0.03 mg/g tamoxifen/body weight at E12.0 (Fig. 5A). Next, we harvested E13.5 Sox9-tdTomato+/ScxGFP+ embryos and micro-dissected their forelimbs at the shoulder level, above the presumable deltoid tuberosity, and at flanking sides of the elbow (Fig. 5Bi,Bii). Finally, the dissected segments were collected into individual tubes, homogenized and then prospectively isolated by FACS (Fig. 5Biii). For each proximal and distal segment, we collected three biological repeats from three separate litters. Each sample contained 7-9% Sox9-tdTomato+/ScxGFP+ progenitors of total cell population, of which 10,000 cells were collected for transcriptome analysis (Fig. 5C). Further information regarding controls, gate settings and instrument configuration is found in Fig. 5D and in Materials and Methods.
Fig. 5.
Comparative transcriptomic analysis highlights regional regulation of superstructure patterning. (A) Sagittal section through the forelimb of E13.5 Sox9-CreERtdTomato;Scx-GFP transgenic reporter embryo following tamoxifen administration at E12.0. Sox9 and Scx co-expressing cells are indicated by white arrows. Dashed white rectangles indicate the proximal and distal regions of interest that were dissected for FACS and transcriptome analysis. (B) Schematics of sample preparation. Labeled forelimbs were dissected (i), the proximal and distal segment cells were isolated (ii) and then sorted independently (iii). (C) A table summarizing cell type distribution among 50,000 collected and FACS-sorted cells. The number and percentage of Sox9-tdTomato+/ScxGFP+ progenitors are indicated by a black arrow. (D) Illustrations of control and gating settings. (i) Live cells were controlled for by DAPI staining. (ii) Single cells were gated according to droplet area (FSC-A) versus width (FSC-W). (iii) Gating settings for Sox9-tdTomato+/ScxGFP+ progenitors resulted in collection of 7-9% of the total living single-cell population (orange rectangle). Colored rectangles indicate populations as listed in C. (E) Heat map showing the clustering of 40 of the most differentially expressed genes between proximal and distal Sox9-tdTomato+/ScxGFP+ progenitors. (F) Using Ingenuity software, these genes were annotated and found to be biologically relevant to limb development and tissue morphology.
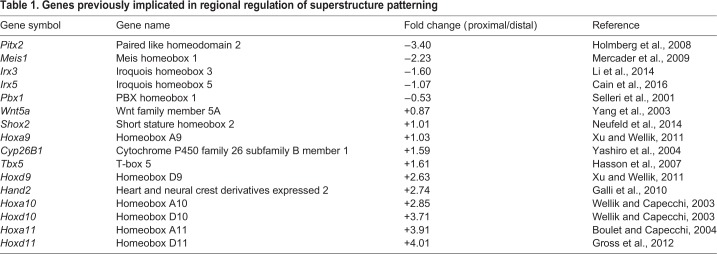
Comparison between the transcriptomes of proximal and distal Sox9-tdTomato+/ScxGFP+ progenitors revealed 561 differentially expressed genes with at least two-fold change in expression levels between segments (Table S2). We further analyzed 40 of these genes that were considered most likely to be involved, based on the difference in expression levels, enrichment in relevant biological function and known roles in embryonic development (Fig. 5E,F). Notably, 16 of these 40 candidate genes (listed in Table 1) have previously been implicated in superstructure development, providing a strong validation for the effectiveness of our strategy.
Table 1.
Genes previously implicated in regional regulation of superstructure patterning
Hoxa11, Hoxd11 and Pbx1 genes regionally regulate distal or proximal superstructure patterning
Our transcriptome analysis revealed that Hoxa11 and Hoxd11 were highly expressed in distal Sox9-tdTomato+/ScxGFP+ progenitors and scarcely expressed in proximal cells. Intriguingly, a previous study has demonstrated that a Hoxa11/Hoxd11 compound mutation (Hox11aadd) resulted in diverse distal forelimb abnormalities, such as the formation of a detached olecranon resembling a sesamoid bone, whereas the proximal limb segment, including the deltoid tuberosity, was not affected (Koyama et al., 2010). Nevertheless, the mechanism underlying the abnormal olecranon development remained to be elucidated. Therefore, we proceeded to study this process in Hox11aadd mutant embryos.
To that end, we crossed double-heterozygous Hox11AaDd mice and harvested embryos at E13.5 and E17.5 (Wellik and Capecchi, 2003). Examination of skeletal preparations from E17.5 control and mutant embryos validated the abnormal development of the olecranon (Fig. 6A,A′). Next, we validated the differential expression of Hoxd11 by fluorescent in situ hybridization (FISH) assay on sagittal sections of E13.5 forelimbs. In agreement with our transcriptome analysis results, Hoxd11 was highly expressed distally in proximity to the elbow, but scarcely expressed in proximal parts of the limb (Fig. 6B). Furthermore, Hoxd11 expression was downregulated in Hox11aadd mutants (Fig. 6B′). The differential expression of Hoxa11 at this stage has been previously demonstrated (Swinehart et al., 2013).
Fig. 6.
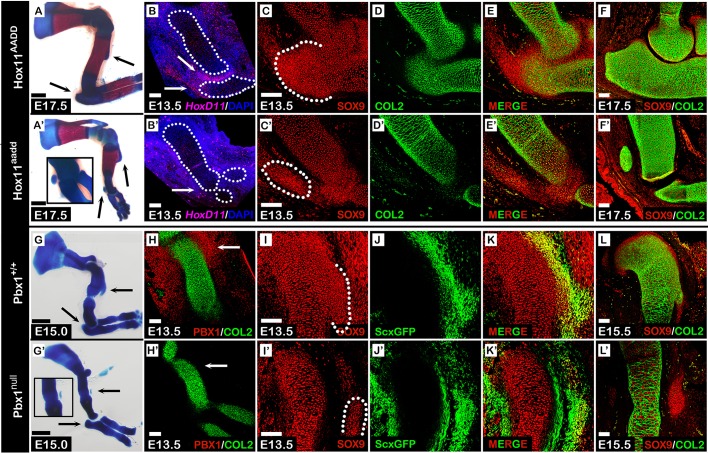
Distal and proximal superstructure patterning is regulated by Hoxa11, Hoxd11 and Pbx1. (A-F′) Hoxa11 and Hoxd11 regionally regulate patterning of the distal olecranon. (A,A′) Skeletal preparations of E17.5 forelimbs from compound Hoxa11 and Hoxd11 mutant and control embryos. Distal olecranon but not proximal deltoid tuberosity developed abnormally in Hox11aadd mutant forelimbs (black arrows and inset). (B,B′) Fluorescent in situ hybridization for Hoxd11 in forelimb sections shows that at E13.5, Hoxd11 is distally expressed in the elbow region (B, white arrows). Expression of Hoxd11 was downregulated in Hox11aadd mutant forelimbs (B′). Long bones are demarcated by dotted lines. (C-F′) Sagittal sections through the elbow of E13.5 (C-E′) and E17.5 (F,F′) Hox11aadd compound mutant and control embryos that were stained against SOX9 and COL2A1. At E13.5, spatial organization of olecranon precursors is abnormal (C,C′). By E17.5, the developing olecranon of mutant embryos has detached from the ulnar shaft (F,F′). (G-L′) Pbx1 regionally regulates patterning of the proximal deltoid tuberosity. (G,G′) Skeletal preparations of E15.0 forelimbs from Pbx1-null and control embryos. Proximal deltoid tuberosity but not distal olecranon developed abnormally in Pbx1 mutant forelimbs (black arrows and inset). (H,H′) Staining against PBX1 and COL2A1 in forelimb sections shows that at E13.5, Pbx1 is proximally expressed at the shoulder region (H, white arrows). Expression of Pbx1 was downregulated in Pbx1null mutant forelimbs (H′). (I-K′) Sagittal sections through the proximal humeri of E13.5 Pbx1;ScxGFP transgenic embryos that were stained against SOX9. Although SOX9 and Scx co-expressing cells are observed in control and mutant embryos (K,K′), their spatial organization was abnormal in Pbx1null mutants (I,I′). (L,L′) Sagittal sections through the proximal humeri of E15.5 Pbx1 mutant and control embryos that were stained against SOX9 and COL2A1. At E15.5, the deltoid tuberosity of mutant embryos is detached from the humeral shaft (L,L′). Scale bars: 500 µm (skeletal preparations); 100 µm (sections).
To study the regulatory roles of Hoxa11 and Hoxd11 in distal patterning of superstructure progenitors, we examined the patterning of the olecranon in control and mutant embryos. For this, we crossed double-heterozygous Hox11AaDd mice and stained forelimb sections from E13.5 double-homozygous Hox11aadd embryos using antibodies against SOX9 and COL2A1, such that the substructure cells were expected to be Sox9+/Col2a1+ and the undifferentiated superstructure progenitors to be Sox9+/Col2a1−. Results showed that the olecranon progenitors failed to organize in the typical pattern observed in control limbs (Fig. 6C-E′). Specifically, the cells were scattered in a proximolateral direction away from the ulnar shaft (Fig. 6C,C′; dotted lines). Sections from E17.5 Hox11aadd mutants illustrated that the olecranon precursors differentiated at a distance from the ulnar shaft, creating a distinct skeletal element resembling a sesamoid bone (Fig. 6F,F′). These results suggest that Hoxa11 and Hoxd11 are necessary for correct localization of the olecranon progenitors and, thus, for their patterning. Moreover, the proper patterning of superstructure progenitors on the proximal side supports our hypothesis that Hoxa11 and Hoxd11 act as regional distal regulators (Fig. S1A,A′).
Another promising candidate gene from our transcriptome analysis was Pbx1. In contrast to Hoxa11 and Hoxd11, Pbx1 was highly expressed in proximal Sox9-tdTomato+/ScxGFP+ progenitors compared with distal cells. Indeed, Pbx1null mutation has been shown to affect only proximal elements, such as the deltoid tuberosity, whereas the distal limb segment, including the olecranon, was not affected (Selleri et al., 2001). We therefore performed the same analysis on Pbx1null;ScxGFP mutants. After validation of the deltoid tuberosity phenotype in skeletal preparations of E15.0 control and mutant embryos (Fig. 6G,G′), we analyzed Pbx1 expression by immunostaining sagittal sections of E13.5 forelimbs against PBX1 and COL2A1. In agreement with the transcriptome analysis results, PBX1 was highly expressed proximally in proximity to the deltoid tuberosity and was less prominent in the distal forelimb (Fig. 6H). As expected, PBX1 expression was completely ablated in Pbx1null mutants (Fig. 6H′).
Next, forelimb sections from Pbx1null;ScxGFP+ E13.5 embryos were stained using antibodies against SOX9. As seen in Fig. 6I-K, the Sox9+/Scx+ deltoid tuberosity precursors were specified in both control and mutant limbs; yet, in the mutant they were organized abnormally and scattered laterally. Finally, examination of sections immunostained against SOX9 and COL2A1 at E15.5 showed that in Pbx1null mutants, the deltoid tuberosity precursors differentiated into a distinct sesamoid-like cartilaginous element parallel to the humeral shaft (Fig. 6L,L′). These results indicate that Pbx1 is necessary for correct localization of the Sox9+/Scx+ deltoid tuberosity precursors and, thus, for their patterning. Importantly, the patterning of superstructure progenitors at the distal region was independent of Pbx1 regulation (Fig. S1B,B′).
Together, these results indicate that superstructure patterning is regulated regionally through early modulation of the spatial organization of Sox9+/Scx+ progenitors by Hoxa11, Hoxd11 and Pbx1.
Both Pbx1 and Pbx2 are involved in patterning of proximal superstructures
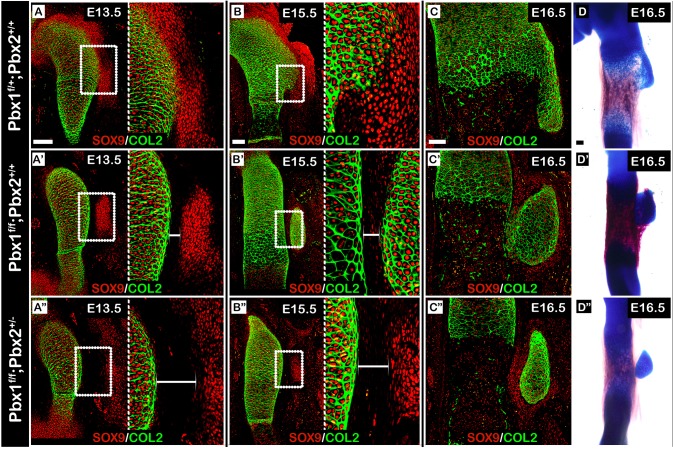
Pbx2 is a paralog of Pbx1 that is expressed throughout the limb mesenchyme. Previously, it was shown that Pbx1 and Pbx2 act in a dosage-dependent manner during proximal limb development (Capellini, 2006). This led us to examine whether Pbx2 co-regulates proximal superstructure patterning with Pbx1. To avoid early embryonic lethality, we ablated one allele of Pbx2 on the background of a limb-specific knockout of Pbx1 (Prx1-Cre;Pbx1fl/fl;Pbx2+/−) and compared the phenotype with both wild-type (Prx1-Cre−;Pbx1+/+;Pbx2+/+) and Pbx1 cKO (Prx1-Cre;Pbx1fl/fl) embryos (Ficara et al., 2008; Selleri et al., 2004). To this end, sections from E13.5, E15.5 and E16.5 control and mutant forelimbs were immunostained using antibodies against SOX9 and COL2A1 (Fig. 7A-C″). At E13.5, we observed abnormal patterning of deltoid tuberosity precursors in both Prx1-Cre;Pbx1fl/fl and Prx1-Cre;Pbxfl/fl;Pbx2+/− mutant embryos. The precursors were laterally scattered (Fig. 7A′,A″), comparable to the deltoid tuberosity precursors of Pbx1null limbs (Fig. 6I′). By E15.5, the deltoid tuberosity precursors of Prx1-Cre;Pbx1fl/fl embryos had fully differentiated, whereas in Prx1-Cre;Pbx1fl/fl;Pbx2+/− we observed delayed differentiation (Fig. 7B-B″). Notably, in Prx1-Cre;Pbx1fl/fl embryos the humerus-deltoid tuberosity boundary was populated by SOX9+ cells that had not been detected in Prx1-Cre;Pbx1fl/fl;Pbx2+/− embryos. Moreover, the gap between the humeral shaft and deltoid tuberosity precursors was roughly twice as large in Prx1-Cre;Pbx1fl/fl;Pbx2+/− limbs than in Prx1-Cre;Pbx1fl/fl limbs. Finally, by E16.5, the deltoid tuberosity of Prx1-Cre;Pbx1fl/fl embryos has attached to the humeral shaft, whereas the deltoid tuberosity of Prx1-Cre;Pbx1fl/fl;Pbx2+/− mutants remained separated (Fig. 7C-C″), as in Pbx1null mutants (Fig. 6L′). These diverging morphologies were further validated by skeletal preparations made of limbs taken from each mutants (Fig. 7D-D″). Taken together, these results suggest that PBX genes act together in regulating the patterning of proximal superstructures, possibly by regulating the level of lateralization in a dose-dependent manner.
Fig. 7.
Pbx1 and Pbx2 coordinately regulate the patterning of proximal superstructures. (A-C″) Sagittal sections through the proximal humeri of E13.5 (A-A″), E15.5 (B-B″) and E16.5 (C-C″) limbs from control (A-C), Prx1-Cre;Pbx1floxed (B-B″) or Prx1-Cre;Pbx1floxed;Pbx2het (C-C″) embryos that were stained against SOX9 and COL2A1. In A-B″, the right side of the panel is an enlargement of the humerus-deltoid tuberosity boundary region demarcated by a white rectangle on the left. At E13.5, spatial organization of deltoid tuberosity precursors in both mutants was abnormal (A′,A″) as deltoid tuberosity precursors were scattered laterally away from the humeral shaft. The gap between the humeral shaft and deltoid tuberosity precursors was twice as large in Prx1-Cre;Pbx1floxed;Pbx2het mutants than in Prx1-Cre;Pbx1floxed mutants (A′,A″, white brackets). At E15.5, whereas cells at the humerus- deltoid tuberosity boundary of Prx1-Cre;Pbx1floxed mutants began to express high levels of SOX9, such expression was not observed in Prx1-Cre;Pbx1floxed;Pbx2het mutants. Moreover, the difference in boundary region size remained consistent (B,B′, white brackets). At E16.5, deltoid tuberosity morphology ranged from attached but aplastic deltoid tuberosity in Prx1-Cre;Pbx1floxed mutants (C′) to a detached deltoid tuberosity in Prx1-Cre;Pbx1floxed;Pbx2het mutants (C″). (D-D″) Skeletal preparations of E16.5 forelimbs further validate these diverging morphologies. Scale bars: 100 µm.
Interaction between global and regional genetic programs fine-tunes patterning of superstructures
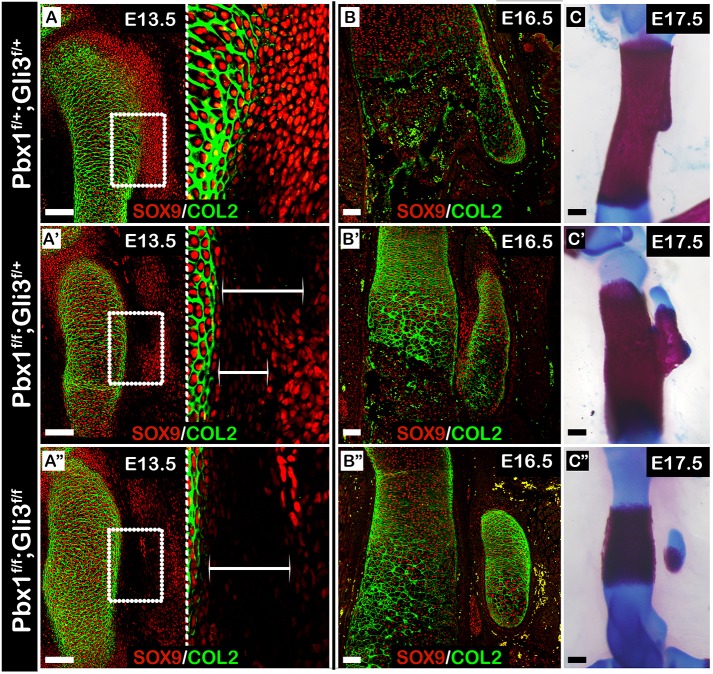
The finding that superstructure patterning can be regulated both globally and regionally led us to hypothesize that these two programs interact with one another. To examine this possibility, we produced a compound mutant mouse carrying limb-specific knockout of Pbx1 and Gli3. Specifically, on the background of a limb-specific knockout of Pbx1 (Prx1-Cre;Pbx1fl/fl) we ablated either one or two alleles of Gli3 (Prx1-Cre;Pbx1fl/fl;Gli3fl/+ or Prx1-Cre;Pbx1fl/fl;Gli3fl/fl, respectively). Then, sections from E13.5 and E16.5 control and mutant forelimbs were immunostained using antibodies against SOX9 and COL2A1 (Fig. 8A-B″). At E13.5, deltoid tuberosity precursors in the Prx1-Cre;Pbx1fl/fl;Gli3fl/fl embryos were laterally scattered and noticeably less condensed than in Prx1-Cre;Pbx1fl/fl;Gli3fl/+ embryos (Fig. 8A-A″). The phenotypic difference between genotypes became more pronounced by E16.5. Whereas ablation of a single Gli3 allele resulted in a dysplastic deltoid tuberosity that was attached to the humeral shaft, ablation of both alleles resulted in a detached deltoid tuberosity (Fig. 8B-B″). These diverging morphologies were further validated by skeletal preparations of limbs from each mutant group (Fig. 8C-C″). Importantly, whereas individual limb-specific knockout of either Prx1-Cre;Gli3fl/fl (data not shown) or Prx1-Cre;Pbx1fl/fl (Fig. 7D′) resulted in dysplastic deltoid tuberosity, the compound Prx1-Cre;Pbx1fl/fl;Gli3fl/fl double knockout mutant displayed a detached deltoid tuberosity, as in both Gli3null (Fig. 2G) and Pbx1null (Fig. 6G′) embryos. These results suggest that, similar to the compound Pbx1/Pbx2 mutation, genetic interactions also occurred between Pbx1 and Gli3 in a dose-dependent manner, highlighting the possibility of interactions between the global and regional regulatory programs.
Fig. 8.
Coordinated regulation by Pbx1 and Gli3 highlights interaction between global and regional genetic programs. (A-B″) Sagittal sections through the proximal humeri of E13.5 (A-A″) and E16.5 (B-B″) limbs from control (A,B), Prx1-Cre;Pbx1floxed;Gli3fl/+ (A′,B′) or Prx1-Cre;Pbx1floxed;Gli3floxed (A″,B″) embryos that were stained against SOX9 and COL2A1. (C-C″) Skeletal preparations of E17.5 forelimbs from control (C), Prx1-Cre;Pbx1floxed;Gli3fl/+ (C′) or Prx1-Cre;Pbx1floxed;Gli3floxed (C″) embryos. In A-A″, the right side of the panel is an enlargement of the humerus-deltoid tuberosity boundary region demarcated by a white rectangle on the left. At E13.5, spatial organization of deltoid tuberosity precursors in both mutants was abnormal (A′,A″) as deltoid tuberosity precursors were scattered laterally away from the humeral shaft. The gap between the humeral shaft and deltoid tuberosity precursors was twice as large in Prx1-Cre;Pbx1floxed;Gli3floxed mutants than in Prx1-Cre;Pbx1floxed;Gli3fl/+ mutants (A′,A″, white brackets). At E16.5, whereas cells at the humerus-deltoid tuberosity boundary of Prx1-Cre;Pbx1floxed;Gli3fl/+ mutants began to express high levels of SOX9, such expression was not observed in Prx1-Cre;Pbx1floxed;Gli3floxed mutants. Moreover, the difference in boundary region size remained consistent (B′,B″). At E17.5, deltoid tuberosity morphology ranged from attached but aplastic deltoid tuberosity in Prx1-Cre;Pbx1floxed;Gli3fl/+ mutants (C′) to a detached deltoid tuberosity in Prx1-Cre;Pbx1floxed;Gli3floxed mutants (C″). Scale bars: 200 µm (skeletal preparations); 100 µm (sections).
The mechanism underlying the abnormal superstructure development
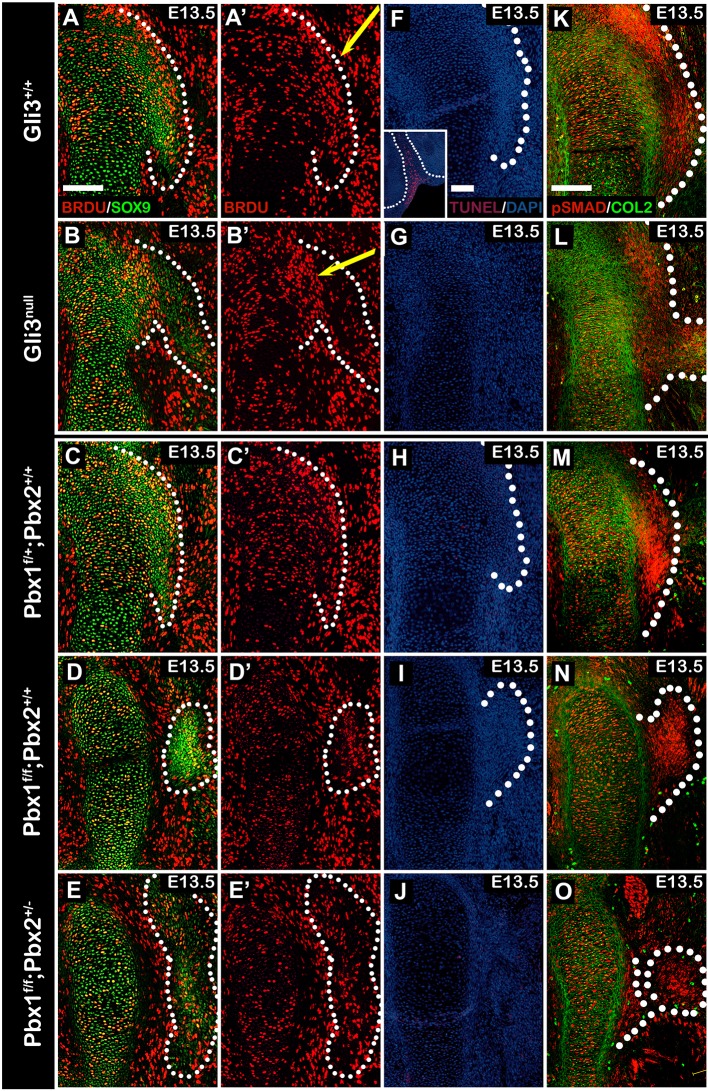
The abnormal superstructure development we observe in the different mutant mouse models could be the result of several possible mechanisms that affect the Sox9+/Scx+ progenitors, including patterning, proliferation, differentiation and cell death. Having already demonstrated abnormal patterning of the Sox9+/Scx+ progenitors in the different mutants, we next sought to study whether proliferation rate contributed to the observed phenotype. To this end, we analyzed incorporation of 5-bromo-2-deoxyuridine (BrdU) into Sox9+/Scx+ cells, focusing on the hypoplastic deltoid tuberosity. Gli3 and Prx1-Cre;Pbx1;Pbx2 pregnant mice were injected subcutaneously with BrdU. Incorporation of BrdU was allowed for a period of 2 h, after which the pregnant mice were sacrificed. Incorporated BrdU was detected by immunostaining using antibodies against BrdU and SOX9 on sections taken from E13.5 Gli3 and Prx1-Cre;Pbx1;Pbx2 control and mutant forelimbs. Next, the total SOX9+ and SOX9+/BrdU+ cells were counted in the region of the greater and deltoid tuberosities and the fraction of proliferating cells was calculated for both control and mutant embryos (Fig. S2). Our results showed that in Gli3 mutants, the center of proliferation appeared to shift inwards from the lateral side of the superstructures to the main shaft (Fig. 9A′,B′; yellow arrows demarcate center of proliferation). However, quantification showed no significant difference in the fraction of proliferating cells. In contrast, we observed a mild but significant reduction of proliferating cells in both Prx1-Cre;Pbx1floxed and Prx1-Cre;Pbx1floxed;Pbx2het mutant embryos (Fig. 9D′,E′) compared with control embryos (Fig. 9C′) (Fig. S2).
Fig. 9.
Superstructure size and shape is independent of proliferation, differentiation or apoptosis. (A-O) Sagittal sections through the proximal humeri of E13.5 Gli3 control (A,A′,F,K) and mutant (B,B′,G,L) embryos or from control (C,C′,H,M), Prx1-Cre;Pbx1floxed (D,D′, I,N) or Prx1-Cre;Pbx1floxed;Pbx2het (E,E′,J,O) embryos. (A-E′) BrdU staining was performed to highlight proliferating cells within the superstructure precursors (demarcated by dotted lines). No reduction in proliferating cells was observed in Gli3 mutants; however, the center of proliferation appeared to shift from the lateral face of the superstructures in control embryos to the main shaft of the mutant embryos (A′,B′; yellow arrows demarcate center of proliferation). In Prx1-Cre;Pbx1floxed (D,D′) and Prx1-Cre;Pbx1floxed;Pbx2het (E,E′) mutant embryos, we observed a mild reduction of proliferating cells in comparison with the control embryos (C,C′). (F-J) TUNEL staining was performed to highlight apoptotic cells within the superstructure precursors (demarcated by dotted lines). No cell death within the superstructure precursors was noticed in either control or mutant embryos. The interdigital space, which undergoes massive cell death at E13.5, was used as control for the staining (F, inset). (K-O) To highlight differentiating cells within the superstructure precursors (demarcated by dotted lines), we performed immunostaining for pSMAD1/5 and counterstained for COL2A1. The results show high pSMAD1/5 activity within all superstructure precursors indicating normal differentiation in both control and mutant embryos. Scale bars: 100 µm.
The detached deltoid tuberosity could be the consequence of apoptosis. To examine the possible involvement of cell death in this phenotype, we performed a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay on sections taken from E13.5 Gli3 and Prx1-Cre;Pbx1;Pbx2 control and mutant forelimbs. As a control, we examined the interdigital space, which undergoes massive cell death at E13.5 (Fig. 9F, inset). Results showed no cell death within the superstructure precursors in either control or mutant embryos (Fig. 9F-J; superstructures are demarcated by dotted lines).
Previously, we showed that BMP4 is necessary for the differentiation of superstructure progenitors (Blitz et al., 2009). To address the possibility that BMP signaling is reduced in our mutants, we examined the activation of SMAD1/5/8 (Retting et al., 2009) in the Sox9+/Scx+ progenitors. Sections taken from E13.5 Gli3 and Prx1-Cre;Pbx1;Pbx2 control and mutant forelimbs that were immunostained using antibodies against pSMAD1/5/8 and COL2A1 revealed different patterns of pSmad1/5/8 activity in control and mutant embryos (Fig. 9K-O), yet the abnormal patterns correlated with the mispatterning of the Sox9+/Scx+ progenitors in these mutants. This finding suggests that the aberrant Smad activation is secondary to the progenitor mispatterning. Overall, we concluded that although cell death did not contribute to the observed phenotypes, reduction in cell proliferation might have had some effect.
DISCUSSION
The musculoskeletal system acts as a system of levers and pulleys to create locomotion. Thus, one way to achieve variation in locomotor strategies is by changing the location of the connection between lever and pulley. Shifting the position of a bone superstructure, to which a muscle is connected by a tendon, along the bone shaft modifies the pulling force vector of that given muscle and, thereby, facilitates different types of locomotive capabilities. Variations in the positioning of superstructures and their effect on locomotion have been well documented (Archer et al., 2011; Milne and O'Higgins, 2012; Polly, 2007; Salton and Sargis, 2008, 2009). However, the mechanism that produces these variations was unknown.
An early step in the development of endochondral bones is mesenchymal condensation and the formation of the cartilaginous template that prefigures the future bone. Here, we identify components of the genetic program that regulate the subsequent step of this process, namely the patterning and condensation of superstructure precursors. Moreover, we demonstrate that the mechanism of superstructure patterning involves both global and regional modules, highlighting the modularity in long bone development. Specifically, by combining comparative transcriptomic analysis with cross-reference to existing literature, we identified a list of candidate genes that might be involved in this genetic program. Further analysis of several of these candidate genes, namely, Gli3, Pbx1, Pbx2, Hoxa11 and Hoxd11, established their involvement in regulating superstructure patterning. Importantly, Gli3 perturbation affected superstructures throughout different parts of the limb skeleton, whereas mutations in Pbx1 and Pbx2 or in Hoxa11 and Hoxd11 affected either proximal or distal superstructures, respectively. Interestingly, we found that the global and regional components of the genetic program are interconnected, as compound mutations of the proximal regulator Pbx1 and the global regulator Gli3 led to increased phenotype severity, compared with mutations in either gene alone.
In addition to these genes, our transcriptomic analysis identified 12 more candidate genes that were reportedly involved in long bone development. Interestingly, many of these genes regulate either proximal or distal limb segments, or alternatively, establish the proximodistal axis itself. For example, we identified additional HOX genes, such as Hoxa9, Hoxd9, Hoxa10 and Hoxd10, which were shown to regulate the patterning of proximal superstructures, such as the deltoid tuberosity (Wellik and Capecchi, 2003; Xu and Wellik, 2011). Other candidate genes, such as Shox2 or Meis1, operate locally either as downstream effectors of HOX genes (Neufeld et al., 2014) or as co-factors of proximal regulators, such as Pbx1 (Mercader et al., 2009), respectively. We also identified genes that control the formation of the proximodistal axis, such as Cyp26B1, which modulates the levels of retinoic acid (Yashiro et al., 2004) or Irx3, Irx5 and Hand2, which modulate the activity of the Shh/Gli3 signaling pathway (Galli et al., 2010; Li et al., 2014). Together, these reports further support the modular model for superstructure patterning and suggest the involvement of at least two hierarchical programs in this process. Higher is the program that governs the establishment of the proximodistal axis and, further downstream, local programs adjust the development of each individual segment. Finally, whereas our transcriptomic analysis was designed to highlight differentially expressed genes along the proximodistal axis, it lacked information regarding globally expressed genes, which could potentially regulate superstructure patterning, such as Gli3, which was expressed ubiquitously in both proximal and distal Sox9+/Scx+ progenitors. Additional evidence for the existence of other global regulatory genes is given by a recent report demonstrating that Tbx3 globally regulates patterning of forelimb superstructures, namely the greater and deltoid tuberosity and olecranon (Colasanto et al., 2016).
Although our work sheds light on the genetic mechanism that regulates superstructure patterning, the cellular mechanism underlying the patterning process is still missing. A clue for its nature is given by the observations that in Gli3, Pbx and Hox11 mutant embryos, superstructure precursors spread abnormally away from the developing bone substructure and display delayed differentiation. This may imply the existence of a mechanism controlling the migration of Sox9+/Scx+ progenitors to a specific condensation site. Once this program is perturbed, cells migrate to the wrong site, where they form an abnormal bony element. Alternatively, it is possible that Sox9+/Scx+ chondroprogenitors are specified from selected cells already present at the designated superstructure locations. In that case, the mechanism would involve activation of a chondrogenic program in specific cell subpopulations by extrinsic or intrinsic signals at specific spatiotemporal positions. In either scenario, it appears that the mechanism that decides where superstructure development should take place involves the Gli3, Pbx and Hox genes. Our finding that ablation of Gli3 specifically in Sox9+ and/or Scx+ cell lineage resulted in hypoplasia of selected superstructures clearly suggests that Gli3 regulates the Sox9+/Scx+ progenitors autonomously. However, the differences we observed in the severity of the phenotype between the Gli3-null and the different conditional alleles may suggest a non-autonomous contribution as well. An alternative explanation for the differences in severity is differences in efficiency between the various Cre lines in targeting the Gli3 locus.
A previous work in which the expression of both Shh and Gli3 was blocked proposed that the function of Shh and Gli3 in limb skeletal patterning is limited to refining autopodial morphology (Litingtung et al., 2002). Our work clearly expands the involvement of Gli3 in skeletal patterning by demonstrating its role as a global regulator of superstructure patterning. Interestingly, both Shh loss of function and Shh/Gli3 double knockout resulted in apparently normal deltoid tuberosity, suggesting that the effect of Gli3, at least in the proximal domain of the limb, is independent of Shh.
The patterning of superstructures involves control over their size. Our finding of reduced progenitor cell proliferation in Prx1-Cre;Pbx1fl/fl and Prx1-Cre;Pbx1fl/fl;Pbx2+/− mutant embryos might suggest that the genetic program regulates superstructure morphology by coordinating patterning and cell proliferation. In contrast, we present evidence that size control does not involve apoptosis of Sox9+/Scx+ progenitors.
Another level of coordination that is required is between the skeleton and the musculature. Interestingly, several reports have demonstrated that mispatterning of specific muscles is coupled with aberrant superstructures. For example, ablation of Tbx3 results in abnormal patterning of stylopod musculature, which attaches to dysplastic olecranon and deltoid tuberosity (Colasanto et al., 2016), whereas in Hox11Aadd or Hox11aaDd compound mutants, abnormal patterning of zeugopod musculature is accompanied by abnormal olecranon patterning (Swinehart et al., 2013). Taken together, these results strongly suggest that muscle and superstructure patterning may be regulated in a coordinated manner.
Previously, we reported that sesamoid bones, which are small auxiliary bones that are highly variant in anatomical location, size and number, also originate from Sox9- and Scx-positive precursors (Eyal et al., 2015; Eyal et al., 2019). Interestingly, we demonstrated that the Sox9- and Scx-positive precursors of some sesamoid bones, such as the lateral fabella, were patterned such that they developed away from the long bone substructure (Eyal et al., 2019). Those results are in line with our current evidence showing the formation of sesamoid-like bones following perturbation in either Gli3, PBX or Hox11 genes. Moreover, they further highlight the degree of plasticity stemming from the modular model, which offers an evolutionary mechanism to induce not only local variations in bone morphology by patterning the superstructures, but also formation of new auxiliary bones without having to rewrite the entire skeletogenic program. We therefore propose that bone superstructures and sesamoid bones be placed under one category of cartilage elements that share a common origin and developmental program.
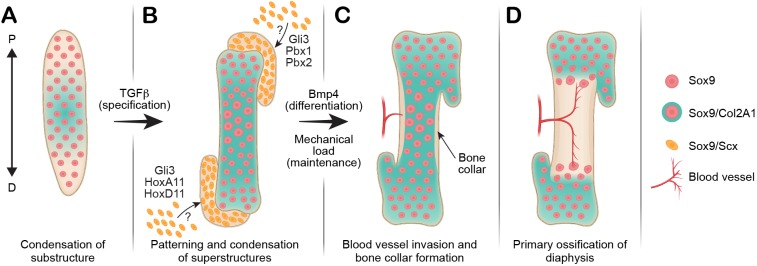
To conclude, our results provide insight into the formation, patterning and development of long bone superstructures and their impact on bone morphology. Furthermore, they provide strong evidence in support of the modular model of skeletogenesis and demonstrate the level of modularity during long bone morphogenesis in terms of cellular origins, genetic regulation, morphology and skeletal assembly of superstructures, namely their ability to attach or detach from the substructure. Specifically, we show that whereas Sox9+ progenitors establish the bone substructure (Fig. 10A), superstructure anlagen are formed and assembled modularly onto the substructure by Sox9+/Scx+ progenitors (Fig. 10B). We show that these unique Sox9+/Scx+ progenitors contribute not only to bone eminences but also to various condyles and sesamoid bones and that their patterning involves both global and regional regulatory modules that include Gli3, Pbx and Hox genes (Fig. 10B). Following their specification and patterning, these superstructures differentiate and become integral to the developing long bones (Fig. 10C,D). Importantly, by performing a comparative transcriptomic analysis, we were able to highlight additional candidate genes that may be implicated in superstructure patterning. Further studies of these molecular players will provide a better understanding of the regulatory network at play in superstructure formation and long bone morphogenesis.
Fig. 10.
A modular model for long bone development and superstructure patterning. (A) Superstructure initiation is preceded by formation of the cartilaginous substructures derived from primary Sox9+ progenitors. (B) Subsequent to substructure differentiation, a secondary wave of de novo specification produces Sox9+/Scx+ superstructure progenitors. This second specification wave occurs in juxtaposition to the substructure at different time points during embryogenesis and is regulated by the TGFβ signaling pathway. The initial spatial patterning of superstructure progenitors is regulated by both global and regional molecular players, such as Gli3 or Pbx and Hox genes, respectively. (C) Following specification, patterning and condensation of the superstructure precursors, they differentiate and become integral to the substructure anlagen, thus producing a complex three-dimensional cartilaginous template that is unique to each long bone. Differentiation of the superstructure is regulated by intrinsic BMP4 signaling and extrinsic mechanical stimuli. (D) Finally, ossification of the substructure, and later of the superstructures, will give rise to the morphology of the mature long bone. D, distal; P, proximal.
MATERIALS AND METHODS
Animals
All experiments involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the Weizmann Institute. For all timed pregnancies, plug date was defined as E0.5. For harvesting of embryos, timed-pregnant females were euthanized by cervical dislocation. The gravid uterus was dissected out and suspended in a bath of cold PBS and the embryos were harvested after removal of the placenta. Tail genomic DNA was used for genotyping by PCR. All experiments were performed in at least three biological repeats, i.e. on three embryos from separate litters.
Sox9-CreERT2 mice (Soeda et al., 2010) were received from the laboratory of Haruhiko Akiyama, Kyoto University, Kyoto, Japan. ScxGFP,Scx-Cre and Sox9-Cre transgenic mice were obtained from Ronen Schweitzer, Shriners Hospital for Children Research Division, Portland, OR, USA.
The generation of Col2A1-CreERT2 (Nakamura et al., 2006), Sox9-CreERT2 (Soeda et al., 2010), ScxGFP (Pryce et al., 2007), Gli3null (Johnson, 1967), Prx1-Cre (Logan et al., 2002), floxed-Gli3 (Blaess et al., 2008), Pbx1null (Selleri et al., 2001), floxed-Pbx1 (Ficara et al., 2008), Pbx2null (Selleri et al., 2004), Hox-a11null (Hsieh-Li et al., 1995), Hox-d11null (Davis and Capecchi, 1994) and Rosa26-tdTomato (Madisen et al., 2010) mice has been described previously.
To create Gli3, Hox11aadd and Pbx1 mutant mice, animals heterozygous for the mutations were crossed; heterozygous embryos were used as a control. To create Prx1-Gli3 or Prx1-Pbx1 mutant mice, floxed-Gli3 or floxed-Pbx1 mice were mated with Prx1-Cre-Gli3 or Prx1-Cre-Pbx1, respectively. To create Sox9-Gli3 or Scx-Gli3 mutant mice, floxed-Gli3 mice were mated with Sox9-Cre-Gli3 or Scx-Cre-Gli3, respectively. To create Prx1-Pbx1flox-Pbx2+/− mutant mice, floxed-Pbx1-Pbx2+/− mice were mated with Prx1-Cre-Pbx1flox mutant mice. To create Prx1-Pbx1-Gli3 mutant mice, floxed-Pbx1-Gli3 mice were mated with Prx1-Cre-Pbx1-Gli3 mutant mice. As a control, Prx1-Cre-negative embryos were used.
For genetic lineage analysis and FACS experiments, either Col2A1-CreERT2 or Sox9-CreERT2 mice were crossed with Rosa26-tdTomato reporter mice. Induction of Cre recombinase was performed at various pregnancy stages by administration of 0.03 mg/g tamoxifen/body weight in corn oil by oral gavage (stock concentration was 5 mg/ml).
Skeletal preparations
Cartilage and bone in whole mouse embryos were visualized, after skinning and disemboweling, by staining with Alcian Blue (Millipore Sigma, A5268) and Alizarin Red S (Millipore Sigma, A5533) and clarification of soft tissue with 3% potassium hydroxide and 100% glycerol (McLeod, 1980).
Paraffin sections
For preparation of paraffin sections, embryos were harvested at various ages, dissected and fixed in 4% paraformaldehyde (PFA)/PBS at 4°C overnight. After fixation, tissues were dehydrated in an ethanol series to 100% and embedded in paraffin. The embedded tissues were sectioned at a thickness of 7 µm using a microtome (LM2235, Leica) and mounted onto slides.
O.C.T-embedded sections
For preparation of O.C.T-embedded sections, embryos were harvested at various ages, dissected and fixed in 1% PFA/PBS at 4°C overnight. Fixed embryos were then dehydrated in 30% sucrose overnight at 4°C. Next, samples were dissected, soaked in O.C.T (Tissue-Tek) for 30-60 min and then frozen in O.C.T. Frozen samples were sectioned at a thickness of 10 µm using a cryostat (CM3050S, Leica) and mounted onto slides.
FISH
FISH on paraffin sections were performed using Hoxd11 digoxigenin (DIG)-labeled RNA probe (Shwartz and Zelzer, 2014). Probe was detected using anti-DIG-POD (1:300, 11207733910, Roche), followed by Cy3-tyramide labeled fluorescent dyes, according to the instructions of the TSA Plus Fluorescent Systems Kit (PerkinElmer). Finally, slides were counter-stained using DAPI (1:1000, D9542, Millipore Sigma).
Immunofluorescence staining
For immunofluorescence staining for SOX9 and COL2A1, 7 μm-thick paraffin sections of embryo limbs were deparaffinized and rehydrated in water. Antigen was then retrieved in 10 mM citrate buffer (pH 6.0), boiled and cooked for 10 min in a microwave oven. In order to block non-specific binding of immunoglobulin, sections were incubated with 7% goat serum, 1% bovine serum albumin (BSA) dissolved in PBT (PBS+0.1% Tween 20) for 60 min at room temperature. Following blockage, sections were incubated overnight at 4°C with primary anti-SOX9 antibody (1:150, AB5535, Millipore Sigma). Then, sections were washed in PBT and incubated with Cy3-conjugated secondary fluorescent antibodies (1:100, 711-165-215, Jackson ImmunoResearch) for 60 min at room temperature. After staining for SOX9, slides were washed in PBT and fixed in 4% PFA at room temperature for 10 min. Then, slides were incubated with proteinase K (Millipore Sigma, P9290), washed and post-fixed again in 4% PFA. Next, sections were washed and incubated overnight at 4°C with primary anti-COL2A1 antibody (1:50, II-II6B3, the Developmental Studies Hybridoma Bank). The next day, sections were washed in PBT and incubated with Cy2-conjugated secondary fluorescent antibodies (1:100; 715-225-150, Jackson ImmunoResearch) for 60 min at room temperature. Slides were mounted with Immu-mount aqueous-based mounting medium (Thermo Fisher Scientific).
For immunofluorescence staining for SOX9 and ScxGFP, 10 μm-thick cryostat sections of embryo limbs endogenously labeled for ScxGFP were used. SOX9 immunofluorescence staining was performed as described above, but with omission of the antigen retrieval step, using primary SOX9 antibody and secondary Cy3-fluorescent antibodies.
For immunofluorescence staining for PBX1 and COL2A1, paraffin sections were used and antigen retrieval and blockage of non-specific binding were performed as described above. Endogenous peroxidase was quenched by incubation in 2% H2O2/PBS for 30 min at room temperature. Staining for PBX1 was performed using primary anti-PBX1 antibodies (1:400, C-4342, Cell Signaling Technology) followed by HRP-conjugated secondary antibodies (1:1000, 711-035-152, Jackson ImmunoResearch) and Cy3-tyramide labeled fluorescent dyes, according to the instructions of the TSA Plus Fluorescent Systems Kit (PerkinElmer). Subsequently, staining against COL2A1 was performed as described above.
For immunofluorescence staining for pSMAD1/5 and COL2A1, 7 μm-thick paraffin sections of embryo limbs were deparaffinized and rehydrated in water. For antigen retrieval, slides were incubated with proteinase K (Millipore Sigma, P9290) for 20 min at room temperature, washed and post-fixed again in 4% PFA. Endogenous peroxidase was quenched by incubation in 2% H2O2/PBS for 30 min at room temperature. Next, staining for pSMAD1/5 was performed using primary anti-pSMAD1/5/9 antibodies (1:200, CST-13820, Cell Signaling Technology) followed by biotin-conjugated secondary antibodies (1:100, 711-065-152, Jackson ImmunoResearch) and HRP-conjugated streptavidin (1:200, NEL750001EA, PerkinElmer). Finally, detection was performed using Cy3-tyramide labeled fluorescent dyes, according to the instructions of the TSA Plus Fluorescent Systems Kit (PerkinElmer). Subsequently, staining against COL2A1 was performed as described above.
BrdU staining and quantification
To examine cell proliferation, pregnant females were injected subcutaneously with 100 µg/g BrdU/body weight dissolved in PBS (stock concentration was 10 mg/ml) (B5002, Millipore Sigma). After 2 h of exposure, pregnant mice were scarified and the embryos were harvested, genotyped, fixated and embedded in paraffin. To detect BrdU-incorporated cells, immunostaining using primary anti-BrdU antibodies (1:200, MCA2060, Bio-Rad) and secondary Cy3-conjugated fluorescent antibodies (1:100; 712-165-153, Jackson ImmunoResearch) was performed on 7 μm-thick paraffin sections. Antigen retrieval using 10 mM citrate buffer (pH 6.0) and counterstaining for SOX9 were performed as described above. Quantification of BrdU-stained cells was performed using sections collected from three embryos from three different litters. From each embryo, at least two sections were used for statistical analysis (n≥6). In each section, the number of Sox9+ and Sox9+/BrdU+ stained cells were counted within the area of the greater and deltoid tuberosities. The fraction of proliferative cells was calculated for each group and the difference between control and mutant embryos was tested using Student's t-test. Statistical significance was determined as P≤0.05.
TUNEL staining for detection of apoptosis
To examine cell death, we performed a TUNEL assay using ApopTag plus peroxidase in situ apoptosis kit (S7101, Millipore Sigma) according to the user guide supplied by the manufacturer, except for the use of DAB as the main peroxidase substrate. As a substitute, we used Cy3-tyramide labeled fluorescent dyes, according to the instructions of the TSA Plus Fluorescent Systems Kit (PerkinElmer). Finally, counterstaining was performed using DAPI (1:1000, D9542, Millipore Sigma).
Tissue clearing for light-sheet fluorescence microscopy
For whole-mount imaging, samples were first cleared and immunostained using the PACT-decal technique (Treweek et al., 2015; Yang et al., 2014). Briefly, either Col2A1-CreERT2- or Sox9-CreERT2 mice were crossed with Rosa26-tdTomato reporter mice. Following tamoxifen administration at E11.5 (Col2A1-CreERT2) or E10.5 (Sox9-CreERT2), as described above, embryos were harvested at E15.5 or E14.5, respectively. Embryos were dissected and fixed in 1% PFA/PBS at 4°C overnight. Next, samples were washed in PBTx (PBS+0.1% Triton X-100+0.01% sodium azide) at room temperature, then embedded into a hydrogel of 4% (wt/vol) acrylamide in 1× PBS with 0.25% thermal initiator 2,2′-azobis[2-(2-imidazolin-2-yl)propane] dihydrochloride (VA-044, FUJIFILM-Wako). The hydrogel was allowed to polymerize at 37°C for 5 h. The samples were removed from the hydrogel, washed in PBTx, and moved to 10% sodium dodecyl sulfate (SDS) with 0.01% sodium azide, shaking at 37°C for 3 days, changing the SDS solution each day. Samples were washed four times with 1× PBTx at room temperature over the course of 24 h and fixed in 4% PFA at room temperature for 10 min. Then, samples were incubated with proteinase K (Millipore Sigma, P9290) for 30 min with shaking at 37°C, washed and post-fixed again in 4% PFA. To detect COL2A1, samples were first incubated with 5% goat serum dissolved in PBTx at 37°C overnight in order to block non-specific binding of immunoglobulin. Next, samples were incubated with primary COL2A1 antibodies (1:150, II-II6B3, the Developmental Studies Hybridoma Bank) in 2.5% goat serum/ PBTx shaking at 37°C for 3 days, changing the antibody solution each day. Samples were washed four times with 1× PBTx at room temperature over the course of 24 h. Next, samples were incubated with secondary Cy3 or Cy2 antibodies (1:150, 715-165-150 or 715-225-150, respectively, Jackson ImmunoResearch) in 2.5% goat serum/PBTx with shaking at 37°C overnight. Samples were washed again with four changes of 1× PBTx, and the refractive index (RI) of the sample was brought to 1.45 by submersion in a refractive index matching solution (RIMS) prepared by dissolving 35 g of Histodenz (Millipore Sigma, D2158) in 0.02 M phosphate buffer (74% wt/vol), and shaking gently at room temperature overnight. Finally, samples were embedded in 1% low gelling Agarose (Millipore Sigma, A9414) in PBS in a glass capillary, submerged in RIMS and stored in the dark at room temperature until imaging.
Light-sheet fluorescence microscopy
The cleared samples were imaged with a Zeiss Lightsheet Z.1 microscope. For each limb, a low-resolution image of the entire limb was taken with the 20× Clarity lens at a zoom of 0.36. Light-sheet fusion of images was done if necessary (Zen software, Zeiss). Tile stitching and 3D-image reconstruction was performed using Arivis Vision4D (Arivis) and Imaris (Bitplane) Software.
FACS
Flow cytometry analysis and sorting were performed on a BD FACS AriaIII instrument (BD Immunocytometry Systems) equipped with a 488, 407, 561 and 633 nm lasers, using a 70 µm nozzle, controlled by BD FACS Diva software v8.0.1 (BD Biosciences), at the Weizmann Institute of Science Flow Cytometry Core Facility. Further analysis was performed using FlowJo software v10.2 (Tree Star).
For collection of cells, Sox9-CreERT2-tdTomato;ScxGFP mice were crossed with Rosa26-tdTomato;ScxGFP reporter mice. Embryos were harvested at E13.5 following tamoxifen administration at E12.0, as described above. Forelimbs were dissected and suspended in cold PBS using 15 ml tubes.
To extract cells from tissues, PBS was replaced with 1 ml pre-heated 0.05% trypsin (0.25% Trypsin EDTA solution A, Biological Industries) diluted in DMEM media (Thermo Fisher Scientific) and incubated for 15 min at 37°C, with gentle agitation every 5 min. Tissues were then dissociated by vigorous pipetting using 1 ml tips. Next, 4 ml of DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution (Biological Industries) was added and cell suspensions were filtered into a 15 ml tube using a syringe and a 40 µm filter net. Finally, tubes were centrifuged at 1000 rpm (90 g) for 7 min, supernatant was removed and cells were resuspended in 1 ml of cold PBS and used immediately for FACS.
Single-stained GFP and tdTomato control cells were used for configuration and determining gate boundaries. Live cells were gated by size and granularity using FSC-A versus SSC-A and according to staining with propidium iodide (PI, 1 µg/ml) and DAPI (1 µg/ml). FSC-W versus FSC-A was used to further distinguish single cells. In addition, unstained, GFP-stained only and tdTomato-stained only cells were mixed in various combinations to verify that the analysis excluded false-positive doublets.
Bulk MARS-Seq
Purified RNA from FACS-isolated Sox9+/Scx+ cell samples was used for library preparation according to a standard MARS-Seq protocol (Jaitin et al., 2014). Library quality was analyzed by a 2200 TapeStation instrument (Agilent Technologies, data not shown). The experiment produced libraries of high quality and sufficient quantity. Libraries were subsequently sequenced by Illumina NextSeq 500.
Bioinformatic analysis
The analysis was performed as described by Köster and Rahmann (2012). Briefly, a unique molecular identifier (UMI) sequence present in the R2 read was inserted in the read name of R1 sequence file using a python script. Cutadapt (Martin, 2011) was used to trim low quality and poly A/T and adapter sequences (-a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -a “A{100}” -a “T{100}” --times 2 -q 20 -m 30). Sequences were mapped using TopHat2 to mouse genome build mm10 (Kim et al., 2013). UMI information was integrated into the BAM files as tags, using a python script. The BAM file was converted to SAM format using Samtools (Li et al., 2009). Duplicate reads were marked and filtered based on having the same UMI and mapping to the same gene, using a python script. UMI counts per gene were calculated using modified HTSeq-count script (Anders et al., 2014) and a RefSeq gtf file (downloaded from igenomes UCSC), which was modified to contain a window surrounding the 3′ UTR.
DESeq2 (Love et al., 2014) was used for normalization and to detect differentially expressed genes based on negative binomial distribution and a generalized linear regression model. The model used contained two factors: olecranon or deltoid tuberosity Sox9+/Scx+ precursors and a batch factor. Genes were considered differentially expressed if the difference in expression between at least two sample types was statistically significant after adjustment with the fdrtool package (adjusted P-value≤0.05) (Strimmer, 2008). After filtering based on expression levels (sum normalized count in all samples greater than 10 and maximum expression level higher than 5), clustering of the standardized normalized counts was carried out using click algorithm (Expander package, Ulitsky et al., 2010). Further analysis was performed using GSEA (Broad institute) and Ingenuity (IPA).
Supplementary Material
Acknowledgements
We thank N. Konstantin for expert editorial assistance, and members of the Zelzer laboratory for their advice and suggestions; R. Schweitzer for providing the ScxGFP, Scx-Cre and Sox9-Cre mice, L. Selleri for providing the Pbx1null and Pbx2null mice, and H. Akiyama for providing the Sox9-CreERT2 mice; and O. Golani from the Weizmann Institute Department of Life Sciences Core Facilities for her guidance and assistance in analyzing and designing the experiments. Light sheet imaging was made possible thanks to the De Picciotto-Lesser Cell Observatory in memory of Wolf and Ruth Lesser of the MICC. We thank C. Vega from the Weizmann Institute Department of Design, Photography and Printing for designing the graphic model.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.E., E.Z.; Methodology: S.E., S. Kult, S.R., S. Krief, N.F., K.M.P., D.L., T.-M.S., Y.A., E.Z.; Software: D.L.; Validation: S.E., S. Kult, S.R., S. Krief, N.F., D.L., T.-M.S., Y.A., D.M.W.; Formal analysis: D.L., T.-M.S.; Investigation: S.E., S. Kult, S.R., S. Krief, N.F., K.M.P.; Resources: N.F., K.M.P., D.M.W.; Data curation: D.L., T.-M.S.; Writing - original draft: S.E., E.Z.; Writing - review & editing: D.L., T.-M.S., D.M.W.; Visualization: S.E., Y.A.; Supervision: E.Z.; Project administration: E.Z.; Funding acquisition: E.Z.
Funding
This study was supported by grants from the National Institutes of Health (R01 AR055580), the European Research Council (310098), the Jeanne and Joseph Nissim Foundation for Life Sciences Research, the Y. Leon Benoziyo Institute for Molecular Medicine, Beth Rom-Rymer, the Estate of David Levinson, the Jaffe Bernard and Audrey Foundation, the Georges Lustgarten Cancer Research Fund, the David and Fela Shapell Family Center for Genetic Disorders, the David and Fela Shapell Family Foundation INCPM Fund for Preclinical Studies, and the Estate of Bernard Bishin for the WIS-Clalit Program. Deposited in PMC for release after 12 months.
Data availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus under accession number GSE129820.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.167882.supplemental
References
- Anders S., Pyl P. T. and Huber W. (2014). HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer M., Beck R., Gott M., Hand S., Godthelp H. and Black K. (2011). Australia's first fossil marsupial mole (Notoryctemorphia) resolves controversies about their evolution and palaeoenvironmental origins. Proc. R. Soc. B 278, 1498-1506. 10.1098/rspb.2010.1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen A. D. and Olsen B. R. (2015). Bone development. Bone 80, 14-18. 10.1016/j.bone.2015.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S., Stephen D. and Joyner A. L. (2008). Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development 135, 2093-2103. 10.1242/dev.015990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz E., Viukov S., Sharir A., Shwartz Y., Galloway J. L., Pryce B. A., Johnson R. L., Tabin C. J., Schweitzer R. and Zelzer E. (2009). Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell 17, 861-873. 10.1016/j.devcel.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz E., Sharir A., Akiyama H. and Zelzer E. (2013). Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 140, 2680-2690. 10.1242/dev.093906 [DOI] [PubMed] [Google Scholar]
- Boulet A. M. and Capecchi M. R. (2004). Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131, 299-309. 10.1242/dev.00936 [DOI] [PubMed] [Google Scholar]
- Cain C. J., Gaborit N., Lwin W., Barruet E., Ho S., Bonnard C., Hamamy H., Shboul M., Reversade B., Kayserili H., et al. (2016). Loss of Iroquois homeobox transcription factors 3 and 5 in osteoblasts disrupts cranial mineralization. Bone Rep. 5, 86-95. 10.1016/j.bonr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini T. D. (2006). Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133, 2263-2273. 10.1242/dev.02395 [DOI] [PubMed] [Google Scholar]
- Cervantes-Diaz F., Contreras P. and Marcellini S. (2017). Evolutionary origin of endochondral ossification: the transdifferentiation hypothesis. Dev. Genes Evol. 227, 121-127. 10.1007/s00427-016-0567-y [DOI] [PubMed] [Google Scholar]
- Colasanto M. P., Eyal S., Mohassel P., Bamshad M., Bonnemann C. G., Zelzer E., Moon A. M. and Kardon G. (2016). Development of a subset of forelimb muscles and their attachment sites requires the ulnar-mammary syndrome gene Tbx3. Dis. Model. Mech. 9, 1257-1269. 10.1242/dmm.025874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P. and Capecchi M. R. (1994). Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development 120, 2187-2198. [DOI] [PubMed] [Google Scholar]
- Eyal S., Blitz E., Shwartz Y., Akiyama H., Schweitzer R. and Zelzer E. (2015). On the development of the patella. Development 142, 1831-1839. 10.1242/dev.121970 [DOI] [PubMed] [Google Scholar]
- Eyal S., Rubin S., Krief S., Levin L. and Zelzer E. (2019). Common cellular origin and diverging developmental programs for different sesamoid bones. Development 146, dev167452 10.1242/dev.167452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficara F., Murphy M. J., Lin M. and Cleary M. L. (2008). Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2, 484-496. 10.1016/j.stem.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A., Robay D., Osterwalder M., Bao X., Bénazet J.-D., Tariq M., Paro R., Mackem S. and Zeller R. (2010). Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6, e1000901 10.1371/journal.pgen.1000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S., Krause Y., Wuelling M. and Vortkamp A. (2012). Hoxa11 and Hoxd11 regulate chondrocyte differentiation upstream of Runx2 and Shox2 in mice. PLoS ONE 7, e43553 10.1371/journal.pone.0043553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P., Del Buono J. and Logan M. P. (2007). Tbx5 is dispensable for forelimb outgrowth. Development 134, 85-92. 10.1242/dev.02622 [DOI] [PubMed] [Google Scholar]
- Holmberg J., Ingner G., Johansson C., Leander P. and Hjalt T. A. (2008). PITX2 gain-of-function induced defects in mouse forelimb development. BMC Dev. Biol. 8, 25 10.1186/1471-213X-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H. M., Witte D. P., Weinstein M., Branford W., Li H., Small K. and Potter S. S. (1995). Hoxa11 structure, extensive antisense transcription, and function in male and female fertility. Development 121, 1373-1385. [DOI] [PubMed] [Google Scholar]
- Jaitin D. A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S. and Tanay A. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776-779. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. R. (1967). Extra-toes: a new mutant gene causing multiple abnormalities in the mouse. Development 17, 543-581. [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez-León J. and Belmonte J. C. I. (2006). The role of TGFβs and Sox9 during limb chondrogenesis. Curr. Opin. Cell Biol. 18, 723-729. 10.1016/j.ceb.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J. and Rahmann S. (2012). Snakemake – a scalable bioinformatics workflow engine. Bioinformatics 28, 2520-2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Koyama E., Yasuda T., Minugh-Purvis N., Kinumatsu T., Yallowitz A. R., Wellik D. M. and Pacifici M. (2010). Hox11 genes establish synovial joint organization and phylogenetic characteristics in developing mouse zeugopod skeletal elements. Development 137, 3795-3800. 10.1242/dev.053447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H. M. (2003). Developmental regulation of the growth plate. Nature 423, 332-336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Lessa E. P., Vassallo A. I., Verzi D. H. and Mora M. S. (2008). Evolution of morphological adaptations for digging in living and extinct ctenomyid and octodontid rodents. Biol. J. Linn. Soc. 95, 267-283. 10.1111/j.1095-8312.2008.01057.x [DOI] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Sakuma R., Vakili N. A., Mo R., Puviindran V., Deimling S., Zhang X., Hopyan S. and Hui C.-C. (2014). Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Dev. Cell 29, 233-240. 10.1016/j.devcel.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Litingtung Y., Dahn R. D., Li Y., Fallon J. F. and Chiang C. (2002). Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979 10.1038/nature01033 [DOI] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N. and Tabin C. J. (2002). Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77-80. 10.1002/gene.10092 [DOI] [PubMed] [Google Scholar]
- Long F. and Ornitz D. M. (2013). Development of the Endochondral Skeleton. Cold Spring Harbor Perspect. Biol. 5, a008334 10.1101/cshperspect.a008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McHenry H. M. and Corruccini R. S. (1975). Distal humerus in hominoid evolution. Folia Primatol. 23, 227-244. 10.1159/000155673 [DOI] [PubMed] [Google Scholar]
- McLeod M. J. (1980). Differential staining of cartilage and bone in whole mouse fetuses by Alcian Blue and Alizarin Red S. Teratology 22, 299-301. 10.1002/tera.1420220306 [DOI] [PubMed] [Google Scholar]
- Mercader N., Selleri L., Criado L. M., Pallares P., Parras C., Cleary M. L. and Torres M. (2009). Ectopic Meis1 expression in the mouse limb bud alters PD patterning in a Pbx1-independent manner. Int. J. Dev. Biol. 53, 1483-1494. 10.1387/ijdb.072430nm [DOI] [PubMed] [Google Scholar]
- Milne N. and O'Higgins P. (2012). Scaling of form and function in the xenarthran femur: a 100-fold increase in body mass is mitigated by repositioning of the third trochanter. Proc. R. Soc. B 279, 3449-3456. 10.1098/rspb.2012.0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E., Nguyen M.-T. and Mackem S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603-2612. 10.1002/dvdy.20892 [DOI] [PubMed] [Google Scholar]
- Neufeld S. J., Wang F. and Cobb J. (2014). Genetic interactions between Shox2 and Hox genes during the regional growth and development of the mouse limb. Genetics 198, 1117-1126. 10.1534/genetics.114.167460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B. R., Reginato A. M. and Wang W. (2000). Bone development. Annu. Rev. Cell Dev. Biol. 16, 191-220. 10.1146/annurev.cellbio.16.1.191 [DOI] [PubMed] [Google Scholar]
- Polly P. D. (2007). Limbs in mammalian evolution. In Fins into Limbs: Evolution, Development and Transformation (ed. Hall B. K.), pp. 245-268. University of Chicago Press. [Google Scholar]
- Pryce B. A., Brent A. E., Murchison N. D., Tabin C. J. and Schweitzer R. (2007). Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev. Dyn. 236, 1677-1682. 10.1002/dvdy.21179 [DOI] [PubMed] [Google Scholar]
- Retting K. N., Song B., Yoon B. S. and Lyons K. M. (2009). BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136, 1093-1104. 10.1242/dev.029926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton J. A. and Sargis E. J. (2008). Evolutionary morphology of the Tenrecoidea (Mammalia) forelimb skeleton. In Mammalian Evolutionary Morphology (ed. Sargis E. J. and Dagosto M.), pp. 51-71. Dordrecht: Springer Netherlands. [Google Scholar]
- Salton J. A. and Sargis E. J. (2009). Evolutionary morphology of the Tenrecoidea (Mammalia) hindlimb skeleton. J. Morphol. 270, 367-387. 10.1002/jmor.10697 [DOI] [PubMed] [Google Scholar]
- Selleri L., Depew M. J., Jacobs Y., Chanda S. K., Tsang K. Y., Cheah K. S., Rubenstein J. L., O'Gorman S. and Cleary M. L. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128, 3543-3557. [DOI] [PubMed] [Google Scholar]
- Selleri L., DiMartino J., van Deursen J., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K. S., Rhee J., Popperl H. et al. (2004). The TALE Homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell. Biol. 24, 5324-5331. 10.1128/MCB.24.12.5324-5331.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz Y. and Zelzer E. (2014). Nonradioactive in situ hybridization on skeletal tissue sections. Skelet. Dev. Repair 1130, 203-215. 10.1007/978-1-62703-989-5_15 [DOI] [PubMed] [Google Scholar]
- Soeda T., Deng J. M., de Crombrugghe B., Behringer R. R., Nakamura T. and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635-644. 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. (2008). fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461-1462. 10.1093/bioinformatics/btn209 [DOI] [PubMed] [Google Scholar]
- Sugimoto Y., Takimoto A., Akiyama H., Kist R., Scherer G., Nakamura T., Hiraki Y. and Shukunami C. (2013). Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140, 2280-2288. 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- Swinehart I. T., Schlientz A. J., Quintanilla C. A., Mortlock D. P. and Wellik D. M. (2013). Hox11 genes are required for regional patterning and integration of muscle, tendon and bone. Development 140, 4574-4582. 10.1242/dev.096693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek J. B., Chan K. Y., Flytzanis N. C., Yang B., Deverman B. E., Greenbaum A., Lignell A., Xiao C., Cai L., Ladinsky M. S. et al. (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 10, 1860-1896. 10.1038/nprot.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Maron-Katz A., Shavit S., Sagir D., Linhart C., Elkon R., Tanay A., Sharan R., Shiloh Y. and Shamir R. (2010). Expander: from expression microarrays to networks and functions. Nat. Protoc. 5, 303 10.1038/nprot.2009.230 [DOI] [PubMed] [Google Scholar]
- Wellik D. M. and Capecchi M. R. (2003). Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301, 363-367. 10.1126/science.1085672 [DOI] [PubMed] [Google Scholar]
- Xu B. and Wellik D. M. (2011). Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc. Natl Acad. Sci. USA 108, 4888-4891. 10.1073/pnas.1018161108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Treweek J. B., Kulkarni R. P., Deverman B. E., Chen C.-K., Lubeck E., Shah S., Cai L. and Gradinaru V. (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945-958. 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Topol L., Lee H. and Wu J. (2003). Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development 130, 1003-1015. 10.1242/dev.00324 [DOI] [PubMed] [Google Scholar]
- Yashiro K., Zhao X., Uehara M., Yamashita K., Nishijima M., Nishino J., Saijoh Y., Sakai Y. and Hamada H. (2004). Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev. Cell 6, 411-422. 10.1016/S1534-5807(04)00062-0 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Eberspaecher H., Lefebvre V. and de Crombrugghe B. (1997). Parallel expression ofSox9 andCol2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377-386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.