ABSTRACT
The sinoatrial node (SAN), the primary cardiac pacemaker, consists of a head domain and a junction/tail domain that exhibit different functional properties. However, the underlying molecular mechanism defining these two pacemaker domains remains elusive. Nkx2-5 is a key transcription factor essential for the formation of the working myocardium, but it was generally thought to be detrimental to SAN development. However, Nkx2-5 is expressed in the developing SAN junction, suggesting a role for Nkx2-5 in SAN junction development and function. In this study, we present unambiguous evidence that SAN junction cells exhibit unique action potential configurations intermediate to those manifested by the SAN head and the surrounding atrial cells, suggesting a specific role for the junction cells in impulse generation and in SAN-atrial exit conduction. Single-cell RNA-seq analyses support this concept. Although Nkx2-5 inactivation in the SAN junction did not cause a malformed SAN at birth, the mutant mice manifested sinus node dysfunction. Thus, Nkx2-5 defines a population of pacemaker cells in the transitional zone. Despite Nkx2-5 being dispensable for SAN morphogenesis during embryogenesis, its deletion hampers atrial activation by the pacemaker.
KEY WORDS: Nkx2-5, Shox2, Sinoatrial node, Cardiac conduction system, Sinus node dysfunction
Summary: Mouse lines in which two distinct pacemaker cell populations can be distinguished reveal a novel role for Nkx2-5 in pacemaking function in the sinoatrial node.
INTRODUCTION
The rhythmic heartbeats in mammals are regulated by a steady pace of electrical impulses generated by the sinoatrial node (SAN), functionally known as the primary cardiac pacemaker (Christoffels et al., 2010; Moorman et al., 1998; Ye et al., 2015a). Anatomically, the SAN is a structurally heterogeneous tissue with a densely packed head domain that extends superiorly and wraps around the right superior vena cava (RSVC), and a loosely packed junction/tail region that extends inferiorly towards the inferior vena cava (IVC) (Boyett et al., 2000; Liu et al., 2007; Opthof, 1988). Interestingly, the SAN junction can maintain pacemaker function even in the presence of a severe hypoplastic SAN head, as shown in Tbx18 mutants (Wiese et al., 2009). In contrast, mice lacking Shox2 in the SAN junction manifest severe sinus node dysfunction associated with a virtual absence of the SAN junction, despite a normal SAN head (Ye et al., 2015b). This observation demonstrates an essential role for the SAN junction in normal pacemaking function. However, whether more than one population of pacemaking cells exist in the SAN remained an unanswered question. Additionally, the mechanism and molecular basis responsible for the distinct function of the SAN head and junction are also unknown.
SAN development has been studied extensively. Gene expression and genetic studies in mice have revealed a complex genetic network involving many genes, including Hcn4, Tbx3, Tbx5, Tbx18, Isl1 and Shox2 (Munshi, 2012). Among them, Shox2, Tbx3, Isl1 and Hcn4 are expressed in the entire developing SAN, with Hcn4 being regarded as a functional marker of the SAN (Moosmang et al., 2001; Santoro and Tibbs, 1999). Nkx2-5, one of the earliest cardiac-specific markers, plays an essential role in heart formation (Lints et al., 1993; Moses et al., 2001). Previously, Nkx2-5 was thought to be excluded from the developing SAN and to be essential for establishing a strict boundary between the SAN domain and the surrounding atrial myocardium by inhibiting Tbx3 and Hcn4, but activating Cx40 (Gja5) expression (Christoffels et al., 2006; Espinoza-Lewis et al., 2011; Kasahara et al., 1998; Mommersteeg et al., 2007). Despite repression of Nkx2-5 by Tbx3 and Shox2 in the SAN head region (Blaschke et al., 2007; Espinoza-Lewis et al., 2009; Wu et al., 2014), Nkx2-5 expression was nevertheless detected in the developing SAN junction (Liang et al., 2013; Wiese et al., 2009; Ye et al., 2015b). Our recent studies demonstrated that, in the developing SAN junction, Shox2 functions to inhibit the transcriptional output of Nkx2-5 through a Shox2-Nkx2-5 antagonistic mechanism (Ye et al., 2015b). However, whether Nkx2-5 is essential for the development of the SAN junction and SAN function is completely unknown.
RESULTS AND DISCUSSION
Patch-clamp recording identifies two distinct populations of pacemaking cells in the developing SAN
While several molecular markers, including Hcn4, Shox2, Isl1 and Tbx3, are expressed in the entire developing SAN, Tbx18 expression is restricted to the SAN head, and Nkx2-5 expression is found in the SAN junction from early embryonic stage to adulthood, indicating the existence of two genetically distinct domains in the SAN (Wiese et al., 2009; Munshi, 2012; Ye et al., 2015a,b; Fig. 1A-C′). Although genetic studies have demonstrated different requirements of these two domains for pacemaking function, the exact cell identity in these two distinct SAN regions has not been studied, likely due to difficulties in determining the precise isolation of cells from each domain. We took advantage of several unique genetically modified mouse lines that can precisely define the SAN head and junction domains. We compounded these alleles to generate mice carrying Nkx2-5Cre/+;Shox2HA−DsRed;R26RYFP alleles. In these mice, the entire SAN is labeled by DsRed expressed from the Shox2HA−DsRed allele (Ye et al., 2015b), while the junction domain is marked by both DsRed and YFP that is activated by the Nkx2-5Cre allele. As a result, the junction cells (DsRed+/YFP+) are distinguishable from the surrounding atrial (YFP+) and SAN head cells (DsRed+) (Fig. S1).
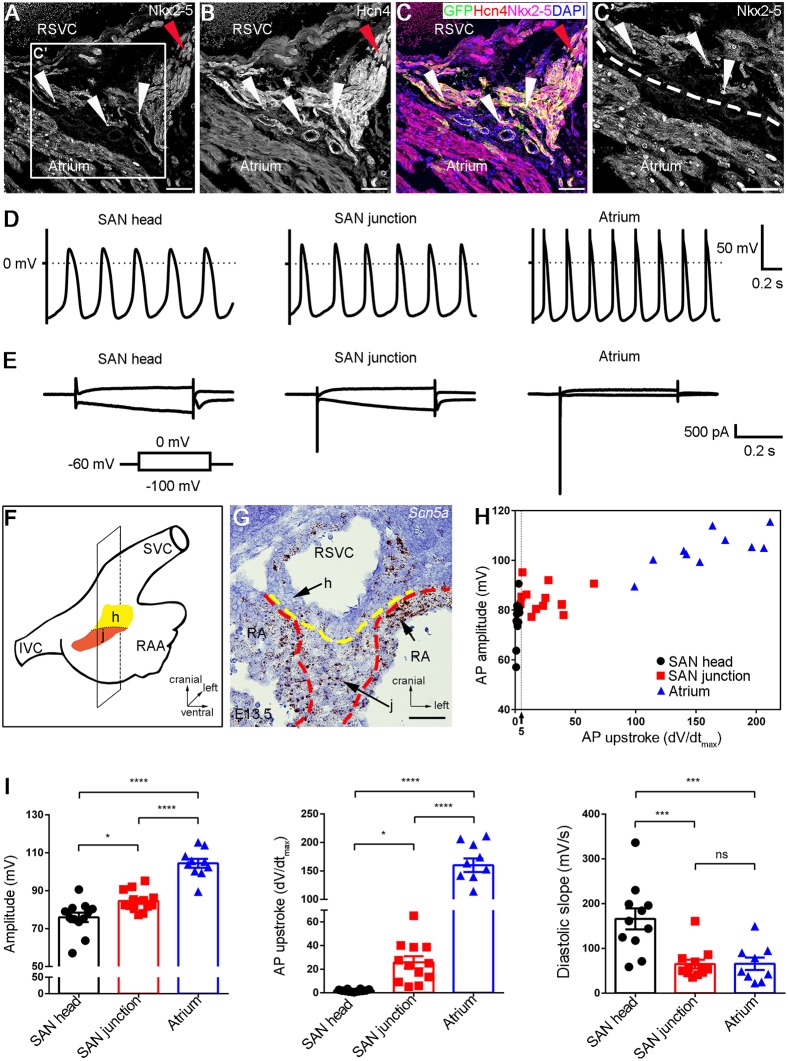
Fig. 1.
AP recordings identify two distinct types of pacemaking cells in the SAN. (A-C,C′) Co-immunofluorescent staining shows the persistent expressions of Nkx2-5 and Hcn4 in the SAN junction of Shox2Cre/+;R26RmTmG mice at adulthood. Red arrowheads indicate the SAN head; white arrowheads indicate the Nkx2-5+ cells in the SAN junction. (D) Sample traces of spontaneous action potentials show typical firing configurations of cells from the SAN head, SAN junction and atrium. (E) Typical examples of the current response in the SAN head, SAN junction and atrium cells upon 0.5 s voltage step from holding the potential of –60 mV to −100 mV or 0 mV. (F) Schematic overview of the section plane on the SAN shown in G. (G) In situ hybridization shows Scn5a expression in the SAN junction and atrial tissue. The expression level in the SAN junction is lower than that in the atrial cells. (H) A two-dimensional diagram shows the AP amplitude and AP upstroke for each type of cell. (I) Summary graphs of the AP amplitude, AP upstroke and diastolic slope in each group. Data are mean±s.e.m. ns, non-significant; *P<0.05; ***P<0.001; ****P<0.0001. h, SAN head; j, SAN junction; RA, right atrium; IVC, inferior vena cava; RAA, right atrial appendage; SVC, superior vena cava. Scale bars: 50 μm.
To understand the developmental status of the electrophysiological properties of a SAN cell, we developed an exo utero electrocardiography (ECG) protocol whereby an ECG recording is conducted on embryos via the placenta to the uterus attachment within the female abdominal cavity. We performed exo utero ECG on wild-type embryos from embryonic day 12.5 (E12.5) to E16.5 and found that the P waves, which accurately reflect atrial depolarization, became detectable at E12.5, and appeared typically from E13.5 onwards (Fig. S2), consistent with a previous report that SAN cells begin to exhibit typical action potential (AP) configurations of pacemaking activity at E12.5 (van Eif et al., 2019). Therefore, we isolated the SAN and adjacent atrial tissues from E13.5 Nkx2.5Cre/+;Shox2HA-DsRed;R26RYFP embryos. Single cells were dissociated from the tissues and plated in culture. Patch-clamp recordings were performed on cultured cells from the SAN head and junction, as well as the atrial tissues, based on corresponding fluorescent markers. The AP properties of the SAN head cells and atrial cells (Fig. 1) exhibited typical configurations characterized previously for pacemaking and atrial cells (Sun et al., 2015; van Eif et al., 2019; Ye et al., 2015b; Zhang et al., 2002). However, the SAN junction cells presented unique AP configurations that were intermediate to those produced by the SAN head and atrial cells (Fig. 1). The AP amplitude in the SAN junction cells was larger in comparison with the SAN head cells, but smaller than that in the atrial cells (Fig. 1). The AP upstroke, which is the maximal rate of the rising, was faster in the SAN junction cells than that in the SAN head cells, but slower in comparison with the atrial cells (Fig. 1). The AP amplitude and the AP upstroke from each cell type are summarized in Fig. 1H, which demonstrates that the SAN junction cells have characteristics of both the SAN head and atrial cells. Statistical analysis of the AP configurations from these three types of cells showed significant differences between each of them (Fig. 1I and Fig. S3), indicating that the SAN cells are electrically heterogeneous. Moreover, compared with the Nkx2-5− head and the atrial cells, the Nkx2-5+ SAN junction cells showed a distinct electrophysiological property manifested by both the hyperpolarization-activated HCN current that shows only in the SAN head cells and the depolarization-activated fast sodium current that is seen in the atrial cells (Fig. 1E,G). Previous studies have shown that the electrical coupling during propagation of APs from the SAN to the atrium creates transitional AP shapes in the cells adjacent to the border zone of these two different cell types (Joyner and Van Capelle, 1986; Verheijck et al., 1998; Zhang et al., 2001). Thus, the SAN junction cells show combinatory electrophysiological properties of the SAN head and atrial cells. This domain appears to act as a transitional zone in which the spontaneous firing arising from the HCN current elicits the activity generated by the fast sodium channel, thus playing a role in both impulse generation and SAN-atrial exit conduction.
scRNA-seq reveals distinct gene expression profiles underlying the cellular heterogeneity of the SAN
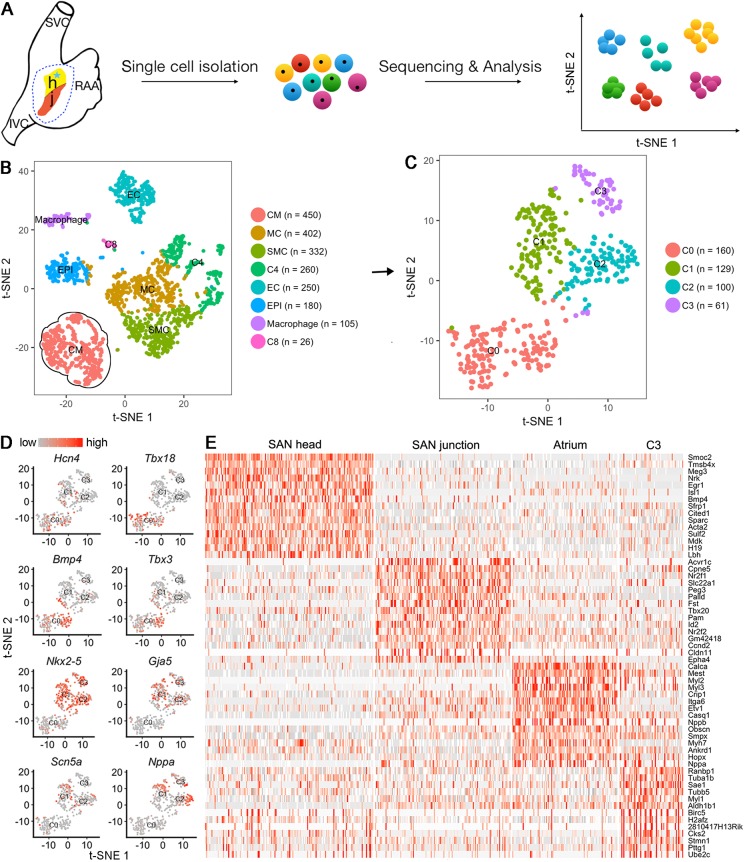
The functional heterogeneity of a cell population is often a reflection of cellular heterogeneity (Tan et al., 2013). Recently, single-cell RNA-sequencing (scRNA-seq) has become a powerful tool for profiling cell-to-cell variability on a transcriptomic scale. We conducted an unbiased survey of the cellular diversity in the SAN, with the surrounding atrial cells as a reference control. The SAN domain, which is highlighted by GFP, was isolated with adjacent atrial tissue from E13.5 Shox2Cre/+;R26RmTmG mice and subjected to scRNA-seq. As quality control of the duplicate samples was mostly coincident (Fig. S4C), we chose one of them for deeper sequencing and further analysis. A total of 3770 individual cells from the SAN and atrium were profiled. After unbiased analysis, 2005 cells were acquired and subjected to significant principal components analysis, resulting in eight distinct clusters, including cardiomyocytes (CM), mesenchymal cells (MC), vascular smooth muscle (VSM), endothelial cells (ECs), epicardial cells (EPIs) and macrophages (Chasseigneaux et al., 2018; DeLaughter et al., 2016; Khazen et al., 2005; Lee et al., 2017; Li et al., 2016; Souders et al., 2009; Tarnawski et al., 2015; Vanlandewijck et al., 2018; Fig. 2B). The clustering results were visualized through the non-linear dimensional reduction algorithm, t-distributed stochastic neighbor embedding(tSNE) (Maaten and Hinton, 2008; Macosko et al., 2015). Because Hcn4, Gja5, Nppa, Nkx2-5 and Tnnt2 genes were all present within the CM cluster (Fig. S4), the 450 cells in the CM cluster were then used for further analysis, which further identified four clusters (C0 to C3) (Fig. 2C). Using Seurat (Butler et al., 2018), we found that, among these four clusters, C0 and C1 exhibited expression of several SAN marker genes, including Isl1, Tbx18, Hcn4, Tbx3, Bmp4 and Smoc2 (Fig. 2D,E, Fig. S4E), as reported previously (Liang et al., 2015; Puskaric et al., 2010; van Eif et al., 2019; Wiese et al., 2009). Thus, C0 and C1 correspond to the SAN cells. C2 showed expression of atrial-specific markers such as Nppa and Itga6 (Lee et al., 2017; Tarnawski et al., 2015; Vedantham et al., 2015), indicating that the C2 population consists of atrial cells. However, in C1, we also found the expression of C2-specific genes, making this group an intermediate between the C0 and C2 populations (Fig. 2D,E). Consistent with the unique AP configurations contributed by both the HCN current and fast sodium current in the SAN junction cells (Fig. 1), C1 cells also expressed Scn5a, which is known to be responsible for fast sodium current. Therefore, we determined that C0 corresponds to the SAN head cells and C1 to the SAN junction cells. In addition, the expression of Gja5 (which encodes Cx40) in C1 would also confer this group of cells with conduction activity (van Weerd and Christoffels, 2016). Although the transcriptome of C3 did not pin-point any specific type of cell, venous valve cells seemed likely. Nevertheless, gene expression profiling supports the existence of a population of SAN cells that exert physiological functions intermediate between the SAN head and atrium in impulse generation and impulse conduction. This is consistent with other data demonstrating that mice lacking the SAN head due to Tbx18 mutation have a normal beating rhythm, but mice lacking the SAN junction exhibit sinus node dysfunction (Wiese et al., 2009; Ye et al., 2015b).
Fig. 2.
scRNA-seq of the SAN and atrial cells. (A) Schematic overview of data collection and analysis. (B) tSNE visualization of the eight clusters on the SAN and atrial cells from Shox2cre/+;R26RmTmG mice. (C) A tSNE plot shows four clusters (C0-C3) resulting from iterative clustering of 450 CM cells. (D) The marker gene expression of the SAN head, SAN junction and atrium on the tSNE plot of the four clusters. (E) Heatmap shows the top 15 marker genes for each cluster. CM, cardiomyocyte; EC, endothelial cells; MC, mesenchymal cells; EPI, epicardial cells; IVC, inferior vena cava; RAA, right atrial appendage; SMC, smooth muscle cells; SVC, superior vena cava. C3, C4 and C8 are cluster numbers 3, 4 and 8.
Nkx2-5 is essential for normal SAN function
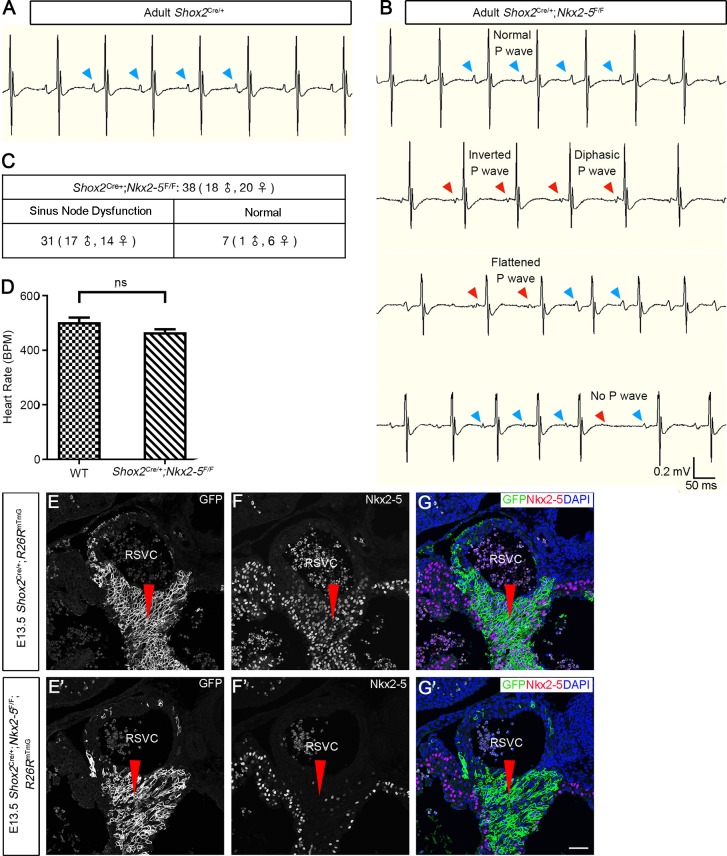
As Nkx2-5 is expressed in the developing SAN junction from E11.5 onwards (Liang et al., 2013; Wiese et al., 2009), we sought to investigate its role in SAN morphogenesis and function. To inactivate Nkx2-5 in the SAN junction specifically (Fig. 3D-F′), we crossed Shox2-Cre knock-in mice that exhibit Cre activity initially in the sinus venosus at E9.0 and later in the developing SAN with Nkx2-5F/F mice (Pashmforoush et al., 2004; Sun et al., 2013). These Shox2Cre/+;Nkx2-5F/F mice survived to adulthood without an obvious phenotype. However, although heartbeat rates appeared comparable with controls, surface ECG identified compromised SAN function in adult mutants (2-6 months old) (Fig. 3B,C); manifested sinus node dysfunction included arrhythmia, inverted P wave and sinoatrial exit block (Fig. 3B), which were also found in 1-month-old young mice (data not shown). Detailed ECG measurements are presented in Fig. S5.
Fig. 3.
Deletion of Nkx2-5 in the SAN junction causes sinus node dysfunction. (A,B) Surface ECG recordings reveal irregular P waves, including an inverted P wave, a diphasic P wave, no P wave and a flattened P wave, in adult Shox2Cre/+;Nkx2-5F/F mice (B, red arrowheads), compared with normal P waves in control mice (A, blue arrowheads). Out of 38 Shox2Cre/+;Nkx2-5F/F mice (18 male and 20 female), 31 (17 male and 14 female) have sinus node dysfunction and 7 (1 male and 6 female) are normal. (C) Heartbeat rates appear comparable in adult Shox2Cre/+;Nkx2-5F/F and control mice. Data are mean±s.e.m. (D-F′) Immunofluorescence shows an absence of Nkx2-5 in the SAN junction (red arrowheads) of E13.5 Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice. ns, non-significant; BPM, beats per minute; RSVC, right superior vena cava. Scale bar: 50 μm.
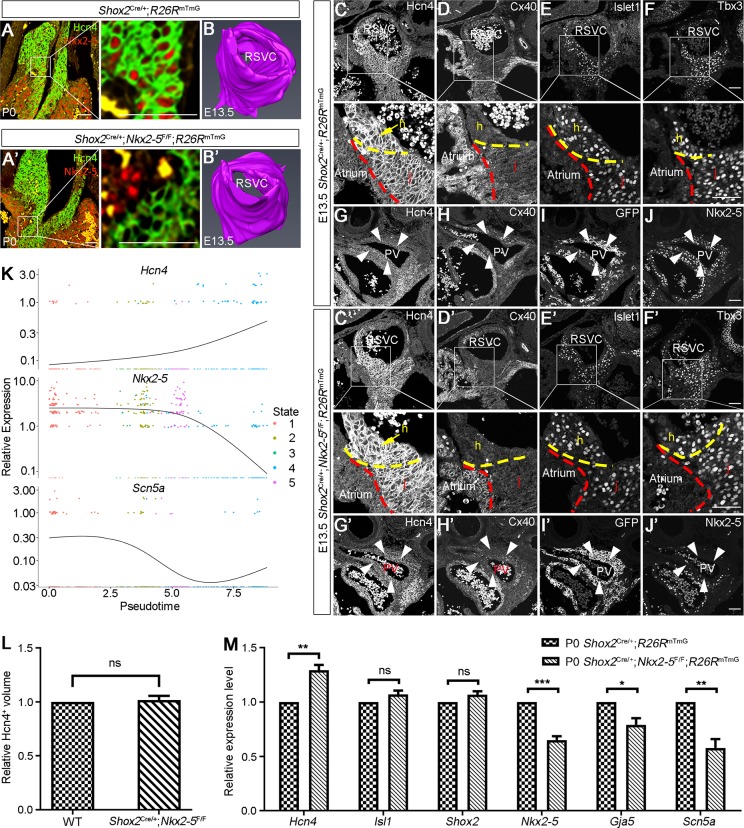
To uncover the morphogenetic and molecular changes underlying the sinus node dysfunction in Shox2Cre/+;Nkx2-5F/F mice, we first assessed SAN morphology using the pan-SAN marker Hcn4. We found comparable structures in P0 controls and mutants (Fig. 4A,A′). Three-dimensional (3D) reconstruction of the SAN from E13.5 controls and mutants also showed similar structures (Fig. 4B,B′,L). However, despite similar expression levels and patterns of the SAN markers Tbx3 and Islet1 in the SAN junction of E13.5 controls and mutants, we did observe an elevated level of Hcn4 and a slightly reduced level of Cx40 in the mutants (Fig. 4C-F′). RT-qPCR analyses further confirmed significantly reduced levels of Nkx2-5, Gja5 and Scn5a in the SAN of P0 mutants compared with controls (Fig. 4M). Downregulation of Scn5a and Gja5 expression could explain the reduced fast sodium current and conduction activity in the SAN junction, thereby contributing to the sinoatrial exit block. The presence of inverted P wave or diphasic P wave is an indication of the retention of the dominant pacemaking site in the left side of the heart (MacLean et al., 1975). We have reported previously that Shox2 and Nkx2-5 are also co-expressed in the developing pulmonary vein (PV) myocardium and that reduced doses of Nkx2-5 lead to ectopic Hcn4 expression and reduced Cx40 in the PV myocardium (Ye et al., 2015b). Similar results were observed in the PV myocardium of Shox2Cre/+;Nkx2-5F/F mice (Fig. 4G-J′), which could be responsible for the inverted and diphasic P waves identified by the surface ECG (Fig. 3; Fig. S5). Our results present unambiguous evidence that Nkx2-5 is dispensable for SAN morphogenesis and differentiation during embryogenesis, but plays an essential role for normal SAN physiological function.
Fig. 4.
Nkx2-5 is dispensable for SAN morphogenesis during embryogenesis. (A,A′) The morphology of the SAN, defined by Hcn4 immunofluorescence, appears comparable between P0 Shox2Cre/+;Nkx2-5F/F;R26RmTmG and Shox2Cre/+;R26RmTmG mice. (B,B′) 3D reconstruction of the SAN (based on Hcn4 immunostaining) in E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice (n=3 for each genotype). (C-F′) Immunofluorescence shows the colocalization of Hcn4, Cx40, Islet1 and Tbx3 in the SAN junction in E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice. Compared with the Shox2Cre/+;R26RmTmG mice (C-F), Nkx2-5 deletion in the SAN junction of Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice (C′-F′) caused visible Hcn4 elevation but Cx40 reduction. Intermediate levels of Hcn4 and Cx40 expression are found in the SAN junction compared with the SAN head and surrounding atrial cells in control mice (enlarged images from C,D). Broken lines indicate the boundaries of the atrium, SAN head and SAN junction. (G-J′) Immunofluorescence shows the expression of Nkx2-5, Hcn4 and Cx40 in the PV myocardium of E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice. Nkx2-5 deletion in Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice (G′-J′) also resulted in elevated Hcn4 and reduced Cx40 expression compared with the controls (G-J). White arrowheads indicate PV myocardium. (K) The dynamic expression of Hcn4, Scn5a and Nkx2-5 along the pseudotime trajectory. (L) Comparison of Hcn4+ volume in the SAN of E13.5 Shox2Cre/+;Nkx2-5F/F;R26RmTmG and Shox2Cre/+;R26RmTmG mice. (M) RT-qPCR assays show downregulated expression of Nkx2-5, Gja5 and Scn5a, and upregulated expression of Hcn4 in the SAN of P0 Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice. ns, non-significant; *P<0.05; **P<0.01; ***P<0.001. h, SAN head; j, SAN junction; PV, pulmonary vein; RSVC, right superior vena cava. Scale bars: 50 μm.
As most SAN-specific markers are present in both the SAN head and junction, but Nkx2-5 is absent in the SAN head, we wondered whether Nkx2-5 expression confers the SAN junction cells unique AP configurations. We therefore set out to conduct patch-clamp recordings on the SAN junction cells with deletion of Nkx2-5. We first performed patch-clamp recording on the GFP-positive SAN cells from E13.5 Shox2Cre/+;R26RmTmG and calculated the proportion of head/junction cells in the whole SAN based on their AP configurations, effectively setting a baseline for calibration. In the 17 recorded cells (with 10 expressing typical head AP configurations and seven manifesting typical junction AP configurations), the proportion of head/junction cells (59% vs. 41%) is consistent with the ratio of head/junction cells defined by scRNA-seq (55% versus 45%; Fig. 2C). Thereafter, we randomly recorded 26 GFP-positive cells from the SAN of E13.5 Shox2Cre/+;Nkx2-5F/F;R26RmTmG mice. Among them, 22 presented head AP patterns and four showed typical junction AP configurations (Fig. S3C). Thus, the majority of the recorded SAN cells (85%) exhibited head AP configurations, suggesting that some of them originated from junction cells. The four cells exhibiting junction AP configurations likely escaped Cre recombination.
To further understand the elicited function of Nkx2-5 in AP configurations, the temporal expression of Scn5a, Hcn4 and Nkx2-5 in the SAN cells from E13.5 Shox2Cre/+;R26RmTmG mice were reconstructed by pseudotime analysis using Monocle2 (Qiu et al., 2017). We found that expression of Nkx2-5 and Scn5a along the pseudotime trajectory gradually decreased, whereas expression of Hcn4 increased (Fig. 4K), strongly suggesting that Nkx2-5 expression is responsible for the generation of the typical SAN junction AP configurations. In the absence of Nkx2-5, the SAN junction cells would stimulate SAN head AP configuration.
Although Nkx2-5 function in the formation of working myocardium has been well defined, its role in SAN formation is controversial. Although mutations in Nkx2-5 lead to conduction system disturbances in humans and mice (Benson et al., 1999; Biben et al., 2000; Jay et al., 2004; Schott et al., 1998), Nkx2-5 was thought to be detrimental to SAN development, at least under overdose conditions (Christoffels et al., 2006; Espinoza-Lewis et al., 2011; Kasahara et al., 1998). Our current studies demonstrate the existence of two distinct domains of the SAN, with the Nkx2-5+ junction cells exhibiting unique electrophysiological properties and transcriptomic profiles intermediate between the SAN head and the adjacent atrial cells. We also provide convincing evidence that, although it is not required for SAN morphogenesis during embryogenesis, Nkx2-5 is essential for normal SAN physiological function. Taken together, our studies define two distinct pacemaker cell populations and reveal a novel role for Nkx2-5 in pacemaking function in the SAN. These results could shed light on the regeneration or directed differentiation of specific pacemaking cell types, and on the development of gene- and cell-based therapies for congenital and acquired cardiac conduction abnormalities.
MATERIALS AND METHODS
Animal models, embryos, histology, immunofluorescence and in situ hybridization
The generation and genotyping protocols of Shox2Cre, Shox2HA-DsRed, Nkx2.5Cre, Nkx2.5F/F, R26RYFP and R26RmTmG mice have been described previously (Barry et al., 2015; Moses et al., 2001; Muzumdar et al., 2007; Pashmforoush et al., 2004; Sun et al., 2013; Ye et al., 2015b). The animal experiments in this study were approved by the Institutional Animal Care and Use Committee at Tulane University.
Embryos or embryonic hearts were harvested from timed pregnant female mice, fixed in 4% paraformaldehyde (PFA) at 4°C overnight, embedded in paraffin wax and sectioned at 8μm for Hematoxylin and Eosin or immunofluorescent staining, as described previously (Ye et al., 2015b). The primary antibodies used in this study were: anti-Hcn4 (ab32675, Abcam; 1:500), anti-Nkx2-5 (sc-8697, Santa Cruz; 1:500), anti-Tbx3 (sc-17871, Santa Cruz; 1:500), anti-Cx40 (sc-20466, Santa Cruz; 1:250), anti-islet1 (ab20670, Abcam; 1:1000) and anti-GFP (sc-9996, Santa Cruz; 1:500). The secondary antibodies used at 1:1000 dilution and all from Life Technologies were: donkey anti-mouse (A21202, A31571), donkey anti-rat (A21208, A21209), donkey anti-rabbit (A31573) and donkey anti-goat (A32849, A11058, A11055).
In situ hybridization was performed with the RNAscope Intro Pack 2.5 HD Reagent Kit Brown-Mm (Advanced Cell Diagnostics, 322371) according to the manufacturer's instructions. Probe information is as below: Scn5a probe (RNAscope Probe - Mm-Scn5a, 429881), Smoc2 probe (RNAscope Probe-Mm-Smoc2, 318541) and Acvr1c probe (RNAscope Probe - Mm-Acvr1c, 429291). All experiments were repeated at least three times.
Surface ECG and whole-cell patch-clamp recordings
Surface ECG recordings of 1-month-old and older mice were conducted as reported previously (Ye et al., 2015b). For exo utero ECG recording of developing embryos, timed pregnant females were subjected to exo utero surgery as described previously (Song et al., 2008). After the opening of the body wall, the uterus was subsequently opened with iridectomy scissors at the opposite site of the placenta. An incision was then made in the extraembryonic membrane to allow the embryo to emerge but remain attached to the uterus via the placenta. Embryos were then positioned properly with styrofoam within the female abdomen, and ECGs were measured with subcutaneous electrode needles.
The SAN head, SAN junction and adjacent atrial tissue of embryonic hearts from Shox2HA-DsRed;Nkx2.5Cre/+;R26RYFP mice at designated age were removed using a fluorescent dissecting microscope. Tissues were dissociated into single cells and were plated onto fibronectin/gelatin-coated coverslips and cultured overnight prior to whole-cell patch-clamp recordings, as described previously (Sun et al., 2015). A coverslip with attached cells was transferred to a recording chamber and perfused with an external solution containing (in mM): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 5.5 glucose and 5 HEPES (pH 7.4), 310-320 mOsm. The temperature during recordings was maintained at 37°C. Cell types (atrial cell, YFP+; SAN head cell, DsRed+; SAN junction cell, DsRed+/YFP+) were determined by fluorescent microscopy (BX51WI, Olympus) through a 40× water-immersion objective and DAGE-MTI camera. Patch-clamp recordings were performed with a Multiclamp 700B amplifier (Molecular Devices). Glass pipettes had open tip resistances of 3-5 MΩ and were filled with an internal solution containing (in mM): 120 K-gluconate, 20 KCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 14 Na2-phosphocreatine and 0.3 Tris-GPT (pH 7.3), 290 mOsm. Only the myocytes with spontaneous action potentials were recorded under both current-clamp and voltage-clamp modes. Signals were filtered at 4-10 kHz and sampled at 50 kHz. Action potentials were characterized by amplitude, maximal upstroke velocity (dV/dtmax), duration at 50% and 90% repolarization (ADP50 and ADP90), the ratio of ADP50 and ADP90, the maximal diastolic potential (MDP), and frequency. Voltage steps were applied to measure different channel-mediated currents.
scRNA-seq
For scRNA-seq, the entire SAN and adjacent atrial tissue, as shown in Fig. 2A, were dissected out from E13.5 Shox2Cre/+;R26RmTmG mice under a stereo fluorescent microscope, subjected to digestion by a cocktail of collagenase I, II and IV, and followed by a brief StemPro Accutase treatment. The suspended cells were processed immediately as follows. Cell count and viability were determined and 5000 cells were loaded for capture onto 10x Genomics Chromium system using single-cell 3′ library and gel bead kit v2. Following capture and lysis, cDNA was synthesized and amplified following the manufacturer's protocol. The amplified cDNAs from each channel of the Chromium System were used to construct an Illumina sequencing library and sequenced on NextSeq 500 with 150 cycle sequencing. Illumina basecall files (*.bcl) were converted to FASTQs using the bcl2fastq2/2.17.1 wrapped in CellRanger/2.1.0. FASTQ files were then aligned to the mm10 mouse reference genome using the CellRanger/2.1.0 software pipeline with default parameters, followed by data analysis in Seurat and Monocle2.
3D reconstruction
For 3D reconstruction, consecutive 10 µm sections were stained using anti-Hcn4 antibodies, imaged, and loaded into Amira 6.0.1, in which subsequent alignment, segmentation, and 3D model generation were performed.
RT-qPCR analyses
For quantitative reverse transcription polymerase chain reaction (RT-qPCR), the SAN was isolated from P0 Shox2Cre/+;R26RmTmG and Shox2Cre/+;Nkx2.5F/F;R26RmTmG mice (N=3 for each genotype), and subjected to RNA extraction (RNeasy Micro Kit; Qiagen). The RNAs were subsequently reversely transcribed into complementary DNAs (cDNAs) (RevertAid First Strand cDNA Synthesis Kit). SYBR green and gene-specific primers were used and transcript levels were examined using a CFX96 Real-Time PCR Detection System. Primer details are as follows: Hcn4 (F, 5′-CTTCTGCTGTGTCACTGGGA-3′; R, 5′-ATACTGCTTCCCCCAGGAGT-3′), Shox2 (F, 5′-ACCAATTTTACCCTGGAACAAC-3′; R, 5′-TCGATTTTGAAACCAAACCTG-3′), Nkx2-5 (F, 5′-TTAGGAGAAGGGCGATGACT-3′; R, 5′-AGGTCCGAGACACCAGGCTA-3′), Scn5a (F, 5′-ATGGCAAACTTCCTGTTACCTC-3′; R, 5′-CCACGGGCTTGTTTTTCAGC-3′), Isl1 (F, 5′-ATGATGGTGGTTTACAGGCTAAC-3′; R, 5′-TCGATGCTACTTCACTGCCAG-3′), Gja5 (F, 5′-GGTCCACAAGCACTCCACAG-3′; R, 5′-CTGAATGGTATCGCACCGGAA-3′), GAPDH (F, 5′-ATCAAGAAGGTGGTGAAGCAG-3′; R, 5′-GAGTGGGAGTTGCTGTTGAAGT-3′). Differences in the RT-qPCR were analyzed using Student's t-test in GraphPad Prism7; results are presented as mean±s.e.m. P<0.05 was considered significant.
Statistical analysis
To ensure scientific rigor and reproducibility, all experiments were repeated at least three times unless specifically indicated. Quantification results are presented as mean±s.e.m., and statistical analysis was conducted using Student's t-test. P<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Kejing Song and Alanna Wanek at the NextGen Sequencing Core in the Tulane Center for Translational Research in Infection and Inflammation for technical assistance with scRNA-seq and initial bioinformatic analyses. We also thank Dr Stryder Meadows for reading and editing the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.C., H.L., C.-L.C., H.H., Y.Z.; Methodology: Y.C., H.L.; Software: H.L., Z.H., Q.T.; Validation: Y.C., H.L.; Formal analysis: H.L., D.L., Z.H., Q.T.; Investigation: H.L., D.L., Y.W., J.X., T.Y., L.W.; Resources: Y.C., C.-L.C.; Data curation: Y.C., H.L., D.L.; Writing - original draft: H.L., Y.W., Q.T.; Writing - review & editing: Y.C., C.-L.C., H.H., Y.Z.; Visualization: H.L.; Supervision: Y.C.; Project administration: Y.C.; Funding acquisition: Y.C.
Funding
We acknowledge financial support from the National Institutes of Health (R01HL136326 to Y.C.). H.H. is supported by the National Institutes of Health (R01DC016324). H.L. was supported in part by a fellowship from Fujian Normal University, and J.X. and L.W. received fellowship from the China Scholarship Council. Deposited in PMC for release after 12 months.
Data availability
scRNA-Seq data have been deposited in GEO under accession number GSE130461.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.178145.supplemental
References
- Barry D. M., Xu K., Meadows S. M., Zheng Y., Norden P. R., Davis G. E. and Cleaver O. (2015). Cdc42 is required for cytoskeletal support of endothelial cell adhesion during blood vessel formation in mice. Development 142, 3058-3070. 10.1242/dev.125260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. W., Silberbach G. M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S., Smalls O., Johnson M. C., Watson M. S. and Seidman J. (1999). Mutations in the cardiac transcription factor NKX2. 5 affect diverse cardiac developmental pathways. J. Clin. Invest. 104, 1567-1573. 10.1172/JCI8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C., Weber R., Kesteven S., Stanley E., McDonald L., Elliott D. A., Barnett L., KÖentgen F., Robb L. and Feneley M. (2000). Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ. Res. 87, 888-895. 10.1161/01.RES.87.10.888 [DOI] [PubMed] [Google Scholar]
- Blaschke R. J., Hahurij N. D., Kuijper S., Just S., Wisse L. J., Deissler K., Maxelon T. and Anastassiadis K. (2007). Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830-1838. 10.1161/CIRCULATIONAHA.106.637819 [DOI] [PubMed] [Google Scholar]
- Boyett M. R., Honjo H. and Kodama I. (2000). The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 47, 658-687. 10.1016/S0008-6363(00)00135-8 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E. and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasseigneaux S., Moraca Y., Cochois-Guégan V., Boulay A.-C., Gilbert A., Le Crom S., Blugeon C., Firmo C., Cisternino S. and Laplanche J.-L. (2018). Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci. Rep. 8, 12272 10.1038/s41598-018-30739-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V. M., Mommersteeg M. T., Trowe M.-O., Prall O. W., de Gier-de Vries C., Soufan A. T., Bussen M., Schuster-Gossler K., Harvey R. P. and Moorman A. F. (2006). Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 98, 1555-1563. 10.1161/01.RES.0000227571.84189.65 [DOI] [PubMed] [Google Scholar]
- Christoffels V. M., Smits G. J., Kispert A. and Moorman A. F. M. (2010). Development of the pacemaker tissues of the heart. Circ. Res. 106, 240-254. 10.1161/CIRCRESAHA.109.205419 [DOI] [PubMed] [Google Scholar]
- DeLaughter D. M., Bick A. G., Wakimoto H., McKean D., Gorham J. M., Kathiriya I. S., Hinson J. T., Homsy J., Gray J. and Pu W. (2016). Single-cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480-490. 10.1016/j.devcel.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J. F., Wang D. and Yang J. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376-385. 10.1016/j.ydbio.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A., Liu H., Sun C., Chen C., Jiao K. and Chen Y. (2011). Ectopic expression of Nkx2. 5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 356, 359-369. 10.1016/j.ydbio.2011.05.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P. Y., Harris B. S., Maguire C. T., Buerger A., Wakimoto H., Tanaka M., Kupershmidt S., Roden D. M., Schultheiss T. M. and O'Brien T. X. (2004). Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Invest. 113, 1130-1137. 10.1172/JCI19846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner R. W. and Van Capelle F. (1986). Propagation through electrically coupled cells. How a small SA node drives a large atrium. Biophys. J. 50, 1157-1164. 10.1016/s0006-3495(86)83559-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H., Bartunkova S., Schinke M., Tanaka M. and Izumo S. (1998). Cardiac and extracardiac expression of Csx/Nkx2. 5 homeodomain protein. Circ. Res. 82, 936-946. 10.1161/01.RES.82.9.936 [DOI] [PubMed] [Google Scholar]
- Khazen W., M'Bika J.-P., Tomkiewicz C., Benelli C., Chany C., Achour A. and Forest C. (2005). Expression of macrophage-selective markers in human and rodent adipocytes. FEBS Lett. 579, 5631-5634. 10.1016/j.febslet.2005.09.032 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Protze S. I., Laksman Z., Backx P. H. and Keller G. M. (2017). Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21, 179-194.e4. 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Li G., Xu A., Sim S., Priest J. R., Tian X., Khan T., Quertermous T., Zhou B., Tsao P. S. and Quake S. R. (2016). Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39, 491-507. 10.1016/j.devcel.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Wang G., Lin L., Lowe J., Zhang Q., Bu L., Chen Y., Chen J., Sun Y. and Evans S. M. (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399-407. 10.1161/CIRCRESAHA.113.301588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N. J., Jiang C., Cao X., Zhao X. and Zhang X. (2015). Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256-3268. 10.1172/JCI68257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints T. J., Parsons L. M., Hartley L., Lyons I. and Harvey R. P. (1993). Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419-431. [DOI] [PubMed] [Google Scholar]
- Liu J., Dobrzynski H., Yanni J., Boyett M. R. and Lei M. (2007). Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc. Res. 73, 729-738. 10.1016/j.cardiores.2006.11.016 [DOI] [PubMed] [Google Scholar]
- Maaten L. v. d. and Hinton G. (2008). Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579-2605. [Google Scholar]
- MacLean W., Karp R. B., KouCHOUKOS N. T., James T. N. and Waldo A. L. (1975). P waves during ectopic atrial rhythms in man: a study utilizing atrial pacing with fixed electrodes. Circulation 52, 426-434. 10.1161/01.CIR.52.3.426 [DOI] [PubMed] [Google Scholar]
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N. and Martersteck E. M. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202-1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg M. T. M., Hoogaars W. M. H., Prall O. W., de Gier-de Vries C., Wiese C., Clout D. E. W., Papaioannou V. E., Brown N. A., Harvey R. P. and Moorman A. F. M. (2007). Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 100, 354-362. 10.1161/01.RES.0000258019.74591.b3 [DOI] [PubMed] [Google Scholar]
- Moorman A. F. M., de Jong F., Denyn M. M. F. J. and Lamers W. H. (1998). Development of the cardiac conduction system. Circ. Res. 82, 629-644. 10.1161/01.RES.82.6.629 [DOI] [PubMed] [Google Scholar]
- Moosmang S., Stieber J., Zong X., Biel M., Hofmann F. and Ludwig A. (2001). Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur. J. Biochem. 268, 1646-1652. 10.1046/j.1432-1327.2001.02036.x [DOI] [PubMed] [Google Scholar]
- Moses K. A., DeMayo F., Braun R. M., Reecy J. L. and Schwartz R. J. (2001). Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31, 176-180. 10.1002/gene.10022 [DOI] [PubMed] [Google Scholar]
- Munshi N. V. (2012). Gene regulatory networks in cardiac conduction system development. Circ. Res. 110, 1525-1537. 10.1161/CIRCRESAHA.111.260026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Opthof T. (1988). The mammalian sinoatrial node. Cardiovasc. Drugs Ther. 1, 573-597. 10.1007/BF02125744 [DOI] [PubMed] [Google Scholar]
- Pashmforoush M., Lu J. T., Chen H., St Amand T., Kondo R., Pradervand S., Evans S. M., Clark B., Feramisco J. R. and Giles W. (2004). Nkx2-5 pathways and congenital heart disease: loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373-386. 10.1016/S0092-8674(04)00405-2 [DOI] [PubMed] [Google Scholar]
- Puskaric S., Schmitteckert S., Mori A. D., Glaser A., Schneider K. U., Bruneau B. G., Blaschke R. J., Steinbeisser H. and Rappold G. (2010). Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet. 19, 4625-4633. 10.1093/hmg/ddq393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A. and Trapnell C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro B. and Tibbs G. R. (1999). The HCN gene family: molecular basis of the hyperpolarization-activated pacemaker channels. Ann. N. Y. Acad. Sci. 868, 741-764. 10.1111/j.1749-6632.1999.tb11353.x [DOI] [PubMed] [Google Scholar]
- Schott J.-J., Benson D. W., Basson C. T., Pease W., Silberbach G. M., Moak J. P., Maron B. J., Seidman C. E. and Seidman J. G. (1998). Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281, 108-111. 10.1126/science.281.5373.108 [DOI] [PubMed] [Google Scholar]
- Song Y., Yan M., Muneoka K. and Chen Y. P. (2008). Mouse embryonic diastema region is an ideal site for the development of ectopically transplanted tooth germ. Dev. Dyn. 237, 411-416. 10.1002/dvdy.21427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souders C. A., Bowers S. L. K. and Baudino T. A. (2009). Cardiac fibroblast: the renaissance cell. Circ. Res. 105, 1164-1176. 10.1161/CIRCRESAHA.109.209809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhang T., Liu C., Gu S. and Chen Y. P. (2013). Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis 51, 515-522. 10.1002/dvg.22397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Yu D., Ye W., Liu C., Gu S., Sinsheimer N. R., Song Z., Li X., Chen C., Song Y. et al. (2015). The short stature homeobox 2 (Shox2)-bone morphogenetic protein (BMP) pathway regulates dorsal mesenchymal protrusion development and its temporary function as a pacemaker during cardiogenesis. J. Biol. Chem. 290, 2007-2023. 10.1074/jbc.M114.619007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. W. B., Jensen K. B., Trotter M. W., Connelly J. T., Broad S. and Watt F. M. (2013). Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development 140, 1433-1444. 10.1242/dev.087551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnawski L., Xian X., Monnerat G., Macaulay I. C., Malan D., Borgman A., Wu S. M., Fleischmann B. K. and Jovinge S. (2015). Integrin based isolation enables purification of murine lineage committed cardiomyocytes. PLoS ONE 10, e0135880 10.1371/journal.pone.0135880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eif V. W. W., Stefanovic S., van Duijvenboden K., Bakker M., Wakker V., de Gier-de Vries C., Zaffran S., Verkerk A. O., Boukens B. J. and Christoffels V. M. (2019). Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 146, dev173161 10.1242/dev.173161 [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M., He L., Mäe M. A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B. and Gouveia L. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475-480. 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- van Weerd J. H. and Christoffels V. M. (2016). The formation and function of the cardiac conduction system. Development 143, 197-210. 10.1242/dev.124883 [DOI] [PubMed] [Google Scholar]
- Vedantham V., Galang G., Evangelista M., Deo R. C. and Srivastava D. (2015). RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 116, 797-803. 10.1161/CIRCRESAHA.116.305913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijck E. E., Wessels A., van Ginneken A. C., Bourier J., Markman M. W. M., Vermeulen J. L. M., de Bakker J. M. T., Lamers W. H., Opthof T. and Bouman L. N. (1998). Distribution of atrial and nodal cells within the rabbit sinoatrial node: models of sinoatrial transition. Circulation 97, 1623-1631. 10.1161/01.CIR.97.16.1623 [DOI] [PubMed] [Google Scholar]
- Wiese C., Grieskamp T., Airik R., Mommersteeg M. T. M., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A. F. M., Kispert A. and Christoffels V. M. (2009). Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 104, 388-397. 10.1161/CIRCRESAHA.108.187062 [DOI] [PubMed] [Google Scholar]
- Wu M., Peng S., Yang J., Tu Z., Cai X., Cai C.-L., Wang Z. and Zhao Y. (2014). Baf250a orchestrates an epigenetic pathway to repress the Nkx2.5-directed contractile cardiomyocyte program in the sinoatrial node. Cell Res. 24, 1201-1213. 10.1038/cr.2014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Song Y., Huang Z., Zhang Y. and Chen Y. (2015a). Genetic regulation of sinoatrial node development and pacemaker program in the venous pole. J. Cardiovasc. Dev. Dis. 2, 282-298. 10.3390/jcdd2040282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Wang J., Song Y., Yu D., Sun C., Liu C., Chen F., Zhang Y., Wang F., Harvey R. P. et al. (2015b). A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521-2532. 10.1242/dev.120220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Holden A. V. and Boyett M. R. (2001). Gradient model versus mosaic model of the sinoatrial node. Circulation 103, 584-588. 10.1161/01.CIR.103.4.584 [DOI] [PubMed] [Google Scholar]
- Zhang Y. M., Hartzell C., Narlow M. and Dudley S. C. Jr (2002). Stem cell-derived cardiomyocytes demonstrate arrhythmic potential. Circulation 106, 1294-1299. 10.1161/01.CIR.0000027585.05868.67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.