Abstract
T cell receptor (TCR)–mediated inhibition of interleukin-7 (IL-7) signaling is important for lineage fate determination in the thymus and for T cell survival in the periphery because uninterrupted IL-7 signaling results in T cell death. The initial event in IL-7 signaling is the transactivation of Janus kinases 1 and 3 (Jak1 and Jak3), which are associated with the cytosolic tails of the IL-7 receptor α chain (IL-7Rα) and the γc subunit, the two cell surface proteins that constitute IL-7R. We found that Jak1 is a highly unstable protein with a half-life of only 1.5 hours, so that continuous Jak1 protein synthesis is required to maintain Jak1 protein in sufficient abundance to support IL-7 signaling. However, we also found that Jak1 protein synthesis was acutely reduced by TCR-responsive microRNAs in the miR-17 family, which targeted Jak1 mRNA (messenger RNA) to inhibit its translation. Thus, this study identifies a molecular mechanism by which TCR engagement acutely disrupts IL-7 signaling.
INTRODUCTION
T cells differentiate in the thymus and then emigrate into the periphery, where they initiate and mediate T cell immune responses. The development of T cells in the thymus and the maintenance of T cells in the periphery require transduction of signals from cell surface T cell antigen receptors (TCRs) and interleukin-7 (IL-7) receptors (IL-7Rs) (1–3). Signaling by either receptor alone is insufficient because mature T cells do not survive in the lymphoid periphery of either major histocompatibility complex–deficient mice (which lack TCR ligands) or IL-7–deficient mice (3). T cells require both TCR and IL-7R signals even though signaling by each receptor alone is sufficient to induce expression of genes encoding prosurvival factors, such as that encoding Bcl-2 (4, 5). Indeed, TCR signals are required to intermittently interrupt IL-7 signaling to prevent it from becoming persistent and toxic to T cells (6). Intermittent interruptions of IL-7R signaling by TCR stimulation are also necessary to maintain T cells during peripheral homeostasis (7) and for lineage fate determination in the thymus (8); however, the molecular mechanisms by which TCR stimulation interrupts IL-7 signal transduction remain to be fully elucidated.
IL-7 is a member of the family of cytokines whose cell surface receptors use the common cytokine receptor gamma chain (γc). γc-Dependent cytokines include IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21, and their cell surface receptors consist of at least two transmembrane proteins: a cytokine-specific receptor chain (such as IL-7Rα for IL-7) and γc (9). The cytosolic tail of the cytokine-specific receptor chain is associated with the protein tyrosine kinase Janus kinase 1 (Jak1), whereas the cytosolic tail of γc is associated with Jak3. γc-Dependent cytokines, such as IL-7, initiate cell signaling by inducing the physical approximation of the IL-7Rα and γc proteins on the cell surface, which causes transactivation of Jak1 and Jak3 in the cytosol. Activated Jak1 and Jak3 then phosphorylate monomeric signal transducer and activator of transcription (STAT) proteins to induce STAT protein dimerization, and the dimeric phosphorylated STAT (pSTAT) molecules translocate to the nucleus to activate gene expression. In this way, γc-dependent cytokines such as IL-7 trigger activation of cytokine-responsive genes in T cells.
The only reported molecular mechanism by which TCR signals impair IL-7 signal transduction involves the TCR-dependent activation of the calcium-sensitive protease calpain, which cleaves the cytosolic tail of γc to dissociate Jak3 from surface IL-7Rs, thus aborting IL-7R signaling (10). The TCR-stimulated desensitization of other cytokine receptors, including those for IL-2, IL-4, and IL-6, is dependent on the TCR-dependent activation of the mitogen-activated protein kinase (MAPK) and calcineurin pathways (11, 12), but the molecular mechanism by which these TCR signaling pathways impair cytokine signaling has not been further elucidated.
We undertook the present study to determine how TCR signaling interrupts IL-7 signal transduction. We found that TCR signaling rapidly reduced the cellular abundance of Jak1 protein, the Jak that is associated with the cytosolic tails of cytokine-specific receptor proteins and that is required for signaling by all γc-dependent cytokine receptors. We found that Jak1 was a highly unstable intracellular protein and that continuous Jak1 protein synthesis was required for cells to maintain sufficient amounts of Jak1 to transduce cytokine signals. Furthermore, we found that TCR signals led to the increased abundance of microRNA-17 (miR-17), which targets Jak1 mRNA (messenger RNA) to block its translation, preventing synthesis of new Jak1 protein. As a result, TCR signaling acutely reduced the cellular abundance of Jak1 protein to impair IL-7 signaling. Thus, this study identifies a previously uncharacterized molecular mechanism by which TCR stimulation acutely disrupts IL-7 signaling.
RESULTS
TCR signaling reduces the abundance of Jak1 protein
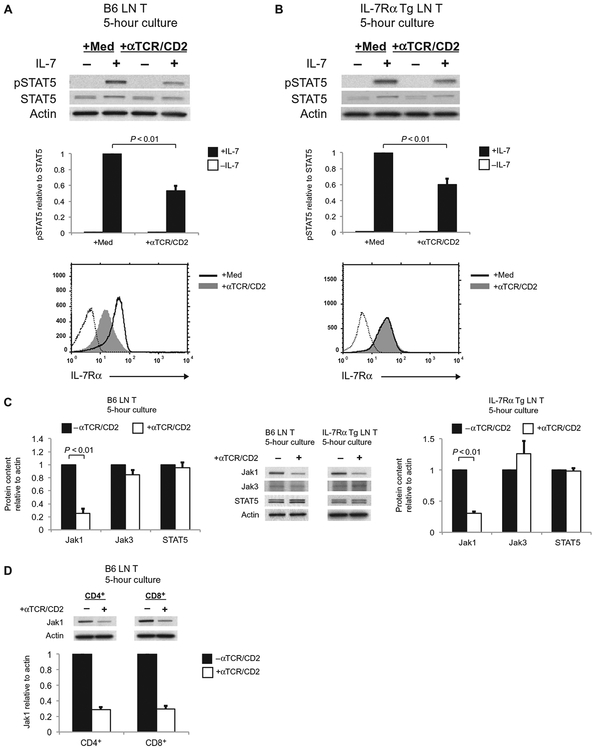
To study how TCR stimulation impaired IL-7 signal transduction, we cultured T cells obtained from mouse lymph nodes (LNs) for 5 hours either alone or with immobilized antibodies against TCR and CD2 (anti-TCR/CD2 antibodies), after which the T cells were assessed for their ability to transduce IL-7R signals in response to short-term (20-min) exposure to IL-7. IL-7 signaling as assessed by detection of STAT5 phosphorylation was quantitatively reduced in TCR-stimulated T cells compared to that in unstimulated T cells (Fig. 1A). Because strong TCR signals can reduce the cell surface abundance of IL-7Rα by reducing expression of the gene encoding IL-7Rα (13), we considered that impaired IL-7 signaling might be a result of the reduced amount of IL-7Rα on the surface of TCR-stimulated cells (Fig. 1A) (13–15). This was not a sufficient explanation, however, because IL-7 signaling as determined by measurement of STAT5 phosphorylation was also impaired in TCR-stimulated T cells from IL-7Rα transgenic (Tg) mice (Fig. 1B) whose cell surface abundance of IL-7Rα was not reduced in response to TCR signaling (Fig. 1B). Consequently, we examined the effect of TCR signaling on the abundance of proteins other than IL-7Rα that are necessary for IL-7 signaling, such as Jak1, Jak3, and STAT5.
Fig. 1.
TCR stimulation impairs IL-7 signaling and reduces Jak1 protein abundance. (A and B) LN T cells from B6 mice (A) and IL-7Rα Tg mice (B) were cultured with medium or with plate-bound anti-TCR and anti-CD2 antibodies (αTCR/CD2) for 5 hours, after which they were left untreated (–) or were treated with IL-7 for 20 min. Top: Cells were harvested, and cell lysates were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and then analyzed by Western blotting with antibodies against the indicated proteins. Middle: The relative amounts of pSTAT5 normalized to those of total STAT5 in the indicated samples were determined by densitometric analysis of the Western blots shown above. Bottom: After the 5-hour culture, the T cells were analyzed by flow cytometry to determine the cell surface expression of IL-7Rα. Dotted histograms show staining with an isotype control antibody. Western blots and flow cytometry plots are representative of three independent experiments. Data in bar graphs are means ± SEM of three independent experiments. (C) LN T cells from B6 mice (left) and IL-7Rα Tg mice (right) were cultured for 5 hours as described for (A), and cell lysates were then analyzed by Western blotting with antibodies specific for the indicated proteins. Jak1, STAT5, and actin were detected by Western blotting analysis of whole-cell lysates, whereas immunoprecipitated samples were analyzed to detect Jak3. Western blots are representative of three independent experiments. Bar graphs show the relative amounts of the indicated proteins, normalized to that of actin, in the unstimulated and TCR-stimulated cells. Data are means ± SEM of three independent experiments. (D) Purified populations of CD4+ and CD8+ T cells from the LNs of wild-type B6 mice were cultured for 5 hours in the absence or presence of αTCR/CD2 antibodies before being analyzed by Western blotting as described for (C). Western blots are representative of two independent experiments, and data in the bar graph are means ± SE of two independent experiments.
TCR signaling did not affect the cellular abundance of either Jak3 or STAT5 proteins, but it reduced the amount of Jak1 protein by more than2.5-fold in both B6 T cells and IL-7Rα Tg T cells, even though cell surface expression of IL-7Rα on IL-7Rα Tg T cells was unaffected by TCR signaling (Fig. 1C). Indeed, TCR signaling reduced the cellular abundance of Jak1 protein in individually purified populations of CD4+ and CD8+ T cells (Fig. 1D). Thus, TCR stimulation impaired IL-7 signaling and reduced Jak1 protein abundance in T cells independently of changes in the surface abundance of IL-7Rα.
TCR signaling inhibits the synthesis of Jak1 protein
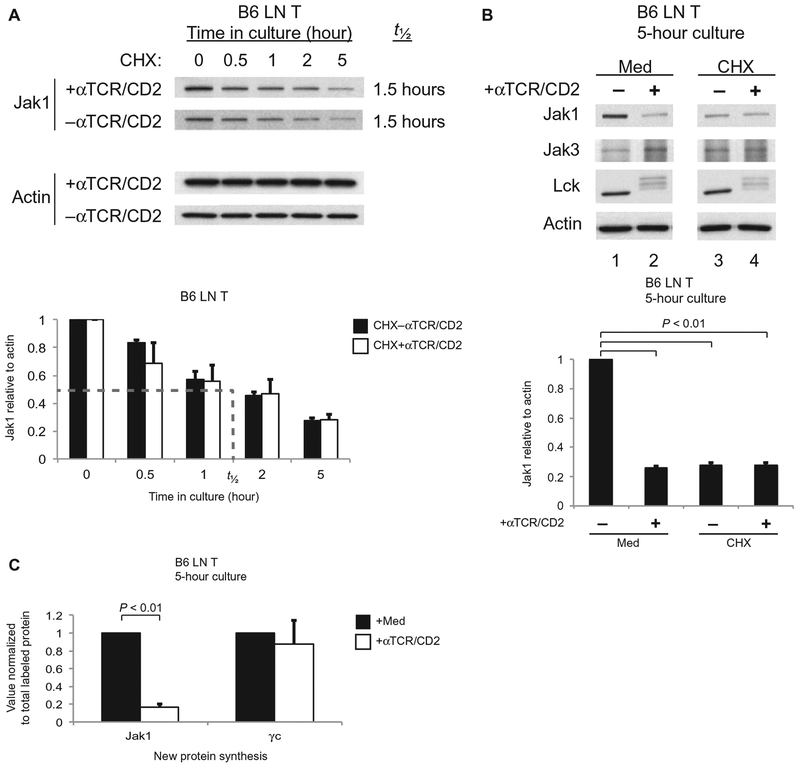
To understand how TCR signaling reduced the abundance of Jak1 protein, we considered that TCR signaling might destabilize Jak1 proteins to accelerate their degradation. To assess this possibility, we examined the effect of TCR signaling on the half-life of Jak1 protein in cells treated with cycloheximide (CHX) to block new protein synthesis (Fig. 2A). We found that Jak1 protein was unexpectedly short-lived in unstimulated T cells, with a half-life of only 1.5 hours, but that it was not further destabilized in response to anti-TCR/CD2 antibodies because Jak1 protein abundance declined with identical kinetics in TCR-stimulated and unstimulated, CHX-treated cells (Fig. 2A). Treatment with anti-TCR/CD2 antibodies induced the appearance of higher–molecular mass (corresponding to phosphorylated) forms of p56Lck (Lck) in both medium- and CHX-treated T cells, suggesting that anti-TCR/CD2 antibodies successfully stimulated TCR signaling in both medium- and CHX-treated T cell populations (Fig. 2B).
Fig. 2.
TCR signaling reduces the abundance of Jak1 by inhibiting the synthesis of new Jak1 protein. (A) LN T cells from B6 mice were pretreated with CHX before being incubated for the indicated times in the absence or presence of αTCR/CD2 antibodies. Top: Cell lysates were analyzed by Western blotting with antibodies specific for the indicated proteins. Bottom: Densitometric analysis of the relative abundance of Jak1 protein, normalized to that of actin, in the indicated samples over time. The dashed line indicates the half-life (t1/2) of Jak1 protein. Data are means ± SEM of three independent experiments. (B) LN T cells from B6 mice were left unstimulated or were stimulated with αTCR/CD2 for 5 hours in the absence or presence of CHX. Top: Samples were analyzed by Western blotting with antibodies against the indicated proteins. Bottom: The relative abundance of Jak1 protein in the indicated samples was determined by densitometric analysis as described for (A). Data are means ± SEM of three independent experiments. Values for Jak1 protein (mean + SE) in TCR-signaled or CHX-treated cells were significantly different from cells in medium only (P < 0.01). (C) LN T cells from B6 mice were left unstimulated or were stimulated with αTCR/CD2 for 5 hours and then were metabolically labeled with [35S]Met/Cys for 30 min. Samples were subjected to immunoprecipitation with anti-Jak1 or anti-γc antibodies. Samples were resolved by SDS-PAGE and quantified for 35S by autoradiography. Densities of bands corresponding to Jak1 and γc were corrected for total TCA-precipitable counts, and the values obtained for TCR-stimulated cells were normalized to those for unstimulated cells, which were set equal to 1.0. Data are means ± SEM of three independent experiments.
These results suggested that TCR signaling did not affect the half-life of Jak1 protein, a conclusion that was also confirmed by our observation that the total abundance of Jak1 protein in T cells pretreated with CHX was unaffected by the presence or absence of TCR signaling during 5 hours of culture (Fig. 2B, lanes 3 and 4); however, CHX alone caused a marked reduction in Jak1 protein abundance because of the short half-life of Jak1 protein (Fig. 2B, lanes 1 and 3). Curiously, the amount of Jak1 protein in medium-treated T cells was reduced by TCR signaling to essentially the same amount as that in CHX-treated, unstimulated T cells (Fig. 2B, lanes 2 and 3), suggesting that TCR signaling might have similar effects to those of CHX in reducing Jak1 protein abundance by inhibiting Jak1 protein synthesis.
To determine the effect of TCR signaling on new Jak1 protein synthesis, we metabolically labeled new proteins synthesized in T cells that had been precultured for the previous 5 hours in the presence or absence of immobilized anti-TCR/CD2 antibodies. After 30 min of metabolic labeling, cell lysates were obtained and specifically immunoprecipitated with anti-Jak1 and anti-γc antibodies to quantify the amount of new Jak1 and γc proteins synthesized, or they were precipitated with trichloroacetic acid (TCA) to quantify total protein synthesis (Fig. 2C and fig. S1). We found that TCR signaling reduced the relative amount of new Jak1 protein synthesized by ~80% compared to that in unstimulated T cells, but TCR signaling did not reduce the relative amount of new γc proteins synthesized (Fig. 2C). We conclude that Jak1 has a short intracellular half-life, so that the overall Jak1 protein abundance in T cells is dependent on continuous synthesis of new protein. Moreover, we conclude that TCR signaling acutely reduces Jak1 protein abundance by specifically inhibiting its synthesis.
Jak1 protein synthesis is regulated by microRNAs
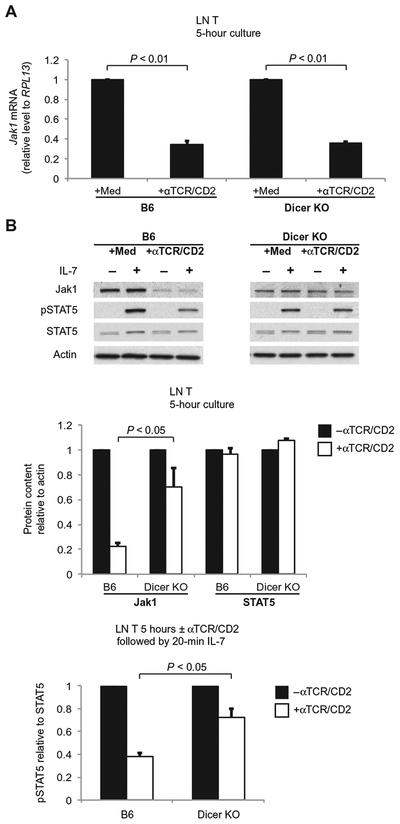
To determine how TCR signaling inhibited Jak1 protein synthesis, we investigated whether TCR signaling might inhibit the transcription of Jak1. To assess this possibility, we quantified Jak1 mRNA abundance by quantitative polymerase chain reaction (PCR) assay and found that the amount of Jak1 mRNA was reduced by TCR signaling (Fig. 3A). A reduction in Jak1 mRNA might have resulted from several different causes, including destabilization of Jak1 mRNA by TCR-induced microRNAs. To assess the latter possibility, we also quantified Jak1 mRNA in T cells from T cell–specific Dicer1 conditional knockout mice, which lack the Dicer nuclease that is required for processing of pre-microRNAs into functional microRNAs (Fig. 3A) (16). We found that Jak1 mRNA abundance was significantly reduced by TCR signaling in T cells from both B6 mice and Dicer-deficient mice, regardless of the presence or absence of microRNAs (Fig. 3A; P < 0.01), suggesting that TCR-stimulated reductions in Jak1 mRNA abundance were independent of microRNA (Fig. 3A).
Fig. 3.
Transcriptional and posttranscriptional effects of TCR signaling on Jak1. (A) LN T cells from B6 mice and Dicer-deficient (Dicer KO) mice were left un-stimulated or were stimulated with αTCR/CD2 for 5 hours. The abundance of Jak1 mRNA in the cells was determined by quantitative reverse transcription PCR (RT-PCR) analysis. Data are means ± SEM of three independent experiments. (B) TCR signaling has substantially less of an effect on Jak1 protein content and IL-7 signal transduction in Dicer KO T cells. LN T cells from B6 mice and Dicer KO mice were cultured for 5 hours with medium or with plate-bound αTCR/CD2 before they were left untreated or were incubated with IL-7 for 20 min. Top: Cell lysates were then analyzed by Western blotting with antibodies against the indicated proteins. Blots are representative of three independent experiments. Middle: Densitometric analysis of the abundance of Jak1 and STAT5, normalized to that of actin, in the indicated cells. Bottom: Densitometric analysis of the relative abundance of pSTAT5 relative to that of total STAT5 in the indicated cells. Data in bar graphs are means ± SEM of three independent experiments and are normalized to values from cells cultured in the absence of αTCR/CD2, which were set equal to 1.0.
If a reduction in Jak1 mRNA abundance was the mechanism by which TCR signaling led to reduced amounts of Jak1 protein, then TCR signaling should reduce the amount of Jak1 protein in both wild-type B6 T cells and Dicer-deficient T cells; however, TCR signaling had markedly different effects in B6 T cells and Dicer-deficient T cells, because TCR signaling substantially reduced Jak1 protein abundance in B6 T cells but had substantially less effect on Jak1 protein abundance in Dicer-deficient T cells (Fig. 3B). These results reveal that the mechanism by which TCR signals acutely reduced the abundance of Jak1 protein was primarily microRNA-dependent and was distinct from the microRNA-independent mechanism by which TCR signals reduced the abundance of Jak1 mRNA. Furthermore, it was the microRNA-dependent reduction in Jak1 protein abundance that impaired IL-7 signaling because, in the absence of microRNAs, TCR stimulation of Dicer-deficient T cells only minimally impaired IL-7–dependent STAT5 phosphorylation (Fig. 3B).
We conclude that TCR signaling can reduce the abundance of Jak1 mRNA but that this is not the major mechanism by which TCR signaling reduces Jak1 protein synthesis to acutely impair IL-7 signaling. Rather, TCR signaling reduces Jak1 protein abundance by a microRNA-dependent mechanism that inhibits translation of Jak1 mRNA and reduces synthesis of new Jak1 protein.
miR-17 directly targets the 3′ untranslated region of Jak1 mRNA
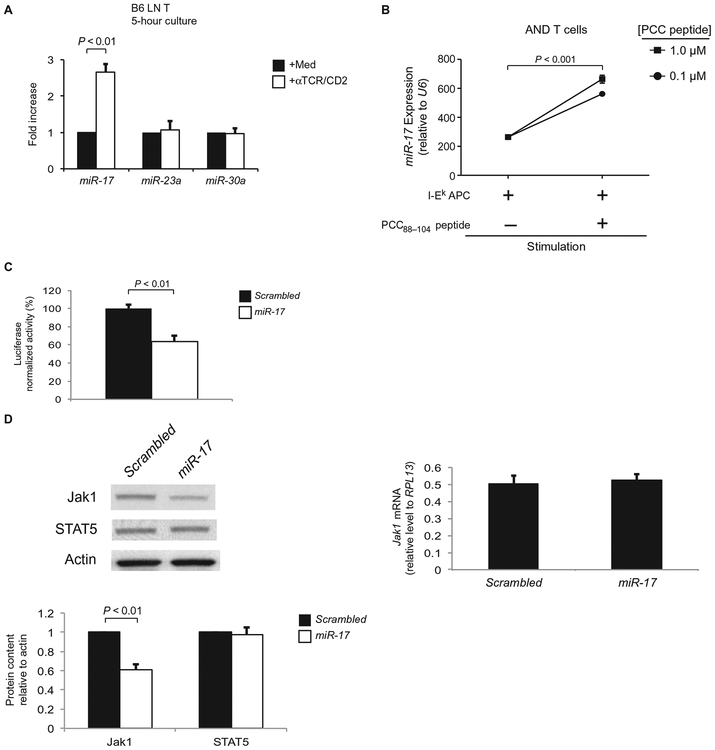
Having determined that the effect of TCR signaling on Jak1 protein abundance was microRNA-dependent, we wanted to identify the responsible microRNA(s). To identify microRNAs with the potential to block Jak1 protein synthesis, we used two different algorithms to analyze the 3′ untranslated region (3′UTR) of Jak1 mRNA for conserved microRNA “seed” regions. Both algorithms identified three microRNA families (mmu–miR-17, mmu–miR-23, and mmu–miR-30) that had imperfect matches with 3′UTR sequences of Jak1 that were predictive of translational inhibition (fig. S2) (16, 17). Although each of these three microRNA families could theoretically inhibit Jak1 translation, only miR-17 abundance was increased by TCR stimulation (Fig. 4A). miR-17 is encoded on murine chromosome 14 together with five other microRNAs that are under the control of a single promoter (referred to as the miR-17~92 cluster) (fig. S3A); thus, the expression of all six members of the miR-17~92 cluster was increased by TCR stimulation (fig. S3B), including the miR-17 family member miR-20a, whose abundance was increased by TCR stimulation to a substantially lesser extent than was that of miR-17 (fig. S3C). These results suggest that miR-17 is the major TCR-responsive microRNA capable of reducing translation of Jak1 mRNA.
Fig. 4.
TCR stimulation increases the abundance of miR-17, which inhibits translation of Jak1 mRNA. (A) LN T cells from B6 mice were left unstimulated or were stimulated with αTCR/CD2 for 5 hours. The abundance of the three microRNA families with potential specificity for Jak1 was then determined by quantitative RT-PCR analysis. Data are means ± SEM of the fold increase in microRNA abundance in response to TCR stimulation from three independent experiments. (B) LN T cells purified from AND Rag2−/− mice were cultured overnight in the presence of I-Ek splenic B cells from B10.A mice (I-Ek APC) and the indicated concentrations of the PCC88–104 peptide. Samples were then analyzed by quantitative RT-PCR. The abundance of miR-17, normalized to that of small nucleolar RNA U6, was then calculated. Data are means ± SEM of quadruplicate cultures for each time point and are representative of two independent experiments. (C) HEK 293T cells were transiently transfected with a dual luciferase reporter in which mRNA for firefly luciferase contained the 3′UTR of Jak1, whereas mRNA for the Renilla luciferase served as an intrinsic control. HEK 293T cells were simultaneously cotransfected with plasmid encoding either miR-17 or scrambled control microRNA. Luciferase activity was measured 48 hours after transfection and is displayed as the activity of firefly luciferase normalized to the activity of Renilla luciferase. Data are means ± SEM of triplicate samples from a single experiment that is representative of three independent experiments. (D) Jurkat cells stably transfected with either miR-17 or scrambled control microRNA were subjected to Western blotting analysis with antibodies against the indicated proteins (left) and to quantitative PCR analysis for Jak1 mRNA abundance (right). Bottom: Densitometric analysis of the abundances of the indicated proteins. Bar graphs show means ± SEM of four independent experiments.
To determine whether miR-17 abundance was increased in T cells stimulated by their physiological ligand, rather than by anti-TCR/CD2 antibodies, we purified LN T cells from AND TCR Tg Rag2−/− mice whose TCR recognizes a complex composed of pigeon cytochrome c (PCC) peptide 88 to 104 and I-Ek (18). Confirming our earlier findings with T cell stimulation by anti-TCR/CD2, stimulation of AND TCR Tg T cells by PCC-bound I-Ek increased the abundance of miR-17 (Fig. 4B).
To determine whether miR-17 targeted the 3′UTR of Jak1 mRNA, we transfected human embryonic kidney (HEK) 293T cells with plasmids encoding luciferase mRNA containing the Jak1 3′UTR. We then compared luciferase activity in cells cotransfected with plasmids encoding either miR-17 or control scrambled microRNA. Indeed, coexpression of miR-17 significantly reduced luciferase protein activity relative to that in cells coexpressing the scrambled microRNA (Fig. 4C; P < 0.01), which demonstrated that miR-17 led to a reduction in the amounts of proteins encoded by mRNAs containing the Jak1 3′UTR. These findings are concordant with an observation in human umbilical vein endothelial cells that miR-17 targets sequences in the 3′UTR of Jak1 mRNA (19). To determine whether miR-17 expression led to reduced Jak1 protein abundance, we stably transfected Jurkat cells (which express endogenous Jak1) with either miR-17 or scrambled microRNA and a green fluorescent protein (GFP) reporter (which enabled identification of cells transfected with microRNA) (Fig. 4D and fig. S4). Compared to control scrambled microRNA, miR-17 significantly reduced Jak1 protein abundance (Fig. 4D; P < 0.01) without affecting Jak1 mRNA abundance (Fig. 4D). We conclude that miR-17 interferes with the translation of Jak1 mRNA without affecting Jak1 mRNA abundance.
miR-17 abundance is increased in constitutively activated T cells from autoimmune mice
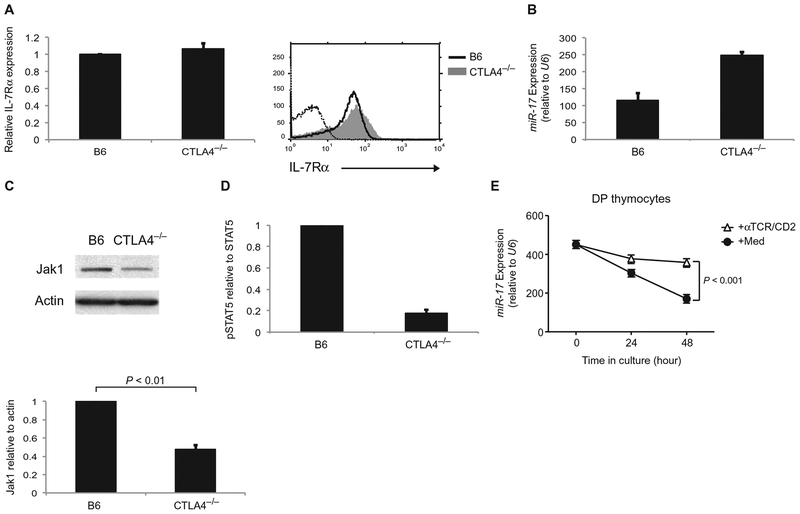
Our results demonstrated that TCR stimulation impaired IL-7 signaling by increasing the expression of miR-17, which led to reduced Jak1 protein synthesis and, as a result, reduced Jak1 protein abundance. On the basis of these findings, we would predict that activated T cells in vivo would have increased miR-17 abundance, reduced Jak1 protein abundance, and impaired IL-7 signaling compared to resting T cells. To assess these predictions, we examined T cells from mice deficient in the inhibitory T cell co-receptor CTLA4 (Ctla4−/− mice), which are spontaneously activated in vivo by the TCR-mediated recognition of endogenous self-antigens and develop autoimmunity (20). We found that the cell surface abundance of IL-7Rα was essentially identical on T cells from Ctla4−/− mice and wild-type B6 mice (Fig. 5A); however, as predicted, miR-17 abundance was increased, whereas Jak1 protein was markedly reduced in Ctla4−/− T cells (Fig. 5, B and C). Accordingly, IL-7 signaling, as determined by measurement of STAT5 phosphorylation, was substantially impaired in Ctla4−/− T cells compared to that in B6 T cells (Fig. 5D and fig. S5). Thus, our assessment of auto-immune T cells in vivo support the conclusion that reductions in Jak1 protein abundance by the TCR-responsive microRNA miR-17 contribute to impaired IL-7 signaling in TCR-stimulated T cells.
Fig. 5.
Examination of T cells from autoimmune Ctla4−/−mice. (A) LN T cells from Ctla4−/− mice and B6 mice were analyzed by flow cytometry to determine the cell surface abundance of IL-7Rα. Left: The relative mean fluorescence intensity of IL-7Rα on Ctla4−/− T cells was expressed relative to that on B6 T cells, which was set equal to 1. Data are means ± SEM of three independent experiments. Right: Representative flow cytometry plots. (B) The abundance of miR-17 in LN T cells from Ctla4−/− mice and B6 mice was determined by quantitative RT-PCR analysis. Data are means ± SEM of triplicate samples from a single experiment that is representative of two independent experiments. (C) Top: LN T cells from Ctla4−/− mice and B6 mice were analyzed by Western blotting with antibodies against the indicated proteins. Bottom: Bar graphs show the relative abundance of Jak1 protein normalized to that of actin. Data are means ± SEM of four independent experiments. (D) LN T cells from Ctla4−/− mice and B6 mice were treated for 20 min with IL-7 before being analyzed by Western blotting with antibodies against pSTAT5 and total STAT5 proteins. The ratio of the abundance of pSTAT5 to that of STAT5 in Ctla4−/− T cells was normalized to that in B6 T cells, which was set equal to1.0. Data are means ± SEM of two independent experiments. (E) Purified DP thymocytes from β2m−/− mice were cultured in the presence or absence of immobilized anti-TCR/CD2 antibodies for the indicated times. Samples were then analyzed by quantitative PCR analysis as described for Fig. 4B. Data are means ± SEM of quadruplicate samples from a single experiment, which is representative of three independent experiments.
TCR signaling maintains miR-17 expression in double-positive thymocytes
The regulation of cytokine signaling by TCR stimulation is important not only for mature T cells in the periphery but also for immature thymocytes during their differentiation in the thymus. Indeed, immature T cells in the thymus go through a developmental checkpoint in which they become CD4+CD8+ [double-positive (DP)] cells, which are refractory to cytokines (21). The lack of responsiveness of these cells to cytokines is partly a result of their constitutively increased amount of suppressor of cytokine signaling 1 protein, which suppresses cytokine signaling (22, 23). Moreover, cytokine-unresponsive DP thymocytes also have constitutively increased amounts of miR-17 (24). Thus, we wanted to determine whether TCR signaling affected miR-17 expression in DP thymocytes similarly to that in mature peripheral T cells. We confirmed that miR-17 was highly abundant in immature DP thymocytes and that TCR stimulation was necessary to maintain miR-17 expression, because miR-17 abundance decreased in the absence of TCR signaling (Fig. 5E). The small reduction in miR-17 abundance in thymocytes stimulated with anti-TCR/CD2 antibodies (Fig. 5E) was a result of DP cells that did not express surface ab TCR complexes and so could not respond to antibody-mediated stimulation. Thus, these results suggest that miR-17 expression is responsive to TCR signaling in immature thymocytes as well as in mature peripheral T cells.
DISCUSSION
Here, we found that TCR stimulation inhibits IL-7 signaling by specifically targeting Jak1, a Jak required for signal transduction by all γc-dependent cytokines. We found that Jak1 protein had a half-life of only1.5 hours, so that continuous synthesis of new Jak1 protein was required to maintain sufficient amounts of Jak1 to support IL-7 signaling. Although TCR signaling did not further destabilize preexisting Jak1 protein or affect its half-life, TCR signaling prevented the synthesis of new Jak1 protein by inhibiting both Jak1 expression and Jak1 mRNA translation. The inhibition of Jak1 mRNA translation was the major mechanism by which TCR signaling rapidly reduced the abundance of Jak1 protein, which was mediated by the TCR-responsive microRNA, miR-17. In contrast, TCR-dependent inhibition of Jak1 transcription was microRNA-independent; however, it did not lead to a rapid reduction in Jak1 protein abundance to acutely disrupt IL-7 signaling. We conclude that IL-7 signaling is limited by Jak1 protein instability and that TCR signals acutely impair IL-7 signal transduction by inducing the expression of a Jak1-specific microRNA, miR-17, which prevents new Jak1 protein synthesis.
The only previously identified mechanism for TCR-mediated interruption of IL-7 signaling involves the TCR-stimulated proteolysis of the cytosolic tail of the γc subunit by the calcium-sensitive protease calpain, which causes Jak3 to dissociate from γc (10). In this mechanism, the enzymatic activity of calpain is activated by cytosolic calcium directly released from intracellular stores by TCR signaling. Because calcium release is a direct consequence of TCR signaling, it would be expected that calpain would be activated regardless of the presence or absence of microRNAs; however, we found little evidence for microRNA-independent disruption of IL-7 signaling in response to TCR engagement, because IL-7 signal transduction was reduced only minimally by TCR stimulation of T cells from Dicer-deficient mice. Nevertheless, because the TCR-dependent targeting of γc (through the action of calpain) (10) and Jak1 (through miR-17) are potentially complementary mechanisms for inhibiting IL-7 signaling, it is possible that TCR-dependent signals use different mechanisms to disrupt IL-7 signaling in different circumstances.
However, why did acute reductions in Jak1 protein abundance result in impaired IL-7 signaling? Our current findings indicate that Jak1 protein is present in T cells in limiting amounts, so that acute reductions in the amount of Jak1 would result in the appearance on the cell surface of “unarmed” IL-7Rα proteins that lack associated Jak1 molecules. Consequently, we think that there are two reasons why acute reductions in Jak1 protein abundance led to impaired IL-7 signaling. First, the number of Jak1-associated IL-7Rα proteins on the cell surface was reduced, so fewer surface receptors could transduce IL-7 signals. Second, unarmed IL-7Rα proteins on the cell surface could have competed with signaling-competent, Jak1-associated IL-7Rα proteins for binding to IL-7. Thus, we think that TCR-dependent reductions in Jak1 protein abundance reduced the number of signaling-competent IL-7Rs on the cell surface and caused these receptors to have to compete with unarmed IL-7Rs for available IL-7.
TCR-mediated interruptions in IL-7 signaling are important for T cell development in the thymus and T cell homeostasis in the periphery (6–8). Because of its fundamental importance for T cells, we do not think that it is surprising that more than one molecular mechanism may have evolved for the TCR to disrupt IL-7 signaling. Indeed, we have identified two mechanisms by which TCR signals targeted Jak1, namely, microRNA-mediated inhibition of Jak1 mRNA translation and microRNA-independent inhibition of Jak1 transcription. Within the 5-hour window of TCR stimulation of mature T cells that we examined, the microRNA-mediated inhibition of Jak1 mRNA translation was substantially more effective in disrupting IL-7 signaling than was inhibition of Jak1 transcription. Nonetheless, we would expect that the effectiveness of inhibition of Jak1 transcription in disrupting IL-7 signaling transduction would increase with longer TCR stimulation times.
Although our study specifically examined IL-7 signaling, Jak1 is important for signaling by all γc-dependent cytokines, so it is reasonable to expect that TCR signaling would similarly disrupt signaling by all γc-dependent cytokines. Thus, our study identifies the TCR-dependent targeting of Jak1 as a regulatory mechanism for interrupting cytokine signaling in T cells.
MATERIALS AND METHODS
Mice
B6 mice were obtained from The Jackson Laboratory. IL-7Rα Tg mice were previously generated in our laboratory (25) and were bred to IL-7Rα–deficient mice (6). Ctla4−/− mice (20), AND TCR Tg mice on the Rag2−/− background (18), B10.A mice, and β2m−/− mice were maintained in our own animal colony. All mice were cared for in accordance with National Institutes of Health guidelines. To construct conditional Dicer-deficient mice, we designed a targeting vector that introduced a loxP site into intron 22 of dcr-1 and an frt/loxP-flanked neomycin resistance cassette into intron 23 of dcr-1. Integration of the targeting vector by homologous recombination into the dcr-1 allele of 129-derived embryonic stem (ES) cells was verified by Southern blotting and PCR analyses. ES cells with the homologously recombined dcr-1 allele were injected into blastocysts to generate mice containing the targeted allele. The neomycin resistance cassette was removed by introduction of the actin-flip transgene, which resulted in the generation of Dicer1fl/wt mice that were then backcrossed and maintained on a C57BL/6 background.
Antibodies
Antibodies with the following specificities were used. For flow cytometry: anti–IL-7Rα (A7R34, eBioscience) and immunoglobulin G (IgG) isotype control (BD Pharmingen); for in vitro stimulation: anti-TCRβ (H57–957, BD Pharmingen) and anti-CD2 (RM2–5, BD Pharmingen); for Western blotting: anti-Jak1 (73/JAK1, BD Pharmingen and Cell Signaling Technology), anti-STAT5 (Cell Signaling Technology), anti-Lck (BD Pharmingen), anti-pSTAT5 (BD Pharmingen), anti-actin (C4, Millipore), and anti-Jak3 (Millipore); and for protein immunoprecipitation: anti-Jak1 (Santa Cruz Biotechnology), anti-Jak3 (Millipore), and anti-γc (Santa Cruz Biotechnology). Biotinylated anti-CD4 (GK1.5, BD Pharmingen) and anti-CD8 antibodies (53–6.7, BD Pharmingen) were used to deplete CD4+ and CD8+ T cells, respectively.
Flow cytometric analysis
Cells were analyzed on an LSR II flow cytometer (Becton Dickinson) with four-decade logarithmic amplification, and data were analyzed with software designed by the Division of Computer Research and Technology (National Institutes of Health).
Cell culture
LN T cells were depleted of B cells with anti-mouse IgG beads (Qiagen) and were purified into CD4+ or CD8+ subpopulations with the Pan T Cell Isolation Kit (Miltenyi). For stimulation, cells were cultured for 5 hours with plate-bound anti-TCRβ (5 μg/ml) and anti-CD2 (5 μg/ml) antibodies, whereas unstimulated cells were cultured only in medium (RPMI 1640 supplemented with 10% fetal calf serum that had been depleted of steroids by charcoal stripping). Where indicated, cultured cells were then exposed to IL-7 (1 ng/ml; PeproTech) for 20 min and then were harvested. In some experiments, cells were pretreated with CHX (10 μg/ml; Sigma) for 10 to 15 min. To monitor new protein synthesis, cells were labeled with [35S]Met/Cys mix (0.5 mCi/10 × 106 cells; PerkinElmer) for 30 min. Where indicated in the figure legends, AND Rag2−/− LN T cells (1 × 106) were stimulated overnight with 2 × 106 B10.A splenic B cells and PCC88–104 peptide (American Peptide Company). Where indicated in the figure legends, DP thymocytes were purified from β2m−/− mice by adherence to immobilized anti-CD8 monoclonal antibody (83-12-5) and then were cultured.
Immunoprecipitations and Western blotting
Cells were solubilized in lysis buffer (1× tris-buffered saline, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing 2 mM EDTA, 1 mM sodium orthovanadate, and protease inhibitors. Immunoprecipitates or whole-cell lysates were resolved by SDS-PAGE on 4 to 20% acrylamide gels (Invitrogen) under reducing conditions and were transferred to nitro-cellulose membranes (Amersham). Membranes were incubated with the appropriate antibodies and horseradish peroxidase–conjugated secondary antibodies. Reactivity was revealed by enhanced chemiluminescence (PerkinElmer). To detect 35S labeling, SDS-PAGE gels were fixed in 10% acetic acid and 20% methanol, soaked in rapid autoradiography enhancer (PerkinElmer), dried, and then exposed to film. TCAwas obtained from Sigma.
Quantitative real-time RT-PCR
Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and reverse-transcribed with oligo dT and SuperScript III reverse transcriptase (Invitrogen). The resulting complementary DNA was subjected to real-time PCR amplification with an SYBR Green PCR Kit (Qiagen). The primers used are shown in table S1. For microRNA analysis, RNA was extracted with the mirVana miRNA Isolation Kit (Applied Biosystems), reverse-transcribed, and then analyzed by real-time PCR amplification with TaqMan MicroRNA Assays (Applied Biosystems). Where indicated, RNA was quantified relative to the housekeeping gene RPL13.
Transfection
Jurkat cells were transfected by electroporation with plasmids encoding either scrambled control microRNA or miR-17 (GeneCopoeia). Stable transfection was achieved by selection in medium containing puromycin (Sigma).
Luciferase assays
To measure luciferase activity, HEK 293T cells were cotransfected Lipofectamine 2000 (Invitrogen) with the Jak1-3′UTR hLuc-hRLuc vector (GeneCopoeia) and with plasmids encoding either scrambled control microRNA or miR-17. Luciferase activity was assessed 48 hours later with the LucPair miR Luciferase Assay Kit (GeneCopoeia).
Statistical analysis
P values were determined by Student’s two-tailed t test.
Supplementary Material
Acknowledgments:
We thank P. Roche, H. Park, and G. Patino-Lopez for helpful discussions and D. Singer, R. Hodes, and H. Park for critically reading the manuscript.
Funding: This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/7/340/ra83/DC1
Fig. S1. Analysis of total protein synthesis in unstimulated and TCR-stimulated cells.
Fig. S2. Imperfect match for microRNAs that potentially bind to the 3′UTR of Jak1 mRNA.
Fig. S3. Effect of TCR signaling on the expression of microRNAs in the miR-17~92 cluster.
Fig. S4. Analysis of GFP abundance in Jurkat cells stably transfected with plasmids encoding GFP and either scrambled control microRNA or miR-17.
Fig. S5. IL-7 signaling is impaired in Ctla4−/− mice.
Table S1. Primers used for real-time PCR analysis.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Akashi K, Kondo M, Weissman IL, Role of interleukin-7 in T-cell development from hematopoietic stem cells. Immunol. Rev 165, 13–28 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Goldrath AW, Bevan MJ, Selecting and maintaining a diverse T-cell repertoire. Nature 402, 255–262 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Jameson SC, Maintaining the norm: T-cell homeostasis. Nat. Rev. Immunol 2, 547–556 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Linette GP, Grusby MJ, Hedrick SM, Hansen TH, Glimcher LH, Korsmeyer SJ, Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity 1, 197–205 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A, In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J. Exp. Med 197, 475–487 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura MY, Pobezinsky LA, Guinter TI, Thomas J, Adams A, Park JH, Tai X,Singer A, IL-7 signaling must be intermittent, not continuous, during CD8+ T cell homeostasis to promote cell survival instead of cell death. Nat. Immunol 14, 143–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Adoro S, Lucas PJ, Sarafova SD, Alag AS, Doan LL, Erman B, Liu X, Ellmeier W, Bosselut R, Feigenbaum L, Singer A, ‘Coreceptor tuning’: Cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat. Immunol 8, 1049–1059 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Singer A, Adoro S, Park JH, Lineage fate and intense debate: Myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol 8, 788–801 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rochman Y, Spolski R, Leonard WJ, New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol 9, 480–490 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi M, Sarin A, Aman MJ, Nakajima H, Shores EW, Henkart PA, Leonard WJ, Functional cleavage of the common cytokine receptor γ chain (γc) by calpain. Proc. Natl. Acad. Sci. U.S.A. 94, 11534–11539 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee IH, Li WP, Hisert KB, Ivashkiv LB, Inhibition of interleukin 2 signaling and signal transducer and activator of transcription (STAT)5 activation during T cell receptor–mediated feedback inhibition of T cell expansion. J. Exp. Med 190, 1263–1274 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Huang H, Guo L, Stonehouse T, Watson CJ, Hu-Li J, Paul WE, Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J. Exp. Med 192, 1125–1134 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alves NL, van Leeuwen EM, Derks IA, van Lier RA, Differential regulation of human IL-7 receptor α expression by IL-7 and TCR signaling. J. Immunol 180, 5201–5210 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Hammerbeck CD, Mescher MF, Antigen controls IL-7R α expression levels on CD8 T cells during full activation or tolerance induction. J. Immunol 180, 2107–2116 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Schluns KS, Kieper WC, Jameson SC, Lefrançois L, Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol 1, 426–432 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Filipowicz W, Bhattacharyya SN, Sonenberg N, Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Fabian MR, Sonenberg N, Filipowicz W, Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem 79, 351–379 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Kaye J, Hsu ML, Sauron ME, Jameson SC, Gascoigne NR, Hedrick SM, Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341, 746–749 (1989). [DOI] [PubMed] [Google Scholar]
- 19.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S, Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115, 4944–4950 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW, Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 270, 985–988 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Yu Q, Park JH, Doan LL, Erman B, Feigenbaum L, Singer A, Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J. Exp. Med 203, 165–175 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong MM, Cornish AL, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, Starr R, Alexander WS, Kay TW, Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity 18, 475–487 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE, Kubo M, Hennighausen L, Feigenbaum L, Singer A, Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol 11, 257–264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K, Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol 9, 405–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A, IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgt: Impact on thymocyte development. J. Exp. Med 200, 797–803 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.