Abstract
Smoking perpetuates in cytologically normal airways a molecular “field of injury” that is pertinent to lung cancer and early detection. The evolution of airway field changes prior to lung oncogenesis is poorly understood largely due to the long latency of lung cancer in smokers. Here, we studied airway expression changes prior to lung cancer onset in mice with knockout of the Gprc5a gene (Gprc5a−/−) and tobacco carcinogen (NNK) exposure and that develop the most common type of lung cancer, lung adenocarcinoma, within 6 months following exposure. Airway epithelial brushings were collected from Gprc5a−/− mice before exposure and at multiple times post-NNK until time of lung adenocarcinoma development and then analyzed by RNA sequencing. Temporal airway profiles were identified by linear models and analyzed by comparative genomics in normal airways of human smokers with and without lung cancer. We identified significantly altered profiles (n = 926) in the NNK-exposed mouse normal airways relative to baseline epithelia, a subset of which were concordantly modulated with smoking status in the human airway. Among airway profiles that were significantly modulated following NNK, we found that expression changes (n = 22) occurring as early as 2 months following exposure were significantly associated with lung cancer status when examined in airways of human smokers. Furthermore, a subset of a recently reported human bronchial gene classifier (Percepta; n = 56) was enriched in the temporal mouse airway profiles. We underscore evolutionarily conserved profiles in the normal-appearing airway that develop prior to lung oncogenesis and that comprise viable markers for early lung cancer detection in suspect smokers.
Introduction
Non-small cell lung cancer (NSCLC) constitutes the most frequently diagnosed type of lung malignancy (1,2). NSCLC has an overall 5-year survival rate of 15% (3). This survival rate is particularly low among advanced stage NSCLC, largely due to late diagnosis (2,3). Although low-dose CT reduces lung cancer mortality, this screening strategy presents high false-positive rates warranting new or complementary strategies for early detection (4). Lung adenocarcinoma is the most common histologic subtype of NSCLC and accounts for the majority of cancer-related deaths in smokers (1, 2). Advances in diagnosis heavily rest on understanding early changes, and thus markers, in the pathogenesis of NSCLC and, particularly, its major histologic subtype lung adenocarcinoma (2).
Although the majority of NSCLCs are causally linked to cigarette smoking, only 15% of smokers develop the disease (5). Also, approximately 50% of NSCLCs are diagnosed in former smokers (6). Following exposure to smoking, cytologically normal airway epithelial cells comprise molecular alterations (e.g., genome-wide gene expression changes) that embody a “field of injury” throughout the lung that is thought to be pertinent to lung oncogenesis in smokers (5, 7, 8). Earlier work revealed that airways of smokers with lung cancer exhibit significant differential expression profiles compared with airways of cancer-free smokers (7). Of note, gene expression changes in the airway field of injury have been translated to viable markers for early lung cancer detection. A 232-gene classifier in the bronchial airway (Percepta) was derived that is able to significantly detect lung cancer in suspect current or former smokers (9). Also, a recent study developed and tested a bronchial gene expression classifier that improved the diagnostic performance of endoscopic bronchoscopy for detection of lung cancer in smokers with indeterminate nodules, particularly in intermediate-risk patients when bronchoscopy was nondiagnostic (10).
Studies reporting diagnostic expression profiles in the airway field of injury of smokers were for the most part cross-sectional. We still do not know how and what airway field expression patterns evolve with time following the onset of smoking to signify the development of a nearby lung cancer. To understand the evolution of the airway field of injury, we sought to survey genome-wide temporal airway gene expression changes by RNA sequencing (RNA-Seq) in vivo. We studied airway expression changes in mice with knockout of the G-protein-coupled receptor Gprc5a (Gprc5a−/−) and nicotine-specific nitrosamine ketone (NNK) exposure, which we recently found to develop the most common type of lung cancer, lung adenocarcinoma, within 6 months following exposure (11). Here, we underscore evolutionarily conserved gene expression profiles in the cytologically normal airway field of injury that progress in vivo following tobacco carcinogen exposure and up to the development of lung tumors. Notably, we find that a subset of these progressive airway expression changes is significantly enriched in cytologically normal airways of human smokers with lung cancer and is indicative of lung malignancy in suspect smokers with indeterminate nodules. Our current study sheds light on evolutionarily conserved early changes in the normal bronchial airway preceding onset of lung cancer.
Materials and Methods
Chemical and reagents
NNK (purity > 99%) and hematoxylin and eosin (H&E) staining reagents were purchased from Sigma-Aldrich.
Animal housing and carcinogenesis experiments
Gprc5a—/— mice (C57BL/6 × 129sv) were generated as described previously (12) and were maintained according to a protocol approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. The mice were housed in a pathogen-free animal facility approved by the American Association for Accreditation of Laboratory Animal Care. Twenty-eight 8-week-old Gprc5a—/— mice (16 females and 12 males) were divided into groups of 5 to 6 mice per time point (1:1, 2:3, or 1:2 male to female ratios) and were injected intraperitoneally with 50 mg/kg of body weight NNK three times per week for 8 weeks (150 mg/kg total per week) dissolved in PBS. Mice were sacrificed at baseline (prior to NNK treatment), immediately after completion of NNK treatment and at 2, 4, and 6 months following treatment. After sacrifice, lungs were excised and inflated by injection with formalin. Lung surface lesions were examined by macroscopic observation. Formalin-fixed lung specimens were then embedded in paraffin (FFPE) and histologic sections were prepared, stained with H&E, and analyzed for lung tumor development by an experienced pathologist (Junya Fujimoto) using previously reported criteria for characterization of murine lung lesions (13).
Collection of mouse tracheal epithelial brushings
Epithelial brushings of the cytologically normal airway from mouse trachea were obtained after mouse sacrifice and based on the method previously described and reported by Sugimoto and colleagues (14). Brushes were made from 60-grit sandpaper-polished polyethylene tube (PE-10 tube; BD Biosciences) with an inserted stainless wire. Brushes (three/mouse) were inserted through an incision into the midsection of the trachea. Initially, brushes were placed in 1 × PBS and cell pellets were stained with pan-cytokeratin to confirm epithelial content and ensure lack of preneoplastic and neoplastic cells. Harvested epithelia from three brushes were collected into 500 mL of Qiazol lysis buffer (Qiagen) and immediately placed on dry ice. Brushes were stored at −80°C until further processing for RNA isolation.
Total RNA isolation from mouse airway brushings
Tubes containing brushes in lysis buffer were vortexed gently to dissociate harvested epithelia and brushes were then removed. Total RNA were purified from the brushings using the miRNeasy Kit from Qiagen according to the manufacturer’s instructions. Purified RNA samples were then cleaned and resuspended in nuclease-free water using the RNeasy MinElute Cleanup Kit from Qiagen according to the manufacturer’s protocol. RNA quantity was measured using the NanoDrop 1000. Quality of RNA samples was quantitatively assessed by computation of RNA integrity numbers (RIN) using the Agilent RNA 6000 Pico Kit (Agilent) following the manufacturer’s protocol.
RNA-Seq
RNA samples with RNA integrity numbers ≥7 were selected for RNA-Seq. Libraries were then prepared from poly-A RNAs using the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) according to the manufacturer’s protocol for barcoded whole-transcriptome libraries. RNA-Seq was performed using the Ion Torrent Proton platform according to the manufacturer’s instructions. On average, we obtained approximately 39.5 million reads per sample. Alignment of reads was performed using the STAR algorithm v2.4.1d (15) and Bowtie2 v. 2.1.0 (16). Transcripts were quantified (mm10) using a modified version of the expectation-maximization (E/M) algorithm (17). Raw sequencing and normalized expression data of the mouse normal-appearing airways (n = 28 samples) are deposited in the gene expression omnibus (GEO) under series GSE102707 (samples GSM2743377 - GSM2743404).
Gene expression analysis
A linear model was used to identify gene expression profiles that were differentially expressed in the mouse airway following NNK exposure (t = 0) relative to baseline. A Benjamini-Hochberg controlled FDR was used to identify significant features based on a q-value cutoff. A linear model was also applied to identify profiles that were differentially expressed at each time point following NNK exposure (t = 2, t = 4, and t = 6) relative to t = 0. All statistical analyses and hierarchical clustering were conducted using the R statistical language environment v3.1.1 and the ilmma package v3.22.7.
Comparative genomics analysis
Comparative genomics analysis was used to analyze mouse airway expression changes in human airway datasets. To analyze changes influenced by NNK (at end of exposure vs. baseline), the previously reported human expression dataset by Spira and colleagues (18) consisting of airway samples from phenotypically healthy current smokers (n = 34) and nonsmokers (n = 23) was interrogated. Orthologues from the murine airway expression profiles were determined, ranked by the t-statistic of the coefficient for smoking status from a linear model, and analyzed by gene set enrichment analysis (GSEA; ref. 19) in the human smoking dataset. Temporal mouse airway expression changes were similarly analyzed by GSEA in the recently reported array dataset by Silvestri and colleagues (10) of bronchial brushings from smokers with suspicion of lung malignancy (n = 253 and n = 90 with and without lung cancer, respectively) and who had undergone endoscopic bronchoscopy for diagnosis. The expression of a recently reported bronchial 232-gene classifier (9) was also analyzed in the temporal post-NNK mouse airway samples.
Detailed methods are available in the Supplementary Methods accompanying the article.
Results
Analysis of temporal genome-wide gene expression changes in the mouse airway following tobacco carcinogen exposure and preceding lung adenocarcinoma
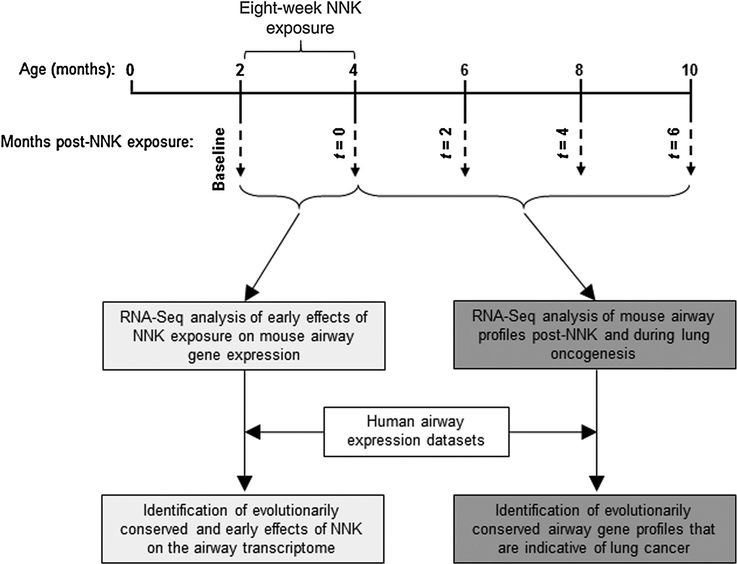
The progression of airway expression changes following smoking and prior to lung cancer onset is poorly understood. This is largely due to the long latency of human lung malignancy and, thus, the difficulty in temporally tracking airway field effects following smoking. To fill this void, we surveyed, by RNA-Seq, genome-wide temporal expression changes in the normal-appearing airway that occur prior to and during oncogenesis of a nearby lung tumor (Fig. 1). We studied Gprc5a−/− mice exposed to the tobacco-specific carcinogen NNK, which we recently found to develop the most common histologic subtype of lung malignancy, lung adenocarcinoma, within 6 months following exposure (Fig. 1; ref. 11). Groups of five to six 8-week-old mice (n = 28 total) were intraperitoneally administered 50 mg/kg bodyweight NNK three times per week for 8 weeks. Airway (tracheal) epithelia were collected by brushing and studied at baseline (prior to NNK exposure) and immediately following the 8-week exposure (t = 0) to understand early effects of tobacco carcinogen exposure on the airway transcriptome (Fig. 1). Mice were followed up at two (t = 2), four (t = 4), and six (t = 6) months following completion of NNK exposure for lung tumorigenesis. Lung tumor burden was significantly increased with time post-NNK particularly at t = 4and t = 6 (P < 0.001 of the ANOVA test, Supplementary Fig. S1). Airway epithelia were also collected every 2 months following completion of NNK treatment (t = 2, t = 4, and t = 6) and studied to understand progressive or lasting field effects following exposure and until tumor development (t = 6; Fig. 1). Comparative genomics analyses were also per formed, using expression datasets of normal airways from human smokers, to determine evolutionarily conserved profiles including those that can diagnose lung cancer in smokers with suspicion of the disease (Fig. 1).
Figure 1.
Identification of global gene expression changes in the normal-appearing airwayduring the development of lung adenocarcinoma. Study design depicting analysis of genome-wide changes in gene expression in the tobacco-carcinogen normal-appearing airway in vivo. Two-month-old Gprc5a−/− were treated intraperitoneally with 50 mg/kg bodyweight nicotine-specific NNK three times per week for 8 weeks. Upper airway epithelia were collected by brushings before NNK treatment (baseline) and at the indicated four time points after completion of NNK exposure as described in the Materials and Methods section. Total RNA was isolated and interrogated by RNA-Seq using the Ion Torrent Proton platform. Gene profiles were statistically analyzed to determine patterns that are impacted early on in the airway by NNK exposure (t = 0 vs. baseline) and those that temporally evolve for 6 months following completion of the treatment and during the development of lung adenocarcinoma. Using publicly available human airway expression datasets, cross-species comparative genomics analysis (see Materials and Methods and Supplementary Methods) was performed to identify genes from the mouse airway expression profiles that are evolutionarily conserved in airways of human suspect smokers, including patients with lung malignancy.
Early evolutionarily conserved expression changes in the mouse airway following tobacco carcinogen exposure
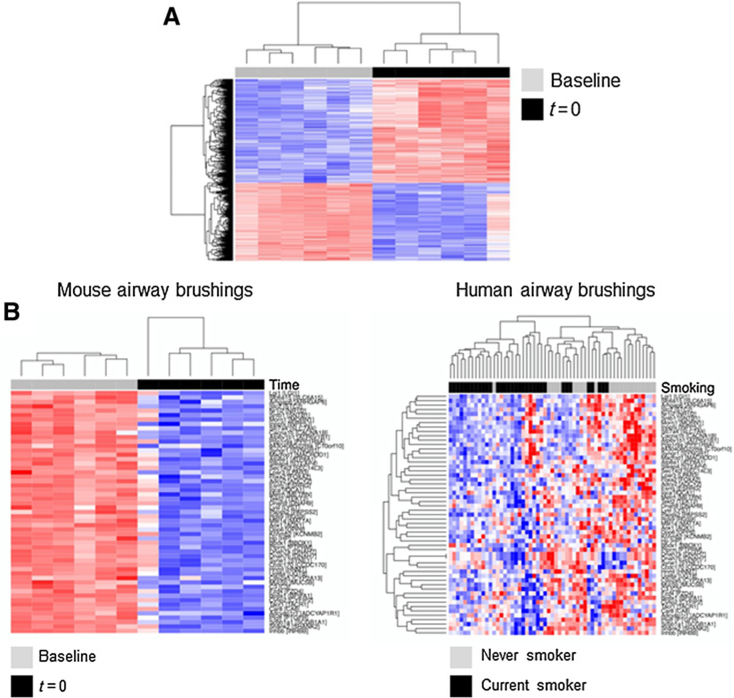
We first compared and contrasted the transcriptome of mouse airways prior to (baseline) and immediately following completion of NNK exposure (t = 0; n = 6 samples each). Using a linear model and a controlled (by the Benjamini-Hochberg method) FDR threshold of 0.1%, we found 926 gene features (529 upregulated and 397 downregulated) that were significantly differentially expressed in the airway following NNK exposure (Fig. 2A; Supplementary Table S1). Hierarchical clustering revealed that the identified profiles clustered airway samples according to NNK exposure status (Fig. 2A). We then sought to determine from the murine airway profiles those that are enriched in cytologically normal airways of human smokers. We interrogated a reported expression dataset of cytologically normal airway (mainstem bronchi) brushings obtained from phenotypically healthy (cancer-free) current smokers (n = 34) and nonsmokers (n = 23; ref. 18). After matching gene features between our study and the human airway dataset, 853 remained (491 upregulated and 362 downregulated by NNK). These orthologous profiles were then ranked according to the t statistic between human smokers and nonsmokers and analyzed by GSEA in the human dataset. Of the 362 downregulated genes in the airway by the tobacco carcinogen, 54 genes were found to be statistically significantly enriched (P < 0.001) within gene features downregulated in airways of human current smokers compared with airways of never smokers (Supplementary Fig. S2; Supplementary Table S2). In addition, the enriched gene features, following hierarchical clustering analysis, not only separately clustered mouse airway samples by NNK exposure status (Fig. 2B, left) but also significantly (P < 0.001 of the Fisher exact test) distinguished airways of human smokers from those of nonsmokers (Fig. 2B, right). Of note, similar effects were found with gene features upregulated by NNK, albeit they did not reach statistical significance (data not shown). These data underscore early expression changes by tobacco carcinogen (NNK) in the mouse airway that are also present in airways of humans smokers and are thus likely to be evolutionarily conserved in their response to smoking.
Figure 2.
Evolutionary conserved effects of NNK tobacco carcinogen exposure on airway gene expression. A, Differentially expressed profiles (n = 926; 529 upregulated and 397 downregulated) between mouse airways at the time of completion of NNK exposure (t = 0) relative to baseline were identified using a linear model and a controlled FDR threshold of 0.1% (see Materials and Methods). B, Using orthologues from the identified genes, GSEA was performed to identify expression profiles that are concordantly (in same direction) enriched in airways of human cancer-free current smokers relative to nonsmokers. Statistically significant leading edge set of 54 downregulated orthologous genes was analyzed by semisupervised clustering in the mouse airway brushings (left). The same genes were assessed by unsupervised clustering in the human airway brushings (right). Columns represent samples and rows denote gene features; red and blue: higher and lower expression, respectively.
Early expression changes in the mouse normal-appearing airway field of injury that signify lung cancer in human smokers
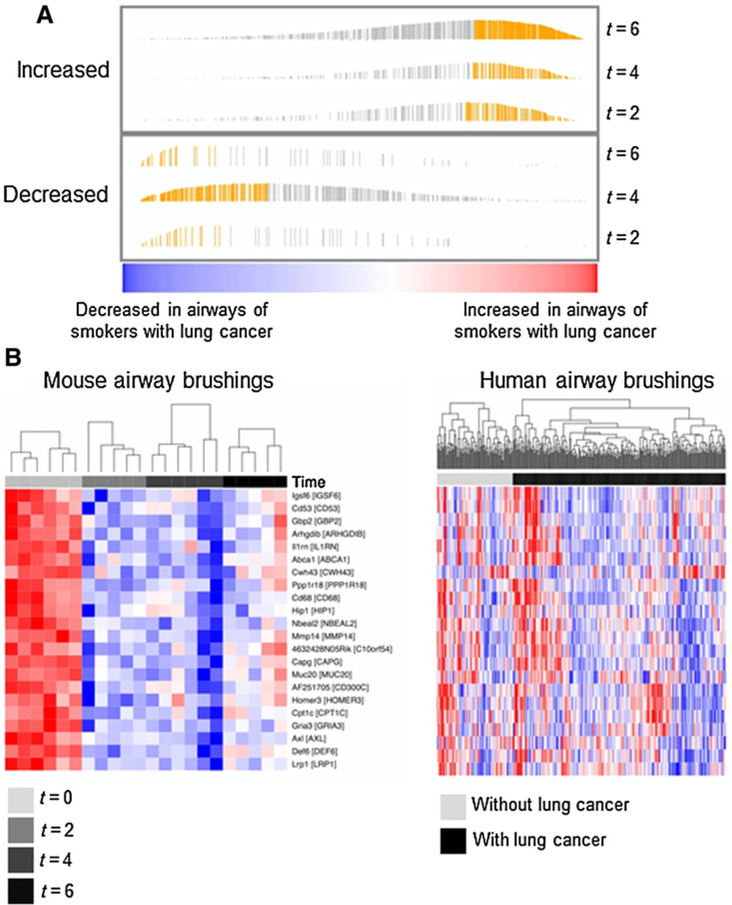
We then sought to pinpoint global gene expression profiles in the airway that progress following tobacco carcinogen exposure and during lung oncogenesis. Using linear models and a controlled FDR threshold of 0.5%, we identified gene profiles in the mouse airway that were differentially expressed at 2 (n = 362), 4 (n = 795), and 6(n = 980) months following NNK relative to the time (t = 0) at completion of carcinogen treatment (Supplementary Table S3). Notably, changes occurring early on at 2 months following NNK (t = 2) were still evident in the airways of mice at both 4 and 6 months following carcinogen treatment (Supplementary Fig. S3). Next, we probed the relevance of the mouse airway field of injury expression changes to the airway in human smokers and in the context of lung cancer. We studied the recently reported expression dataset by Silvestri and colleagues (10) comprised of cytologically normal airway (mainstem bronchi) brushings obtained from smokers who had undergone bronchoscopy for suspected lung cancer (with lung cancer: n = 253; without: n = 90). After matching orthologous mouse and human features, genes were ranked according to the t statistic between airways of human smokers with and without lung cancer and based on a linear model incorporating final diagnosis of lung cancer as well as other clinicopathologic variables including chronic obstructive pulmonary disease status, age, and gender (Supplementary Methods). Following GSEA, we identified subsets within gene features differentially expressed at each time point following NNK exposure in the mouse airway that were overall enriched and concordantly (i.e., in same direction) modulated in airways of smokers with lung cancer relative to cancer-free smokers (Fig. 3A). We paid particular attention to orthologous profiles that were induced at t = 2, the earliest time point we had sampled following NNK. Of these early changes, we identified a profile of 22 orthologous genes (Supplementary Table S4) that not only persisted (downregulated) at later time points (4 and 6 months) following NNK (Fig. 3B, left), but were also significantly (P < 0.001) and concordantly enriched (downregulated) in the airways of human smokers with lung cancer relative to cancer-free smokers (Fig. 3B, right).
Figure 3.
Temporal mapping of the molecular airway field of injury upon following tobacco carcinogen exposure and during in vivo development of lung adenocarcinoma. A, Murine genes changing in the normal-appearing airway at two (t = 2), four (t = 4), and six (t = 6) months following completion of NNK exposure and relative to the time at the end of the treatment (t = 0) were statistically identified as described in the Materials and Methods section and Supplementary Methods. The identified temporal mouse profiles were analyzed by comparative genomics and GSEA in the reported array dataset by Silvestri and colleagues (10), comprised of bronchial brushings from smokers with suspicion of lung malignancy (253 with and 90 without lung cancer). Enrichment plots generated by GSEA of ranked gene expression are depicted demonstrating enriched up-and down-regulated murine genes post NNK and at the indicated time points. Bar heights correspond to enrichment scores and yellow bars correspond to genes in the leading edge subset that were concordantly enriched in airways of human smokers with lung cancer relative to cancer-free smokers. B, Concordantly enriched genes that were differentially expressed at the earliest time point (t = 2) following NNK exposure were interrogated to derive a leading edge set consisting of 22 downregulated genes. The leading edge set was then analyzed by hierarchical semisupervised clustering in mouse (left) and human smoker (right) airway brushings. Columns, samples; rows, gene features (red, higher; blue, lower expression).
We then sought to interrogate the relevance of the identified early downregulated airway field of injury gene expression changes in the mouse (at t = 2, n = 68 genes) to profiles in lung tumors themselves. We interrogated by GSEA orthologous features of the 68-gene profile we found to be downregulated in the mouse normal airway at t = 2in publicly available expression datasets of lung adenocarcinomas/NSCLCs and paired normal lung tissues (20–22). The profile was also assessed in six NNK-exposed Gprc5a−/− lung tumors (that developed in 5 Gprc5a−/− mice at 4–7 months following NNK exposure) and adjacent normal lung tissues that we also surveyed by RNA-Seq (See Supplementary Methods). We found that the 68-gene profile was significantly (all P < 0.05) suppressed in human lung adenocarcinomas/NSCLCs (Supplementary Fig. S4A–S4C) as well as in mouse lung tumors relative to their respective normal lung tissues (Supplementary Fig. S4D). These findings point to early changes in the evolution of the molecular airway field of injury following tobacco carcinogen exposure that persist even after tobacco cessation and may signify the ensuing development of a cancer in the lung epithelial field.
Cross-species temporal analysis of a human bronchial genomic classifier
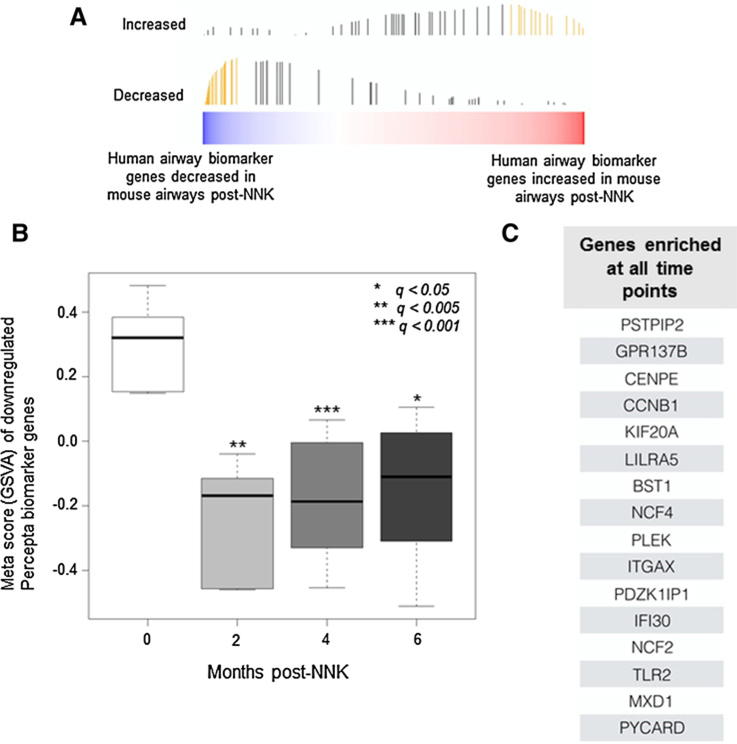
A recent report studying cytologically normal airways in suspect lifetime smokers who have undergone diagnostic bronchoscopy derived bronchial gene expression profiles that are significantly indicative of lung cancer status (114 upregulated and 118 downregulated genes in airways of smokers with lung cancer; ref. 9). Here, we sought to begin to understand the nature of development and progression of this human bronchial gene signature prior to lung cancer onset. We cross-species examined the human signature (9) in our temporal murine airway field of injury RNA-Seq dataset. Following gene set variation analysis (GSVA), we found that 56 of the 118 downregulated genes were significantly (q < 0.005) and concordantly reduced in the mouse airway field of injury (Fig. 4A). Collectively, when summarized as a metascore, these conserved markers were significantly downregulated in the mouse airway field of injury following NNK exposure, a subset of which was suppressed as early as 2 months following NNK treatment (Fig. 4B). Among these leading edge genes, 16 were found alternatively by GSEA to be statistically significantly enriched in the mouse airway at all tested time points following NNK exposure (Fig. 4C). Pathway analysis of the leading edge set revealed significant modulation of molecular and biological processes associated with the inflammatory response as well as leukocyte migration, suggesting early aberration of immune signaling in the tobacco carcinogen-exposed airway field (Supplementary Table S5). These findings point to airway field genes that are not only indicative of lung cancer in smokers with indeterminate nodules, but are also induced or evolve prior to tumor onset and, thus, may serve as high-potential surrogate markers to predict lung cancer in high-risk smokers.
Figure 4.
Human to mouse cross-species analysis of a human bronchial genomic classifier for lung cancer detection. A, A recently reported human bronchial 232-gene classifier (9) was analyzed by GSVA (see Supplementary Methods) in the identified mouse airway profiles that were found to be modulated in vivo following NNK exposure (bar heights correspond to running enrichment score calculated in GSVA and yellow bars correspond to the leading edge subset). B, Meta scores were plotted based on GSVA of a 56-gene leading edge set from the Percepta classifier that is downregulated in airways of human smokers with lung cancer relative to cancer-free smokers and concordantly decreased in the temporal mouse airway brushings. Meta scores were derived and plotted for conserved markers, which were concordantly downregulated at 2, 4, or 6 months after NNK treatment. Statistically significant differences among the different time points were based on a q-value cutoff (*, < 0.05; **, < 0.005; ***, < 0.001). C, GSEA (see Materials and Methods) of the 232-gene human classifier was performed to identify, from the human biomarker, a gene set that was concordantly enriched in the mouse airway samples across all time points.
Discussion
The nature of the development and progression of the molecular airway field of injury following tobacco is poorly understood. In this study, we sought to determine temporally progressive airway field effects that can elucidate quintessential airway expression signatures during tobacco carcinogen-induced lung tumorigenesis. Following RNA-Seq analysis, we identified evolutionarily conserved airway field changes that are not only modulated early on by the tobacco carcinogen NNK, but are also prevalent and enriched in smoking-exposed cytologically normal airway epithelia of human smokers relative to airway epithelia of human nonsmokers. This airway whole-transcriptome survey also revealed expression profiles that are temporally modulated in vivo following tobacco carcinogen exposure prior to and during lung tumor development. By crossspecies comparative analysis, we found that a subset of these early post-tobacco murine airway expression changes was significantly enriched in airways of human suspect smokers with lung cancer relative to those without the malignancy. In addition, a recently derived airway expression classifier for lung cancer among suspect smokers (9) was found to be enriched in the identified mouse orthologous airway profiles. Furthermore, genes found to be suppressed in the mouse normal airway as early as 2 months post-NNK were overall concordantly decreased in mouse and human lung tumors relative to normal lung tissue, pointing to the probable roles of these early deregulated airway profiles in lung oncogenesis. Our study highlights airway expression changes that occur and progress early on during tobacco carcinogen-associated lung cancer development. These changes, otherwise difficult to survey (e.g., longitudinally) in humans, may epitomize the early development of nearby lung lesions within this mutagenized field of injury and, thus, may serve as a low hanging fruit of biomarkers for early detection.
Earlier work from our group and others characterized gene expression changes in the airway field of injury of human smokers and derived markers that are indicative/ diagnostic of lung cancer among suspect smokers (5, 7, 9, 10, 20, 23, 24). These studies were largely cross-sectional, that is, they compared, following diagnostic bronchoscopy, airway profiles of smokers with lung cancer relative to those without the malignancy at a specific point in time. Because these studies were not aligned with time, the question remains on how airway expression patterns change following tobacco carcinogen exposure and prior to lung cancer onset. Specifically, it is not clear whether airway expression profiles that are differentially expressed in smokers with lung cancer relative to cancer-free smokers are related, phenotypically, to the already developed tumor or whether these airway profiles are a consequence that emerged sometime after nearby tumor development. This is particularly significant in light of the long latency of lung cancer development in humans (25). In this context, temporal tracking of tobacco-induced and progressive airway field effects, presumed to recapitulate the genotype of nearby lung tumors that emerged from the field, are arduous to characterize and thus have remained elusive. We sought to fill this void by surveying airway expression changes that occur with time following tobacco carcinogen exposure in vivo. We employed RNA-Seq and a lung cancer model comprising knockout of the lung-specific tumor suppressor Gprc5a and that we previously showed to develop lung adenocarcinomas following NNK exposure (11, 26). Our first goal here was to characterize effects of NNK exposure on the airway transcriptome by RNA-Seq analysis of airway brushings pre- and post-NNK (t = 0). We first pinpointed murine airway expression profiles that were not only altered by tobacco carcinogen but were also evolutionarily conserved in cancer-free human smokers relative to airways in nonsmokers. Biological processes (gene ontology annotated) of a number of gene products identified in those profiles include response to oxidative stress (Hspb6, Txnrd3, Hopx, Bcl2l11, Cat), inflammation (Mgst2, Ccl28, Tgfb1, Il7ra, Il1rn, Tlr2, Itgb2) as well as immune regulation (Cybb, Tnfrsf19, Csf2ra, Csf2rb, Csf2rb2, Stk39, Tlr7, Elf4, Cd93, Fcer1g), and those genes were similarly modulated in both NNK-exposed mouse airway and human smokers. Of note, previous studies have demonstrated modulation of oxidative stress, inflammation, and immune reaction related pathways in the molecular airway field of injury (2). These findings suggest that the effects of NNK on the mouse airway transcriptome recapitulate genome-wide effects of cigarette smoke in human airways. It is noteworthy that the results presented herein showed that the murine NNK-modulated genes that were evolutionarily conserved and enriched in human smokers (relative to nonsmokers) were particularly downregulated (compared with nonsmokers). This is in agreement with previous reports showing prevalence of dampened gene expression in the airway field of injury in smokers with early-stageNSCLC (20). Our findings warrant further studies that can potentially survey these profiles as possible early molecular markers of smoking exposure.
By serial analysis of the airway transcriptome up to the time of lung adenocarcinoma development, our study next underscored airway expression changes that develop and/ or progress following completion of tobacco carcinogen exposure and during lung oncogenesis. Our study design enabled temporal tracking of changes in airway gene expression before, immediately after exposure to NNK, and at various time points following exposure to the tobacco carcinogen. Notably, from these temporal airway expression profiles, we identified by cross-species comparative analysis those that were significantly enriched and concordantly modulated in airways of human smokers with lung cancer relative to brushings from cancer-free smokers. In retrospect, our analysis thus identified, from gene profiles previously reported to be dissimilar between airways of smokers with cancer relative to cancer-free smokers (10), profiles that likely emerged following tobacco exposure and prior to lung cancer development. It is noteworthy that mouse airway field of injury profiles that were downregulated, but not elevated, post-NNK were suppressed in cytologically normal airways of human smokers with lung cancer relative to cancer-free smokers. We also showed that these early downregulated profiles in the mouse normal airway field of injury (at t = 2) were also modulated in the same direction (downregulated) in both human and mouse lung tumors themselves relative to normal lung tissues. It is plausible that these progressive airway profiles are not only biomarkers for lung malignancy but also causally related to ensuing lung tumor development within the mutagenized airway field of injury. It is reasonable to surmise that downregulated expression of the identified genes in the airway field of injury may be, in part, mediated by methylation, a known molecular genome-wide mechanism in smoking-associated lung oncogenesis (27). It is important to note that upon completion of 8 weeks of NNK exposure, we compared gene signatures after 2, 4, and 6 months to the onset signature (t = 0), thereby excluding direct/immediate effects post-NNK and those possibly occurring during carcinogen exposure. By virtue of this analytic design, it is thus plausible that our study centered on airway field of injury profiles that are induced or progressed following cessation of smoking activity, thereby signifying the human former smoker phenotype and the capacity to elucidate the significance of residual effects of smoking in lung cancer development. Indeed, despite reduction in relative lung cancer risk in former smokers, the risk never returns to baseline (6). Also, former smokers constitute up to half of patients diagnosed with lung cancer (6). Future studies are warranted to determine the precise relevance of profiles in our study to former smokers (as compared with current smokers).
Further analysis revealed that a significant fraction of the identified temporal airway field of injury profiles play crucial roles in the host immune response. For example, immunoregulatory receptors C10orf54 and Cd300c can affect T-cell/natural killer cell-mediated ligand-receptor interactions or inflammatory responses (28, 29). Other previously reported functions of the profiles identified include Th1 responses (Def/SLAT; ref. 30), evasion of immunosurveillance (Mmp14; ref. 31), immune infiltration, tumor proliferation, progression, and metastases (Lrpl; ref. 32), cancer cell survival (loss of Abcal; ref. 33), while some have emerged as risk markers for lung cancer (Il1rn; ref. 34). Of note, our temporal RNA-Seq analysis demonstrated that the immune marker Cd68 and the tyrosine receptor kinase Axl were downregulated across all surveyed time points post-NNK exposure. Cd68 plays a crucial role in tumor-associated macrophage infiltration, and its expression was found to be significantly associated with favorable prognosis in NSCLC (35). The Axl kinase was found to influence immunosurveillance of tumor cells (36). To further support the significance of immune-related functions of the identified temporal airway field of injury profiles, several of the concordantly enriched Per-cepta (9) biomarker orthologues were shown to possess immunomodulatory roles (Supplementary Table S5). These findings suggest that the identified early airway field of injury changes (i.e., 68 downregulated genes at t = 2) comprise early immunomodulatory effects. Although expression of immune markers (e.g., the immune check-pointPD-L1; ref. 37) or immunotherapeutic response (38) has been shown to be associated with specific genomic features (e.g., mutation burden), the interplay between the molecular airway field of injury and markers of the immune response is still extremely poorly understood. Emerging and ongoing efforts have been put into understanding the clinical ramifications of the dynamic interplay between tumors and the host immune response (39). In lung cancer, compromised antitumor immunity is associated with poor prognosis (35), and immune therapies that recuperate the host immunity have proven successful (40, 41). In this context, it is intriguing to suggest that modulation of the host immune response occurs very early on following smoking and in the pathogenesis of lung cancer (42), thus providing a framework for immune-based prevention of this fatal disease.
Our study is not without limitations. We acquired epithelial brushings from mice following euthanasia (14) given the difficulty of obtaining such samples from mice that continue to survive over a long course. Thus, true longitudinal assessment of temporal expression patterns in the same variables (mice) was limited by mice euthanasia. Yet, our study underscored global expression patterns that denoted temporal progression of the airway field of injury in vivo, findings that warrant future confirmation. In addition, although our study employed the nicotine-specific and derived carcinogen NNK, cigarette smoke comprises other potentially carcinogenic constituents, including polycyclic aromatic hydrocarbons among others (43). Considering this, cigarette smoke exposure may exhibit additional or disparate effects on the airway transcriptome. Nonetheless, earlier work showed that NNK is one of the most prevalent carcinogenic constituents in cigarette smoke (43). In addition, in our current study, mice were not exposed to a single dose of NNK; rather, they were exposed to NNK continuously over a course of 2 months to attempt to mimic prototypical chronic cigarette/nicotine exposure in human smokers. Also, our cross-species genomic analysis demonstrated that NNK exposure in the mouse does recapitulate known transcriptomic effects of cigarette smoke in human airways. An additional factor to consider in our study is the specificity of the mouse model (Gprc5a−/−) employed to lung adenocarcinoma (11,44). It is reasonable to surmise that the temporally progressive airway field of injury profiles presented in this study are most likely relevant to the pathogenesis of lung adenocarcinomas. Considering that lung adenocarcinomas constitute the major histologic subtype (~65%) among NSCLCs (5), our study’s findings are applicable to understanding the pathogenesis of asignificant fraction of lung malignancies. Adding to this, our group previously reported that protein expression of human GPRC5A itself was significantly decreased in the normal-appearing airway field of injury of smokers with NSCLC relative to cancer-free smokers (45). Also, we recently found, by whole-exome sequencing, that lung adenocarcinomas from tobacco carcinogen-exposed Gprc5a−/− mice exhibit activating mutations in Kras, the most commonly mutated oncogene in smoker lung adenocarcinomas (11). Thus, it is conceivable to construe that the tobacco carcinogen-exposed Gprc5a−/− mouse is useful to interrogate the temporal evolution of at least a significant molecular fraction (e.g., Kras mutant) of smoking-associated lung adenocarcinomas. Future studies are warranted to probe the evolution of airway field effects following tobacco carcinogen and during oncogenesis of other lung cancer subtypes. Along these lines, Xiong and colleagues (46) identified evolutionary conserved early airway expression alterations in a mouse model of lung squamous cell carcinoma and that are modulated in response to chemopreventive agents.
In conclusion, by surveying transcriptome-wide alterations in cytologically normal airways in vivo, we delineated evolutionarily conserved expression changes that occur early on during tobacco-associated lung oncogenesis and that are otherwise difficult to ascertain during the long latency of lung cancer development in human smokers. We underscored evolutionarily conserved airway expression profiles that are modulated by NNK in the mouse and that are significantly and concordantly enriched in airways of human smokers relative to nonsmokers. We also identified murine airway profiles that temporally progress in the epithelial field following NNK exposure and that are significantly enriched in airways of human smokers with lung cancer relative to cancer-free smokers, as well as in both murine and human lung tumors relative to normal lung tissues. Our findings point to early evolutionarily conserved changes in the normal-appearing exposed airway that embody the development of nearby lung lesions within the mutagenized field and thus serve as minimally invasive biomarkers and targets for early detection and prevention.
Supplementary Material
Acknowledgments
This work was supported in part by the Lung Cancer Discovery award LCD-303708 from the American Lung Association (to H. Kadara), the American University of Beirut Collaborative Research Stimulus award 103319 (to H. Kadara), and the NCI grant R01CA205608 (to H. Kadara).
Footnotes
Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerprevres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A.E. Spira has ownership interest (including patents) in and is a consultant/advisory board member for Veracyte Inc. No potential conflicts of interest were disclosed by the other authors.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadara H, Scheet P, Wistuba II, Spira AE. Early events in the molecular pathogenesis of lung cancer. Cancer Prev Res 2016;9:518–27. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 4.National Lung Screening Trial Research Team, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadara H, Wistuba II. Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc 2012;9:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong L, Spitz MR, Fueger JJ, Amos CA. Lung carcinoma in former smokers. Cancer 1996;78:1004–10. [DOI] [PubMed] [Google Scholar]
- 7.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361–6. [DOI] [PubMed] [Google Scholar]
- 8.Steiling K, Ryan J, Brody JS, Spira A. The field of tissue injury in the lung and airway. Cancer Prev Res 2008;1:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitney DH, Elashoff MR, Porta-Smith K, Gower AC, Vachani A, Ferguson JS, et al. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med Genomics 2015;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, et al. Abronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med 2015;373:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimoto J, Nunomura-Nakamura S, Liu Y, Lang W, McDowell T, Jakubek Y, et al. Development of Kras mutant lung adenocarcinoma in mice with knockout of the airway lineage-specific gene Gprc5a. Int J Cancer 2017;141:1589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao Q, Fujimoto J, Men T, Ye X, Deng J, Lacroix L, et al. Identification of the retinoic acid-inducible Gprc5a as a new lung tumor suppressor gene. J Natl Cancer Inst 2007;99:1668–82. [DOI] [PubMed] [Google Scholar]
- 13.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res 2004;64:2307–16. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto K, Kudo M, Sundaram A, Ren X, Huang K, Bernstein X, et al. The alphavbeta6 integrin modulates airway hyperresponsiveness in mice by regulating intraepithelial mast cells. J Clin Invest 2012;122:748–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res 2006;34: 3150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcrip-tome. Proc Natl Acad Sci U S A 2004;101:10143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadara H, Fujimoto J, Yoo SY, Maki Y, Gower AC, Kabbout M, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst 2014;106:dju004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007;8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phos-phoinositide 3-kinase/AKT signaling cascade. Cancer Sci 2012; 103:1640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med 2010;2:26ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadara H, Shen L, Fujimoto J, Saintigny P, Chow CW, Lang W, et al. Characterizing the molecular spatial and temporal field of injury in early-stage smoker non-small cell lung cancer patients after definitive surgery by expression profiling. Cancer Prev Res 2013;6:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thrift AP, Whiteman DC. Can we really predict risk of cancer? Cancer Epidemiol 2013;37:349–52. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto J, Kadara H, Men T, van Pelt C, Lotan D, Lotan R. Comparative functional genomics analysis of NNK tobacco-carcinogen induced lung adenocarcinoma development in Gprc5a-knockout mice. PLoS One 2010;5:e11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013;339: 1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res 2014;74:1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lankry D, Rovis TL, Jonjic S, Mandelboim O. The interaction between CD300a and phosphatidylserine inhibits tumor cell killing by NK cells. Eur J Immunol 2013;43:2151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becart S, Charvet C, Canonigo Balancio AJ, De Trez C, Tanaka Y, Duan W, et al. SLAT regulates Th1 and Th2 inflammatory responses by controlling Ca2+/NFAT signaling. J Clin Invest 2007;117:2164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenlee KJ, Werb Z, Kheradmand F. Matrix metalloprotei-nases in lung: multiple, multifarious, and multifaceted. Physiol Rev 2007;87:69–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staudt ND, Jo M, Hu J, Bristow JM, Pizzo DP, Gaultier A, et al. Myeloid cell receptor LRP1/CD91 regulates monocyte recruitment and angiogenesis in tumors. Cancer Res 2013;73:3902–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith B, Land H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep 2012;2:580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lind H, Zienolddiny S, Ryberg D, Skaug V, Phillips DH, Haugen A. Interleukin 1 receptor antagonist gene polymorphism and risk of lung cancer: a possible interaction with polymorphisms in the interleukin 1 beta gene. Lung Cancer 2005;50:285–90. [DOI] [PubMed] [Google Scholar]
- 35.Parra ER, Behrens C, Rodriguez-Canales J, Lin H, Mino B, Blando J, et al. Image analysis-based assessment of PD-L1 and tumor-associated immune cells density supports distinct intra-tumoral microenvironment groups in non-small cell lung carcinoma patients. Clin Cancer Res 2016;22:6278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gay CM, Balaji K, Byers LA. Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer 2017;116:415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadara H, Choi M, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol 2017;28:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321–30. [DOI] [PubMed] [Google Scholar]
- 40.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spira A, Disis ML, Schiller JT, Vilar E, Rebbeck TR, Bejar R, et al. Leveraging premalignant biology for immune-based cancer prevention. Proc Natl Acad Sci U S A 2016;113:10750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hecht SS. Approaches to chemoprevention of lung cancer based on carcinogens in tobacco smoke. Environ Health Perspect 1997; 105 Suppl 4:955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadara H, Fujimoto J, Men T, Ye X, Lotan D, Lee JS, et al. AGprc5a tumor suppressor loss of expression signature is conserved, prevalent, and associated with survival in human lung adenocarcinomas. Neoplasia 2010;12:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujimoto J, Kadara H, Garcia MM, Kabbout M, Behrens C, Liu DD, et al. G-protein coupled receptor family C, group 5, member A (GPRC5A) expression is decreased in the adjacent field and normal bronchial epithelia of patients with chronic obstructive pulmonary disease and non-small-cell lung cancer. J Thorac Oncol 2012;7:1747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong D, Pan J, Zhang Q, Szabo E, Miller MS, Lubet RA, et al. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer che-mopreventive agents. Oncotarget 2017;8:18885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.