Abstract
Background
The aim of this study was to elucidate the involvement of cPLA2-AA-COX-2 pathway factors and their potential role in lung cancer early diagnosis and prognosis.
Material/Methods
We selected 80 lung cancer patients as the cancer group, and 30 normal patients were selected as the normal group. Serum contents of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 were measured, and mRNA levels of COX-2, cPLA2, COX-1, and mPGES in serum were determined. Spearman’s P-test was used to analyze the correlation between expression of PGI2 and mPGES in serum and the clinical characteristics of these lung cancer patients. The factors affecting the prognosis lung cancer were analyzed by COX regression model.
Results
The serum contents of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 in the cancer patient group were significantly higher (p<0.05) than in the normal group; after treatment, the serum contents of these factors were significantly decreased (p<0.05). However, distant metastasis had a significant effect on serum contents of mPGES and PGI2 (p<0.05), but not on the other factors. The mRNA levels of COX-2, cPLA2, COX-1, and mPGES in cancer patients were significantly higher than in normal patients. In addition, the 5-year survival rate of patients with high expression of mPGES and/or PGI2 was lower than that of the low expression group. Cox regression analysis showed that the expression of mPGES and PGI2 had statistical significance in predicting the prognosis of lung cancer.
Conclusions
The cPLA2-AA-COX-2 pathway is closely associated with lung cancer. These findings are important for clinical diagnosis of lung cancer.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; Cyclooxygenase 2; Diagnosis; Group IV Phospholipases A2; Prognosis
Background
Lung cancer is a well-known leading cause of cancer-related death among both men and women around the world [1]. Lung cancer can be broadly divided into 2 major types: small cell lung cancer (SCLC, ~15%) and non-small cell lung cancer (NSCLC, ~85%). Among NSCLC cases, there are 2 main histological subtypes – adenocarcinoma and squamous cell carcinoma – in which there exist specific DNA mutations that allow further stratification [2]. The survival rate of lung cancer patients is very low, although in recent years there have been some significant developments in the clinical treatment of late-stage lung cancer. Most patients (~75%) have late-stage disease (stages III or IV) at the time of diagnosis, which leads to low survival rate [3]. According to a 2014 report from the UK Office for National Statistics, patients who were diagnosed with stage IV cancers only had 1-year survival rates of 15–19%, compared to 1-year survival rates of 81–85% for those who had stage I cancers [4]. Therefore, early detection of lung cancer is a key factor to improve patient survival rate [5]. One solution is to identify biomarkers that can be used for early diagnosis.
In recent years there have been reports that some key factors in the cPLA2-AA-COX-2 pathway are involved in lung cancers. The cPLA2-AA-COX-2 pathway is an important signaling pathway in inflammation, and increased risk of cancer development has been reported to be associated with the presence of inflammation [6,7]. The cPLA2-AA-COX-2 pathway has the following 3 steps: (1) arachidonic acid (AA) is first released from membrane glycerophospholipids by PLA2, (2) AA is converted to the intermediate PGH2 by COX-1 or COX-2, and (3) PGH2 is isomerized to prostaglandins such as PGE2 by prostaglandin E synthase (PGES) [8,9]. It has been reported that phospholipase A2 (PLA2) is associated with certain types of cancer, including colorectal, gastrointestinal, and prostate carcinomas [10–13]. Other studies showed that overexpression of cyclooxygenase-2 (COX-2) is a tumor initiating and promoting event for some solid tumors, including lung cancers [14–16]. A number of clinical studies have used COX-2 inhibitors in lung cancer drug development [17–19]. These previous studies indicate that the close connection between the cPLA2-AA-COX-2 pathway and lung cancer may be beneficial in lung cancer diagnosis and treatment.
As mentioned above, early detection of lung cancer is key to improving patient survival. It was reported that surgical resection can offer good prognosis in patients with stage I tumors, leading to 5-year survival rates of 70–90% [20]. However, early diagnosis of lung cancer remains a challenging problem. To help advance the identification of biomarkers for early detection of lung center, in the present study we evaluated the expressions of key factors in the cPLA2-AA-COX-2 pathway, including COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2, and assessed their correlation with lung cancer patient characteristics and distant metastasis. These findings may help to establish a sound basis to further evaluate them more quantitively as potential biomarkers for early detection and prognosis of lung cancer.
Material and Methods
Patient selection and serum sample preparation
We divided subjects into 2 groups: one group consisted of patients with lung cancer (n=80) and the other group (control) consisted of healthy people who came to the hospital for a regular physical examination (n=30). Eighty lung cancer patients being treated in the Affiliated Yantai Yuhuangding Hospital of Qingdao University from 1 Jan 2011 to 31 Dec 2012 were selected as the lung cancer group. Among these 80 patients, 49 were male and 31 were female, and the age range was 36–78 years. In terms of lung cancer types, 34 out of the 80 patients had adenocarcinoma, 15 had squamous cell carcinoma, 20 had small cell lung cancer, and 11 had other types of lung cancer. In terms of disease stage, 52 patients had no distant metastasis, and the other 28 had distant metastasis to other organs, including the liver, bones, and brain. All these 80 patients have complete clinical examination and treatment records. Based on each patient’s examination and tumor diagnosis results, doctors chose specific treatment methods, including radiotherapy, chemotherapy, immunotherapy, or other treatment methods. The control group consisted of 30 randomly selected healthy people (19 males, 11 females, age range 30–80 years).
To prepare the serum samples to be used in experimental analysis, 10-ml vein blood samples were collected from all subjects in both groups after fasting. For the lung cancer patients, blood samples were collected again after 12 months of anti-cancer treatment. The blood samples were centrifuged at low temperature, and the supernatant (serum) was harvested and kept at −20°C before being used in the experimental analysis.
The proposed experimental plan and procedures were reviewed and approved by the Ethics Committee of the hospital. All 80 lung cancer patients and 30 healthy subjects provided signed consent to participate. The patients were followed up by telephone or outpatient follow-up for 3~5 years. The follow-up started on the second day after treatment. The deadline was the end of follow-up or the day of death or loss of patients to follow-up. The survival time was calculated as the second day after treatment.
Serum content of cPLA2-AA-COX-2 pathway factors measured by ELISA
To evaluate the expression levels of the cPLA2-AA-COX-2 pathway factors, serum contents of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 were determined by ELISA assay. All the ELISA assay kits used in this study were purchased from Winter Song Boye Biotechnology Company (Beijing, China), which included the kits for COX-2 (YX-031526H), cPLA2 (YX-191615H), COX-1 (DG-031525H), mPGES (YX-131607H), PGE2 (DG-160707H), and PGI2 (YX-160711H). All the experimental procedures in the ELISA assays were performed strictly following the manufacturer’s instructions.
mRNA levels of cPLA2-AA-COX-2 factors determined by qRT-PCR
To measure the mRNAs levels of these cPLA2-AA-COX-2 pathway factors, the total RNA in the serum samples was extracted using RNAprep Pure Kit (Cat. No. DP433, TIANGEN, Beijing, China) following the manufacturer’s protocol. The concentration of total RNA was quantified by the optical density at 260 nm. Then, 1000 ng of the total RNA was applied as a template for reverse transcription reaction using a reverse transcription kit (Applied Biosystems, Waltham, MA, USA), following the manufacturer’s instructions. β-actin was selected as internal reference. The sequences of primers used in qRT-PCR are listed in Table 1, and the qRT-PCR was performed using Mastercycler® nexus X2 (Eppendorf, Hamburg, Germany), with the following cycling parameters: 95°C for 15 min, then 30 cycles of 95°C for 15 s, 60°C for 30 s, and 55°C for 60 s, and finally a single cycle at 55°C for 10 min. For β-actin, the cycling parameters were 94°C for 5 min, then 30 cycles of 94°C for 40 s, 55°C for 40 s, and 72°C for 40 s, and a final single cycle at 72°C for 10 min. We added 2 μL loading buffer to 10 μL of each qRT-PCR product, and applied 1.5% agarose gel containing 0.05 μg/mL ethidium bromide at 70 V for 50 min. The relative level of mRNA was calculated with the 2−ΔΔCt method [21] using β-actin mRNA as an internal reference.
Table 1.
Sequences of primers used in qRT-PCR.
| Primer sequence | |
|---|---|
| COX-2 | F: 5′-CTGGCGCTCAGCCATACAGC-3′ |
| R: 5′-GGCCCTCGCTTATGATCTGTC-3′ | |
| cPLA2 | F: 5′-CAGTGGGCTCACATTTAACCT-3′ |
| R: 5′-CTTCCCGATCAAACAGG-3′ | |
| COX-1 | F: 5′-CCGGATGCCAGTCAGGATGATG-3′ |
| R: 5′-CTAGACAGCCAGATGCTGACTG-3′ | |
| mPGES | F: 5′-TGGAGACCATCTACCCCTT-3′ |
| R: 5′-ACCTTGAAGGAGTCAGGCATGAG-3′ | |
| β-actin | F: 5′-GTGGACATCCGCAAAGAC-3′ |
| R: 5′-AAAGGGTGTAACGCAACTA-3′ |
Statistical data analysis
Statistical analyses were carried out using SPSS, version 19.0 (IBM Corp., Armonk, NY). All data are presented in the form of mean ± standard deviation (SD). For comparison between 2 groups, statistical analysis was performed with the 2-tailed t test. For comparison between more than 2 groups, one-way analysis of variance (ANOVA) was applied. If ANOVA showed a significant difference, post hoc least significant difference (LSD) testing was used for paired comparison. For correlation analysis, the Spearman correlation coefficient method was used. Cox regression analysis was used to assess prognostic factors of lung cancer patients. Kaplan-Meier method was used to analyze survival curves. In all statistical analyses, statistical significance was assumed at p<0.05, and very significant difference was assumed at p<0.01.
Results
Expression of cPLA2-AA-COX-2 pathway factors
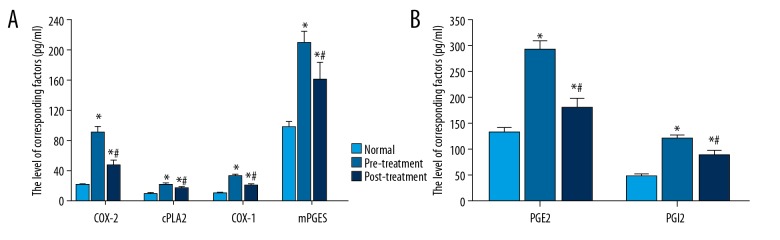
The serum contents of the cPLA2-AA-COX-2 pathway factors, COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2, from the normal (control) group and lung cancer patient group were measured and compared. The measurements for the lung cancer patient group were done both before (pre-treatment) and after the treatment (post-treatment). As shown in Figure 1, a statistically significant difference was found between the lung cancer patient group and the control group (p<0.05) for both pre-treatment and post-treatment results. Figure 1 shows that the expression of these factors was significantly decreased after the treatment compared to the pre-treatment results (p<0.05).
Figure 1.
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 in serum of the normal patient group and in lung cancer patients before and after treatment. The data are presented by mean ±SD (standard deviation). * Indicates a statistically significant difference was found between the lung cancer patient group and the normal patient group (p<0.05). # Indicates a statistically significant difference was found in the comparison between before treatment and after treatment (p<0.05).
Expression of cPLA2-AA-COX-2 pathway factors in different lung cancer types
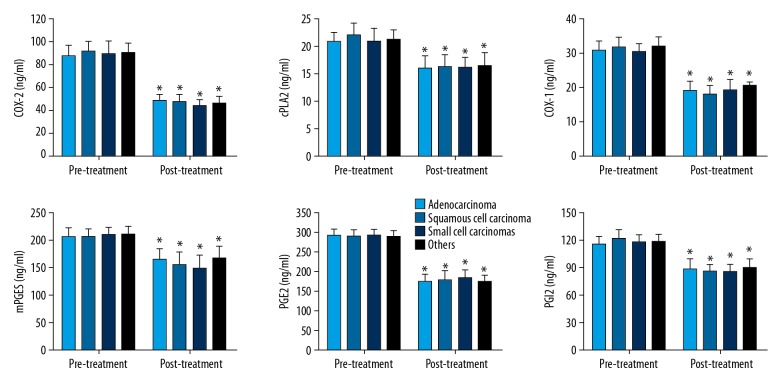
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 in serum of lung cancer patients in different lung cancer type groups was compared between pre-treatment and post-treatment measurements, which are summarized in Figure 2. In all 4 lung cancer type groups (adenocarcinoma, squamous cell carcinoma, small cell lung cancer, and other types), the expressions of all these factors was greatly reduced, and the differences were all statistically significant (p<0.05). However, in the pre-treatment or post-treatment measurements, no significant difference was found between the different lung cancer types.
Figure 2.
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 before and after treatment in serum of lung cancer patients in different lung cancer type groups. The data are presented by mean ±SD (standard deviation). * Indicates a statistically significant difference was found between the before and after treatment comparison (p<0.05).
Expression of cPLA2-AA-COX-2 pathway factors in different lung cancer types
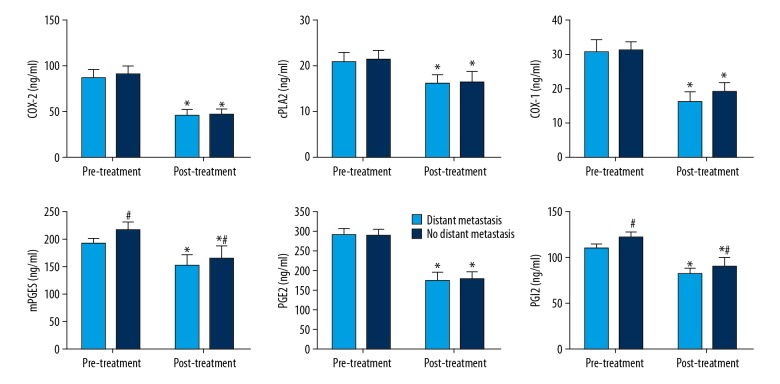
The influence of distant metastasis on expression of these cPLA2-AA-COX-2 pathway factors was also evaluated. As seen in Figure 3, before treatment there was no significant difference in their expression. After treatment, the expressions of these factors were all decreased significantly different compared to the pre-treatment measurements. In the post-treatment measurements, there was no significant difference observed for COX-2, cPLA2, COX-1, and PGE2 between patients with metastasis and patients without metastasis; however, for mPGES and PGI2, the expression in patients without distant metastasis was significantly higher than that in patients with distant metastasis.
Figure 3.
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 before and after treatment in serum of lung cancer patients with or without distant metastasis. The data are presented by mean ±SD (standard deviation). * Indicates a statistically significant difference was found between the before and after treatment comparison (p<0.05). # Indicates a significant different compared to the lung cancer patient group with distant metastasis (p<0.05).
Expression of cPLA2-AA-COX-2 pathway factors in males vs. females
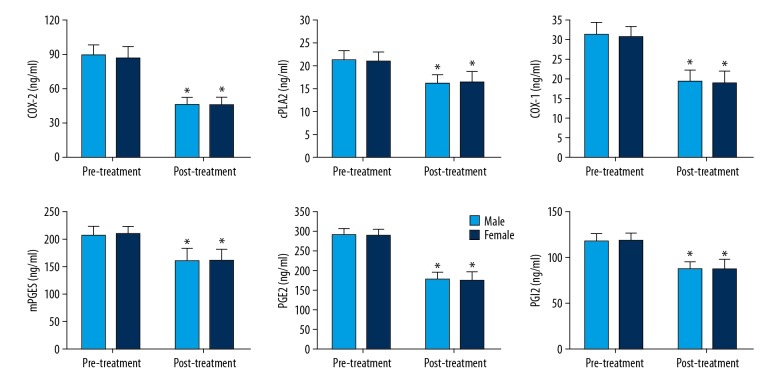
The correlation of the expression of the cPLA2-AA-COX-2 pathway factors with sex was also studied (Figure 4). After treatment, their expression in both sexes was greatly reduced, which was significant for all 6 factors. However, when we compared the differences in males vs. females, no significant difference was observed, either before treatment or after treatment.
Figure 4.
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 before and after treatment in serum of male and female lung cancer patients. The data are presented by mean ±SD (standard deviation). * Indicates a statistically significant difference was found between the before and after treatment comparison (p<0.05).
Expression of cPLA2-AA-COX-2 pathway factors in different age groups
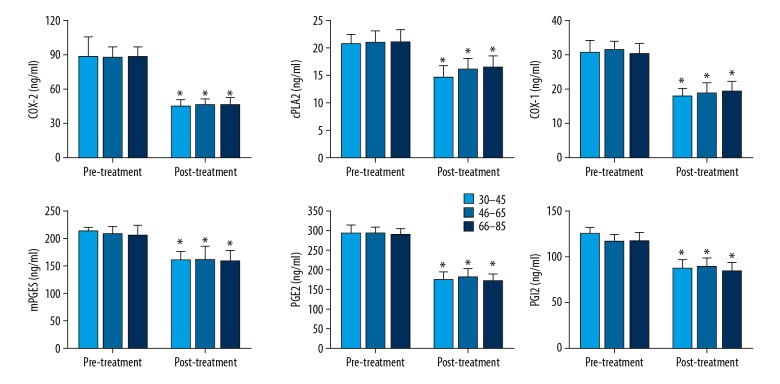
The correlation of the expression of the cPLA2-AA-COX-2 pathway factors with patient ages was also studied (Figure 5). The patients were divided into 3 groups according to age: 30–45, 46–65, and 66–85. After treatment, their expression in patients in all age groups was greatly reduced and was significant for all 6 factors. However, when comparing the difference in each age group, no significant difference was observed, either before treatment or after treatment.
Figure 5.
Expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 before and after treatment in serum of lung cancer patients in different age groups. The data are presented by mean ±SD (standard deviation). * Indicates a statistically significant difference is found between the before and after treatment comparison (p<0.05).
Correlation of expression of cPLA2-AA-COX-2 pathway factors and patient characteristics
The above results showed that the expression of all 6 factors of cPLA2-AA-COX-2 pathway was not correlated with patient age, sex, or lung cancer type. In terms of distant metastasis, the expression of COX-2, cPLA2, COX-1, and PGE2 was not correlated with distant metastasis, but the expression of mPGES and PGI2 was correlated with distant metastasis. These findings were summarized in Table 2.
Table 2.
Serum levels of mPGES and PGI2 in different patient groups.
| Character | mPGES (ng/ml) | PGI2 (pg/ml) | p Value | |
|---|---|---|---|---|
| Sex | Male | 215.21±15.36 | 121.31±7.47 | >0.05 |
| Female | 206.79±15.88 | 116.82±7.18 | ||
| Age group | 30–45 | 211.58±8.99 | 124.69±7.20 | >0.05 |
| 46–65 | 195.93±4.32 | 109.09±4.55 | ||
| 66–85 | 185.64±10.73 | 107.98±7.82 | ||
| Lung cancer type | Adenocarcinoma | 204.56±17.58 | 115.31±8.42 | >0.05 |
| Squamous cell carcinoma | 205.28±14.96 | 120.36±10.02 | ||
| Small cell lung cancer | 208.41±15.17 | 116.31±8.64 | ||
| Other | 209.32±17.35 | 117.22±7.85 | ||
| Distant metastasis | Yes | 214.78±13.10 | 121.21±6.65 | <0.05 |
| No | 191.08±8.62 | 108.51±5.67 |
Serum mRNA levels of COX-2, cPLA2, COX-1, and mPGES with or without distant metastasis
The qRT-PCR experiment (Figure 6) showed that the mRNA levels of COX-2, cPLA2, COX-1, and mPGES in serum of lung cancer patients, either with or without distant metastasis, were much higher than in the normal control group, and the differences were very significant (p<0.01). However, when comparing the mRNA levels of COX-2, cPLA2, COX-1, and mPGES in serum between lung cancer patients with distant metastasis and those without distant metastasis, there was a significant difference only in mPGES mRNA (p<0.05), not in the mRNA of the other 3 factors.
Figure 6.
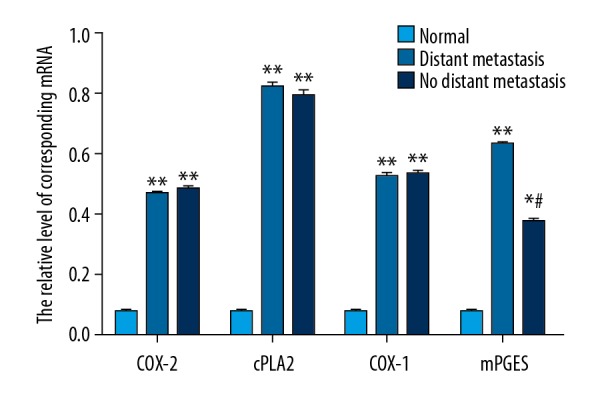
mRNA levels of COX-2, cPLA2, COX-1, and mPGES in serum of the normal control group and lung cancer patient group with or without distant metastasis, determined by RT-PCR. The data are presented by mean ±SD (standard deviation). ** Indicates a very significant difference compared to the normal control group (p<0.01), and # indicates a significant different compared to the lung cancer patient group with distant metastasis (p<0.05).
Relationship between serum mPGES, PGI2, and survival rate in patients with lung cancer
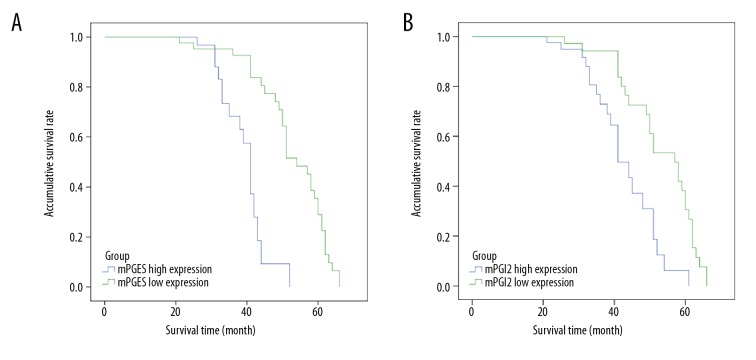
The study subjects were followed up for 20–70 months (Figure 7). Survival curve analysis showed that the 5-year survival rate of patients who had high expression of mPGES was 0% and that of the group with low expression was 28.9%, with very significant difference between those with high expression vs. low expression of mPGES (p=0.001). The 5-year survival rate was 8.0% in the high expression group and 32.7% in the low expression group, and the difference between the 2 groups was very significant (p=0.004).
Figure 7.
(A, B) Postoperative survival curve of lung cancer patients with high and low expression of mPGES and PGI2, respectively.
Significance of mPGES and PGI2 expression in lung cancer in predicting prognosis of lung cancer
We used a COX proportional risk model to analyze factors that might influence the prognosis of lung cancer, including sex, age, type of lung cancer, metastasis type, and expression levels of mPGES and PGI2. As shown in Table 3, the expression of mPGES and PGI2 was statistically significant in predicting the prognosis of lung cancer (p<0.05).
Table 3.
The results of COX proportional risk model analysis.
| Items | B | P | Exp(B) | 95.0% CI |
|---|---|---|---|---|
| Sex | 0.050 | 0.883 | 1.051 | 0.540–2.046 |
| Age | 0.001 | 0.969 | 1.001 | 0.958–1.406 |
| Lung cancer type | 0.103 | 0.515 | 1.109 | 0.812–1.514 |
| Metastasis type | 0.503 | 0.245 | 1.654 | 0.708–3.861 |
| mPGES | −1.068 | 0.025 | .344 | 0.135–0.875 |
| PGI2 | −0.594 | 0.043 | .552 | 0.249–1.223 |
B – partial regression coefficient; Exp(B) – eelative risk estimate
Discussion
Lung cancer diagnosed at an early stage is more likely to respond to effective treatment, and early detection enhances the probability of higher survival with less morbidity [5]. Also, early detection of lung cancer can significantly improve patient quality of life. However, early detection of lung cancer still remains a significant clinical challenge [5]. The results presented here show that the expression of COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 are all significantly increased in lung cancer patients, independent of demographic characteristics and lung center histological types. The present findings show the possibility of using these key factors in the cPLA2-AA-COX-2 pathway as biomarkers in early detection of lung cancer.
We also demonstrated that the serum contents of mPGES and PGI2 and mRNA level of mPGES are highly correlated with distant metastasis of lung cancer. This is of biomedical importance because it shows that their mRNA level can be used as potential biomarkers in predicting distant metastasis in clinical lung cancer treatment. Furthermore, after anti-cancer treatment, the expression levels of all 6 factors – COX-2, cPLA2, COX-1, mPGES, PGE2, and PGI2 – are all significantly decreased compared to their expression levels before lung cancer treatment, indicating a high correlation between their expression levels and lung cancer treatment status. These findings may help improve lung cancer prognosis.
An advantage of using plasma biomarkers is that they can be assessed by less costly and non-invasive methods, and thus have high potential to be used in the early diagnosis of various diseases [22]. The research presented here is just the initial step in evaluating the potential use of these key factors in the cPLA2-AA-COX-2 pathway as biomarkers for early detection in lung cancer clinical diagnosis, and also have potential implications for improving prognosis in lung cancer treatment. We plan to perform more detailed and systematic research on this topic to more quantitatively determine the correlation of expression of these key factors and lung cancer stages, as well as lung cancer post-treatment status. Towards this end, the present study has laid a very sound basis for further research.
Our study demonstrates that the expression of COX-1, COX-2, cPLA2, mPGES, PGE2, and PGI2 is greatly enhanced in lung cancer patients, which indicates the cPLA2-AA-COX-2 pathway is closely associated with lung cancer. We also found that the mRNA level of mPGES and PGI2 is highly correlated with distant metastasis of lung cancer; therefore, their mRNA levels can be used to predict distant metastasis.
Conclusions
We found a significant decrease in expression of COX-2, PLA2, COX-1, mPGES, PGE2, and PGI2 after treatment, which suggests that the cPLA2-AA-COX-2 pathway can serve as a new method to evaluate prognosis in lung cancer. These findings are important for clinical diagnosis of lung cancer, especially in the early stages, and provide a sound basis for designing new drugs for lung cancer therapy.
Footnotes
Source of support: This work was supported by grants from the Shandong Medical and Health Science and Technology Development Project (2018WS025), Yantai Key Research and Development Program (2019YD011), Science and Technology Development Plan of Yantai (2015ws004, 2016ws013) and the Natural Science Foundation of Shandong province (ZR2016HL26)
Conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters S, Maringe C, Coleman MP, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: A population-based study, 2004–2007. Thorax. 2013;68:551–64. doi: 10.1136/thoraxjnl-2012-202297. [DOI] [PubMed] [Google Scholar]
- 4.Broggio J, Bannister N. Cancer survival by stage at diagnosis for England. Newport: UK Office for National Statistics; 2016. [Google Scholar]
- 5.Blandin Knight S, Crosbie PA, et al. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7:170070. doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crusz SM, Balkwill FR. Inflammation and cancer: Advances and new agents. Nat Rev Clin Oncol. 2015;12:584–96. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Karin M. Roles of tumor suppressors in regulating tumor-associated inflammation. Cell Death Differ. 2014;21:1677–86. doi: 10.1038/cdd.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norregaard R, Kwon TH, Frokiaer J. Physiology and pathophysiology of cyclooxygenase-2 and prostaglandin E2 in the kidney. Kidney Res Clin Pract. 2015;34:194–200. doi: 10.1016/j.krcp.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasrallah R, Hassouneh R, Hebert RL. PGE2, kidney disease, and cardiovascular risk: Beyond hypertension and diabetes. J Am Soc Nephrol. 2016;27:666–76. doi: 10.1681/ASN.2015050528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Huang CJ, Yu GZ, et al. Expression of group IIA phospholipase A2 is an independent predictor of favorable outcome for patients with gastric cancer. Hum Pathol. 2013;44:2020–27. doi: 10.1016/j.humpath.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Zhao X, Wu Z, et al. Polymorphisms in arachidonic acid metabolism-related genes and the risk and prognosis of colorectal cancer. Fam Cancer. 2013;12:755–65. doi: 10.1007/s10689-013-9659-2. [DOI] [PubMed] [Google Scholar]
- 12.Mirtti T, Laine VJ, Hiekkanen H, et al. Group IIA phospholipase A as a prognostic marker in prostate cancer: Relevance to clinicopathological variables and disease-specific mortality. APMIS. 2009;117:151–61. doi: 10.1111/j.1600-0463.2008.00002.x. [DOI] [PubMed] [Google Scholar]
- 13.Menschikowski M, Hagelgans A, Nacke B, et al. Epigenetic control of group V phospholipase A2 expression in human malignant cells. Tumour Biol. 2016;37:8097–105. doi: 10.1007/s13277-015-4670-x. [DOI] [PubMed] [Google Scholar]
- 14.Scheff RJ, Schneider BJ. Non-small-cell lung cancer: Treatment of late stage disease: Chemotherapeutics and new frontiers. Semin Intervent Radiol. 2013;30:191–98. doi: 10.1055/s-0033-1342961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokouchi H, Kanazawa K, Ishida T, et al. Cyclooxygenase-2 inhibitors for non-small-cell lung cancer: A phase II trial and literature review. Mol Clin Oncol. 2014;2:744–50. doi: 10.3892/mco.2014.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith WL, Langenbach R. Why there are two cyclooxygenase isozymes. J Clin Invest. 2001;107:1491–95. doi: 10.1172/JCI13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou LC, Huang F, Xu HB. Does celecoxib improve the efficacy of chemotherapy for advanced non-small cell lung cancer? Br J Clin Pharmacol. 2016;81:23–32. doi: 10.1111/bcp.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou YY, Hu ZG, Zeng FJ, Han J. Clinical profile of cyclooxygenase-2 inhibitors in treating non-small cell lung cancer: A meta-analysis of nine randomized clinical trials. PLoS One. 2016;11:e0151939. doi: 10.1371/journal.pone.0151939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Shen P, Zhang XC, et al. Efficacy and safety profile of celecoxib for treating advanced cancers: A meta-analysis of 11 randomized clinical trials. Clin Ther. 2014;36:1253–63. doi: 10.1016/j.clinthera.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Kondo T. Inconvenient truth: Cancer biomarker development by using proteomics. Biochim Biophys Acta. 2014;1844:861–65. doi: 10.1016/j.bbapap.2013.07.009. [DOI] [PubMed] [Google Scholar]