Abstract
The lack of tools to identify causative variants from sequencing data greatly limits the promise of Precision Medicine. Previous studies suggest one-third of disease alleles alter splicing. We discovered that splicing defects cluster in diseases (e.g. haploinsufficient genes). We analyzed 4,964 published disease-causing exonic mutations using a Massively Parallel Splicing Assay (MaPSy) that showed 81% concordance rate with patient tissue splicing. ~10% of exonic mutations altered splicing, mostly by disrupting multiple stages of the spliceosome assembly. We present the first large-scale characterization of exonic splicing mutations using a novel technology that facilitates variant classification that keeps pace with variant discovery.
Introduction
Human genetic disorders occur in ~8% of the population1. Significant technological advancements in the past decade have made it possible to detect all sequence variations in individual genomes in a cost effective manner. Combined with capture technologies, targeted sequencing of the entire protein-coding regions of the human genome (exomes) has been increasingly used for routine diagnostics in Mendelian disorders2,3. Unfortunately, the tremendous progress that has been made in variant detection has outpaced the capacity to characterize sequence variations. Recent deep sequencing of human exomes detected ~14,000 single nucleotide variants (SNVs) per individual, 47% of which were predicted to be deleterious by one or more in silico prediction tools, but there were very little agreements (<1%) between all the commonly used methods4.
Large-scale sequencing has identified many loss-of-function variants thought to cause severe genetic disorders in asymptomatic individuals5,6. These variants could represent annotation or sequencing errors, partial penetrance or recessive alleles carried by asymptomatic individuals. This uncertainty illustrates the urgency for better characterization of sequence variation. While it is difficult to predict the effect of a single nucleotide variant on protein function, the characterization of splicing mutations is a tractable problem. Splicing mutations are easily detected and quantified. They are deleterious and one-third of hereditary disease alleles are predicted to confer some degrees of missplicing7. While some of these mutations disrupt canonical splice-sites, others disrupt the multitude of enhancers and silencers that can modulate splice-site usage. Any change in exonic sequence may therefore disrupt or create cis-acting elements that facilitate exon recognition and cause aberrant splicing. Here, we present a novel parallel splicing reporter system to characterize 4,964 published disease-causing exonic mutations for effects on splicing. The present study identified allelic splicing imbalance caused by these exonic mutations and provided insights into the determinants and mechanisms of splicing aberrations.
Massively Parallel Splicing Assays
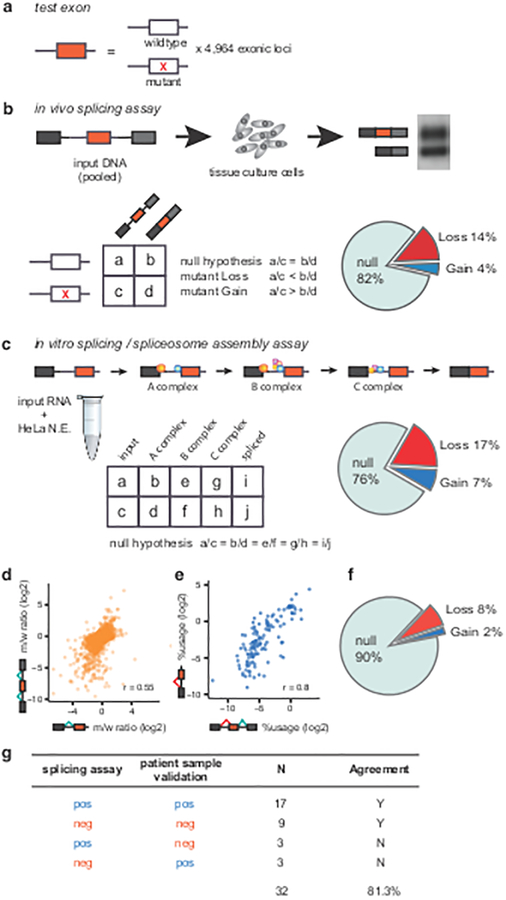
We developed a Massively Parallel Splicing Assay (MaPSy) to screen a panel of 4,964 exonic disease mutations (5K panel) reported in the Human Gene Mutation Database8 (HGMD) for splicing defects. One library was designed to evaluate the effects of the mutations on splicing in vivo via transfection in tissue culture cells. The second library was comprised of RNA substrates designed to evaluate the mutations’ effects on splicing in vitro via incubation in cell nuclear extract. Solid phase oligonucleotide synthesis technology and PCR was used to manufacture the in vivo library and the template for the in vitro library (Fig. 1). Each reporter in the library contains a 170mer genomic fragment of either the mutant or wildtype (reference) sequence consisting of the exon, at least 55 nts of upstream intron and 15 nts of downstream intron (Fig. 1a)9. The allelic ratio for each mutant/wildtype pair was determined from allelic counts obtained from deep sequencing of the input libraries, the output spliced fractions and the RNA pools isolated from different in vitro spliceosomal intermediates (Fig. 1b,c). The most common outcome of disrupted splicing in vivo is exon skipping, whereas most pre-mRNAs with splicing mutations in vitro remain unspliced. While changes in transcription or stability may account for an altered allelic ratio in the spliced fraction in vivo, the in vitro assay is a direct measure of splicing. Despite significant differences in processing and substrate design, general agreements were observed between allelic splicing ratios obtained from the two assays (Fig. 1d, Pearson’s r = 0.55). Approximately 10% of the exonic mutations in the 5K panel altered splicing in both systems (Fig. 1f, >1.5 fold change, two-sided Fisher’s exact test, adjusted with 5% FDR) and thus were regarded as unambiguous splicing changes and classified as exonic splicing mutations (ESM). We also performed MaPSy on a control panel of common SNPs, which disrupted splicing at a significantly lower level (8/228 or 3%, P = 9.94 × 10−5, two-sided Fisher’s exact test, Supplementary Table 1). Additionally, cryptic 3’ splice-site usage was identified in both assays (Fig. 1e, Pearson’s r = 0.8). Although most bona fide cryptic splicing events (74%) were caused by the creation of an AG, a significant number of disease alleles caused dramatic shifts in the usage of an existing AG (Supplementary Fig. 1).
Figure 1. Massively Parallel Splicing Assay (MaPSy) on the 5K panel.
a, The panel consists of 4,964 mutant and wildtype pairs. b, The panel is incorporated into three exons in vivo library. Allelic ratios of both input and output were determined by deep sequencing. The result of RT-PCR from output RNA (spliced species) is shown (Supplementary Figure 2f). Splicing aberrations were found in 18% of mutants. c, Allelic ratios were determined in spliceosomal intermediates, ~24% species disrupt splicing in vitro. N.E.: nuclear extract d, Allelic splicing ratios in vivo versus in vitro. e, Cryptic splice-site usage in vivo versus in vitro. f, Exonic splicing mutations identified in ~10% of the 5K panel. g, Summary of MaPSy validations in patient samples.
MaPSy was found to be robust (Pearson’s r = 0.85 – 0.89 between allelic splicing ratios from experimental replicates, Supplementary Fig. 2a–d). In order to assess the validity and relevance of splicing aberrations detected by MaPSy, we performed RTPCR validations in RNAs extracted from patient samples consisting of lymphoblastoid cell lines, fibroblasts, whole-blood and post-mortem brain tissues (Supplementary Fig. 3a–f, Supplementary Table 2). The validation samples were chosen solely based on availability. In addition, we searched the literature for follow-up studies involving the mutations in the 5K panel that include RNA splicing analyses in patient tissue samples. The summary of validations can be found in Supplementary Table 2. Overall, ~81% (26/32) of MaPSy-detected ESMs were validated in patient tissue samples (Fig. 1g). Furthermore, we compared the splice-site usage in 19 different cell lines that are part of ENCODE data set with wildtype (reference) splicing in our 5K panel. Exons that splice best in the 5K panel also has the highest average splice-site usage in ENCODE cell lines, while exons that splice worst in the 5K panel also has the lowest average splice-site usage in ENCODE data (Supplementary Fig. 3g).
Non-uniform distribution of Splicing Mutations
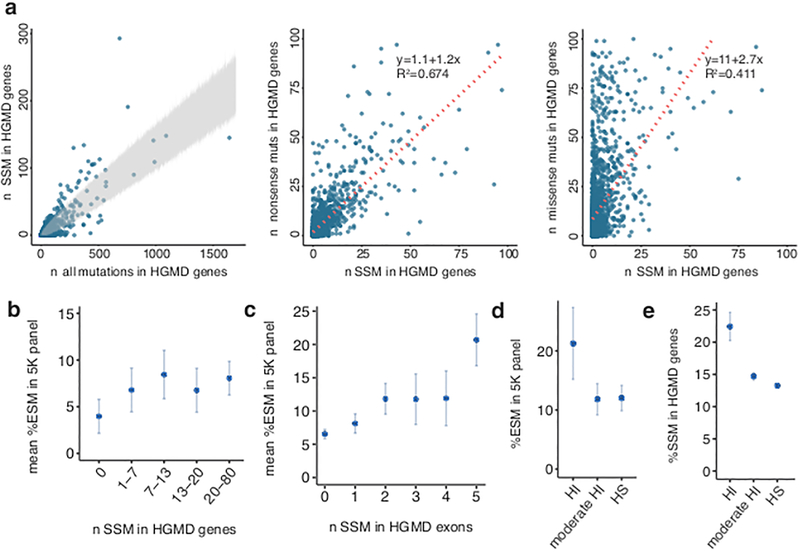
Some exons appeared to have a higher fraction of splicing mutations than others (e.g. exon 8 of MLH1 and exon 18 of BRCA1, adjusted P = 2.26 × 10−3 and 4.18 × 10−6, respectively, two-sided binomial test). Interestingly, the set of (mostly) intronic splice-site mutations (SSM) are also not distributed uniformly in disease genes. Analyses of 2,314 disease gene loci identified 64 genes that are predisposed to SSM (Fig. 2a left, Supplementary Table 3)8. SSM often result in exon skipping. Not surprisingly, SSM and nonsense mutations in human disease transcripts were positively correlated as they both result in the loss-of-function of the proteins. This correlation was not observed between missense mutations and SSM (Fig. 2a, middle, right). We found ESM to be more abundant in genes that are also enriched with SSM (Fig. 2b, P = 3 × 10−6, Kruskal-Wallis, Online Methods). This effect was more pronounced at the level of the individual exons (P = 2.1 × 10−34, Kruskal-Wallis, Fig. 2c, Online Methods). Moreover, disease mutations with autosomal dominant inheritance showed a two-fold ESM enrichment in haploinsufficient genes, as compared to haplosufficient genes (P = 0.002, Kruskal-Wallis, Fig. 2d). This finding agrees with splicing mutations acting mainly via a loss-of-function mechanism and further confirms the utility of MaPSy in identifying deleterious ESM (Supplementary Fig. 4). The same enrichment was also observed in SSM reported at HGMD (P = 0.02, Kruskal-Wallis, Fig. 2e)10. Recently, the Exome Aggregation Consortium (ExAC) identified 3,230 genes that are depleted of protein-truncating variants (PTV) in 60,706 humans6, thus showing evidence for extreme selective constraint. Because PTV and splicing mutations often share the same loss-of-function mechanism, we examined the disease ESM occurrence in PTV-intolerant genes (pLI >= 0.9)6 in comparison to other genes. Indeed, we found a three-fold excess of ESM in PTV-intolerant genes (n = 92) as compared to PTV-tolerant (n = 66) genes in the 5K panel that are causing dominant disease traits (adjusted P = 0.005, Kruskal-Wallis, Supplementary Fig. 5a)6. These findings suggest that ESM and SSM are enriched in haploinsufficient genes, in which the loss of one functional copy likely leads to a disease phenotype.
Figure 2. Prevalence of splicing mutations in disease genes.
a, Left: Splice-site mutations (SSM) versus all exonic mutations in the Human Gene Mutation Database (HGMD8) with region of 99.9% confidence interval shown in gray. Middle, right: Number of SSM versus nonsense (middle), and SSM versus missense (right) in all disease genes. b, Mean of exonic splicing mutation (ESM) percentage in each gene is plotted against roughly equal bins of percent SSM in HGMD genes (n = 708). c, Mean of ESM percentage in each exon versus number of SSM per exon (n = 2,048). d, Percent ESM in haploinsufficient (HI, n = 174), moderate HI (n = 567) and haplosufficient (HS, n =874) genes in autosomal dominant diseases in the 5K panel10. e, Percent SSM in HGMD with autosomal dominant inheritance in HI (n = 1,383), moderate HI (n = 14,059) and HS (n = 59,901) genes10. Error bars in b,c represent standard error of the mean. Error bars in d,e represent 95% confidence intervals.
Random Forest Classification of Exonic Splicing Mutations
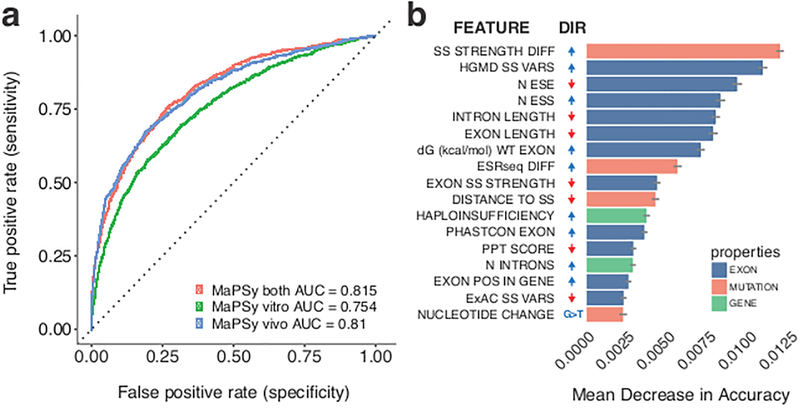
Various genomic and sequence features have been reported to affect splicing10–14 While most of these studies were only done in a few substrates, MaPSy enables direct comparisons of splicing performance of thousands of exons in vivo and in vitro (Supplementary Fig. 2e). Many of these features (e.g. differential GC content between exons and introns, density of exonic splicing silencers (ESSs)) were confirmed with MaPSy (Supplementary Fig. 6a)11,13. We used Random Forest classification (Online Methods) on the ESM dataset generated with MaPSy to further understand the different contributions of the various genomic and sequence features that may lead to ESM15. Performance of the Random Forest model was measured by mean area under the curve (AUC = 0.81, 0.755 and 0.816 for in vivo, in vitro and combined, respectively) (Fig. 3a). The in vivo assay performed better than in vitro, but combining the two assays resulted in further increase in sensitivity to detect ESM. Measures of feature importance were calculated as mean decrease in accuracy (MDA). Each feature was categorized as the property of the mutation, the exon or the gene (colored salmon, blue and green, respectively, Fig. 3b). It was surprising that the majority of the top predictors for the ESM that are not within the splice-site regions (~76%) were exon level features, rather than some properties of the nucleotide substitutions (e.g. exon splicing enhancer (ESE) disruption and ESS creation). In other words, some exon properties (e.g. low ESE density and high ESS density) sensitize an exon to ESM (adjusted P = 1.8 × 10−12, 7.8 × 10−18, Kruskal-Wallis, ESE and ESS density, respectively, Supplementary Fig. 6b). In addition, the Random Forest model suggests that genes with many introns pose a greater risk for ESM. We found that PTV-intolerant genes6 also contain more introns than average disease genes (P < 2.2 × 10−16, Mann-Whitney test), similar to ESM and SSM-enriched genes (Supplementary Fig. 5b).
Figure 3. Random forest classification of exonic mutations that disrupt splicing.
a, Classification performance of the random forest model was calculated as the area under the curve (AUC) in receiver operating characteristic (ROC) analysis. b, The order of variable importance by mean decrease in accuracy. Error bars indicate standard deviations. The directions (DIR) of change that promote exonic splicing mutations (ESM) are indicated, positive directions are colored blue, and negative directions are colored red. Variables include differences in splice-site strength35 and hexamer splicing scores11 (SS STRENGTH DIFF, ESRseq DIFF), sum of the effects of splice-site variants at Human Gene Mutation Database (HGMD) and Exome Aggregation Consortium (ExAC) datasets (HGMD SS VARS, ExAC SS VARS)6,8, numbers of exon splicing enhancers (ESE) and exon splicing silencers (ESS) in the exon (N ESE, N ESS), free-energy estimate (dG (kcal/mol) WT EXON)31, exon conservation (EXON PHASTCON), number of introns (N INTRONS) and relative exon position in the gene (EXON POS IN GENE). PPT: Polypyrimidine track.
RNA binding protein motifs in the 5K panel
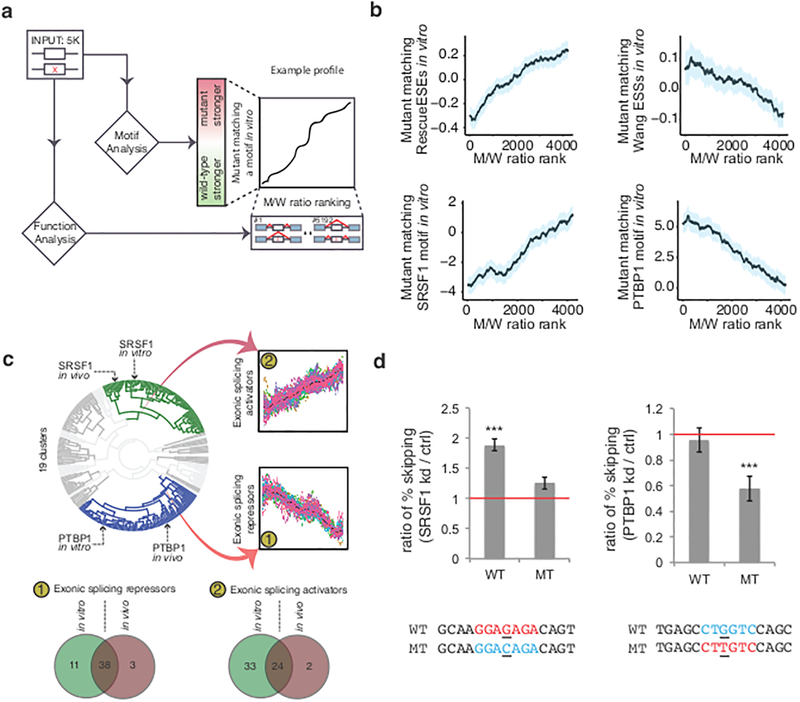
Presumably, most mutations that alter splicing act by disrupting the binding site of an activator or by creating a binding site for a repressor. The loss or gain of previously characterized exonic enhancer and silencer elements was compared to splicing performance in MaPSy12,16–19 (Fig. 4a). A positive correlation was observed between gains of known exonic enhancing elements and relative splicing performance (i.e. m/w ratio, adjusted P = 7.75 × 10−25, linear regression, Fig. 4b, see Online Methods). In contrast, a negative correlation was observed between gains of known exonic silencing elements and splicing (adjusted P = 0.0001, linear regression, Fig. 4b).
Figure 4. Detection of RNA binding protein (RBP) motifs that affect splicing.
a, All mutant/wildtype (M/W) pairs were examined for difference in position-weight-matrices agreement with 155 RBP motifs and known exonic cis-elements. b, Motif profiles show clear trends in agreement with previously defined functions. Shaded blue regions indicate 95% confidence intervals. c, Clustering of data shows similar function of RBP motifs in vivo and in vitro. The mean values from each bin are colored black. d, Left: In the absence of SRSF1, the mutant (MT) that disrupts the SRSF1 binding motif had a modest but not a significant increase in exon skipping, while the wildtype (WT) exon with the SRSF1 motif had a two-fold increase in exon skipping. Right: the splicing phenotype of a mutation that creates a PTBP1 binding motif were rescued (~0.5 fold less of skipping event) when PTBP1 was knocked down, but not the wildtype exon. Three stars on top of the bar indicate statistical significance (P < 0.001, two-sided Cochran-Mantel-Haenszel test). Error bars indicate standard deviation. kd: knockdown; ctrl: control.
To predict which trans-acting factors’ binding events were affected by exonic mutations, we recorded the effect of thousands of point mutations with the predicted change of the binding affinity of 155 human RNA binding proteins (RBP)20. Briefly, mutant-wildtype pairs were ranked from lowest to highest degree of inclusion of the mutant allele relative to the wildtype allele. The predicted changes in binding affinity were compared to the observed gain or loss of splicing activity (i.e. m/w ratio)21. SRSF1, a well-characterized exonic splicing activator22,23, showed a positive correlation with splicing (adjusted P = 3.34 ×10−27, linear regression, Fig. 4b), while Polypyrimidine Tract Binding Protein 1 (PTBP1), a known exonic splicing repressor, correlated negatively with splicing (adjusted P = 3.26 × 10−21, linear regression, Fig. 4b)24,25. As the presence of a RBP motif does not necessarily result in a binding event20,26, it is necessary to validate the relationship between the loss/gain of protein binding with the loss/gain of splicing. An ESM in exon 20 of COL1A2 (NM_000089.3:c.1045G>T) was predicted to create a PTBP1 motif. If PTBP1 binding was responsible for splicing repression, depletion of PTBP1 would be predicted to relieve the splicing defect. We found that in the absence of PTBP1, a rescue of splicing (i.e. ~0.5 fold less skipping) was observed in the mutant, but not in the wildtype exon (P = 4.19 × 10−5, two-sided Cochran-Mantel-Haenszel χ2 test, Fig. 4d right and Supplementary Fig. 7a). An ESM predicted to function by disrupting SRSF1 binding in exon 8 of MLH1 (NM_000249.3:c.595G>C) was also selected for similar analysis. In the absence of SRSF1, the wildtype exon had a significant increase in skipping event (P = 0.0002, two-sided Cochran-Mantel-Haenszel χ2 test, Fig. 4d left and Supplementary Fig. 7b), but not the mutant exon (P = 0.07, two-sided Cochran-Mantel-Haenszel χ2 test). This result demonstrates how motif prediction can identify mutations where the gain of PTBP1 binding or the loss of SRSF1 binding can lead to the ESM phenotype.
Clustering the functional profiles of human RBP motifs in the 5K panel (see Online Methods) resulted in 19 clusters, with the two largest clusters matching the profile of exonic splicing enhancers and repressors (Fig. 4c). The method was robust; >90% of all motifs that functioned as silencers or enhancers in vivo segregated into the same category in vitro (P = 1×10−16 and 1.5×10−10, one sided Fisher’s Exact Test for Venn diagram overlap exonic splicing repressors and activators, respectively, Fig. 4c, Supplementary Fig. 8e). Overall, 38 motifs corresponding to 35 RBP consistently behaved as exonic repressors and 24 motifs corresponding to 25 RBP behaved as exonic activators in both assays. Comparing the degree of predicted intronic binding with splicing performance suggests that most exonic repressors enhance splicing when bound in the introns (57%, Supplementary Fig. 8c) and most exonic activators repress splicing when bound in the introns (77%, Supplementary Fig. 8d). These findings reinforce the notion that splicing factors behave in highly position-dependent manners7,27.
Mechanistic signatures of splicing mutants
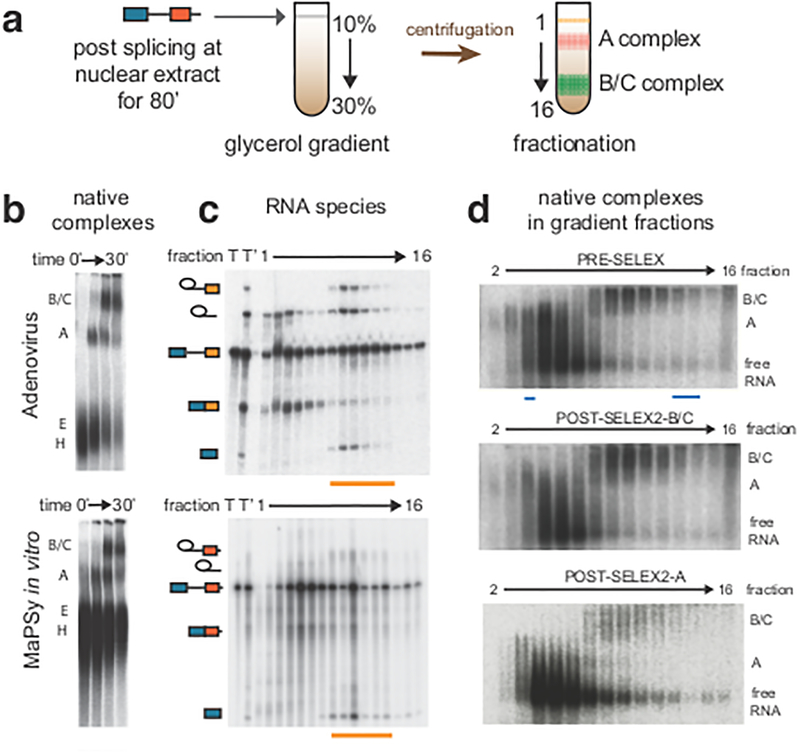
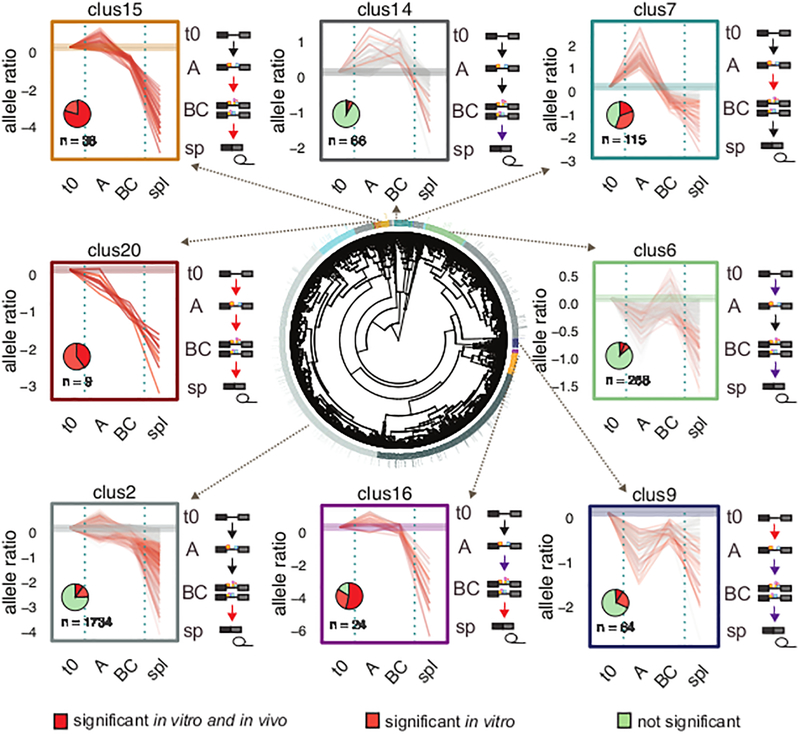
During the development of the in vitro splicing assay in the 1980s, techniques were developed to isolate the biochemical intermediates in the stepwise assembly of the spliceosome28. Spliceosome is assembled from A through B to C complex on the model Adenovirus substrate, as previously described29,30. Consistent with catalysis occurring in the C complex, chemical intermediates of splicing co-migrated with the C complex during glycerol gradient centrifugation (Fig. 5a). This same procedure was implemented on the 5K panel of mixed library substrates. Although each library member is the same length, greater heterogeneity in complex mobility was observed (Fig. 5b). Despite this increased heterogeneity, distinct splicing complexes were effectively partitioned as the splicing intermediates and final products were found to segregate into the same fractions as seen in the control (orange underlines, Fig. 5c). Furthermore, each stage of spliceosome assembly contains a distinct composition of library species that could be further enriched by a SELEX approach (Fig. 5d, Supplementary Fig. 9a). For example, extracting RNA from the B/C fraction and repeating the spliceosome assembly assay returned a clear bias towards B/C complex (Fig. 5d middle), while reassembly of the A fraction resulted in a bias toward the A complex (Fig. 5d bottom). By utilizing glycerol gradient centrifugation coupled to next-generation sequencing, the allelic ratio of each locus was determined at the different stages of the spliceosome assembly: pre-assembly (t0), A, B/C and spliced. In general, RNA species that were enriched in the early A complex were underrepresented in the spliced fraction, suggesting that the species that were being blocked from transitioning to the catalytic B/C complex were accumulating in the A complex. Conversely, RNA species that were enriched in the B/C complex were also enriched in the spliced fraction, suggesting that spliceosomes at this stage were mostly committed to splicing (Supplementary Fig. 9b). Clustering the 5K panel by allelic ratios in the different spliceosomal fractions showed distinct patterns of disruptions. Most mutations affected multiple transitions of the spliceosome (red arrows indicating major disruptions, purple arrows indicating minor disruptions, Fig. 6, Supplementary Fig. 9c). We found that mutations in the same exon were more likely to cluster together (P = 0.008, permutation test). This result suggests that an exon disrupted by splicing mutations will tend to fail at the same stage of the spliceosome assembly, a behavior consistent with the finding that exon properties are strong predictors of ESM (Fig. 3b). Remarkably, the allelic ratio profiles in the different assemblies seem to represent mechanistically distinct scenarios of splicing disruption. For example, mutants in cluster 20 are strongly inhibited in each step of the spliceosome assembly (Fig.6). Interestingly, cluster 20 is comprised of mutations that are likely to trigger structural rearrangements (average ΔΔG = 1.95 kcal/mol, adjusted P = 0.014, permutation test)31. They are single substitutions that, on average, were predicted to trigger the formation of four new basepairs that contribute to a more closed RNA secondary structure. Cluster 15 contained mutations in weakly defined exons (low differential GC and high numbers of ESS, adjusted P = 0.008 and 0.014, respectively, permutation test) and flanked with highly conserved introns (adjusted P = 0.006, permutation test). The splicing progression of these mutants were stalled in A and B/C, all of which significantly altered splicing in vitro, ~80% of which also significantly altered splicing in vivo. Exons in cluster 15 and 20 are also frequent targets of disease-causing SSM8, consistent with the finding that disease-causing ESM and SSM are often coenriched in the same exons. In contrast, mutations in cluster 14 were associated with strongly defined exons (high differential GC, low numbers of ESS, adjusted P = 0.001 and 0.002, respectively, permutation test) and rarely disrupted splicing (Fig. 6). Mutants in cluster 7 were found in exons with strong splice-sites (adjusted P = 0.01, permutation test), and their respective wildtype exons were strong splicers both in vivo and in vitro (adjusted P = 0.0008 for both assays, permutation test). The splicing progressions of these mutants were mainly inhibited in the A complex. While mutations in cluster 15,16 and 20 represent ESMs with the most severe splicing phenotypes, ESMs in cluster 7 and 14 have modest effects on splicing (Supplementary Fig. 10). It remains to be determined whether these distinct modes of splicing disruptions are associated with the degree of severity or other aspects of disease phenotypes. We predict that mechanism via structural changes (e.g. cluster 20) is likely to be independent of tissues and cell-types, since they seem more independent from trans-acting factors that may vary across tissues and cell-types. While mutational mechanisms that involve trans-acting factors recognizing exonic binding motifs (e.g. cluster 15) are more likely to be tissue and cell-type dependent.
Figure 5. Isolation of spliceosomal intermediates.
a, After MaPSy in vitro, splicing reaction was loaded to 10–30% glycerol gradient, followed by fractionation. Different spliceosome stages were retrieved in different fractions. b, Spliceosomal complexes (B/C, A, E, H) visualized in native gels for control (top) and heterogeneous library substrates (bottom). c, RNA splicing intermediates migrate to the same fractions in control and library substrates (orange underlines). Total RNA pre (T) and post (T’) splicing are indicated. d, Reassembly of purified B/C and A fractions (middle and bottom), compared to the assembly of original input (top). Fractions used for SELEX are underlined (cyan).
Figure 6. Clustering of allelic ratios provides exonic splicing mutation (ESM) mechanistic insights.
The result of the hierarchical clustering of allelic ratios in spliceosomal fractions is shown (center plot) with representative clusters shown in different colors. The individual panels surrounding the center plot show allelic ratios of each mutant/wildtype (m/w) pairs in the different fractions (t0, A, BC and spliced (spl,sp)) for the corresponding clusters. Each pair is colored according to its ESM classification (dark red for significance in both assays, orange for significance in vitro, and gray for negative pairs). The complete profile of all clusters can be found in Supplementary Fig. 9c. Pie charts in individual panels indicate the proportion of ESM classifications. Spliceosome stages are depicted at the right of the individual panels. Major disruptions in assembly transitions are indicated with red arrows and minor disruptions are indicated with purple arrows.
Each mutation in the 5K panel represents a variant reported in a patient/family in the last four decades. We have established the first large-scale collection of the effect of exonic mutations on splicing and created a public web-server that enables the visualization of the MaPSy results (see URLs, Supplementary Fig. 11).
Discussion
The need for better characterization of sequence variation is ever more urgent with the increasing number of rare variants being discovered from many large-scale sequencing efforts6,32. Previous studies had tested the effect of random k-mers in enhancing or silencing splicing11,33,34. We present the results of a survey of 4,964 single point mutations’ effects on splicing using MaPSy, a novel dual parallel splicing system. We further characterized the splicing aberrations by their stages of disruptions in the spliceosome assembly. We found ~10% (513/4,964) of exonic disease alleles disrupt splicing in vivo and in vitro. In contrast, only 3% (7/228) of common SNPs altered splicing in both assays. It is interesting that in diseases that are more frequently caused by splicing mutations, more exonic mutations were also found to disrupt splicing. This likely reflects disease processes that occur through loss-of-function mechanisms. We found that exonic features play a large role in forming ESM. We also identified 24 exonic RBP motifs that are associated with increased splicing and 38 RBP motifs that are associated with weaker splicing.
MaPSy has certain limitations; particularly, only mutations in exons of fewer than 100 nucleotides in length can be evaluated due to the current limitation in oligo synthesis technology. Given that the average length of internal exons is around 130 nucleotides, half of human exons are not accessible for splicing characterization using MaPSy. We also cannot rule out the presence of other influences (e.g. flanking splice-sites, different transcription efficiencies, tissue-specific effects), all of which are not preserved in MaPSy. It is intriguing that some features that have been previously shown to be predictors for SSM but are not present in MaPSy (e.g. flanking intron length, number of introns) were also identified as predictors for ESM (Fig. 3b)14. These findings, together with the high concordance rate with splicing phenotypes in corresponding patient tissue samples, suggest that despite these limitations, MaPSy contains most of the critical elements required for splicing in native conditions and thus it is a powerful tool to characterize sequence variation for splicing aberrations.
In conclusion, MaPSy facilitates the first large-scale identification and characterization of ESM. The system effectively translates to 5K implementations of basic mutational approaches and can be further adapted to other mutation panels, thus accelerating the efforts to characterize all sequence variation.
Online Methods
Library design and synthesis.
Nonsynonymous mutations classified as disease-causing (DM) were downloaded from Human Gene Mutation Database8 (HGMD, accessed in May 2012) and mapped to GRCh37/hg19 human reference sequence. Mutations were mapped to internal exons with <= 100 nucleotides in length and selected for those that fit into 170 nucleotide genomic windows that include 15 nucleotides downstream introns and >= 55 nucleotides upstream introns (n = 4,964). The mutant and wildtype versions of the 170-mer genomic fragments were flanked with 15-mer common primers and designed into a 200-mer oligo library. Solid-phase oligonucleotide synthesis was performed by Agilent Technologies and used to generate in vivo and in vitro reporters.
MaPSy in vivo.
In vivo splicing reporter design includes Cytomegalovirus (CMV) promoter, Adenovirus (pHMS81)36 exon with part of its downstream intron, 200-mer oligo library, exon16 of ACTN1 with part of intron15 and bGH PolyA signal sequence (Supplementary Fig. 12). Common sequences (everything except the 200-mer library) were concatenated by overlapping PCR and cloned with TOPO TA (Invitrogen) to generate 5’ common fragment and 3’ common fragment. Each cloned fragment was PCR amplified and equimolar amounts of common fragments and the oligo library were concatenated in a single PCR reaction and purified/size selected twice with 0.4:1 ratio of Agencourt AMPure beads (Beckman Coulter). The resulting in vivo reporters were transfected to human embryonic kidney hek293T cells (ATCC) in three cell culture replicates using Lipofectamine 2000 (Invitrogen) in 6-wells plate. RNA was extracted 24 hours post transfection using TRIzol (ThermoFisher) and DNase treated. Random 9-mers were used to generate cDNA with SuperScript III Reverse Transcriptase (Invitrogen) followed by PCR (GoTaq, Promega). All PCR reactions were kept in lowest possible cycles (15–20 cycles). Input reporters and spliced species were sequenced in Illumina HiSeq2500 (100bp paired-end).
MaPSy in vitro
In vitro splicing reporter has similar design as in vivo reporters, but excluding the ACTN1 exon and T7 promoter was used (Supplementary Fig. 12). In vitro reporters were obtained via transcription in vitro using T7 RNA Polymerase (Stratagene) and internally labeled with [α−32P]UTP (Perkin Elmer) and capped with G(5’)ppp(5’)G (New England Biolabs). The resulting RNA was gel purified and used for splicing reaction in 40% HeLa-S3 (NCCC) nuclear extract for 80 min at 30°C as previously described37. Pools of input and spliced RNAs were converted to cDNA (SuperScript III, Invitrogen) and made into Illumina library (NEBNext kit, New England Biolabs) for deep sequencing. For glycerol gradient fractionation, 120 μl of splicing reaction was treated with 0.5 mg/ml heparin for 5 min at 30°C and was loaded to 3.75 ml of 10%−30% glycerol gradient and centrifuged at 38,000 rpm at 4°C for 2.5 h. After centrifugation, gradient was fractionated from top to bottom in 16 equal volumes and analyzed with 2.1% native agarose (UltraPure LMP Agarose, Invitrogen) or 8% denaturing polyacrylamide gel (29:1 crosslinking). The in vitro MaPSy was done in two experimental replicates. Gels were visualized with Typhoon PhosphorImager (GE Healthcare). Unspliced RNAs bound to different complexes were extracted from relevant fractions, converted to cDNA (SuperScript III, Invitrogen), reattached to T7 promoter sequence by PCR, gel purified, and used as template for subsequent in vitro transcription to make pre-mRNA substrates for the next round of SELEX (Supplementary Fig. 9a). RNA pools recovered from each purification step were converted to cDNA, PCR amplified, and analyzed with deep sequencing (Illumina HiSeq2500, 100bp paired-end).
Library species alignment and counting.
We generated “reference genomes” for both in vivo and in vitro libraries, with each wildtype (reference) and mutant species treated as their own “chromosomes”. Paired-end reads were mapped using STAR aligner38. For input alignment, we do not allow for split-reads and only uniquely mapped reads with a maximum of 10 mismatches were allowed. We used the same settings as input alignment for output alignment, with the exception that we allowed for split reads. Since the 5K panel may include more than one mutation in an exon, the requirement for calling a wildtype can be more stringent than the requirement for calling each of the mutants, given that calling the wildtype species would require the read pair to span all mutation positions in the same exon, while calling the mutant species would only require the read pair to span the respective mutant position. Thus, we also require all mapped reads to span all mutation positions in order to ensure balance of detection between wildtype and mutant species.
Allelic imbalance analyses.
The allelic ratios for MaPSy analyses were calculated as follows:
in which mo is count of mutant spliced species, mi is count of mutant input, wo is count of wildtype spliced species and wi is count of wildtype input. To assess for statistical significance, two-sided fisher’s exact test was used and the resulting p-values were adjusted to account for multiple comparisons using p.adjust function in R (method=“fdr”). Significance level of < 0.05 and allelic ratio >= 1.5 fold change were used to call ESM.
Splicing efficiency analyses.
To compare splicing performance between individual species, the following splicing index was calculated for each species:
where spli is the count for spliced output for species i and inpi is the count for the input for species i, and n is the number of species in the library pool.
MaPSy validation in patient samples.
Tissue samples (n = 13) were obtained from University of Utah School of Medicine (Salt Lake City, UT), Washington University School of Medicine Alzheimer’s Disease Research Center (St. Louis, WA), Ohio State University (Columbus, OH), National Institute of Child Health and Human Development (Bethesda, MD) and Coriell Repository. Ethical approvals were granted from local institutional review boards, and informed consents were obtained from all participants. RNAs were extracted using PAXgene kit for whole-blood samples, RNAeasy kit (Qiagen) for post-mortem brain samples, or Trizol (Life Technologies) for all other samples, using the respective manufacturers’ protocols. SuperScript III Reverse Transcriptase (Invitrogen) was used to generate cDNA with random 9-mers, followed by PCR (GoTaq, Promega). PCR primers were designed in exons flanking the mutant exon. In the case of patients with nonsense mutations for which we have lymphoblastoid cell lines or fibroblasts available, the cells were also treated with 10 μg/ml of cyclohexamide for 3 h prior to RNA extraction.
MaPSy validation in ENCODE data.
We downloaded 46 whole-cell RNA-Seq Long PolyA+ ENCODE data sets of 19 different cell lines (accession numbers: see Supplementary Table 4). Reads were mapped to hg19 using STAR38 aligner with default parameters. Each STAR output generates a splice-junction file, which was used to calculate percent usage in each splice junction as follows:
Results from multiple runs of the same cell lines were collapsed. Hg19 positions of 3’ splice-site (ss), 5’ss, upstream 5’ss and downstream 3’ss of all wildtype exons in the 5K panel were retrieved and were binned into four groups of increasing splicing performance in MaPSy. Average percent usage at both splice sites were plotted in each bin and compared.
HGMD mutation analyses.
Disease-causing splicing and coding sequence mutations were selected from HGMD (n=77,943). The mutations were classified as splicing, missense, or nonsense mutations and the numbers of all classes of mutations were determined for each gene. The total number of mutations was plotted against the total number of SSM in a gene (Figure 2a). Weighted random sampling was then used to construct a 99.9% confidence interval that capitulates the expected number of SSM given the total number of mutations within a gene. Using the proportion of total SSM to total mutations in the HGMD as a weight for random sampling, the proportion of SSM given the total mutations in each gene was simulated 1,000 times. Genes falling outside the simulated values represent genes that have more (above the confidence interval) or fewer (below the confidence interval) SSM than expected (P <0.01) based on the distribution of mutations types within the dataset. Haploinsufficiency scores were obtained from published data10. HGMD genes were binned as haploinsufficient genes (haploinsufficiency (HI) score = 1), moderate haploinsufficient genes (HI score in between 0.7 and 1) and haplosufficient genes (HI score <= 0.7).
Random Forest classification.
We used R implementation of Random Forest15, a non-parametric ensemble learning method, to model the contribution of various genomic, sequence and functional features on the likelihood that an exonic mutation will impact splicing. Each tree in the forest is constructed with a different bootstrap sample from the original data set, with approximately two-thirds of the bootstrap sample being used for the construction of the kth tree and the remaining one-third of the bootstrap sample (out-of-bag data) is used for cross-validation. The results from all trees are then averaged to provide unbiased estimates of predicted values, error rates and measures of variable importance. Default parameters were used to build the Random Forest model, with the exception of specifying number of trees to 1,000. Since variable importance measures may vary depending on the parameters of the algorithm and both the degree of correlation as well as the scale of the variables can influence them, we opted to use two different methods for feature selection and measures of importance. The first method created shuffled copies of all the features (shadow features) and trained a Random Forest classifier using the Supplementary set while iteratively removing irrelevant features (those with z scores less than the maximum z scores of their respective shadow features). This was done until all features were either confirmed or rejected, using the Boruta39 package in R. For the second method, we generated the null distribution of the variable importance measures by permuting the response variable so that the relationship between response and predictor variables was destroyed. This was done with 1,000 runs of Random Forest, and the empirical p-values for importance measures were calculated by counting the number of times in which each importance measure in the original data was lower or equal to the respective importance measure in the permuted data. Features that are selected in both methods with significance level < 0.05 were used for the final Random Forest model.
Random Forest predictor variables.
Splice-site strength was computed using downloaded Perl scripts from MaxEntScan35 package, which uses a Maximum Entropy approach on large datasets of splice-sites in humans, taking into account both adjacent and non-adjacent dependencies. The splice-site models assign log-odd ratios to 9 basepairs sequences (−3 to +6 positions) for the 5’ splice-site scores and 23 basepairs sequences (−20 to +3 positions) for 3’ splice-site scores. “SS VARS” is the sum of wildtype-mutant Δ splice-site scores of all SSM at HGMD8 and ExAC6 datasets in each exon. ESE and ESS were downloaded from published data11,16,40. “ESRseq DIFF” was computed as the wildtype-mutant difference in hexamer splicing scores11. Haploinsufficiency score was obtained from a previous study that developed haploinsufficiency prediction model using a large deletion dataset (Wellcome Trust Consortium Controls)10. Polypyrimidine track (PPT) score was computed as previously described41. “EXON POS IN GENE” was calculated as exon number divided by total number of exons in the gene (values between 0 and 1). The free-energy estimate (ΔG) was computed using ViennaRNA package31 version 1.8.5, using default settings with -d2 -noLP options.
Motif analyses.
RBP, ESE and ESS motifs were obtained from published sources11,21. ESE and ESS hexamers were mapped and counted in each mutant and wildtype exons of the 5K panel. Contribution of known splicing elements in MaPSy splicing was evaluated by plotting the mutant-wildtype difference in ESE and ESS counts against mutant/wildtype splicing ratio in sliding windows (size = 1,000, step = 1). RBP motifs were mapped to the exons and upstream introns of the 5K panel using matchPWM function from Bioconductor package42 with default settings (minimum score = 0.8). We computed the maximum matchPWM score percentiles of all spanning n-mers at the mutation positions that overlap the exonic motif maps and calculated the mutant – wildtype difference for each mutation position (n = length of motif). In vitro and in vivo splicing profiles of exonic motifs were generated by plotting the mean of the maximum score differences in rolling windows of increasing mutant allele inclusion of spliced species (i.e. m/w ratio, window size = 1,000, step =1). Intronic motif maps of wildtype species (n=2,086) were used to calculate intronic motif density for each RBP (Supplementary Fig. 8a). Wildtype splicing profiles of intronic motifs were generated by plotting the mean motif density in rolling windows of increasing splicing efficiency (window size = 200, step = 1). In vitro and in vivo profiles were combined and fitted using smooth.spline function in R43. Bayesian Information Criterion was used to determine the optimal number of clusters using mclust function from the mclust R package44. Profiles were clustered based on the coefficient values from spline fitting using hclust function in R (Fig. 4c, Supplementary Fig. 8b).
RBP binding motif validation.
We ordered siRNA for human PTBP1 from ThermoScientific (s11436) and siRNA for human SRSF1 from Dharmacon as previously described23. For control siRNA, AllStar Negative control siRNA (Qiagen) was used. Minigenes were synthesized by Synbio Technologies, Inc. HeLa cells (ATCC) were plated 24 h prior to transfection. For PTBP1 knockdown, 7.5 μl of Lipofectamine RNAiMax (Invitrogen) was used to transfect siRNA for PTBP1 (20 nM final concentration) in 6-wells plate for 48 h according to the manufacture’s protocol (Invitrogen). This is followed by a second transfection with 3.75 μl Lipofectamine 3000 (Invitrogen) and the same siRNA in Opti-MEM medium (Life Technologies) and 500 ng DNA in 100 μl pure DMEM (Invitrogen). RNA was extracted 24 hours later with Trizol according to the manufacture’s protocol (Ambion), followed by DNase treatment and RT-PCR as described above. For SRSF1 knockdown, 1.5 μl of Lipofectamine 3000 (Invitrogen) was used to transfect siRNA for SRSF1 (20 nM final concentration) in OptiMEM medium (Life Technologies) and 500 ng DNA in 100 μl pure DMEM (Invitrogen). After 72 hours, RNA was harvested, followed by DNase treatment and RT-PCR. Knockdown efficiencies were evaluated with Western Blot, using anti-SRSF1 (sc-33652, SantaCruz), anti-PTBP1 (32–4800, Thermo Fisher) and anti-GAPDH (sc-47724 and FL-335, SantaCruz). All experiments were done in two cell culture replicates.
Functional SELEX analysis.
The allele ratios were calculated as follows:
where mie is minor allele count in enriched pool, mii is minor allele count in input, mje is major allele count in enriched pool and mji is major allele count in input. Minor allele is the allele that splices less efficiently in comparison to the respective major allele, which differ by one nucleotide. All analyses were performed in R. Hierarchical clustering was performed on all m/w pairs that were recovered in all purified fractions (n=4,873) using hclust function with complete linkage method and Euclidean distances. Bayesian Information Criterion plots were generated for k=1 to k=50 using mclust package to estimate the optimal number of clusters. The resulting clusters were visualized and the tree was cut using cutree function (k=32). To determine the significance of the observation that mutations in the same exons were more often clustered together, we permuted the exon assignment in the 32 clusters 10,000 times and obtained the χ2 distribution of the permuted data. P-value was obtained by counting the number of times the statistics of the permuted data exceeds or equal to that of the original data, divided by the number of permutations. To examine whether certain genomic features may act as “signatures” of the identified clusters, we plotted the distribution of each feature in the different clusters and significance was determined by the mean difference in two-sided t-statistics on the actual data and permuted data 10,000 times, using the flip function, followed by flip.adjust (method=“fdr”) to account for multiple testing45.
Data availability statement
The data generated from this study (raw allelic counts and allelic ratios from each M/W pairs from MaPSy experiments with the corresponding genomic positions, variant allele and HGMD accession numbers) are available at http://fairbrother.biomed.brown.edu/ESM_browser/.
Supplementary Material
Acknowledgments
We thank Karina Villanueva for generating the list for SNPs used in this study and Alexander Leblang for compiling the variants to make the oligo library. We thank Melissa Jurica and Melissa Moore for suggestions and protocols for in vitro spliceosome assembly assay and nuclear extract preparation. We thank Arun Janssens for contacting investigators for patient samples. We thank Amanda Toland (Ohio State University), Joan Marini (NIH/NICHD) and Alison Goate (Washington University Alzheimer’s Disease Research Center) for contributing patient samples for validation. R.S. was supported by Postdoctoral Fellowship from Center for Computational Molecular Biology (CCMB), Brown University. C.R. was supported by Graduate Research Fellowship from National Science Foundation (NSF). This work was supported by National Institutes of Health (NIH) grants R01GM095612 (to W.F.), R01GM105681 (to W.F.) and R21HG007905 (to W.F.) and by SFARI award 342705 (to W.F.). Part of this research was conducted using computational resources and services at the Center for Computation and Visualization, Brown University and Genomics Core Facility, Brown University.
Footnotes
URLs
Visualization of MaSPy results: http://fairbrother.biomed.brown.edu/ESM_browser/.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Baird PA, Anderson TW, Newcombe HB & Lowry RB Genetic disorders in children and young adults: a population study. Am JHum Genet 42, 677–93 (1988). [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312, 1870–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamshad MJ et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12, 745–55 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Tennessen JA et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue Y et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 91, 1022–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim KH, Ferraris L, Filloux ME, Raphael BJ & Fairbrother WG Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proceedings of the National Academy of Sciences of the United States of America 108, 11093–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenson PD et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 21, 577–81 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Taggart AJ, DeSimone AM, Shih JS, Filloux ME & Fairbrother WG Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nature Structural & Molecular Biology 19, 719–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang N, Lee I, Marcotte EM & Hurles ME Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 6, e1001154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke S et al. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Research 21, 1360–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairbrother WG, Yeh RF, Sharp PA & Burge CB Predictive identification of exonic splicing enhancers in human genes. Science 297, 1007–13 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Amit M et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1, 543–56 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Mort M et al. MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol 15, R19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L Random forests. Machine Learning 45, 5–32 (2001). [Google Scholar]
- 16.Wang Z et al. Systematic identification and analysis of exonic splicing silencers. Cell 119, 831–45 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ke S, Zhang XH & Chasin LA Positive selection acting on splicing motifs reflects compensatory evolution. Genome Res 18, 533–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith PJ et al. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15, 2490–508 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Zhang XH & Chasin LA Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18, 1241–50 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray D et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray D et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 27, 667–70 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Long JC & Caceres JF The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417, 15–27 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Rahman MA et al. SRSF1 and hnRNP H antagonistically regulate splicing of COLQ exon 16 in a congenital myasthenic syndrome. Sci Rep 5, 13208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen H, Kan JL, Ghigna C, Biamonti G & Green MR A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. Rna 10, 787–94 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterne-Weiler T, Howard J, Mort M, Cooper DN & Sanford JR Loss of exon identity is a common mechanism of human inherited disease. Genome Research 21, 1563–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Xiao SH & Manley JL Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev 12, 2222–33 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim KH & Fairbrother WG Spliceman--a computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics 28, 1031–2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padgett RA, Grabowski PJ, Konarska MM, Seiler S & Sharp PA Splicing of messenger RNA precursors. Annu Rev Biochem 55, 1119–50 (1986). [DOI] [PubMed] [Google Scholar]
- 29.Konarska MM & Sharp PA Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46, 845–55 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Das R & Reed R Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5, 1504–8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz R et al. ViennaRNA Package 2.0. Algorithms Mol Bioi 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacArthur DG et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Ma M, Xiao X & Wang Z Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19, 1044–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenberg AB, Patwardhan RP, Shendure J & Seelig G Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163, 698–711 (2015). [DOI] [PubMed] [Google Scholar]
Online Method References
- 35.Yeo G & Burge CB Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Journal ofComputationaI Biology 11, 377–94 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Gozani O, Patton JG & Reed R A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO Journal 13, 3356–67 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichert V & Moore MJ Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions. Nucleic Acids Res 28, 416–23 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kursa MB, Jankowski A & Rudnicki WR Boruta - A System for Feature Selection. Fundamenta Informaticae 101, 271–286 (2010). [Google Scholar]
- 40.Fairbrother WG et al. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32, W187–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin CL et al. RNA structure replaces the need for U2AF2 in splicing. Genome Res 26, 12–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasserman WW & Sandelin A Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5, 276–87 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Chambers JM & Hastie T Statistical models in S, xv, 608 p. (Wadsworth & Brooks/Cole Advanced Books & Software, Pacific Grove, Calif., 1992). [Google Scholar]
- 44.Fraley C & Raftery AE Model-based clustering, discriminant analysis, and density estimation. Journal of the American Statistical Association 97, 611–631 (2002). [Google Scholar]
- 45.Pesarin F Multivariate permutation tests : with applications in biostatistics, xxvi, 408 p. (J. Wiley, Chichester; New York, 2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated from this study (raw allelic counts and allelic ratios from each M/W pairs from MaPSy experiments with the corresponding genomic positions, variant allele and HGMD accession numbers) are available at http://fairbrother.biomed.brown.edu/ESM_browser/.