Abstract
Injectable drug delivery systems that autonomously detect, propel towards, and ultimately treat the cancerous tissue, are the future of targeted medicine. Here, we developed a drug delivery system that swims autonomously towards cancer cells, where it releases a therapeutic cargo. This platform is based on viable bacteria, loaded with nanoparticles that contain the chemotherapeutic-antibiotic drug doxorubicin. The bacteria ferry across media and invade the cancer cells, increasing their velocity in the presence of nutrients that are present within the tumor microenvironment. Inside the cancer cells, doxorubicin is released from the nanoparticles, destroying the bacterial swimmer (antibiotic activity) and executing the therapeutic activity against the cancer cells (chemotherapeutic activity). This mode of delivery, where both the carrier and the cancer cell are destroyed, supports implementing nanoswimmers in drug delivery.
Keywords: autonomous swimmers, nanomedicine, nanotechnology, nanoparticle, cancer, bacteria, targeted drug delivery
Introduction
Nanotechnologies have become important clinical tools, enabling therapeutic precision and functionality that cannot be attained using systems of a larger scale. Newly developed nanotechnologies have the potential to revolutionize diagnosis and care, with more than 40 nanomedicines already approved for clinical use [1].
The application of nanotechnology has proven to be especially advantageous in cancer therapy. Anti-cancer drugs, such as doxorubicin and paclitaxel, have harsh side effects when used systemically, including vomiting, alopecia and cardiotoxicity [2]. By using nanoparticles that accumulate in the tumor, there is an increase in the effective therapeutic dose at the disease site and reduced adverse side-effects caused by systemic application [3].
Liposomes, vesicles with a lipid bilayer membrane which surrounds an aqueous core, are common nanoscale drug carriers [4]. This structure enables loading hydrophilic drugs into the aqueous core and hydrophobic drugs into the lipid bilayer. The liposome membrane can carry a neutral, positive, or negative charge, by enriching the membrane with cationic or anionic lipids. The liposomal corona can be decorated with targeting moieties to increase accumulation at the target site [5], or, alternatively, creating a stealth polyethylene glycol corona, to disguise the particle from the immune system [6].
While most nanomedicines are dependent on blood flow trafficking to reach their target site, adding a propulsion modality that will actively drive the nanomedicines to the disease site, will improve targeting [7].
Several strategies have been employed to physically drive nanomedicines towards diseased tissues [8–10], including the use of magnets positioned above the disease site or ultrasonic waves that propel drug-loaded microbubbles towards tumors [11, 12].
Micro- and nano-swimmers are systems that propel autonomously, or under an external force, towards a target site. In a recent study, bio-hybrid materials with a sperm-like structure were developed to self-propel in media by using filaments composed of a rigid head and elastic tail, covered with contractile cardiomyocytes [13]. Adapting autonomous swimmers for drug delivery has been suggested, however, shown only recently [14–16].
Since the 1800's, bacteria have been used as vehicles for therapeutic applications [16–22]. Most bacteria propel using flagella, 10 μm long and 20 nm in diameter filaments that extend from the bacterial body [23, 24]. Flagella rotation can be either clockwise or counterclockwise, for forward or backward swimming [25]. Bacteria use their aptitude of motility to survive and move towards favorable, or flee from unfavorable, microenvironments. They source their power from nutrients in their environment by converting chemical energy to mechanical energy. Bacteria use chemoreceptors that sense favorable nutrient-rich environments, and then navigate towards these sites, in a process called chemotaxis [22]. This motility enables them to overcome diffusion resistances, which is beneficial for improving tissue invasion [26].
In particular, bacteria favor nutrients found in the tumor microenvironment [21, 27, 28]. Pre-clinical and clinical studies have demonstrated that bacteria accumulate preferentially in cancerous tissue [29–31]. In-vivo experiments have demonstrated that bacteria are attracted to the cancerous tissue by a unique set of nutrients that are secreted metabolically in the tumor microenvironment, including – serine, aspartate, ribose, leucine and arginine [20–22, 27, 28, 32–35]. The extracellular matrix of the tumor microenvironment is used by some types of bacteria as anchoring points for propagating within the tissue [36].
Because of their easily manipulated genetics, bacteria can be engineered to overcome toxicity and systemic implications when injected into the human body. Salmonella with a modified lipid A (strain vnP20009) is non-toxic but colonizes in tumors upon injection [1, 37–40].
To invade cells, bacteria either secrete a set of proteins that puncture the host cell, causing cytoskeletal rearrangement, resulting in bacterial uptake. Otherwise, invasion can occur when the bacteria binds to receptors on the host cell, and is then taken up via cytoskeletal-mediated rearrangements of the cell plasma membrane around the bacteria [26, 41].
This study examines the potential of bacteria as carriers of nanomedicine for targeted delivery of cancer drugs. We loaded bacteria with nanomedicines, to autonomously target malignant cells.
Materials and Methods
Hydrogenated soy phosphatidylcholine, HSPC, was purchased from Lipoid GmbH (Ludwigshafen, Germany); 1,2-dioleoyl-3-trimethylammonium-propane, DOTAP; 1,2-distearoyl-sn-glycero-3-phospho-(1'-rac-glycerol), DSPG; 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine-B sulfonyl) ammonium salt, 18:1 Liss Rhod PE, were purchased from Avanti Polar Lipids (USA); cholesterol (Ch), sulforhodamine-B, ampicillin (amp) and doxurubicin-HCl were purchased from Sigma-Aldrich.
Bacterial strains and growth conditions
Salmonella enterica subspecies enterica serovar Typhimurium LT2 (ATCC 700720) were transformed with pGFP plasmid (Clontech, CA, USA) by electroporation using MicroPulser electroporator (Bio-Rad Laboratories, Ca, USA). Transformants were selected on Luria-Bertani (LB) agar plates (1.5% w/v) supplemented with ampicillin (100μg/mL) and stored at -80 °C in LB medium supplemented with 30% glycerol (%v/v).
Escherichia coli (ATCC 53323), JM109 (F' traD36 proA+ proB+ lacIq Δ(lacZ)M15 Δ (lacproAB)supE44 hsdR17 recA1 gyrA96 thi1endA1 relA1 e14λ) was grown on LB medium. Both strains were examined in motility assays. Salmonella Typhimurium LT2 was further examined as carrier in in vitro experiments.
Motility assay
Bacteria glycerol stocks were streaked on LB plates supplemented with 100 μg/mL ampicillin (amp100) and grown at 37 °C overnight (O/N). A single colony was inoculated in fresh LB media and grown O/N at 37 °C, at 250 rpm in a TU400 orbital incubator shaker (MRC, Holon, Israel). This was used as a starter to inoculate fresh LB media the following day at a ratio of 1:10 (starter:media). The culture was grown at 37 °C until an OD600~0.5-0.7 (mid-log phase) was reached; then, centrifuged and washed three times with potassium phosphate buffer (0.1 M at the appropriate pH). The desired glucose concentration was added afterwards. Bacteria were observed in bright field using an inverted Nikon Eclipse TS100 microscope (NU, USA). Velocity was measured as distance/time, where distance was measured using data analysis software NIS-Elements D4.10.00.
Effect of doxorubicin on bacteria
The antibacterial effect of DOX on bacteria was examined adding increasing concentrations of DOX to Salmonella in suspension (OD600=1). DOX concentration was at range 1.2ng/mL up to 12mg/mL. The viability of Salmonella cells was evaluated by live cell count. Decimal dilutions of bacterial suspension were seeded in 20μl droplets on LBamp100 plates. The plates were incubated O/N at 37°C. The next-day bacterial colonies were counted in each of the DOX dilutions. Colony forming unit (CFU)/mL was determined according to: Σ colonies / (Σ volume of droplets X Σ dilutions), cfu/mL.
Invasion of Salmonella into 4T1 cells
1×105 cells/well (2 mL) of triple negative breast cancer mouse cell-line (4T1; ATCC CRL2539) were seeded in 6-well plates in a RPMI 1640 medium (Sigma-Aldrich), supplemented with 10% heat inactivated Fetal Bovine Serum (Biological Industries, Beit-Haemek, Israel), 2 mM of L-Glutamine Solution, 10 U/mL of penicillin G sodium salt and 0.01 mg/mL of streptomycin sulfate for 24 hours at 37 °C in a 5% CO2 humid atmosphere. The following day, the 4T1 cells were washed, to remove remnants of antibiotics, and Salmonella at multiplicity of infection (MOI) 100:1 was added for 3h. The 4T1 cells were washed with PBS and gentamycin sulfate at concentration of 20μg/mL (Biological industries, Israel) to remove external bacteria. Cells were incubated with Hoescht 33342 (Sigma-Aldrich) and Dil (life technologies, Israel) to stain the nucleus and the cell’s membrane, respectively. Images were obtained using Confocal Laser Scanning Microscopy Zeiss LSM 700 and ZEN imaging software integrated to a three dimensional (Z-stack) image.
Liposome preparation
Liposomes were prepared using the ethanol injection method [42]. In short: phospholipids were dissolved in preheated absolute ethanol at 65 °C and injected into preheated (65 °C) 10% phosphate buffer, resulting in the formation of a milky dispersion of multi-lamellar vesicles (MLV). The MLV dispersion was downsized by stepwise extrusion through Nucleopore etched polycarbonate membranes with 400, 200, 100, 80 and 50 nm (Whatman, USA) pores; 5 cycles of extrusion at each pore-diameter, using a Lipex extruder (Transferra Nanosciences, BC Canada). After extrusion, non-entrapped materials were removed by dialysis (12-14 kDa MWCO) against PBS. Neutral liposomes were composed of HSPC:Ch in molar ratio of 4:1, positively charged liposomes were composed of HSPC:Ch:DOTAP at molar ratio of 4:1:1.5 and negatively charged liposomes were composed of HSPC:Ch:DSPG at molar ratio of 4:1.5:1. Total concentration of all components in all formulations was 50 mM. Particle size distribution, mean particle diameter and zeta potential of the liposomes were measured using dynamic light scattering (nano ZSP, Malvern Instruments, Southborough, MA), Figure 4.
Figure 4. Liposome charge affects bacterial viability.
(A) Light and electron microscopy images of bacteria incubated with anionic (left) or cationic (right) liposomes (red, in the upper overlay images). Incubating the bacteria with the cationic liposomes induced significant defects in the bacterial structure. (B) Salmonella were incubated with cationic and anionic liposomes. Bacteria exposed to anionic (-40mV) liposomes showed a mild decrease in viability, while bacteria exposed to cationic liposomes (+25mV), underwent a significant decrease in viability. Non treated bacteria were used as control. (C) Effect of lipid composition on the liposomal surface charge. Liposomes were composed of: Hydrogenated soy phosphatidylcholine (HSPC), 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP, cationic) or 1,2-distearoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (DSPG, anionic), and cholesterol (ch), the zeta potential was measured using ZSP instrument.
Rhodamine encapsulation-passive loading
Rhodamine was dissolved in the 10% PBS phase. Phospholipids were dissolved in EtOH and added to the aqueous solution. Non-encapsulated rhodamine was removed by dialysis against PBS.
Doxorubicin encapsulation – active loading
Phospholipids were dissolved in EtOH, and then added to 120 mM ammonium sulfate solution. Liposomes were formed by stepwise extrusion as described above. Non-entrapped ammonium sulfate was removed by dialysis (12-14 kDa MWCO) against 10% sucrose. Doxorubicin (DOX) was dissolved in 10% sucrose in concentration of 12 mg/mL. The liposomes and DOX were mixed in ratio of 1:2 (final DOX concentration 6 mg/mL) and stirred for 1h at 60 °C. Free drug was removed by three sequential dialysis steps against 10% sucrose.
Bacteria loaded with liposomes
Incubation
We exploited the time interval in which bacteria split and divide, the membrane opens and liposomes can enter the cell. 1mL of bacteria suspension in O.D600~ 0.5 (mid-log phase) were incubated with 200 μl liposomes for 1h.
Electroporation
Bacteria glycerol stocks (E.coli or Salmonella) were inoculated in LB media and grown at 37°C, at 250 rpm, in orbital shaker, O/N. This was used as a starter to inoculate fresh LB media the following day in ratio of 1:10 (starter:media). The culture was grown at 37°C to OD600~0.5. Then the culture was cooled for 30min on ice and centrifuged at 2,500g for 10 min at 4°C. The media was discarded and the pellet was re-suspended in an equal volume of ice-cold 10% (%v/v) glycerol. Afterwards, the centrifugation step was repeated and the pellet was re-suspended with ice-cold glycerol (half of the initial volume). Two additional centrifugation steps were performed (resuspension with 10mL glycerol and 1mL glycerol, respectively). The final suspension was divided into Eppendorf's, immediately frozen in liquid nitrogen and stored at -80°C for further use.
50μl of competent bacteria and 50μl of liposomes were mixed and transferred into ice-cold cuvettes. After electroporation by MicroPulser electroporator the mixture was re-suspended in LB media and incubated for 1h at 37°C.
The morphology of Salmonella incubated with different types of liposomes was examined by scanning electron microscopy (SEM, Zeiss Ultra-Plus FEG-SEM, Germany).
Viability assay
The viability of 4T1 cells exposed to various treatments was determined using the methylene blue (MB) survival assay [43]. 104 4T1 cells/well (200 μl) were seeded in 96-well plates in a RPMI 1640 medium, supplemented with 10% heat inactivated Fetal Bovine Serum, 2 mM of L-Glutamine solution, 10 U/mL of penicillin G sodium salt and 0.01 mg/mL of streptomycin sulfate for 24 hours at 37 °C, in a 5% CO2 humid atmosphere. The cells were washed with PBS and then exposed to different treatments: (1) DOXIL (liposomal DOX); (2) Free DOX; (3) bacteria; (4) bacteria loaded with DOXIL (“BADOX”) (5) Control (10% sucrose) (20μl/well). The experiment was performed in hexaplicate. All drug treatments were 2 ug/mL. Following a 24h incubation at 37°C cells were incubated with gentamycin sulfate (20μg/mL) for 20 minutes to kill the bacteria that didn’t enter the cells. Then the cells were washed three times with PBS and fixed with 2.5% glutaraldehyde (Sigma-Aldrich) (50μl/well) for 15min. Fixed cells were rinsed with de-ionized water (DDW), then once with borate buffer (0.1M, pH=8.5), vacuum drained and stained with MB (Merck, Germany) (100μl of 1% w/v solution in 0.1M borate buffer, pH=8.5) for 1h at RT. Stained cells were rinsed thoroughly with DDW to remove non bound dye. The MB bound to the fixed cells was extracted by incubation with 200μl 0.1M HCl for 1h at 37°C (Frutarom, Haifa, Israel). Optical density (OD) of the dye in each well was determined by Tecan infinite 200Pro (Switzerland) at 620nm. The percentage of living cells was calculated with respect to the untreated controls that were processed simultaneously.
To ensure the media is not depleted of nutrients, the media was refreshed every 3 hr.
Results
In this study we tested the ability of bacteria to act as carriers of drug-loaded nanoparticles to cancerous tissue.
Increased bacterial velocity in tumor conditions
In 1924, Otto Heinrich Warburg observed that compared to normal cells, cancer cells rely mostly on aerobic glycolysis, known as the Warburg Effect [44, 45]. Due to this effect, tumor cells are dependent on high glucose concentrations in the tumor microenvironment. High glucose consumption by cancer cells, through aerobic glycolysis pathways, leads to increased lactate production corresponding to an acidic tumor microenvironment [46].
We examined the effect pH and glucose concentration have on Salmonella and E.coli motility, imitating healthy and tumor microenvironments.
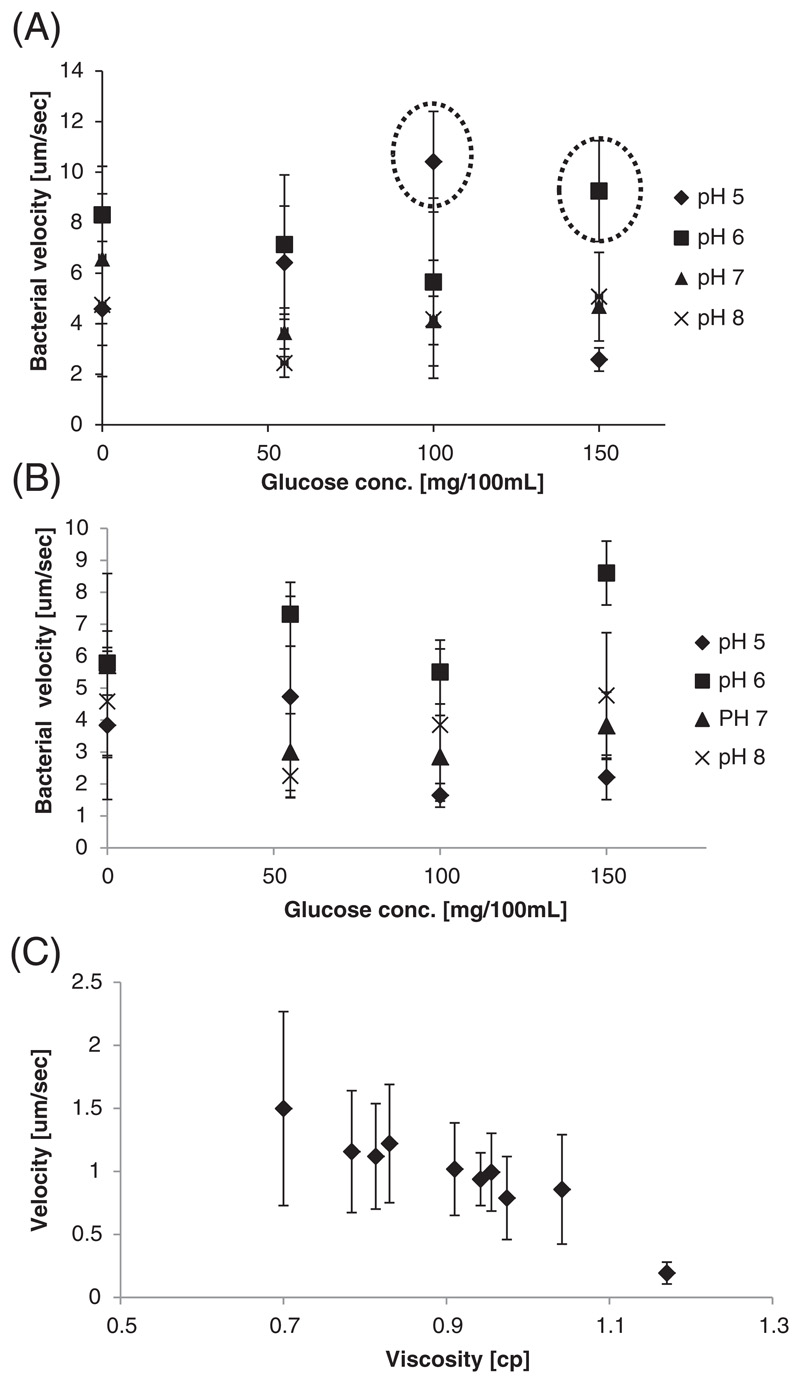
The velocities of both types of bacteria were dependent on the environmental conditions, generally favoring tumoral glucose and pH conditions, Figure 2. At pH 6, the highest bacterial velocity was obtained for all glucose concentrations, maximizing at a glucose concentration of 150 mg/dL (Fig. 2B). At pH 5 there was a decrease in the bacterial velocity. In general, the highest bacterial propulsion velocities were found at pH 6-7, the pH ranges of the tumor microenvironment [47, 48]. For both bacteria types, the higher velocity was obtained in high glucose concentrations, reflecting the abundance of glucose and nutrients and the pH of the tumor microenvironment [27, 28, 35].
Figure 2. Bacterial velocity depends on the glucose concentration, pH and viscosity.
The velocity of (A) Salmonella and (B) E. coli were measured under different glucose concentrations and pH environments. In general, Salmonella were faster swimmers than E. coli, and in both cases the velocity increased in pH and glucose concentrations reported to exist in the tumor microenvironment (emphasized here with the dashed line). (C) Velocity of bacteria was measured in media of increasing viscosity. As the viscosity increased a decline in the bacterial velocity was recorded. Velocity was measured as distance/time, by data analysis software NIS-Elements D 4.10.
The viscosity of tissue varies from organ to organ, and also depends on the disease state [49, 50]. As expected, the bacterial velocity decreased as the viscosity of the environment increased.
In summary, Salmonella was more motile compared to E. coli, therefore we chose to continue our research using this bacterial strain.
Loading liposomes into bacteria
We sought to develop a method for loading liposomes into bacteria, without compromising the bacterial motility. For this, we compared two modes of loading: incubation and electroporation. For incubation, bacteria were incubated with 100-nm PEGylated liposomes over 4 hours. Extremely low levels of liposomal uptake (<5%) were noticed in the bacteria post incubation, Figure 3. We then tested electroporation as an alternative mode of liposomal loading. Electroporation creates transient pores in the bacterial membrane by the application of brief electrical pulses. Using electroporation we detected liposomes in 62% of the bacteria (Fig. 3). The uptake of the liposomes by bacteria was visualized using GFP-Salmonella that were loaded with rhodamine-labeled liposomes.
Figure 3. Loading liposomes into bacteria using incubation and electroporation, reduces bacterial velocity.
(A) Bacteria were incubated with liposomes before and after electroporation. The liposomal uptake after electroporation reached 62% of the bacteria, compared to less than 5% of the bacteria incubated with the liposomes.
(B) Interestingly, a slight decrease in velocity was recorded after loading the bacteria with the nanoparticles.
Liposomes inside Salmonella. (C) A confocal image of Salmonella loaded with doxorubicin-containing liposomes. Salmonella (green fluorescent protein); doxorubicin (red); overlay; and bright-field. (D) Bacteria are loaded by electroporation with 100-nm liposomes. Confocal microscopy images show rhodamine-labeled liposomes (red) inside Salmonella stained with Hoescht (blue).
After determining the optimal loading method, the effect of loading nanoparticles into bacteria was addressed. We compared Salmonella velocity before and after being loaded with liposomes. A slight decrease in the bacterial velocity was measured after loading the bacteria with liposomes (Fig. 3).
Bacterial loading and viability are affected by the surface charge of the nanoparticles
The effect of the liposomal surface charge on the bacterial viability was investigated. Cationic and anionic liposomes were synthesized and loaded into bacteria by electroporation. Bacterial disruption was observed after being loaded with the cationic liposomes, while bacteria loaded with anionic liposomes maintained their integrity (Fig. 4). A viability assay confirmed these observations; compared to the untreated control, bacteria treated with negatively-charged liposomes exhibited reduced growth of 20%, while the cationic liposomes induced a growth reduction of 50% (Fig. 4).
The antibacterial activity of doxorubicin
The antibacterial activity of doxorubicin against Salmonella was examined as a mode for destroying the bacterial carrier after reaching the target site. The rationale of this approach is that the drug loaded into the nanoparticles will kill the bacteria and subsequently the invaded cancer cell.
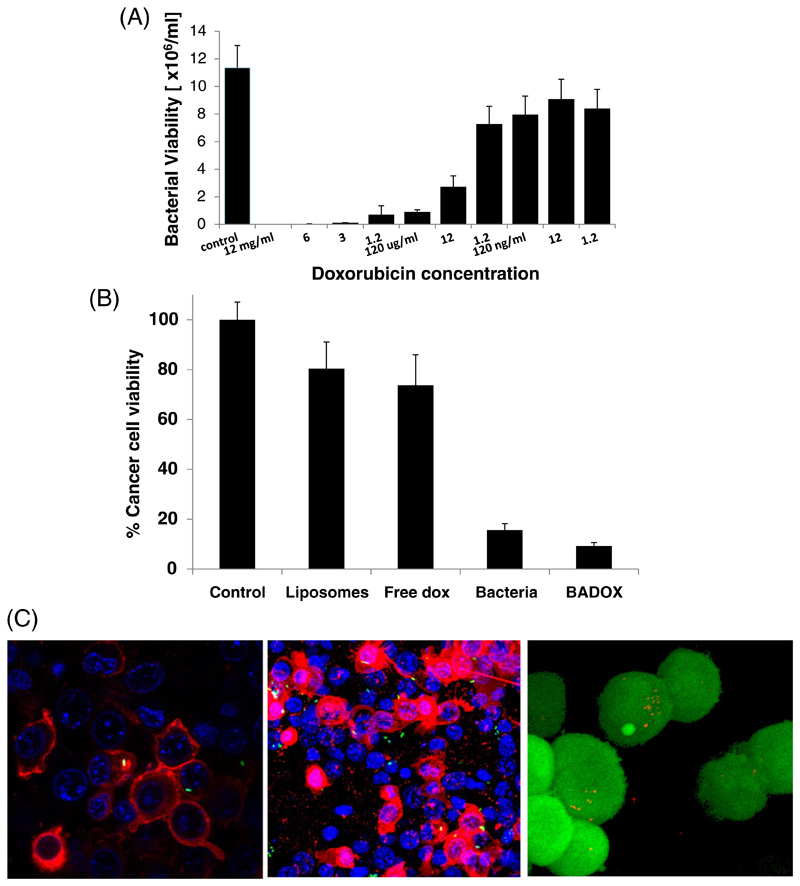
Doxorubicin was added to Salmonella at increasing concentrations and the bacterial viability was determined. A concentration-dependent toxicity of doxorubicin to Salmonella was found (Fig. 5), supporting our approach for using the antibiotic as part of the dual therapeutic mode of action. The IC50 of doxorubicin to the bacteria was estimated to be ~5 μg/mL.
Figure 5. The anti-bacterial and anti-cancer activity of doxorubicin nanoswimmers.
The anti-bacterial activity of doxorubicin was tested on Salmonella (A). Salmonella was found to be sensitive to doxorubicin at increasing concentrations with an IC50 of ~5 ug/mL.
(B) Salmonella were loaded with liposomes that contain doxorubicin. The bacterial nano-swimmers were incubated with triple-negative breast cancer cells (4T1). The viability of viability of the cancer cells, after being incubated for 24 hrs with different treatments, was recorded. The most significant decrease in cell viability was noticed in BADOX (P-value bacteria-BADOX, control-lipo, BADOX-lipo< 0.001).
Salmonella invade cancer cells (C). Confocal microscopy image of bacterial invasion into triple-negative 4T1 cancer cells (left), B16 melanoma cells (middle), HeLa-KB cells (right); nucleus with blue, membrane in red, bacteria are green (GFP) invades the cell; for HeLa bacteria in red and cells in green.
Nanoparticle-loaded bacteria invade and kill cancer cells
We examined the ability of bacteria loaded with liposomes to invade cancer cells. Salmonella was loaded with fluorescently-labeled 100-nm liposomes. The bacteria were added to triple-negative breast cancer cells (4T1) in culture. Three hours later, 30% of the cancer cells were invaded by the nanoparticle-loaded Salmonella (Fig. 5).
After demonstrating the ability of liposome-loaded Salmonella to invade cells, the capacity of Salmonella loaded with liposomes containing doxorubicin ("BADOX") to kill cancer cells was investigated. 4T1 cells were exposed to different treatments: (1) media alone (control) (2) liposomal doxorubicin (3) free doxorubicin (4) bacteria (5) bacteria loaded with liposomal doxorubicin (BADOX). 24 hours later we found that cancer cells treated with BADOX had the lowest viability compared to any other treatment group (Figure 5). However, despite the mathematical significance, the prominent cell death is due to the bacteria itself. To reduce bacterial toxicity, mutant bacterial strains without lipid-A have been described [51].
In summary, our data indicates that Salmonella loaded with nanomedicines can propel across media, and penetrate cancer cells, to induce a therapeutic effect.
Discussion
Autonomous swimmers are an evolving scientific area with many potential medical applications. Our research harnesses the self-propelling abilities of bacteria, loaded with a nanomedicine, to treat cancer. This system is designed to release the anti-cancer drug at the disease site after self-destruction of the swimmer. Specifically, we loaded 100-nm liposomes, that contained doxorubicin, into motile bacteria. The bacteria sense and invade the cancer cells, where the drug is released internally. Due to the dual-activity of the drug, being an antibiotic chemotherapeutic, once released from the liposomes, the doxorubicin destroyed the carrier and killed the invaded cancer cell.
In order to construct the nanoswimmer platform, we chose Salmonella over E. coli, for its higher velocity in the conditions of the tumor microenvironment, and its ability to target tumors and invade triple-negative cancer cells [20–22, 27–36]. We found that the bacterial motility is pH- and glucose-dependent, favoring the tumor conditions [52, 53].
We compared two approaches for loading nanoparticles into the bacteria – incubation and electroporation. Electroporation, which creates transient pores in the bacterial membrane, was more efficient than incubation. Moreover, confocal microscopy indicated that post electroporation the liposomes were loaded into the bacterial body, and not fused to the bacterial external membrane.
The charge of liposome membrane has a major effect on the bacterial viability. Using electron microscopy, we found that cationic liposomes disrupted the bacterial envelope. This anti-bacterial activity of the cationic liposomes (but not of the anionic liposomes) is owed to binding of the negatively-charged bacterial envelope and intra-bacterial nucleic acids to the positively-charged liposomal lipids [54–58].
Doxorubicin, an antibiotic-chemotherapy, has an antibacterial effect against Salmonella. We show that doxorubicin kills bacteria in a concentration-dependent manner. In tumors, doxorubicin is released from the liposomes by a metabolic switch, freeing the drug to perform its therapeutic activity. Specifically, the secretion of ammonia by tumor cells, imbalances the osmotic gradient that is used to captures the doxorubicin inside the liposomes [59, 60]. Once released from the liposomes, doxorubicin destroys the bacterial carrier and kills the invaded cancer cell. This resulted in an enhanced on-site therapeutic activity.
To conclude, this study demonstrates the potential of bacteria-mediated targeted drug delivery as a new approach for cancer treatment. By exploiting natural bacteria characteristics that are attracted to the cancerous tissue, we developed a bacteria-based nanoswimmer that carries liposomal drugs into cancer cells followed by self-destruction of the bacterial vehicle and release of the drugs.
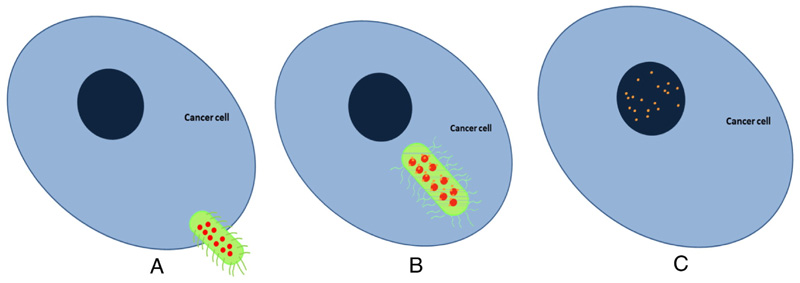
Figure 1. Therapeutic nano-swimmers.
(A) Viable bacteria, with the capacity to autonomously swim in media, are transformed into carriers of anti-cancer agents. The bacteria are loaded with nanoparticles that contain the anti-cancer agent and antibiotic doxorubicin. (B) Cancer cells secret a unique set of nutrients that are strong attractants to the bacterial swimmer. The drug-loaded bacteria swim towards and invade the cancer cell. (C) Inside the cell, the drug releases from the nanoparticle, killing the bacteria and destroying its envelope. Following bacterial death the doxorubicin executes its activity against the invaded cancer cell.
Bacteria are shown in green, liposomes in red and doxorubicin in orange.
References
- [1].Duncan R, Gaspar R. Nanomedicine(s) under the microscope. Mol Pharm. 2011;8:2101–2141. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- [2].Lammers T, Hennink WE, Storm G. Tumour-targeted nanomedicines: principles and practice. Br J Cancer. 2008;99:392–397. doi: 10.1038/sj.bjc.6604483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- [4].Barenholz Y. Doxil(R)--the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- [5].Torchilin VP. Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat Rev Drug Discov. 2014 doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Klibanov AL, Maruyama K, Torchilin VP, Huang L. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990;268:235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- [7].Messager L, Burns JR, Kim J, Cecchin D, Hindley J, Pyne AL, Gaitzsch J, Battaglia G, Howorka S. Biomimetic Hybrid Nanocontainers with Selective Permeability. Angew Chem Int Ed Engl. 2016;55:11106–11109. doi: 10.1002/anie.201604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kievit FM, Zhang M. Surface engineering of iron oxide nanoparticles for targeted cancer therapy. Acc Chem Res. 2011;44:853–862. doi: 10.1021/ar2000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sinha R, Kim GJ, Nie S, Shin DM. Nanotechnology in cancer therapeutics: bioconjugated nanoparticles for drug delivery. Molecular Cancer Therapeutics. 2006;5:1909–1917. doi: 10.1158/1535-7163.MCT-06-0141. [DOI] [PubMed] [Google Scholar]
- [10].Dvirv T, Banghart MR, Timko BP, Langer R, Kohane DS. Photo-Targeted Nanoparticles. Nano Letters. 2009;10:250–254. doi: 10.1021/nl903411s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gao W, Kagan D, Pak OS, Clawson C, Campuzano S, Chuluun-Erdene E, Shipton E, Fullerton EE, Zhang L, Lauga E, Wang J. Cargo-towing fuel-free magnetic nanoswimmers for targeted drug delivery. Small. 2012;8:460–467. doi: 10.1002/smll.201101909. [DOI] [PubMed] [Google Scholar]
- [12].Schamel D, Mark AG, Gibbs JG, Miksch C, Morozov KI, Leshansky AM, Fischer P. Nanopropellers and their actuation in complex viscoelastic media. ACS Nano. 2014;8:8794–8801. doi: 10.1021/nn502360t. [DOI] [PubMed] [Google Scholar]
- [13].Williams BJ, Anand SV, Rajagopalan J, Saif MT. A self-propelled biohybrid swimmer at low Reynolds number. Nature communications. 2014;5 doi: 10.1038/ncomms4081. 3081. [DOI] [PubMed] [Google Scholar]
- [14].Mercado-Lubo R, Zhang Y, Zhao L, Rossi K, Wu X, Zou Y, Castillo A, Leonard J, Bortell R, Greiner DL, Shultz LD, Han G, et al. A Salmonella nanoparticle mimic overcomes multidrug resistance in tumours. Nature communications. 2016;7 doi: 10.1038/ncomms12225. 12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Felfoul O, Mohammadi M, Taherkhani S, de Lanauze D, Zhong Xu Y, Loghin D, Essa S, Jancik S, Houle D, Lafleur M, Gaboury L, et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat Nanotechnol. 2016 doi: 10.1038/nnano.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].MO Din, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, Hasty J. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hess H, Vogel V. Molecular shuttles based on motor proteins: active transport in synthetic environments. Journal of biotechnology. 2001;8:67–85. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- [18].Hiratsuka Y, Miyata M, Tada T, Uyeda TQ. A microrotary motor powered by bacteria. Proc Natl Acad Sci U S A. 2006;103:13618–13623. doi: 10.1073/pnas.0604122103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kojima M, Zhang Z, Nakajima M, Fukuda T. High efficiency motility of bacteria-driven liposome with raft domain binding method. Biomedical microdevices. 2012;14:1027–1032. doi: 10.1007/s10544-012-9711-2. [DOI] [PubMed] [Google Scholar]
- [20].Kasinskas RW, Forbes NS. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnology and bioengineering. 2006;94:710–721. doi: 10.1002/bit.20883. [DOI] [PubMed] [Google Scholar]
- [21].Zhao M, Yang M, Ma H, Li X, Tan X, Li S, Yang Z, Hoffman RM. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647–7652. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- [22].Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67:3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- [23].Tuxhorn J, Daise T, Dentler WL. Regulation of flagellar length in Chlamydomonas. Cell motility and the cytoskeleton. 1998;40:133–146. doi: 10.1002/(SICI)1097-0169(1998)40:2<133::AID-CM3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- [24].Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. Journal of Bacteriology. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pontier-Bres R, Prodon F, Munro P, Rampal P, Lemichez E, Peyron JF, Czerucka D. Modification of Salmonella Typhimurium motility by the probiotic yeast strain Saccharomyces boulardii. PLoS One. 2012;7:e33796. doi: 10.1371/journal.pone.0033796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011;18:457–466. doi: 10.1038/cgt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bettegowda C, Dang LH, Abrams R, Huso DL, Dillehay L, Cheong I, Agrawal N, Borzillary S, McCaffery JM, Watson EL. Overcoming the hypoxic barrier to radiation therapy with anaerobic bacteria. Proceedings of the National Academy of Sciences. 2003;100:15083–15088. doi: 10.1073/pnas.2036598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao M, Yang M, XM Li, Jiang P, Baranov E, Li S, Xu M, Penman S, Hoffman RM. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].King I, Bermudes D, Lin S, Belcourt M, Pike J, Troy K, Le T, Ittensohn M, Mao J, Lang W, Runyan JD, et al. Tumor-targeted Salmonella expressing cytosine deaminase as an anticancer agent. Hum Gene Ther. 2002;13:1225–1233. doi: 10.1089/104303402320139005. [DOI] [PubMed] [Google Scholar]
- [30].Pawelek JM, Low KB, Bermudes D. Bacteria as tumour-targeting vectors. The lancet oncology. 2003;4:548–556. doi: 10.1016/s1470-2045(03)01194-x. [DOI] [PubMed] [Google Scholar]
- [31].Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57:4537–4544. [PubMed] [Google Scholar]
- [32].Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao M, Yang M, Li X-M, Jiang P, Baranov E, Li S, Xu M, Penman S, Hoffman RM. Tumortargeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Griffith TS, Brunner T, Fletcher SM, Green DR, Ferguson TA. Fas ligand-induced apoptosis as a mechanism of immune privilege. Science. 1995;270:1189–1192. doi: 10.1126/science.270.5239.1189. [DOI] [PubMed] [Google Scholar]
- [36].Berg HC, Turner L. Movement of microorganisms in viscous environments. Nature. 1979;278:349–351. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- [37].Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, Freezer LJ, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jia L-J, Wei D-P, Sun Q-M, Huang Y, Wu Q, Hua Z-C. Oral delivery of tumor-targeting Salmonella for cancer therapy in murine tumor models. Cancer Science. 2007;98:1107–1112. doi: 10.1111/j.1349-7006.2007.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen G, Wei D-P, Jia L-J, Tang B, Shu L, Zhang K, Xu Y, Gao J, Huang X-F, Jiang W-H, Hu Q-G, et al. Oral delivery of tumor-targeting Salmonella exhibits promising therapeutic efficacy and low toxicity. Cancer Science. 2009;100:2437–2443. doi: 10.1111/j.1349-7006.2009.01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bermudes D, Low B, Pawelek J. Tumor-targeted Salmonella. Highly selective delivery vectors. Advances in experimental medicine and biology. 2000;465:57–63. doi: 10.1007/0-306-46817-4_6. [DOI] [PubMed] [Google Scholar]
- [41].Velge P, Wiedemann A, Rosselin M, Abed N, Boumart Z, Chaussé AM, Grépinet O, Namdari F, Roche SM, Rossignol A, Virlogeux-Payant I. Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis. MicrobiologyOpen. 2012;1:243–258. doi: 10.1002/mbo3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schroeder A, Avnir Y, Weisman S, Najajreh Y, Gabizon A, Talmon Y, Kost J, Barenholz Y. Controlling liposomal drug release with low frequency ultrasound: mechanism and feasibility. Langmuir. 2007;23:4019–4025. doi: 10.1021/la0631668. [DOI] [PubMed] [Google Scholar]
- [43].Schroeder A, Honen R, Turjeman K, Gabizon A, Kost J, Barenholz Y. Ultrasound triggered release of cisplatin from liposomes in murine tumors. J Control Release. 2009;137:63–68. doi: 10.1016/j.jconrel.2009.03.007. [DOI] [PubMed] [Google Scholar]
- [44].Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochemische Zeitschrift. 1924;152:319–344. [Google Scholar]
- [46].Alfarouk KO, Verduzco D, Rauch C, Muddathir AK, Adil HH, Elhassan GO, Ibrahim ME, David Polo Orozco J, Cardone RA, Reshkin SJ, Harguindey S. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience. 2014;1:777–802. doi: 10.18632/oncoscience.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- [48].Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- [49].Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- [50].Lekka M, Laidler P, Gil D, Lekki J, Stachura Z, Hrynkiewicz AZ. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur Biophys J. 1999;28:312–316. doi: 10.1007/s002490050213. [DOI] [PubMed] [Google Scholar]
- [51].Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J, Lin SL, et al. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17:37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- [52].Demir M, Salman H. Bacterial thermotaxis by speed modulation. Biophys J. 2012;103:1683–1690. doi: 10.1016/j.bpj.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Toley BJ, Forbes NS. Motility is critical for effective distribution and accumulation of bacteria in tumor tissue. Integrative biology : quantitative biosciences from nano to macro. 2012;4:165–176. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Miteva M, Andersson M, Karshikoff A, Otting G. Molecular electroporation: a unifying concept for the description of membrane pore formation by antibacterial peptides, exemplified with NK-lysin. FEBS letters. 1999;462:155–158. doi: 10.1016/s0014-5793(99)01520-3. [DOI] [PubMed] [Google Scholar]
- [55].Nicolas P. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS journal. 2009;276:6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x. [DOI] [PubMed] [Google Scholar]
- [56].Papo N, Shai Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides. 2003;24:1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- [57].Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends in Biotechnology. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- [58].Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Silverman L, Barenholz Y. In vitro experiments showing enhanced release of doxorubicin from Doxil(R) in the presence of ammonia may explain drug release at tumor site. Nanomedicine. 2015;11:1841–1850. doi: 10.1016/j.nano.2015.06.007. [DOI] [PubMed] [Google Scholar]
- [60].Haran G, Cohen V, Bar LK, Barenholz Y. Transmembrane ammonium sulfate gradients in liposomes produce efficient and stable entrapment of amphipathic weak bases. Biochim Biophys Acta, Biomem. 1993;1151:201. doi: 10.1016/0005-2736(93)90105-9. [DOI] [PubMed] [Google Scholar]