Abstract
It is increasingly appreciated that drug response to different cancers driven by the same oncogene is different and may relate to differences in re-wiring of signal transduction. We aimed to study differences in dynamic signaling changes within mutant KRAS (KRASMT), non-small cell lung cancer (NSCLC), colorectal cancer (CRC) and pancreatic adenocarcinoma (PDAC) cells. We used an antibody-based phosphoproteomic platform to study changes in 50 phosphoproteins caused by seven targeted anticancer drugs in a panel of 30 KRASMT cell lines and cancer cells isolated from 10 patients with KRASMT cancers. We report for the first time significant differences in dynamic signaling between CRC and NSCLC cell lines exposed to clinically relevant equimolar concentrations of the pan-PI3K inhibitor pictilisib including a lack of reduction of p-AKTser473 in CRC cell lines (P = 0.037) and lack of compensatory increase in p-MEK in NSCLC cell lines (P = 0.036). Differences in re-wiring of signal transduction between tumor types driven by KRASMT cancers exist and influence response to combination therapy using targeted agents.
Keywords: Context specificity, KRAS, Signal transduction, PI3K pathway, Clinical trial design
Introduction
There are multiple examples of targeted anticancer drugs that are clinically effective in targeting different cancers driven by the same oncogene including the recent report of the activity of TRK inhibitors across a wide range of tumors driven by TRK fusions (1). However, there is emerging evidence related to context specificity where differences in signaling in different tumor types driven by the same oncogene can result in disparate clinical outcomes; for example, the BRAF inhibitor, vemurafenib, causes clinical responses in patients with V600E mutant BRAF-driven melanoma (2) but not colorectal cancer (3). This has been attributed to differences in EGFR signaling between the two tumor types (4).
Mutations in the oncogene KRAS are seen across a wide range of solid tumors, such as pancreatic cancer (97%), colon cancer (40%) and non-small cell lung cancer (30%). Multiple treatment approaches to target have been proposed including post-targeting, post-translational modification (farnesyl transferase inhibitors), combinatorial inhibition of down-stream pathways (5) or directly targeting the mutated KRAS protein (6).
There have been detailed studies of down-stream signaling patterns of KRAS-mutated (KRASMT) mutated non-small cell lung (NSCLC), pancreatic ductal carcinoma (PDAC) and colorectal cancer (CRC) to define patterns of signaling, feedback loops and optimal combination therapy (7–10). In the light of emerging evidence of context specificity of drug response in cancer (3,4) and the reports that patients with KRASMT NSCLC and CRC respond differentially when treated with combinations of MEK and PI3K pathway inhibitors (11,12) we aimed to study differences in dynamic signaling patterns in three diseases with frequent KRAS mutations ie NSCLC, PDAC and CRC.
We chose to study differences in signaling patterns in 30 KRASMT cell lines (10 NSCLC, 10 CRC and 10 PDAC) and 10 KRASMT cells isolated from patients with malignant effusions. It is known that differences in the type of KRAS mutations between these tumor types do occur, for example, G12C mutations occur more frequently in KRAS mutated NSCLC compared to CRC (5). The different KRAS mutations in our cell line panel are listed in Supplementary Table S1. An antibody-based platform was used to screen changes in 50 phosphoproteins which are relevant to KRAS signaling and are related to targets of the drugs used as probes (Figure 1). We exposed the cell lines to clinically relevant concentrations of 7 targeted anticancer drugs: AZD5363 (13) (AKT inhibitor), everolimus (m-TOR inhibitor), gefitinib (EGFR inhibitor), luminespib/NVP-AUY922 (14) (HSP90 inhibitor), pictilisib/GDC-0941 (15) (PI3K inhibitor), trametinib (MEK inhibitor), vemurafenib (BRAF inhibitor). We chose these drugs as tools as they were known to inhibit signaling nodes related to KRAS signaling and/or where KRAS mutations were known to effect sensitivity to the drug. Further, all these drugs had been used in the clinic and it was possible to use concentrations of the drugs that were clinically relevant. We used equimolar drug concentrations across cell lines and patient samples rather than individual GI50 concentrations across all samples as we were focussed on using clinically relevant concentrations and it was not possible to determine GI50 of drugs in cells isolated from ascites of patients as we did not establish cell lines from these samples. We planned to validate any interesting findings in more detailed experiments.
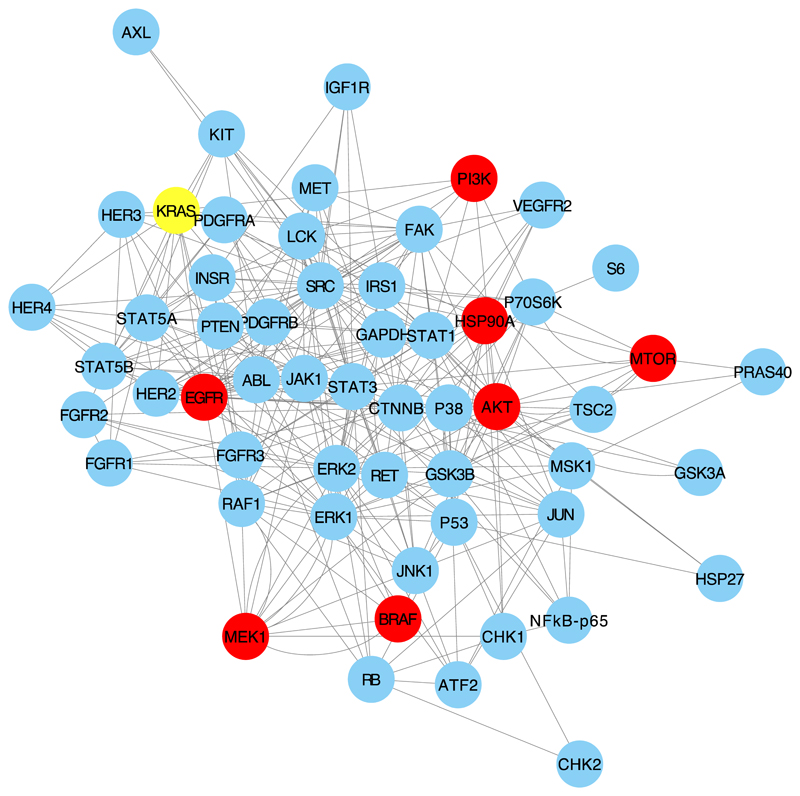
Figure 1.
The figure shows the network of interactions between the phosphoproteins studied in this project and the targets of the drugs used and KRAS. Targets of the drugs are shown as red nodes.
Materials and Methods
Cell lines, tissue culture and drugs
All cell lines were obtained from ATCC (LGC Standards, Teddington, UK), Public Health England (Salisbury, UK), Sigma-Aldrich (St. Louis, MO, USA) or from The Francis Crick Institute’s Cell Services (London, UK). All drugs were sourced from Selleck Chemicals (Stratech, Cambridge UK). Details of cell lines, culture media and concentrations of drugs used are available in the Supplementary Data.
Baseline mutations and m-RNA expression
Data related to whole exome sequencing baseline mRNA for 16,831 genes were extracted from the Cancer Cell Line Encyclopaedia (CCLE) database. Data related to mutations and baseline mRNA expression in 26 and 28 out of 30 cell lines, respectively (mutational data for COLO678, LIM-2099, LIM-1899 and PANC-1 cell lines were not present in the CCLE database, and mRNA data were unavailable for LIM-1899 and LIM-2099). The data file 'CCLE_Expression_Entrez_2012-09-29.gct' and annotation file ‘CCLE_Expression.Arrays.sif_2012-10-18.txt’ were downloaded from the CCLE website. The data file was filtered in R Studio to only the cell lines used in this project, and then the values for each gene were median centered using the base R package. This data file was then loaded into the Broad Institute’s GENE-E software (version 3.0.204) which was used to create Figure 2 using default settings. Pearson’s correlation between global mRNA expression is indicated by a blue-white-red color scale, normalized to the minimum correlation between cell lines seen (0.8367) and a hypothetical perfect correlation of 1.
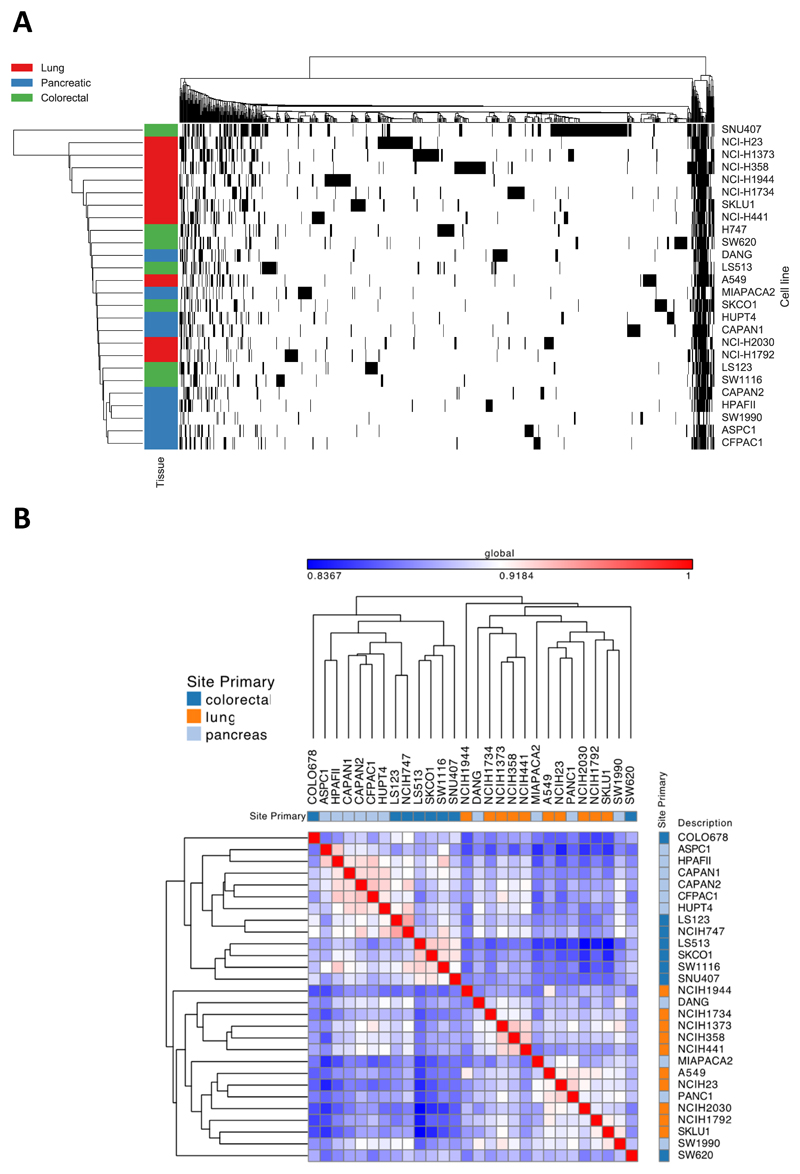
Figure 2.
Baseline characteristics of cell line panel
A) Heat map of mutations in the cell line panel, using agglomerative average linkage clustering with hamming distance. Black = a mutated gene. Cell lines are labelled on the left with their tissue type. B) GENE-E clustered heat map of global Pearson correlations between basal mRNA, with tissue type annotation. The minimal global Pearson correlation between cell lines is 0.8347, reflecting similarities in expression of the majority of the 16,381 genes analyzed, for example, due to house-keeping gene expression. Pearson correlations are indicated by a blue-white-red color scale normalized to this minimum correlation of 0.8347 and maximized to a perfect correlation of 1. Overall, the three tissue types are mixed throughout the correlation matrix: the cell lines do not cluster together with other cell lines of the same tissue in the dendrogram. The highest correlations between mRNA profiles (denoted by a pink-red color in the heat map) are seen between the following cell lines: ASPC1, HPAFII, CAPAN1, CAPAN2, CFPAC1 and HUPT4, all of which are pancreatic. However, the remaining pancreatic cell lines are scattered throughout the dendrogram and have a comparatively low correlation with this cluster of cell lines.
Isolation of cancer cells from CRC and NSCLC serous effusions
Up to 1000 ml of ascites or pleural fluid was collected from the patient and immunomagnetically-separated using previously published methods (16). Pleural and ascitic fluid were used in the study after the investigators had obtained written, informed consent. The tissue collection protocols were approved by the institutional review board and conducted in accordance with the Declaration of Helsinki.
Quantification of phosphoproteins: Luminex magnetic bead suspension array
MILLIPLEX MAP Akt/mTOR phosphoprotein kit, MILLIPLEX MAPK/SAPK signaling kit, MILLIPLEX MAP RTK phosphoprotein kit (48-611MAG, 48-660MAG, HPRTKMAG-01K respectively, MerckMillipore, Billerica, MA, USA) were combined with the following singleplex magnetic bead sets to produce three multiplex Luminex assays: phospho-NFkB, phospho-SRC, phospho-STAT3, phospho-STAT5 A/B, total HSP27 and GAPDH (46-702MAG, 46-710MAG, 46-623MAG, 46-641MAG, 46-608MAG, 46-667MAG, MerckMillipore). Bio-Plex Pro phospho-PDGFRa, phospho-PDGFRb and Akt (Thr308) (171-V50017M, 171-V50018M, 171-V50002, Bio-Rad, Watford, Herts, UK) were combined into a triplex assay. Ten ug of protein was loaded per well and manufacturers’ protocols were followed throughout.
Additionally, an in-house multiplex Luminex assay was created utilizing a range of antibodies from Cell Signaling Technology (Danvers, MA, USA), targeting proteins of interest. These were conjugated to Luminex MagPlex Microspheres (MC100XX-01, Luminex, Austin TX, USA) via an xMAP Antibody Coupling Kit (40-50016, Luminex). A second set of antibodies, targeting phosphorylated versions of the antibodies from the first set, were biotinylated (Biotin Type A conjugation kit, ab102865, Abcam, Cambridge, UK). This home-grown assay followed the Millipore protocol; however, during the optimization process it was found that small amounts of unbound biotin in the secondary antibody mix were binding directly to protein, increasing background. To offset this as much as possible the secondary antibody was added directly post-primary incubation and unbound protein was only washed off post-secondary antibody incubation.
Phosphoprotein levels were measured on the Luminex 200 system utilising xPONENT v3.1 software.
We attempted to quality control our Luminex platform. Three test cell lines (A2780, HT29 and NCI-H520) were run in triplicate and each repetition was run across five separate 96-well plates. All plates were run in a single sitting to avoid inter-daily fluctuations. Baseline phospho-protein levels were measured and the coefficient of variation (CV) per analyte was calculated. There was considerable variation in the assays with the CV being >30% in at least one of the three cell lines in 12 of the 55 analytes tested. The CV of phosphoproteins of interest such as p-MEK and p-AKT was <10% and <20%, respectively, in all three cell lines tested during the validation (Supplementary Table S2).
Each Millipore Mapmate kit provided positive and negative lysate controls for each analyte (as listed in the manufacturer’s manual). These were all run once at the start of the project. A protein concentration titration was also carried out to assess the appropriate amount of protein to use in each assay. Positive controls for the home-grown bead set were found via a literature search and were used in the optimization process.
The assay platform continues to be in development and would not meet the standards required for clinical decision-making. However, as samples of each experiment including untreated controls and samples treated with drugs were analyzed in the same run in order to reduce chances of variability affecting results, we run the untreated control sample for each cell line in each individual experiment thrice and calculated a mean and standard deviation for each phosphoprotein. Following GAPDH normalization only values for each phosphoprotein that were two standard deviations above or below the corresponding untreated control were considered to be significant. Normalizing the values of each phosphoprotein to its total protein in addition to normalization by loading 10 ug protein per well and normalizing phosphoprotein values to GAPDH would improve the quality and validity of the results. This was not done as it would require double the quantity of protein needed to analyse patient samples which we did not have. Other factors taken into consideration for not measuring total protein and normalizing all phosphoproteins to individual total proteins is that this would very significantly increase the cost and complexity of the screening assay.
Interpretation of phosphoproteomic data
All phosphoproteomic data were normalized to GAPDH. Importantly, for each cell line, three samples of control and one sample for each drug treatment was set up. A standard deviation was calculated for each control and if the drug treated sample had a value more than 2 standard deviations above it was classified as ‘increased’ and if 2 standard deviations below it was classified as ‘decreased’. If it was within 2 standard deviations above or below the control it was considered unchanged. We chose to use dichotomous, increased, decreased or no change outputs as we had not validated the linearity of the absolute changes in phosphoprotein in our assays. We ran only controls and did not treat samples in triplicate because of the cost but repeated specific experiments in triplicate to validate any interesting changes seen.
For the purpose of analysis by logistic regression, only individual analytes which were considered to be significantly increased or decreased (2 standard deviations above or below the mean control) compared to control were used. These phosphorylation changes in cell lines from one tumor type were compared to the other two groups e.g. NSLC Vs CRC and PDAC, CRC Vs NSCLC and PDAC, PDAC Vs NSCLC and CRC using the generalized linear models function in R to compute logistic regression (RStudio, V1.1.383, RStudio,Inc). False discovery rate was corrected for via the Benjamini-Hochberg (BH) procedure (RStudio) and only changes that were significant after BH correction are mentioned in Figure 3.
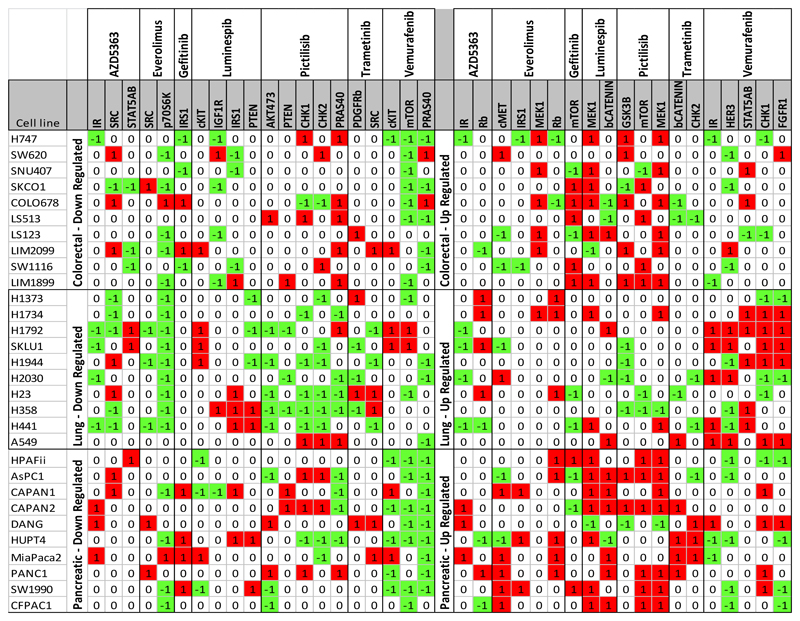
Figure 3.
Results of phosphoproteomic screen
Changes in phosphorylation in cell lines exposed to targeted anticancer drugs. For each analyte, phosphorylation changes had to be first separated out into ‘up regulated vs not’ and ‘down-regulated vs not’ giving binary ‘1/0’ categories. Logistic regression compared the number of ‘1s’ in one tumor type to the ‘1s’ in the other two tumor types combined, repeated for each tumor/drug/analyte configuration and was corrected for multiple testing. The color green and symbol -1 denotes that the levels of the phosphoprotein were 2 standard deviations below the control; the color red and symbol 1 denote levels of phosphoprotein that were 2 standard deviations above control and cells with no color and symbol 0 denote values between 2 standard deviations above and below control. Only changes that were significantly different between any of the three tumor types by logistic regression have been depicted.
Quantification of p-AKT and AKT by ELISA
Cells were lysed and phospho/Total AKT levels were measured using a phospho (ser473)/Total AKT whole cell lysate kit (Meso Scale Discovery, Rockville, MD, USA, K15100D-1) following manufacturers guidelines.
Cytotoxicity and combination studies
Growth inhibition was assessed via standard 72-hour sulforhodamine B assays by previously described methods (17).
Results
Differences between mutations and baseline m-RNA signatures of cell lines
Data related to mutations and baseline m-RNA signatures were available in 26 and 28 of the 30 cell lines, respectively. Unbiased hierarchical clustering did not reveal that KRAS-mutant cell lines of different tumor types of origin significantly clustered together based on either mutation, Figure 2A or in m–RNA expression, Figure 2B. The data has also been fitted to each tumor type/tissue type and represented in Supplementary Figure 1 and Supplementary Figure 2.
Phosphoproteomic screen
Interestingly, p-MEK levels increased in more than 50% of cell lines in cells exposed to the PI3K inhibitor, pictilisib, AKT inhibitor, AZD5363, EGFR inhibitor, gefitinib, BRAF inhibitor, vemurafenib, and HSP90 inhibitor, luminespib. p-ERK levels were increased in more than 50% of cell lines upon exposure to the BRAF inhibitor, vemurafenib, AKT inhibitor, AZD5363, and EGFR inhibitor, gefitinib (Supplementary Table S3). These results suggest increased activation of MEK is a relatively common re-wiring event across multiple different drugs in the cell line panel tested.
There were similarities between changes caused by drugs targeting a defined signaling pathway, for example, more than 50% of all the 30 cell lines showed reduction of p-S6 when exposed to pictilisib, AZD5363 and everolimus all targeting the PI3K pathway. There were differences in phosphorylation of proteins in drugs targeting the PI3K pathway, for example, p-AKT levels were increased in more than 50% of cell lines exposed to AZD5353 but not pictilisib or everolimus. The increase in phosphorylation of AKT despite inhibition of the target is known to be due to change in conformation of the drug target.
We then tested to see if changes in each analyte were different in the three different tumor types when exposed to an individual drug using logistic regression and correcting for multiple testing (Figure 3). A total of 39 changes in phosphorylation was considered significantly different across different tumor types (Table 1).
Table 1. Significant differences in phosphoprotein changes between tumor types.
| Drug (target) | NSCLC vs CRC + PDAC | CRC vs NSCLC + PDAC | PDAC vs NSCLC + CRC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Increased | Not increased | Decreased | Not decreased | Increased | Not increased | Decreased | Not decreased | Increased | Not increased | Decreased | Not decreased | |
| AZD5363 (AKT) | RB | IR SRC |
STAT5 | IR | ||||||||
| Everolimus (m-TOR) | SRC S6K |
MEK | RB | C-MET IRS1 | ||||||||
| Gefitinib (EGFR) | m-TOR | IRS1 | ||||||||||
| Luminespib (HSP90) | MEK | PTEN | IGF1R IRS1 |
B-Catenin | C-KIT | |||||||
| Pictilisib (PI3K) | m-TOR MEK |
CHK1 CHK2 PTEN |
GSK3B | PRAS40 AKT |
||||||||
| Trametinib (MEK) | PDGFRB SRC |
B-Catenin CHK2 |
||||||||||
| Vemurafenib (KRAS) | FGFR1 HER3 IR STAT5 |
m-TOR | CHK1 | PRAS40 C-KIT |
||||||||
Changes in phosphorylation of proteins that were increased or decreased upon exposure to different drugs but were significantly different from cells derived from different tumor types i.e. NSCLC, CRC and PDAC upon logistic regression corrected for multiple testing.
Interestingly, a significantly lower number of CRC cell lines showed a reduction in phosphorylation of AKTSer473 when exposed to pictilisib at a clinically relevant, equimolar concentration compared to NSCLC and PDAC cell lines (P = 0.037). Conversely, p- AKTSer473 levels were reduced in NSCLC cell lines exposed to pictilisib compared to PDAC and CRC, although this was not statistically significant. Further, when exposed to pictilisib, significantly fewer NSCLC cell lines showed an increase in phosphorylation of MEK compared to CRC and PDAC cell lines (P = 0.036). We went on to further validate the findings of differential phosphoprotein changes between NSCLC, CRC and PDAC cell lines when exposed to pictilisib.
We analyzed differences between phosphoprotein changes caused by all seven drugs by unbiased clustering and there were no significant differences between types of KRAS mutation (Supplementary Note).
Validation of findings related to dynamic signaling changes of the pan-PI3K inhibitor, pictilisib
We chose 9 cell lines (3 each of CRC, PDAC and NSCLC) from the original panel to validate our results, exposing them for both 1 hr. We conducted the experiments in triplicate allowing more detailed assessment of fold change compared to threshold based binary representations used for logistic regression in the initial 1hr screen. The patterns of non-reduction of p-AKT in CRC cell lines and no increase in p-MEK levels in the NSCLC cell lines were recapitulated at 1 hr of exposure to pictilisib and technically validated our findings in the larger screen (Figure 4A). We also used a further assay (ELISA) to quantify p-AKTSer473 and total AKT at the 1 hr time-point and the CRC cell lines exposed to pictilisib showed a lesser reduction of p-AKTSer473 normalized to total AKT compared to NSCLC cell lines (Supplementary Figure 3).
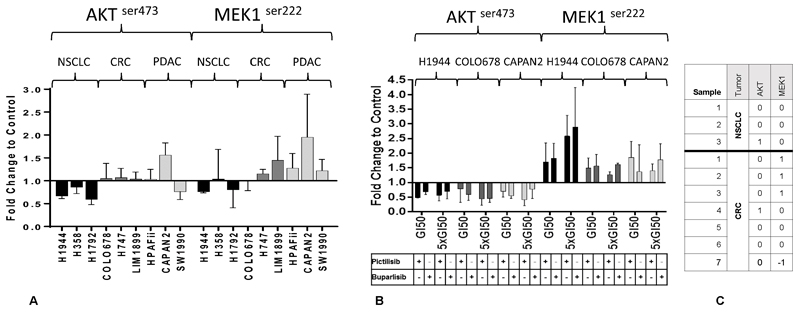
Figure 4.
Validation of findings in phosphoproteomic screen
A) Changes in phosphorylation caused by the pan-PI3K inhibitor, pictilisib. Nine cell lines were exposed to pictilisib in 3 separate experiments and the phosphorylation of p-MEK and p-AKT was measured to confirm findings in the initial screen. B) Changes in phosphorylation caused by equitoxic concentrations of PI3K inhibitors pictilisib and buparlisib for 1 hr at GI50 and x 5 GI50 concentration for 1 hr. The phosphoprotein changes caused by both inhibitors are concordant. C) Changes in phosphorylation in 10 samples of cancer cells isolated from patients with KRASMT cancers exposed to pictilisib. 1, -1 indicate changes more than 2 standard deviations above or below control, respectively and 0 indicates changes between 2 standard deviations above or below the control. None of the NSCLC samples showed a significant increase in p-MEK while 3/7 CRC samples did. Significant reductions in p-AKT levels were not seen in NSCLC or CRC samples. The concentrations of drugs used for the cell lines are detailed in the Supplementary Data. The patient-derived cell lines were exposed to a concentration of pictilisib of 96.3 nM.
The histograms in A and B represent means and the error bars represent standard deviation.
We also determined the GI50 of pictilisib and another pan-PI3K inhibitor, buparlisib, exposing the 9 cell lines to increasing concentrations of pictilisib and buparlisib for 1 hr to study changes in p-AKT and p-MEK. Changes in phosphorylation caused by pictilisib were concordant with that of buparlisib, suggesting that phosphoprotein changes were not due to the off-target effects of pictilisib (Figure 4B). It was, however, noted that p-MEK levels increased in the NSCLC cell line N1944 in Figure 4B as increasing while this was not the case in Figure 3 and Figure 4A. The concentration of pictilisib used in experiments 3 and 4A was 96.3 nM, while concentrations of pictilisib used in Figure 4B were GI50 and x5 GI50, which was considerably higher (Supplementary Table 4) and this may account for this difference.
We studied p-MEK and p-AKT levels in cancer cells isolated from serous effusions (pleural effusions and ascites) in 10 patients with KRAS mutant cancers (3 NSCLC and 7 CRC) exposed to pictilisib for 1 hr as we had done in the 30 cell line screen. We found the increase in the p-MEK levels was significantly raised in 3/7 samples of cells isolated from patients with CRC as opposed to 0/3 samples of cells isolated form NSCLC. No significant reduction of p-AKT was seen in either CRC or NSCLC samples (Figure 4C). The number of samples were not sufficient for statistical analysis however the experiment provided proof of concept that it is possible to conduct ex-vivo assays on cancer cells freshly isolated from cancer patients.
Discussion
To our knowledge, this is the first report to study the differences in dynamic signaling patterns in KRASMT cell lines in an attempt to study context specificity of tumors of origin.
Antibody-based phosphoprotein platforms have been used to study signaling output in cancer cells exposed to drugs, with reverse-phase protein arrays being one of the more established platforms (18). We used the antibody-bead based Luminex platform (19). The drugs used as probes were anticancer drugs that were either licensed (everolimus, gefitinib, trametinib, vemurafenib) or in phase II studies (AZD5363, luminespib and pictilisib). We chose to use clinically relevant concentrations of these drugs at human Cmax and exposed the cell lines and patient samples to these concentrations for 1 hr. Most phosphorylation changes occur early and so we chose a 1-hr time-point to reflect this, as well as the fact that concentrations surrounding Cmax could be maintained for 1 hr. Our aim was to study differences in signal transduction at early time-points. We acknowledge that rewiring of signal transduction may change at later time-points which we did not study. Further, we attempted to standardize the treatment conditions by exposing cells to drugs at 80% confluence. The cells were exposed to drugs for 1 hour so the drugs are unlikely to cause major differences in cell density due to cell death or growth inhibition, however it was not possible to rule out the effects of contact inhibition on signaling in individual cell lines. We did not establish cell lines form each patient sample and cells derived from ascites or pleural effusions were EpCam separated and exposed to drugs for 1 hr in suspension. It was not possible to ascertain the contribution of differences in tissue culture conditions i.e adherent vs suspension in the results while comparing cell lines and patient samples. The importance of PI3K signaling in KRASMT CRC, NSCLC and PDAC has been described previously (20). Co-existence of activating PIK3CA mutations with KRAS mutations in the general population of NSCLC, CRC and PDAC is rare (21,22). However, the co-existence of PIK3CA mutations can significantly affect signaling output drug resistivity to signal transduction inhibitors and we were careful while choosing our panel of cell lines to exclude cell lines with known activating mutations in PIK3CA.
KRAS mutations are found in approximately 90% of PDAC, 40% of CRC and 30% of adenocarcinoma subtype of NSCLC(5). Within NSCLC, a glycine to cysteine substitution is more common (5). Interestingly, within NSCLC there are differences in outcomes for patients depending on the type of KRAS mutation. While evaluating differences in KRAS mutations in NSCLC, the investigators found cell lines with KRASGly12Cys and KRASGly12Val had decreased growth factor-dependent AKT activation suggesting that there are differences in signaling patterns within different KRASMT cell lines (24). Further, analysis of known mutations (not exclusively KRAS) of the cell lines and baseline m-RNA expression available in public databases did not reveal the cell lines clustered significantly to tissue of origin, highlighting the importance of studying post-transcriptional differences such as re-wiring of signal transduction.
Importantly, we found that in KRASMT CRC cell lines, exposure to the pan-PI3K inhibitor, pictilisib had a compensatory increase in p-MEK levels as well as differentiated themselves by not having a reduction in p-AKT levels. In contrast NSCLC cell lines did not have a compensatory increase in p-MEK levels. This differential phosphorylation of MEK by PI3K inhibitors in KRASMT cell NSCLC and CRC cell lines has not been described previously.
We went on to validate specific changes in cells seen in response to pictilisib by repeat experiments in triplicate. We also confirmed that these changes in p-MEK and p-AKT occurred when a second pan-PI3K inhibitor was used, which suggested that the phosphoprotein changes are not due to the off-target effects of pictilisib. Interestingly, in cancer cells isolated from patients with KRASMT, NSCLC did not show activation in p-MEK, while some of the samples isolated from patients with KRASMT CRC did. The number of samples studied did not allow robust statistical analysis however shows proof of concept that such tissue can be used to study changes in phosphoproteins.
The context specificity of oncogenes to induce cancer has been described (25). In the setting of KRASMT cancers, there is increasing evidence that tissue of origin also dictates metabolism of branched chain amino acids (26). The signaling patterns within cancers cells are influenced by surrounding stromal cells as shown in KRAS mutant cancer cells such PDAC (27). It is possible that stromal differences in diverse organs lead to context specificity of KRAS mutated cancers. The findings in the current study of signaling in cancer cells did not take stromal influences into consideration and could be considered as a weakness in the experimental design. However, our findings are of particular significance as they suggest inherent differences in signaling output in KRAS mutant cancer cells sans stroma and may in part be responsible for context specificity of response to signaling inhibitors seen in clinical trials. Our study was confined to cell lines that had known KRAS mutations. However, it was not possible to answer the interesting question relating to context-specificity of signaling extending to non-KRAS mutant cell lines.
Context specificity is a critical facet that influences the precision medicine paradigm. There is already evidence that this is clinically important to drug response, as has been exemplified by BRAF inhibitors (3,4). Our findings suggest there are inherent differences in dynamic signaling output within KRASMT cancer cells, which could influence drug response and should be taken into consideration while designing clinical trials. Finding new treatment paradigms for KRASMT cancers remains an urgent and unmet need.
Supplementary Material
Financial Support
The authors acknowledge institutional funding from Cancer Research UK as part of the Experimental Cancer Medicine Centre initiative [Ref: C12540/A25128]; a Cancer Therapeutics Unit award [Ref: C2739/A22897] and a Cancer Therapeutics Centre award [Ref: C309/A25144]. The authors also acknowledge the National Institute for Health Research (NIHR) Biomedical Centre initiative awarded to The Institute of Cancer Research and The Royal Marsden Hospital NHS Foundation Trust [Ref: IS-BRC-1215-20021]. U Banerji is a recipient of an NIHR Research Professorship award [Ref: RP-2016-07-028].
Disclosures
Three of the seven drugs used in the screen (luminespib, pictilisib and AZD5363) were discovered in collaborations with The Institute of Cancer Research. Elizabeth A Coker, A Stewart, S Pölsterl, A Georgiou, AR Minchom, S Carreira, FI Raynaud, JS de Bono, B Al-Lazikani and U Banerji are employees of The Institute of Cancer Research. None is named on the patent or receives royalties related to the development of these drugs.
Footnotes
Previous presentation of data: This work has been presented, in part, as an oral presentation at the TAT Congress, Paris, March 2018 [Abstract #430] and as an oral presentation at the AACR Annual Meeting, Washington DC, April 2017 [Abstract #996].
References
- 1.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–9. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373:726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 5.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, et al. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep. 2014;8:1475–83. doi: 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 10.Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–51. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res. 2015;21:730–8. doi: 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 12.Tolcher AW, Khan K, Ong M, Banerji U, Papadimitrakopoulou V, Gandara DR, et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin Cancer Res. 2015;21:739–48. doi: 10.1158/1078-0432.CCR-14-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies BR, Greenwood H, Dudley P, Crafter C, Yu DH, Zhang J, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther. 2012;11:873–87. doi: 10.1158/1535-7163.MCT-11-0824-T. [DOI] [PubMed] [Google Scholar]
- 14.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, et al. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–60. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 15.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–38. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carden CP, Stewart A, Thavasu P, Kipps E, Pope L, Crespo M, et al. The association of PI3 kinase signaling and chemoresistance in advanced ovarian cancer. Mol Cancer Ther. 2012;11:1609–17. doi: 10.1158/1535-7163.MCT-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerji U, Walton M, Raynaud F, Grimshaw R, Kelland L, Valenti M, et al. Pharmacokinetic-pharmacodynamic relationships for the heat shock protein 90 molecular chaperone inhibitor 17-allylamino, 17-demethoxygeldanamycin in human ovarian cancer xenograft models. Clin Cancer Res. 2005;11:7023–32. doi: 10.1158/1078-0432.CCR-05-0518. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Ling S, Hegde AM, Byers LA, Coombes K, Mills GB, et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol. 2016;43:476–83. doi: 10.1053/j.seminoncol.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart A, Banerji U. Utilizing the Luminex Magnetic Bead-Based Suspension Array for Rapid Multiplexed Phosphoprotein Quantification. Methods Mol Biol. 2017;1636:119–31. doi: 10.1007/978-1-4939-7154-1_9. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheffler M, Bos M, Gardizi M, Konig K, Michels S, Fassunke J, et al. PIK3CA mutations in non-small cell lung cancer (NSCLC): genetic heterogeneity, prognostic impact and incidence of prior malignancies. Oncotarget. 2015;6:1315–26. doi: 10.18632/oncotarget.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB, et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 2010;70:6804–14. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–53. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–5. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165:1818. doi: 10.1016/j.cell.2016.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.