Summary
Hereditary thrombocytopenias can be subclassified based on mode of inheritance and platelet size. Here we report a family with autosomal dominant (AD) thrombocytopenia with normal platelet size. Linkage analysis and whole exome sequencing identified the R1026W substitution in ITGA2B as the causative defect. The same mutation has been previously reported in 7 Japanese families/patients with AD thrombocytopenia, but all of these patients had macrothrombocytopenia. This is the first report of a family with AD thrombocytopenia with normal platelet size resulting from mutation in ITGA2B. ITGA2B mutations should therefore be included in the differential diagnosis of this latter disorder.
Keywords: Hereditary thrombocytopenia, linkage analysis, whole exome sequencing, ITGA2B, autosomal dominant thrombocytopenia
Introduction
Hereditary thrombocytopenias (HT) are a heterogeneous group of bleeding disorders characterized by varying degrees of thrombocytopenia and a wide spectrum of clinical manifestations. Recent advances in molecular genetics have improved our understanding of the pathophysiology of these disorders. HT can be subclassified based on the mode of inheritance as well as platelet size (Balduini and Savoia 2012). Here we report a family with autosomal dominant (AD) thrombocytopenia characterized by normal platelet size and absence of other phenotypic abnormalities. Linkage analysis and whole exome sequencing identified the genetic defect as mutation in the ITGA2B gene resulting in an R1026W substitution.
Methods
Sample Collection and DNA preparation
A 4-generation family with 10 affected individuals (Fig. 1A) was studied. Following enrollment on a research protocol approved by the University of Michigan Institutional Review Board, genomic DNA was prepared from the individuals indicated in Fig. 1A as described in the Supplemental Methods.
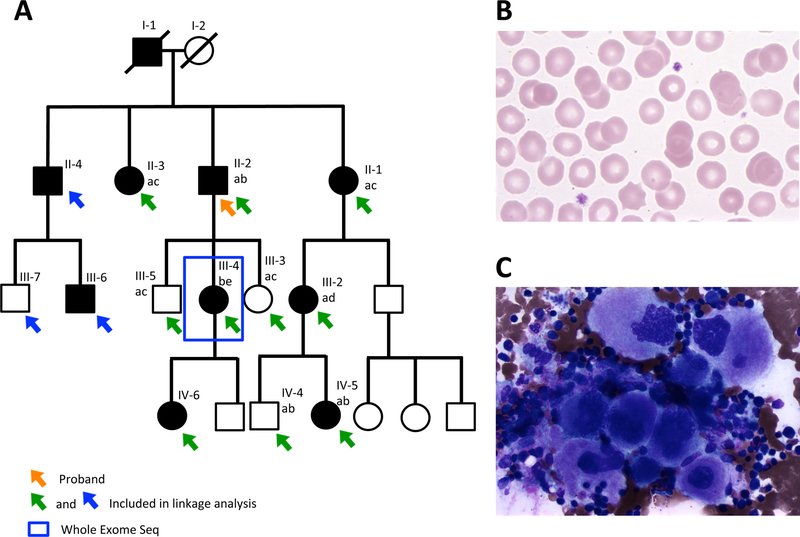
Figure 1. Family with autosomal dominant thrombocytopenia.
(A) Affected individuals are indicated by filled symbols. Arrows indicate individuals included in the linkage analysis, with green or blue arrows denoting collection of blood sample or buccal swab, respectively. The blue box indicates the individual whose DNA was subjected to whole exome sequencing. The proband is indicated by an orange arrow. Genotyping for a microsatellite, ~3Mb upstream of ANKRD26 was performed on 9 family members (6 affected and 3 unaffected), ruling out ANKRD26 as the causative gene in this family. Each genotype is indicated by a different letter (a, b, c, d, e). Affected patients did not share a common microsatellite genotype. (B) Peripheral blood smear from family member II-2 demonstrates thrombocytopenia with uniform, normal-sized platelets. Original magnification x500. (C) Bone marrow aspirate from family member III-4 demonstrates abundant (slightly increased in number) megakaryocytes, 5–10% of which were small with nuclear hypolobation with no other bone marrow abnormalities. Original magnification x500.
Microsatellite Genotyping
Genotyping for a microsatellite located ~3 megabases (Mb) 5’ of ANKRD26 (Chromosome 10:30,059,574–30,059,640) was performed by polymerase chain reaction (PCR with primers MS F1 and MS R1 (Table S1), as described in the Supplemental Methods.
Sanger Sequencing
PCR and Sanger Sequencing were performed as described in the Supplemental Methods. PCR primers are listed in Table S1.
Linkage Analysis
DNA samples from the 13 individuals highlighted with a green or blue arrow in Fig. 1A were genotyped on the Infinium HumanCoreExome v24.1 BeadChip (Illumina, San Diego, CA), which yielded ~530K autosomal single nucleotide polymorphisms (SNPs). After applying multiple filters for quality control, parametric linkage analysis was performed using an AD inheritance model on a range of marker sets and parameters (See Supplemental Methods).
Whole-Exome Sequencing and Analysis
Genomic DNA from family member III-4 (Fig. 1A) was sonicated and subjected to exome capture using a NimbleGen SeqCap EZ Exome Enrichment Kit v3.0 (Roche, Madison, WI, USA) and to 100 base pair paired-end sequencing on an Illumina HiSeq 2000 at the University of Michigan Sequencing Core (UMSC). This sample was one of 733 samples sequenced at the UMSC for which pooled variant calling was performed. The details of the sequencing and downstream analysis are included in the Supplemental Methods.
Imputation and Haplotype Analysis
In a previous report (Kunishima, et al 2011), the disease haplotype at the ITGA2B locus was defined using a set of 11 SNPs. The disease haplotype was defined in this study to determine if the ITGA2B mutation occurs on a different haplotype in our family (see Supplemental Methods). The candidate risk allele was also examined for segregation with the disease haplotype (see Supplemental Methods).
Results and Discussion
This family (Fig. 1A) is of European ancestry and exhibits AD thrombocytopenia with normal platelet size and appearance on peripheral smear (Fig. 1B and Table S2) and normal mean platelet volume of 10.2 fl for the proband (normal range 9–12.2 fl). Bone marrow evaluation of individual III-4 demonstrated a slightly increased number of megakaryocytes, 5–10% of which appeared small with nuclear hypolobation (Fig. 1C). The bone marrow was otherwise normal and metaphase cytogenetics revealed a normal karyotype.
Given the lack of acute leukaemia history or phenotypic abnormalities in this AD thrombocytopenia family with normal platelet size, ANKRD26-related thrombocytopenia, also known as thrombocytopenia 2, was suspected. Thrombocytopenia 2 results from mutations in the 5’UTR of ANKRD26 leading to loss of transcription factor binding (Bluteau, et al 2014, Pippucci, et al 2011). The 5’UTR of ANKRD26 was Sanger sequenced in family members II-2 and III-4, and no mutation was identified. To rule out a disease-causing mutation in ANKRD26 outside of the 5’UTR, genotyping for a microsatellite, ~3Mb upstream of the gene was performed in 9 family members (6 affected and 3 unaffected) (Fig. 1A), excluding the ANKRD26 locus as the cause of thrombocytopenia in this family (logarithm of the odds [LOD] score minus 15.6).
To map the disease-causing gene, we performed linkage analyses on 9 affected and 4 unaffected family members (details described in Methods). All parameter combinations and marker sets highlighted two consensus linkage regions: Chr17:37,222,473–48,110,703 (LOD score 3.31) and Chr13:50,246,074–78,461,133 (LOD score 2.62).
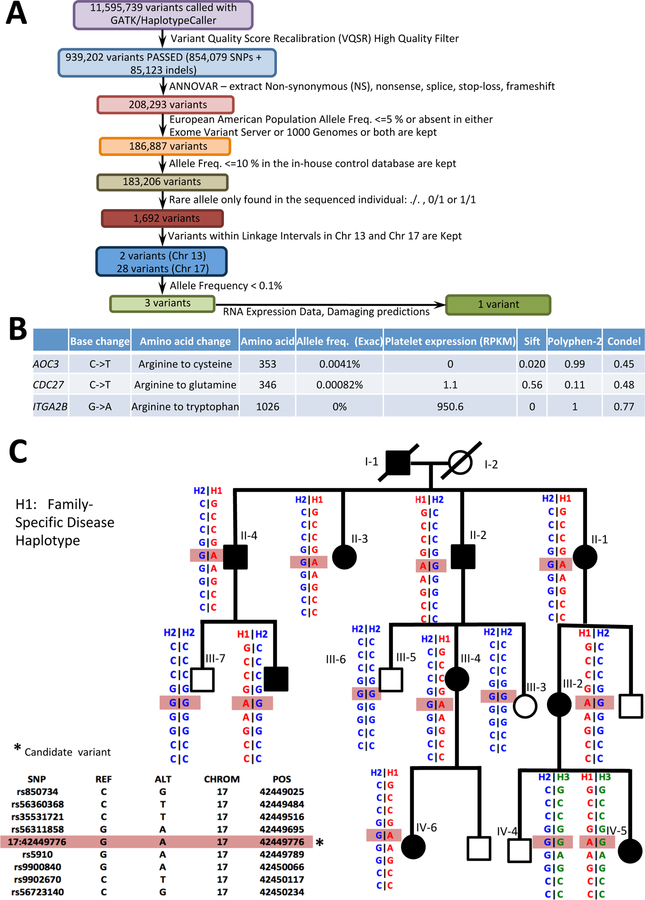
Whole exome sequencing was performed on genomic DNA obtained from family member III-4, with an average coverage of 45.96X (95.95% of the target sequence covered at least 10X). Non-synonymous, nonsense, splice-site, stop-loss and frameshift variants within the identified linkage peaks were subjected to a variety of filters (see Methods and Fig. 2A). Three variants (one in each of AOC3, CDC27 and ITGA2B) passed all filters (Fig. 2B). AOC3 is not expressed in platelets (Rowley, et al 2011), and the CDC27 variant was predicted to be benign/tolerated (Fig. 2B). Therefore, the leading candidate variant was a G to A substitution in ITGA2B, resulting in an Arginine (R) to Tryptophan (W) substitution at amino acid 1026 (p.R1026W). This variant was predicted to be damaging and has been reported previously in AD macro-thrombocytopenia (Kashiwagi, et al 2013, Kunishima, et al 2011), further supporting its identification as the causative mutation in this family. Sanger sequencing showed that the ITGA2B variant was present in heterozygous form in all affected family members and was absent in all unaffected family members.
Figure 2. Sequence analysis.
(A) Sequence analysis pipeline. (B) Three variants passed all the applied filters. RPKM refers to reads per kilobase of transcript per million mapped reads and indicates the expression level of the genes in human platelets (Rowley, et al 2011). Multiple tools (SIFT, Polyphen2, and CONDEL) were utilized to predict the pathogenicity of the variants. (C) The ITGA2B haplotype segregates with the disease.
Homozygous or compound heterozygous loss of function mutations in ITGA2B result in Glanzmann thrombasthenia, a bleeding disorder characterized by normal platelet count but abnormal platelet function (Nurden, et al 2011). In contrast, the ITGA2B R1026W mutation (referred to as R995W in some reports) is thought to result in constitutive activation of the αIIbβ3 receptor (Kunishima, et al 2011) and has been previously reported in 7 Japanese families with AD thrombocytopenia (Kashiwagi, et al 2013, Kunishima, et al 2011). The common geographic origin of these latter families suggested the possibility of a single founder allele, although a mutation in another gene tightly linked to ITGA2B could not be excluded. There are many examples where the true causative mutation is in linkage disequilibrium with many other variants, some of which reside at a distance or even in another gene. For example, before ANKRD26 was identified as the causative gene for thrombocytopenia 2, the disease was initially attributed to mutations in one of two nearby genes, ACBD5 or MASTL (Pippucci, et al 2011.).
The identical disease-associated ITGA2B haplotype was previously reported for 4 of the Japanese families (Kunishima, et al 2011). We determined the ITGA2B haplotype in our Caucasian family, confirmed that it segregates with the disease (Fig. 2C), and demonstrated that the ITGA2B R1026W mutation arose on a different haplotype compared to the previous reports, strongly supporting at least 2 independent origins for this same point mutation. This R to W substitution represents a C to T transition at a CpG site, a known hot spot for human mutations (Rahbari, et al 2016). Taken together, these results suggest that R1026W confers a unique gain of function, consistent with a previous report (Kunishima, et al 2011).
In contrast to our family (European ancestry), all previously reported families (Japanese ancestry) with HT due to the ITGA2B R1026W substitution exhibit macrothrombocytopenia (thrombocytopenia with large platelet size) (Kashiwagi, et al 2013, Kunishima, et al 2011). The lack of macrothrombocytopenia in our family could be the result of a modifier gene(s) difference in these disparate genetic backgrounds. Strain differences in mice have been demonstrated to contribute to variations in platelet size in Gray Platelet Syndrome (Tomberg, et al 2016), analogous to this observation in humans.
A different substitution at the same position of ITGA2B (R1026Q, previously designated R995Q) was reported to result in thrombocytopenia with an mean platelet volume of 10.3 μm3 (control range of 8.6 +/-1 μm3) in one patient (Hardisty, et al 1992, Peyruchaud, et al 1998). Point mutations in ITGB3 have also been reported to result in an identical phenotype (Ghevaert, et al 2008, Gresele, et al 2009). All of these mutations cluster on both sides of the transmembrane domains of αIIbβ3 (Rao and Coller 2014). Recently, rare variants in GP1BB were also found to be associated with AD macrothrombocytopenia (Sivapalaratnam, et al 2017).
To our knowledge, this is the first report of a family with AD thrombocytopenia with normal platelet size, resulting from a mutation in ITGA2B. Mutations in ITGA2B should therefore be included in the differential diagnosis of patients with this disorder.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Health Grants K08 HL128794 (RK), R01 HL124232 (JAS), R01 HL141399 (KD) R35 HL135793 (DG), and by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000433. RK is a recipient of the American Society of Hematology Scholar Award. KD is the recipient of the University of Michigan Charles Woodson Accelerator Award. DG is a Howard Hughes Medical Institute investigator. The authors would like to thank all family members for participation in this research.
Footnotes
Disclosure of conflicts of interest
The authors declare no competing conflicts of interest.
References
- Balduini CL & Savoia A (2012) Genetics of familial forms of thrombocytopenia. Hum Genet, 131, 1821–1832. [DOI] [PubMed] [Google Scholar]
- Bluteau D, Balduini A, Balayn N, Currao M, Nurden P, Deswarte C, Leverger G, Noris P, Perrotta S, Solary E, Vainchenker W, Debili N, Favier R & Raslova H (2014) Thrombocytopenia-associated mutations in the ANKRD26 regulatory region induce MAPK hyperactivation. J Clin Invest, 124, 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghevaert C, Salsmann A, Watkins NA, Schaffner-Reckinger E, Rankin A, Garner SF, Stephens J, Smith GA, Debili N, Vainchenker W, de Groot PG, Huntington JA, Laffan M, Kieffer N & Ouwehand WH (2008) A nonsynonymous SNP in the ITGB3 gene disrupts the conserved membrane-proximal cytoplasmic salt bridge in the alphaIIbbeta3 integrin and cosegregates dominantly with abnormal proplatelet formation and macrothrombocytopenia. Blood, 111, 3407–3414. [DOI] [PubMed] [Google Scholar]
- Gresele P, Falcinelli E, Giannini S, D’Adamo P, D’Eustacchio A, Corazzi T, Mezzasoma AM, Di Bari F, Guglielmini G, Cecchetti L, Noris P, Balduini CL & Savoia A (2009) Dominant inheritance of a novel integrin beta3 mutation associated with a hereditary macrothrombocytopenia and platelet dysfunction in two Italian families. Haematologica, 94, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty R, Pidard D, Cox A, Nokes T, Legrand C, Bouillot C, Pannocchia A, Heilmann E, Hourdille P & Bellucci S (1992) A defect of platelet aggregation associated with an abnormal distribution of glycoprotein IIb-IIIa complexes within the platelet: the cause of a lifelong bleeding disorder. Blood, 80, 696–708. [PubMed] [Google Scholar]
- Kashiwagi H, Kunishima S, Kiyomizu K, Amano Y, Shimada H, Morishita M, Kanakura Y & Tomiyama Y (2013) Demonstration of novel gain-of-function mutations of alphaIIbbeta3: association with macrothrombocytopenia and glanzmann thrombasthenia-like phenotype. Mol Genet Genomic Med, 1, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima S, Kashiwagi H, Otsu M, Takayama N, Eto K, Onodera M, Miyajima Y, Takamatsu Y, Suzumiya J, Matsubara K, Tomiyama Y & Saito H (2011) Heterozygous ITGA2B R995W mutation inducing constitutive activation of the alphaIIbbeta3 receptor affects proplatelet formation and causes congenital macrothrombocytopenia. Blood, 117, 5479–5484. [DOI] [PubMed] [Google Scholar]
- Nurden AT, Fiore M, Nurden P & Pillois X (2011) Glanzmann thrombasthenia: a review of ITGA2B and ITGB3 defects with emphasis on variants, phenotypic variability, and mouse models. Blood, 118, 5996–6005. [DOI] [PubMed] [Google Scholar]
- Peyruchaud O, Nurden AT, Milet S, Macchi L, Pannochia A, Bray PF, Kieffer N & Bourre F (1998) R to Q amino acid substitution in the GFFKR sequence of the cytoplasmic domain of the integrin IIb subunit in a patient with a Glanzmann’s thrombasthenia-like syndrome. Blood, 92, 4178–4187. [PubMed] [Google Scholar]
- Pippucci T, Savoia A, Perrotta S, Pujol-Moix N, Noris P, Castegnaro G, Pecci A, Gnan C, Punzo F, Marconi C, Gherardi S, Loffredo G, De Rocco D, Scianguetta S, Barozzi S, Magini P, Bozzi V, Dezzani L, Di Stazio M, Ferraro M, Perini G, Seri M & Balduini CL (2011) Mutations in the 5’ UTR of ANKRD26, the ankirin repeat domain 26 gene, cause an autosomal-dominant form of inherited thrombocytopenia, THC2. Am J Hum Genet, 88, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, Dominiczak A, Morris A, Porteous D, Smith B, Stratton MR, Consortium UK & Hurles ME (2016) Timing, rates and spectra of human germline mutation. Nat Genet, 48, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AK & Coller BS (2014) Hereditary quantitative platelet disorders. In: Williams Hematology, 9th Edition. McGraw-Hill Education, New York, NY. [Google Scholar]
- Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA & Weyrich AS (2011) Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood, 118, e101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivapalaratnam S, Westbury SK, Stephens JC, Greene D, Downes K, Kelly AM, Lentaigne C, Astle WJ, Huizinga EG, Nurden P, Papadia S, Peerlinck K, Penkett CJ, Perry DJ, Roughley C, Simeoni I, Stirrups K, Hart DP, Tait RC, Mumford AD, BioResource N, Laffan MA, Freson K, Ouwehand WH, Kunishima S & Turro E (2017) Rare variants in GP1BB are responsible for autosomal dominant macrothrombocytopenia. Blood, 129, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomberg K, Khoriaty R, Westrick RJ, Fairfield HE, Reinholdt LG, Brodsky GL, Davizon-Castillo P, Ginsburg D & Di Paola J (2016) Spontaneous 8bp Deletion in Nbeal2 Recapitulates the Gray Platelet Syndrome in Mice. PLoS One, 11, e0150852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.