Abstract
Spatial structuring is important for the maintenance of natural ecological systems1,2. Many microbial communities, including the gut microbiome, display intricate spatial organization3–9. Mapping the biogeography of bacteria can shed light on interactions that underlie community functions10–12, but existing methods cannot accommodate hundreds of species found in natural microbiomes13–17. Here we describe metagenomic plot-sampling by sequencing (MaP-Seq), a culture-independent method to characterize the spatial organization of a microbiome at micron-scale resolution. Intact microbiome samples are immobilized in a gel matrix and cryo-fractured into particles. Neighboring microbial taxa in the particles are then identified by droplet-based encapsulation, barcoded 16S rRNA amplification and deep sequencing. Analysis of three regions of the mouse intestine revealed heterogeneous microbial distributions with positive and negative co-associations between specific taxa. We identified robust associations between Bacteroidales taxa in all gut compartments and showed that phylogenetically clustered local regions of bacteria were associated with a dietary perturbation. Spatial metagenomics could be used to study microbial biogeography in complex habitats.
The local spatial organization of the gut microbiome influences various properties including colonization10,17–19, metabolism11, host-microbe and inter-microbial interactions20 and community stability1,21,22. However, current microbiome profiling approaches such as metagenomic sequencing require homogenization of input material which means that underlying spatial information is lost. While imaging techniques can reveal spatial information, they rely on hybridization with short DNA probes of limited spectral diversity, yielding data with low taxonomic resolution and often require extensive empirical optimization15,23. Bacteria are densely packed in communities, limiting identification and analysis of individual cells using visual methods8. Although imaging approaches can simultaneously profile simple synthetic communities composed of a small number of cultivable species (e.g. 6 in ref. 17)16,17, they are challenging to scale to complex and diverse natural microbiomes. An unbiased method for high-taxonomic resolution and micron-scale dissection of natural microbial biogeography is needed to better study the role of the gut microbiome in health and disease.
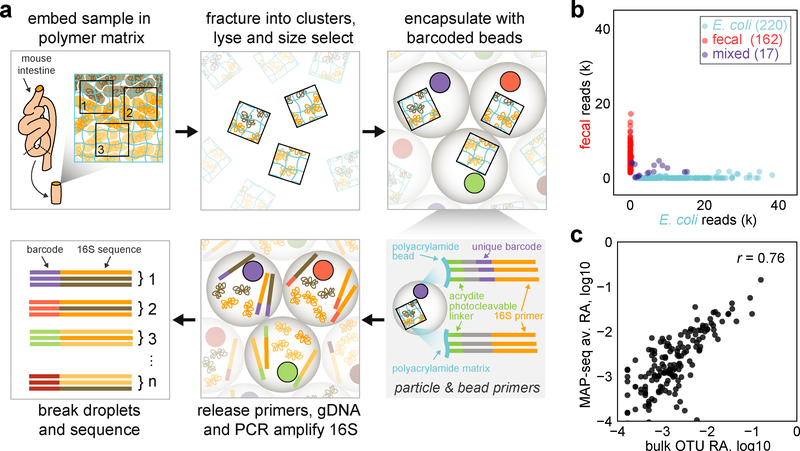
In macroecology, plot sampling is used to study the spatial organization of large ecosystems, which are otherwise impractical to fully characterize. By surveying many smaller plots from a larger region, one can tractably delineate local distributions of species and statistically infer fundamental properties of global community organization and function. Inspired by this approach, we developed Metagenomic Plot-sampling by sequencing (MaP-seq), a multiplexed sequencing technique that analyzes microbial cells in their native geographical context to statistically reconstruct the local spatial organization of the microbiome (Fig. 1a).
Fig. 1: Metagenomic Plot-sampling by sequencing (MaP-seq) and quality control.
a) Schematic of the MaP-seq technique for micron-scale plot-sampling of microbiome samples. b) MaP-seq profiling of a mixture of clusters prepared from homogenized fecal bacteria or E. coli. The number of reads (k, thousands of reads) for each of n=399 barcodes belonging to either the E. coli OTU or fecal OTUs is displayed as a scatter plot; blue dots: E. coli cluster, red dots: fecal cluster, purple dots: mixed cluster. c) Correlation between OTU relative abundance (RA) measurements obtained by standard bulk 16S sequencing of the same homogenized fecal community compared to MaP-seq OTU RA measurements averaged across individual homogenized fecal clusters (n=162 clusters with <10% E. coli reads); all RA are plotted on log10 scale, n=152 OTUs with greater than 0.01% RA are displayed, r indicates Pearson correlation.
To perform MaP-seq, an input sample is first physically fixed by immobilizing the microbiota via perfusion and in situ polymerization of an acrylamide polymer matrix that also contains a covalently linked reverse 16S rRNA amplification primer. The embedded sample is then fractured via cryo-beadbeating, subjected to cell lysis, and passed through nylon mesh filters for size selection to yield cell clusters or particles of desired and tunable physical sizes (i.e. by utilizing different mesh filter sizes). Resulting clusters contain genomic DNA immobilized in their original arrangement, preserving local spatial information. Next, a microfluidic device is used to co-encapsulate these clusters with gel beads, each containing uniquely barcoded forward 16S rRNA amplification primers. Primers are photocleaved from the beads and clusters, genomic DNA is released from clusters by triggered degradation of the polymer matrix within droplets, and PCR amplification of the 16S V4 region is performed. Droplets are then broken apart, and the resulting library is subjected to deep sequencing. Sequencing reads are filtered and grouped by their unique barcodes, which yield the identity and relative abundance of bacterial operational taxonomic units (OTUs) within individual cell clusters of defined size-scales (Methods, Supplementary Figs. 1–4).
To rigorously test the feasibility of this spatial metagenomics approach, we first generated separate cluster communities from either homogenized mouse fecal bacteria or E. coli (Methods, Supplementary Fig. 5) and profiled them with MaP-seq. The resulting data revealed that the majority of detected barcodes mapped uniquely to their respective initial communities with minimal mixing (Fig. 1b, 4.3% mixed) and negligible contamination introduced during sample processing (<0.2% of reads). In addition, the average abundance of taxa across individual fecal clusters obtained by enzymatic lysis and droplet PCR displayed good correlation with standard mechanical cell lysis and bulk 16S PCR measurements (Fig. 1c, Pearson correlation r = 0.76). A replicate community mixing experiment with new particles of a smaller size confirmed technical performance of the approach (Supplementary Fig. 5g–h). Together, these results indicate that MaP-seq accurately measures bacterial identity and abundance within individual spatially-constrained cell clusters.
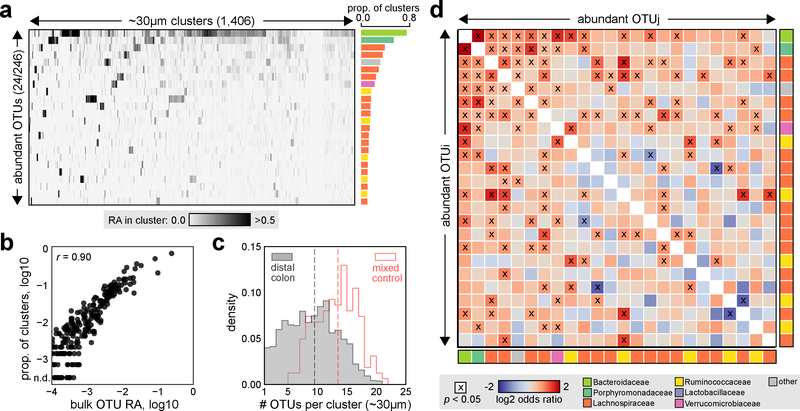
To explore the utility of spatial metagenomics in complex communities, we applied MaP-seq to the mouse colonic microbiome. We generated and characterized cell clusters (~30 μm median diameter) from a segment of the distal colon (including both epithelium and digesta) of a mouse fed a plant-polysaccharide diet, yielding 1,406 clusters passing strict quality filtering across two technical replicates (Fig. 2a, Methods, Supplementary Fig. 6a). 236 total OTUs were identified with their prevalence across clusters highly correlating to bulk abundance obtained by standard 16S sequencing, implying that more abundant taxa are also physically dispersed over more space (Fig. 2b, Pearson correlation r = 0.90). The spatial distribution of taxa across clusters appeared mixed (median 9 OTUs per cluster), but some clusters contained only a few OTUs indicating spatial aggregation or clumping in a fraction of the community (Fig. 2c). Moreover, this observed distribution of OTUs per cluster was significantly lower than clusters of the same size generated from homogenized fecal bacteria, which serve as a control for a well-mixed community (Mann–Whitney U test, p < 10−26). These results suggest that at the scale of tens of microns, individual taxa in the gut microbiome are neither fully mixed nor highly structured, but rather are heterogeneously distributed in mixed patches. Peristaltic mixing across the gut likely acts to decrease strong spatial segregation between taxa, but nevertheless the weak but significant spatial structuring observed could play an important role in the maintenance of high microbial diversity observed in the healthy gut1,22
Fig. 2: Spatial organization of the mouse distal colon microbiota.
a) MaP-seq profiling of ~30 μm median diameter distal colon clusters. Raw relative abundance (RA) data from MaP-seq is displayed as a heatmap; columns represent individual clusters (n=1,406), and rows represent abundant and prevalent OTUs (>2% RA in >10% of all clusters; 24 of 246 detected OTUs) aggregated from two technical replicate datasets of the same sample. Shading denotes the RA of individual OTUs in each cluster (linear scale); OTUs are sorted by decreasing prevalence (proportion of clusters OTU is >2% RA), and clusters are clustered by Euclidean distance. The prevalence of each OTU across clusters is displayed to the right as a bar plot, and each bar is colored by the OTUs assigned taxonomy at the family level (legend in d). b) Correlation between OTU RA measurements obtained by standard bulk 16S sequencing of the same sample compared to OTU prevalence (>2% RA across n=1,406 clusters); n.d. indicates not detected >2% RA in any clusters, n=219 OTUs greater than 0.01% RA as measured by bulk 16S sequencing are displayed, r indicates Pearson correlation. c) Histogram of the number of OTUs per cluster (OTUs >2% RA), shown for homogenized fecal clusters which serve as a mixed control (red outline, n=162) and distal colon clusters (grey, n=1,406) of the same size. Dotted lines indicate median value for each group. d) For each abundant and prevalent OTU pair (OTUi,j; n=24 OTUs) spatial associations were calculated across n=1,406 clusters, shading indicates log2 odds ratio, x denotes statistically significant association (Fisher’s exact test, two-sided, p < 0.05, FDR = 0.05); colored boxes represent OTU taxonomy at the family level.
We next explored whether these observed spatial distributions reflect specific associations between individual taxa that may result from processes such as positive or negative interspecies interactions (e.g., cooperative metabolism24; contact-dependent killing20) or local habitat filtering11. Across abundant and prevalent OTUs (>2% abundance in >10% of clusters, n=24), we assessed whether their pairwise co-occurrences were detected more or less frequently than expected in comparison to a null model of independent, random assortment of OTUs (Methods, Fisher’s exact test, p < 0.05, FDR = 0.05). Application of this strategy to the cluster mixing control experiment confirmed our ability to accurately detect positive and negative spatial associations that are expected (Supplementary Fig. 5f). Out of 276 possible pairwise combinations of taxa in the murine colon, we detected 75 statistically significant associations between diverse taxa, the majority of which were positive (72/75) but relatively weak in magnitude (Fig. 2d, Supplementary Fig. 6b–c). The strongest co-occurrence was a positive association between abundant Bacteroidaceae and Porphyromonadaceae taxa from the Bacteroidales order (odds ratio 3.9, p < 10−23). In addition, a small number of negative associations were observed, which could reflect antagonistic processes such as production of inhibitory factors or competitive exclusion.
The number of detected associations increased as more of the dataset is sampled, implying that detection of weaker relationships between less abundant taxa can be improved by analyzing more clusters (Supplementary Fig. 6d). Nonetheless, the detected associations showed good correspondence between technical replicates (Supplementary Fig. 6e). Importantly, despite high inter-host microbiome variability, the nature of the associations (i.e., sign, magnitude, and number) and some strong associations could be recapitulated in MaP-seq profiling of a second co-housed mouse, such as the co-occurrence of Bacteroidales taxa (Supplementary Fig. 7). This characterization implies that individual taxa in the colon are organized in distinct and reproducible spatial relationships.
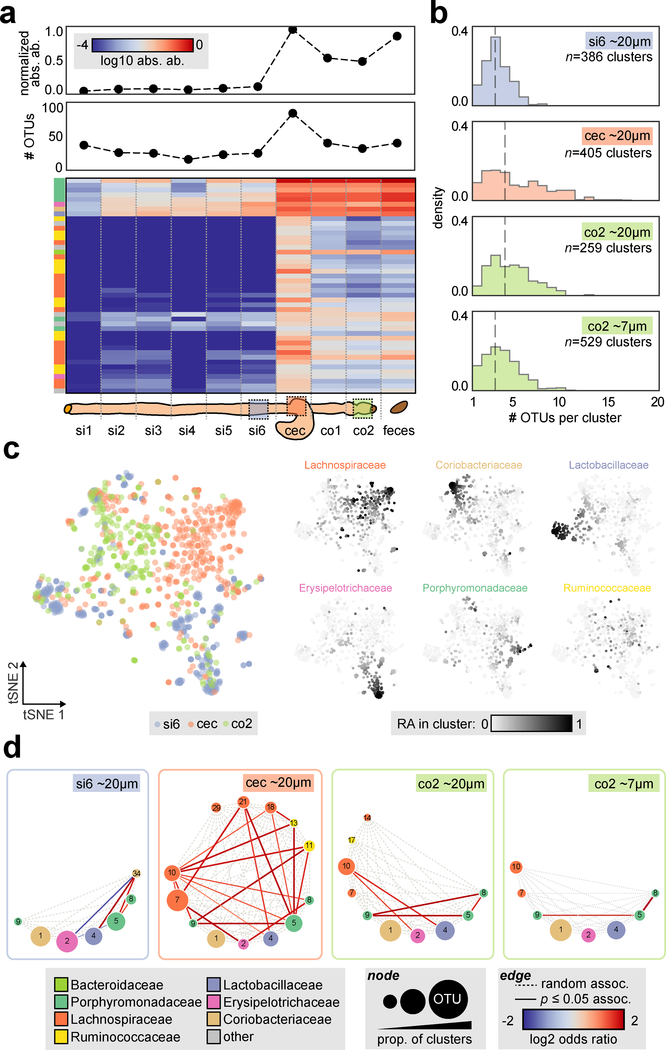
To further investigate how the spatial organization of the microbiota is influenced by their environmental context, we applied spatial metagenomics along the gastrointestinal (GI) tract. The mammalian GI tract is composed of distinct anatomical regions with different pH levels, oxygen concentrations, host-derived antimicrobials and transit times that together influence the local microbiota assemblage9. We first performed an adapted 16S community profiling approach along the murine GI tract that could also infer absolute OTU abundances25 (Fig. 3a, Supplementary Fig. 8, Methods). This new mouse cohort (2 co-housed mice) shared only ~20% of OTUs with the previous group (Supplementary Fig. 8b), illustrating the significant inter-animal microbiome heterogeneity inherent to such studies. This further highlights challenges for other spatial profiling techniques such as 16S FISH imaging where probes must be designed in advance, in comparison to MaP-seq, which can be applied to measure diverse bacteria without advance specification. Analysis of microbiota in absolute abundance across the intestine revealed increased bacterial density (~16 fold higher) and species richness in the large intestine compared to the small intestine, with the cecum harboring the highest bacterial density and number of OTUs. We chose three separate GI regions that exhibited distinct microbiota assemblages for characterization by MaP-seq: the ileum (si6), cecum (cec) and distal colon (co2). Given the high degree of species mixing previously observed at ~30 μm, we used smaller sized clusters (~20 μm median diameter) to capture higher-resolution spatial associations.
Fig. 3: Survey of spatial organization in the mouse gastrointestinal tract.
a) Top: absolute abundance within gut intestinal regions calculated from spike-in sequencing (arbitrary units, normalized to the maximum value) and number of OTUs (i.e. alpha diversity, number of OTUs >0.1% relative abundance). Bottom: absolute abundance of abundant OTUs (>1% of maximum OTU absolute abundance in any sample) is shown below as a heatmap (log 10 scale); OTUs are clustered by Bray-Curtis dissimilarity. b) Histogram of the number of OTUs per cluster (OTUs >2% RA). The number of clusters aggregated from two technical replicates is indicated (si6 ~20 μm n=386, cec ~20 μm n=405, co2 ~20 μm n=259, co2 ~7 μm n=529), and dotted line indicates median value. c) tSNE visualization of n=1,050 ~20 μm clusters from si6, cec and co2 sites utilizing Bray-Curtis dissimilarity of OTU relative abundances (subsampled to 314 reads across all clusters, number of clusters indicated above). On the left, each cluster is colored by site of origin; on the right each cluster is colored by the relative abundance of the six most abundant families within each cluster (linear scale). d) Pairwise spatial associations for abundant and prevalent OTUs visualized as a circular graph; the number of clusters utilized is subsampled to the lowest number across the samples (n=259 clusters). Nodes indicate OTUs, sizing is proportional to the prevalence of OTUs across clusters and color represents OTU taxonomy at the family level, dotted edges denote all possible associations and shaded edges denote statistically significant associations (Fisher’s exact test, two-sided, p < 0.05, FDR = 0.05).
We first assessed the distribution of OTUs per cluster to compare the spatial organization of taxa in the three regions (Fig. 3b). ~20 μm clusters displayed lower numbers of OTUs per cluster than ~30 μm clusters (median 3–4 OTUs per cluster). The ileum possessed significantly fewer OTUs per cluster than the cecum or distal colon (Mann–Whitney U test, p < 10−18 and p <10−14 respectively). In comparison, the cecum and colon displayed similar OTU distributions, while the cecum harbored more clusters with a large number of OTUs. This suggests that GI regions with more diverse microbiota also exhibit higher spatial diversity at microscopic scales. Colonic clusters of an even smaller size-scale (~7 μm) were also profiled, and contained a significantly lower number of OTUs per cluster compared to ~20 μm clusters as may be expected (Mann–Whitney U test, p < 10−6).
To understand how the local spatial organization of the microbiome may vary within and across different gut regions, we visualized the cell clusters data across the three gut regions using t-distributed Stochastic Neighbor Embedding (tSNE, utilizing Bray-Curtis distance of OTU relative abundance within clusters), as well as the abundance of prevalent bacterial families in cell clusters across the resulting manifold (Methods, Fig. 3c, Supplementary Fig. 9). While some cell clusters from the ileum, cecum and distal colon separately projected into distinct groups, other clusters from each site projected more broadly across the manifold. Interestingly, a subset of cell clusters from the cecum projected into a dense group and are compositionally dominated by Lachnospiraceae, which were generally not present in clusters from the ileum or distal colon. When cell clusters from a second co-housed mouse were added to the tSNE analysis, they were distributed in a similar manner to clusters from the first mouse across the manifold and displayed a similar cecum-specific Lachnospiraceae group, further strengthening these results (Supplementary Fig. 10). Our observations suggest that the spatial distribution of some taxa at different GI regions may have distinct local organizations from one another while other taxa may have similar local organization along the GI tract.
Next, we explored whether these different spatial distributions reflect distinct spatial co-associations between taxa at each GI site (Fig. 3d). The ileum harbored a network of positive and negative associations between the few taxa present. On the other hand, the cecum exhibited a dense network of positively co-associated taxa, primarily between abundant Lachnospiraceae, Ruminococcaceae, and Porphyromonadaceae. Similar to the cecum, the distal colon displayed only positive associations, including strong groupings between three abundant Porphyromonadaceae (OTUs 5,8,9). Profiling the colon at an even smaller size-scale (~7 μm) confirmed strong positive associations between a subset of these three taxa, indicating that this spatial co-occurrence is robust at short, local length-scales. Species from these abundant Bacteroidales taxa often contain diverse carbohydrate-active enzymes26 and are known to engage in cooperative metabolic cross-feeding24,27, which could promote these spatial co-associations.
While the spatial association networks revealed by MaP-seq differed across the three GI regions, some common co-associations (or lack of associations) were observed. For example, a positive association between Lachnospiraceae (OTU 10) and Lactobacillaceae (OTU 4) was found in both the cecum and colon; on the other hand, Coriobacteriaceae (OTU 1), an abundant taxon at all sites, lacked co-associations with other taxa and was thus randomly assorted at all sites. Together, the differing spatial architectures observed across GI sites suggest that regional environmental factors can variably shape some local spatial structuring of the microbiota, while conserved spatial patterns across sites are more likely the result of robust ecological interactions not affected by environmental variations.
We further investigated whether MaP-seq could identify individual taxa with unique or altered spatial patterns. While the cecum harbored the densest community and the highest degree of species mixing of the three sites (Fig 3a–b), we hypothesized that specific taxa may self-aggregate to a higher degree than others, for example by uniquely utilizing a specific metabolite11. Assessing the aggregation of abundant taxa revealed a Lachnospiraceae (OTU 7; putatively of the genus Dorea, 60% confidence by RDP) that clustered two-fold greater than the average clustering metric value of all taxa (Supplementary Fig. 11a). To validate this finding with an orthogonal approach, we performed 16S FISH on GI sections from the same murine sample using previously validated probes that targeted Lachnospiraceae (Erec482) as well as two other abundant taxa for which FISH probes were available but were predicted not to cluster at a similar degree (Coriobacteriaceae: Ato291, Lactobacillaceae: Lab148; Methods). Strikingly, imaging confirmed that while Lachnospiraceae were distributed across the cecum, they also formed large clustered aggregates that appeared to exclude other bacteria (Supplementary Fig. 11–12). Importantly, this result highlights that individual taxa in the gut can organize in unique and spatially varying micron-scale structures that can be revealed by MaP-seq.
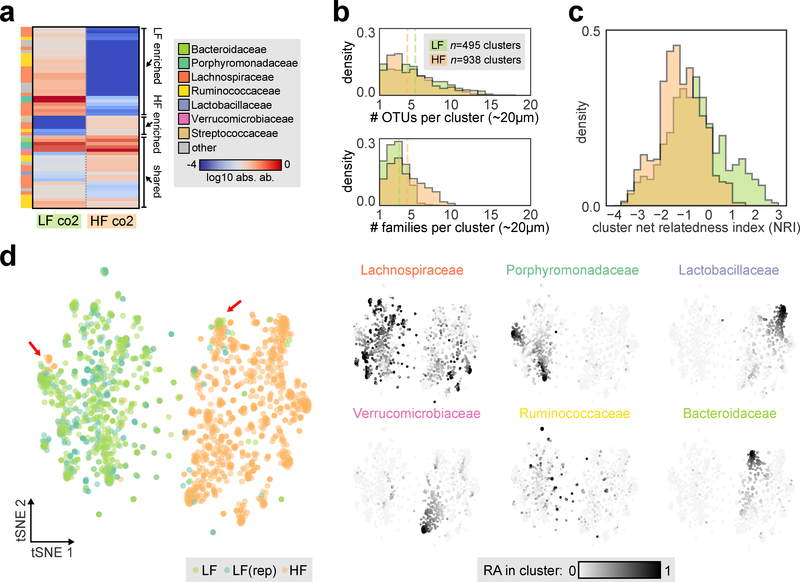
Having established the local spatial organization across the GI tract of mice fed a standard plant-polysaccharide diet, we next sought to understand the extent to which diet might influence spatial structuring. Diet is known to play a major role in shaping the variation of gut microbiota across individuals28,29. While diet shifts can rapidly alter microbiota composition within days30, the detailed ecological mechanisms underlying these community-scale changes are not well understood. We thus took co-housed mice and split them into two cohorts where one was maintained on the plant-polysaccharide based diet (LF, same as in the previous cohorts) and one was switched to a high fat, high sugar diet (HF, commonly utilized in dietary-induced obesity studies) to assess microbiota changes associated with these two diets representing distinct macronutrient profiles. After 10 days on the two diets, a considerable loss of species richness in the cecum and colon was observed in HF-fed mice compared to LF-fed mice (Fig. 4a, Supplementary Fig. 13).
Fig. 4: Spatial organization in the colon after dietary perturbation.
a) Absolute abundance of dominant OTUs (>1% of maximum OTU absolute abundance in any sample) in the distal colon of co-housed mice fed a low fat, plant-polysaccharide diet (LF) or high fat diet (HF) for 10 days is shown as a heatmap (log 10 scale). Labels on right indicate LF enriched, HF enriched and shared OTUs. b) Top: histogram of the number of OTUs per cluster (OTUs >2% RA). Bottom: histogram of the number of distinct families per cluster (families >2% RA). For both plots, green indicates LF clusters and orange indicates HF clusters, dotted line indicates median value, and the number of clusters aggregated from two technical replicates is indicated (LF co2 n=495, HF co2 n=938). c) Histogram of net relatedness index (NRI) calculated for each cluster containing at least two OTUs, green indicates LF clusters and orange indicates HF clusters. d) tSNE visualization of clusters utilizing Bray-Curtis dissimilarity of OTU relative abundances (subsampled to 121 reads across all clusters). Left, cluster colored by site of origin; LF (green), HF (orange), number of clusters indicated above. In addition a biological replicate from an adjacent colonic segment of the same LF mouse is shown (LF(rep), dark green, n=359 clusters). Red arrows indicate examples of cluster configurations observed in both diet conditions. Right, each cluster is colored by the relative abundance of the six most abundant families within each cluster (linear scale).
To determine if a dietary shift could alter the spatial organization of the microbiota, which could contribute to the observed loss of species diversity, we performed MaP-seq on distal colon samples from mice fed the LF or HF diet. We found that the distribution of unique OTUs per ~20 μm cluster was similar between both diets (Fig. 4b, top). This implies that species distributions at the local ~20 μm scale is governed by factors that are either common to or not affected by the two diets, for example spatial autocorrelation of bacterial growth. However, assessing diversity at the higher taxonomic family-rank revealed significantly higher diversity in HF clusters (HF: average 4.0 families/cluster, LF: average 2.7 families/cluster; Mann–Whitney U test, p < 10−22, Fig. 4b, bottom), indicating that while both LF and HF clusters contained similar numbers of OTUs, taxa within individual HF clusters were more phylogenetically diverse. Furthermore, positive co-associations were more frequently observed between diverse taxa in HF diet than in LF diet, which in contrast had co-associations mostly between Porphyromonadaceae or Lachnospiraceae (Supplementary Fig. 14). Interestingly, our observation of increased bacterial mixing at higher taxonomic levels has also been documented in mice fed with a plant-polysaccharide deficient diet (compared to a LF plant-polysaccharide rich diet) using confocal imaging with 16S FISH probes of limited phylum-level specificity6, which further highlights the utility of examining spatial organization at a higher taxonomic resolution that is achievable by MaP-seq.
Understanding the phylogenetic distribution of an ecosystem can provide important insights into ecological processes underlying community assembly31,32. To better quantify possible changes in phylogenetic diversity between the two diets, we calculated the net relatedness index (NRI) of clusters, a standardized effect size of the mean phylogenetic distance of taxa present within clusters against a null model of random sampling from the local species pool within each sample (Methods)31. For each microbiota cluster, a positive NRI value indicates phylogenetic clustering of its taxa, whereas a negative NRI indicates phylogenetic over-dispersion. While most clusters had NRI values near 0, suggesting random phylogenetic distributions, both LF and HF diets showed a subset of clusters with high negative NRI values suggesting a high degree of phylogenetic over-dispersion. Interestingly, NRI values in LF clusters were overall significantly higher compared to HF values (Mann–Whitney U test, p < 10−18), driven by a subset of LF clusters with positive NRIs not observed in HF clusters (Fig. 4c, Supplementary Fig. 15). The phylogenetic clustering observed in this subset of LF clusters suggests that ecological habitat filtering due to factors associated with the LF diet (e.g. complex plant polysaccharides) may be important in shaping in the formation of these clusters at length-scale of ~20 μm (assuming that more phylogenetically similar taxa also have more similar phenotypes). A possible explanation for the loss of species diversity when transitioning from a LF to a HF diet could thus be the loss of this LF-specific local niche, which stably hosts these closely related taxa. Indeed, the same taxa (predominantly Lachnospiraceae OTUs) that are abundantly found in LF clusters with high NRI values are those that are almost completely lost on HF diet (Supplementary Fig. 15b).
Next, to compare the taxa spatial organization across the two diets, we visualized clusters using tSNE as before (Fig. 4d, Supplementary Fig. 16). Cell clusters from the two diets each formed highly distinct groups with minimal overlap, indicating that the spatial organization in the distal colon was significantly altered by the dietary shift. Despite this overall separation, we observed examples of cluster configurations that were shared between the two diets. For example, HF clusters were observed in a predominantly LF region marked by high abundance of a Porphyromondaceae taxa (OTU 5), and LF clusters were observed in a predominantly HF region marked by high abundance of a Bacteroidaceae taxa (OTU 6) (Supplementary Fig. 16d). These shared cluster regions could represent spatial niches that may be independent of the diet (e.g. mucus layers secreted by the host). Taken together, MaP-seq profiling of a diet perturbation enabled mechanistic analysis of ecological processes underlying community shifts and loss of diversity.
Spatial metagenomics enables the high-throughput characterization of microbial biogeography through plot sampling of co-localized nucleic acids at tunable length scales. Our approach could be applied to multiple perturbations in the gut e.g., diet, antibiotics, fecal microbiota transplantation, in other microbiota such as skin, or genitalia, or diverse environmental niches including soils or biofilms. MaP-seq enables in-depth analysis of these processes at previously inaccessible and ecologically meaningful local length scales within individual microbiomes. A variety of established spatial ecology tools and emerging computational and analytical approaches could be applied to this new type of high-dimensional microbiome dataset, which will require more rigorous evaluation and further development. Our approach could be modified to capture metagenomic information by introduction of additional capture primers or use of random priming or tagmentation strategies, which could enable profiling of interactions between bacteria and eukaryotes (e.g. epithelial cells or fungi). Plot sampling of biological structures at microscopic scales opens up new directions of research that employ spatial ecology tools to study these complex systems.
Online methods
Materials and reagents
All primers and FISH probes were ordered from Integrated DNA Technologies. Primers containing any modifications were HPLC purified by the manufacturer. Photocleavable primers were protected from unnecessary light exposure throughout.
Animal procedures
All mouse procedures were approved by the Columbia University Medical Center Institutional Animal Care and Use Committee (protocol AC-AAAR1513) and complied with all relevant regulations. 6–8 week-old female C57BL6/J mice were obtained from Taconic (colonic analysis, Fig. 2) or Jackson (analysis across GI tract, Fig. 3; dietary perturbation, Fig. 4) and fed a plant-polysaccharide based diet (LabDiet 5053). Mice were allowed to adjust to the animal facility for two weeks prior to all studies, and all mice analyzed within each cohort were co-housed within the same cage. Dietary perturbation was performed by splitting four co-housed mice into two cages; one cage received the same plant-polysaccharide based diet and one cage received high fat diet (Teklad TD.06414).
Microfluidic device fabrication
Devices were fabricated utilizing standard SU-8 soft lithography. Silanized SU-8 silicon wafer molds were fabricated by FlowJEM with a feature height of ~40 μm. PDMS (Dow Corning Sylgard 184) was mixed for 5 minutes at a ratio of 10:1 base to curing agent, degassed under house vacuum for 30 minutes, and poured over the wafer. The PDMS mixture was cured at 80 °C for 1 hour, allowed to cool to room temperature and removed from the wafer. Individual devices were cut from the PDMS slab and ports were punched utilizing a 1 mm biopsy punch (World Precision Instruments 504646). A PDMS device and pre-cleaned glass slide (Fisher 12–550-A3) were then treated in a plasma cleaner (Harrick Plasma PDC-32G) for 40 seconds, the device was bonded to the slide and cured at 80 °C for 30 minutes. Devices were then treated to make channel surfaces fluorophilic33; briefly, Aquapel solution was injected to the device, left for 30 seconds, flushed with air, FC-40 (Sigma-Aldrich F9755) and air again, and baked at 80 °C for 10 minutes.
Uniquely barcoded bead design and construction
We designed custom barcoded hydrogel beads containing one of 884,736 unique barcoded primers per bead and a partial sequencing adapter and 16S V4 primer 515f34,35. Theoretically, around ~17,500 clusters can be captured per sample with a 1% multiple barcoding rate36. Barcoded primer sequences were constructed via a split-and-pool primer extension strategy36,37 with three barcode extension rounds. Each barcode position contained 96 possible sequences, and each set of barcodes was selected such that each had at least 3 bp hamming distance from the other barcodes in each set (allowing for 1 bp error correction). The first barcode position was 7–9 bp in length (allowing for dephasing of reads to improve sequencing quality) while the second and third positions were 8 bp in length.
Construction of the barcoded beads followed procedures from Zilionis et al.38 with minor modification for our barcoding scheme. Briefly, acrylamide beads (6% w/w acrylamide, 0.18% w/w N,N’-methylenebisacrylamide [Sigma-Aldrich 146072], 20 μM acry_pc_pe1 [see Supplementary Table 1) were generated using a custom microfluidic droplet device. Resulting beads were ~20–25 μm in diameter. Batches of ~20 million beads were then subjected to three rounds of primer extension using the three sets of 96 barcode sequences (pe1, pe2, and pe3 primer extension sets, see Supplementary Table 2). For each round, beads and primers were distributed into wells of a 96 well PCR microplate and primers were annealed to the beads by incubation. A Bst polymerase reaction master mix (NEB M0537L) was then distributed to each well and incubated to allow for extension. Finally, the reaction was quenched with EDTA and pooled for cleanup steps. The beads were then subjected to denaturing of the extension primers by sodium hydroxide and washing, and the extension protocol was repeated. These procedures were automated on a Biomek 4000 liquid handling robot where possible. After the final extension step, a primer targeted to the terminal 515f primer sequence (515f_RC, see Supplementary Table 1) was annealed, and an Exo1 enzymatic cleanup (NEB M0293L) was utilized to remove extension intermediates. Resulting barcoded beads were subjected to a final denaturing and washing step and stored at 4 °C in TET (10 mM Tris HCl [pH 8.0], 1 mM EDTA, 0.1% Tween-20).
Sample fixation and in situ polymerization
Intact tissue segments (from the colon, cecum or small intestine as noted) were obtained by dissection and immediately fixed in methacarn solution (60% methanol, 30% chloroform, 10% acetic acid) for 24 hours39. The fixed tissue was trimmed with a sterile razor into segments no larger than 3 mm in length, and segments containing digesta were selected. Thus, all input samples for MaP-seq analysis contained undisturbed epithelial tissue and lumenal digesta contents. The trimmed sample was then incubated in phosphate buffered saline (PBS) for 5 minutes and was permeabilized in PBS with 0.1% v/v Triton-X 100 for 5 minutes. Next, a matrix embedding solution40,41 containing a reverse sequencing primer with 16S V4 primer 806rB35,42 and acrydite and photocleavable linker groups was prepared on ice by mixing concentrated stocks of the following components in order: 1X PBS, 10% w/w acrylamide (Sigma-Aldrich A9099), 0.4% w/w N,N′-Bis(acryloyl)cystamine (BAC, Alfa Aesar 44132–03), 5 μM acry_pc_pe2_816r (see Supplementary Table 1), 0.01% w/w 4-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl (Sigma-Aldrich 176141), 0.2% w/w tetramethylethylenediamine (Sigma-Aldrich T7024) and 0.2% w/w ammonium persulfate (Sigma-Aldrich A3678). The BAC crosslinker enables gel degradation upon exposure to reducing conditions. The sample was dabbed dry with a sterile Kimwipe and placed in a PCR tube with excess matrix embedding solution (~50 μL per segment) and incubated on ice for 5 minutes. Excess embedding solution was removed by pipetting and replaced, and the sample was subsequently incubated on ice for >1 hour for perfusion. Excess embedding solution was removed, and samples were placed in a 37 °C incubator in an anaerobic chamber (Coy Laboratory Products) for >3 hours. Gel-embedded samples were removed, excess polymer matrix was trimmed from the sample with a sterile razor, and the sample was washed twice with PBS and once with TET and stored in TET at 4 °C.
Sample fracturing, lysis and size-selection
Samples were placed in a stainless-steel vial (Biospec 2007) along with a 6.35 mm stainless steel bead (Biospec 11709635ss), and were sealed with a silicone rubber plug cap (Biospec 2008). The vial was placed in liquid nitrogen for >2 minutes, vigorously shaken to dislodge the sample from the vial wall, and quickly transferred to a bead beater (Biospec 112011) and subjected to beating for 10 seconds. PBS was added to the vial and vortexed; clusters in PBS were removed and washed twice with PBS via centrifugation at 15K RPM for 1 minute (Eppendorf 5424). Next, embedded cells were lysed43; clusters were resuspended in 500 μL lysis buffer (10 mM Tris-HCl [pH 8.0], 1 mM ethylenediaminetetraacetic acid [EDTA], 100 mM NaCl) with 75 U/μL lysozyme (Epicentre R1810M) and were incubated at 37 °C for 1 hour. Clusters were then resuspended in 500 μL digestion buffer (30 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5% Triton X-100, 800 mM guanidine hydrochloride [Sigma-Aldrich G9284]) with 0.1 μg/μL proteinase K (Epicentre MPRK092), and were incubated at 65 °C for 15 minutes. Finally, clusters were incubated at 95 °C for 5 minutes to inactivate proteinase K and washed three times with TET.
Samples were next subjected to size-selection. Clusters were first passed through a 40 μm cell strainer (Fisher 22–363-547) to remove large particulate matter. Next, nylon mesh filters (Component Supply Company, 7 μm: U-CMN-7-A, 15 μm: U-CMN-15-A, 31 μm: U-CMN-31-A) were cut to size using a ½” hole punch and two filter punches were placed in a holder (EMD Millipore SX0001300) for each size. Clusters were passed through the 31 μm filter, 15 μm filter, and 7 μm filter sequentially using a 3 mL syringe (BD 309657); for each filter, clusters were passed through three times, and retained clusters on filters were washed once with TET. Clusters were washed off the 15 μm filter (large, ~30 μm median diameter) and 7 μm (medium, ~20 μm median diameter) or collected from the pass-through from the final 7 μm filter (small, ~7 μm median diameter). The concentration of clusters was quantified by counting on a hemocytometer (INCYTO DHC-N01) and stored at 4 °C in TET for processing within ~2 days.
Co-encapsulation of beads and clusters
A microfluidic co-encapsulation strategy was utilized with three syringe pumps (Harvard Apparatus Pump 11 Elite) and observed under a microscope (Nikon Eclipse Ti2). First, 300 μL of HFE-7500 (3M) with 5% w/w surfactant (RAN Biotechnologies 008-FluoroSurfactant) was loaded into a 1 mL low dead volume syringe (Air-Tite Products A1), the syringe was fitted with a needle (BD 305122) and polyethylene tubing (Scientific Commodities Inc., BB31695-PE/2) and primed on a syringe pump.
30 μL of packed barcoded beads were then removed and washed twice with wash buffer (WB, 10 mM Tris HCl [pH 8.0], 0.1 mM EDTA, 0.1% Tween-20) and twice with bead buffer (10 mM Tris HCl [pH 8.0], 0.1% Tween-20, 50 mM KCl, 10mM fresh DTT [utilized to degrade clusters within droplets]) by addition of buffer and centrifugation at 15K RPM for 1 minute. After the 4 washes, remaining buffer supernatant was removed with a gel-loading tip (Fisher 02–707-139). ~5 μL of packed beads were loaded into polyethylene tubing and primed with a 1 mL syringe (BD 309626) backfilled with 500 μL HFE-7500. The tubing was protected from light with a black tubing sheath (McMaster-Carr 5231K31) and primed on a syringe pump with needle facing upwards.
Next, a cluster stock was vortexed for 1 minute, ~2,500 clusters were removed, washed three times in WB, and the remaining buffer was removed as above. A 45 μL encapsulation mix was prepared (25 μL NEBNext Q5 Hot Start HiFi PCR Master Mix [NEB M0543L], 4 μL Nycoprep Universal [Accurate Chemical & Scientific Corp. AN1106865), 5 μL 10% w/v Pluronic F-127 [Sigma-Aldrich P2443], 1.25 μL 20 mg/mL BSA [NEB B9000S], 9.75 μL nuclease-free water) and clusters were resuspended in the mix and vortexed for >10 s. A 1 mL low dead volume syringe was backfilled with 500 μL HFE-7500, and the encapsulation mix was added directly into the tip of the syringe. A needle and polyethylene tubing were fitted to the syringe, protected from light with a black tubing sheath, and primed on a syringe pump with needle facing upwards.
Tubing was connected for the carrier, bead and cluster encapsulation mix channels to a new microfluidic device. Pumps were primed for the carrier, beads and cluster encapsulation mix channels in order and once stable bead packing was observed set to final flow rates of 2 μL/min for carrier, 0.3 μL/min for beads, and 2.7 μL/min for cluster encapsulation mix. Once stable droplet formation was observed, polyethylene tubing was connected to the outlet port and emulsion was collected in a PCR tube (Axygen PCR-02-L-C) prefilled with 10 μL of 30% w/w surfactant in HFE-7500 and 50 μL of mineral oil. Under these conditions, generated droplets were ~35–45 μm in diameter with bead occupancy of ~25–50% (packed bead ordering enables loading beating expected Poisson encapsulation statistics44) and extremely low cluster occupancy of <0.1% (cluster aggregation and channel clogging is a limiting factor at higher concentrations).
Emulsion PCR, library preparation and sequencing
The carrier phase underneath the emulsion was removed and replaced with 30 μL of 30% w/w surfactant in HFE-7500 to ensure droplet stability during PCR cycling. Tubes were placed on ice under a 365 nm UV light (Ted Pella Blak-Ray) and exposed for 10 minutes to release amplification primers. The emulsion was then subjected to PCR cycling (10 °C for 2 h, 98 °C for 30 s; 30 cycles of: 98 °C for 10 s, 55 °C for 20 s, 65 °C for 30 s; 65 °C for 2 m) with heated lid off. Coalesced droplet fraction, if present, was removed by pipetting and the carrier phase and mineral oil were removed. Droplets were broken by addition of 20 μL 1H,1H,2H,2H-perfluoro-1-octanol (Sigma-Aldrich 370533), and brief centrifugation in a microfuge tube. The aqueous phase was extracted and passed through a 0.45 μm spin column (Corning 8162) and subjected to an Exo1 cleanup by adding 50 uL of 1X Exo1 buffer with 1U/uL Exo1 (NEB M0293L) and incubating at 37 °C for 30 minutes. The mixture was then subjected to a 1X SPRI bead cleanup (Beckman Coulter A63881) per the manufacturer’s protocol with addition of 1X volume beads and elution in 20 μL of 10 mM Tris-HCl (pH 8.0).
The resulting products were then subjected to a second PCR to add sample indexes and Illumina P5 and P7 adapters. 10 μL of cleanup product was used as template for a 50 μL reaction with 1X NEBNext Q5 Hot Start HiFi PCR Master Mix, 0.5 μM of each of the indexing primers (p5_X, p7_X, see Supplementary Table 3), and 0.1X SYBR Green I (Invitrogen S7567). The PCR (98 °C for 30s, cycle: 98 °C for 10 s, 68 °C for 20 s, 65 °C for 30 s; 65 °C for 2 m) was run on a real-time PCR machine (Bio-Rad CFX96) to stop reactions during exponential amplification (typically ~10 cycles). Products were assessed on an agarose gel (2% E-gel, Thermo Fisher G501802) to confirm the expected ~490 bp amplicon and were subjected to a 1X SPRI bead cleanup as above. Resulting libraries were quantified via fluorometric quantitation (Thermo Fisher Q32854), pooled, and were subjected to sequencing with an Illumina MiSeq 500 cycle v2 kit (read1: 254 bp, read2: 254 bp) at 12 pM loading concentration with 20% PhiX spike in.
Sequence filtering and 16S analysis
For MaP-seq data, a custom python script was utilized to demultiplex reads based on barcode identity and strip primer sequences from reads. Reads were merged and filtered using USEARCH 9.2.6445 with maximum expected errors of 1. The resulting sequences were then dereplicated, de-novo clustered with a minimum cluster size of 2, and reads were mapped to OTUs at 97% identity46. Taxonomy was assigned to OTUs using the RDP classifier47. This yielded an OTU table consisting of individual barcodes (i.e., putative clusters) as samples.
Cluster mixing quality control experiment
Two bacterial communities were assembled; the first contained a single strain (e.g. E. coli NEB-beta), the second contained homogenized fecal bacteria. E. coli is not expected in the mouse gut at high abundances48. To generate homogenized fecal bacteria, fecal pellets were subjected to bead beating (Biospec 1001) with 0.1 mm glass beads in PBS for 1 minute and passed through a 40 μm cell strainer. The two communities were fixed in methacarn, resuspended in approximately equal volume matrix embedding solution to fixed pellet volume and subjected to cluster generation as per the MaP-seq protocol above. The homogenized fecal clusters therefore contained lower cell densities compared to the actual fecal samples, given dilution in embedding solution. The resulting size-selected clusters were then mixed in equal quantity and subjected to encapsulation and sequencing.
Analysis of MaP-seq data
An overview of all MaP-seq datasets generated in this study can be found in Supplementary Table 5. The resulting dataset contained a large number of barcodes/clusters with varying numbers of reads. A conservative threshold cutoff for considering real clusters was set as the total number of reads in a sample divided by 2,500 (i.e., the number of clusters that were utilized as input during microfluidic encapsulation, and assuming an equal read distribution for each cluster). Reactions yielding an extremely low number of clusters passing this threshold (i.e., <50) were conservatively excluded as they may represent failed encapsulation or amplification reactions.
Clusters were first pre-processed to remove a small number of clusters displaying highly similar OTU abundance profiles within a single technical replicate that appeared to represent technical artifacts (i.e., clusters encapsulated into droplets containing multiple barcoded beads or beads erroneously containing multiple barcodes) which could confound association detection. The pairwise Pearson correlation of all clusters was calculated, and highly correlated sets of clusters (r>0.95) dominated by a single technical replicate and large in size (>90% belonging to a single technical replicate, clusters constitute >1% of the overall dataset) were removed. These artifacts constituted a low amount of the overall dataset. For analysis of presence or absence of species within a cluster, a 2% relative abundance threshold within clusters was utilized, given observation of a small amount of background read-through across clusters and to ensure that at least 2 reads (and not singletons) were required to denote presence of a speciess.
To determine pairwise associations, prevalent and abundant OTUs within filtered clusters (>2% relative abundance in >10% of clusters) were identified, and 2 by 2 contingency tables of appearance (>2% relative abundance) were calculated for all pairs of OTUs. Fishers exact test was then used to calculate the probability of pairs occurring more or less together than expected (i.e. a null model of random assortment of the two species, assuming equiprobable occupancy at all sites), and resulting p-values were adjusted via the Benjamini-Hochberg procedure (FDR = 0.05).
For t-distributed Stochastic Neighbor Embedding (tSNE) analysis49, reads for each cluster were subsampled to the lowest number for all clusters in the dataset (as specified in the text) since raw relative abundance values were analyzed (i.e. not utilizing a 2% relative abundance threshold as in other analyses). Bray-Curtis distance between taxa relative abundances within clusters was calculated, and this resulting distance matrix was utilized as the input for tSNE analysis.
The Net Relatedness Index (NRI) was calculated as previously described31 adapting code from the relatedness_library.py script from Qiime 1.9.150 which implements the same calculation as in phylocom 4.251. Briefly, species presence and absences across clusters were defined using the same 2% relative abundance threshold, and clusters containing only one OTU were omitted from analysis. OTU sequences were aligned and a neighbor-joining tree was constructed using MUSCLE 3.8.3152. The NRI was calculated as a standardized effect size for each cluster: NRI = −1 * (MPDcluster – MPDnull)/sd(MPDnull), where MPDcluster denotes the mean phylogenetic distance (MPD), and MPDcluster & sd(MPDnull) indicate the mean MPD, and the standard deviation of the MPD over 1000 iterations of a null mode. The null model, calculated for each cluster, was random draws for the number of OTUs present in the sample (i.e. preserving cluster OTU richness) from the sample pool (i.e. any OTU observed at least once in any cluster in the sample) without replacement. The null model therefore preserves the OTU richness of each cluster but randomizes the OTUs present from the set of OTUs occurring in the sample.
Bulk 16S sequencing and spike-in for absolute abundance calculation
An overview of all bulk 16S sequencing datasets generated in this study can be found in Supplementary Table 6. The bulk sequencing protocol followed our established spike-in sequencing pipeline25. Briefly, genomic DNA (gDNA) extraction was performed using a custom liquid handling protocol on a Biomek 4000 robot based on the Qiagen MagAttract PowerMicrobiome DNA/RNA Kit (Qiagen 27500–4-EP) but adapted for lower volumes. Samples were subjected to bead beating for a total of 10 minutes. For samples processed with the spike-in sequencing approach for absolute abundance calculation, the sample added was weighed on an analytical balance, and 10 uL of a frozen spike-in strain concentrate (Sporocarcina pasteurii, ATCC 11859, an environmental bacterium not found in the gut microbiome) was added during gDNA preparation. Resulting gDNA was subjected to amplification and sequencing of the 16S V4 region following a dual indexing scheme53 but utilized updated 515f and 806rB primers as in the MaP-seq technique. A 20 μL PCR amplification was performed (1 M forward and 1 μM reverse barcoded primers, 1 μL prepared gDNA, 10 μL NEBNext Q5 Hot Start HiFi Master Mix, 0.2X final concentration SYBR Green I). The PCR (98 °C for 30 s; cycle: 98 °C for 20 s, 55 °C for 20 s, 65 °C for 60 s, 65 °C for 5 m) was run on a real-time PCR machine to stop reactions during exponential amplification. Amplicon products were quantified and pooled, the expected 390 bp product was gel-extracted, and paired-end sequencing was performed with an Illumina MiSeq 300 cycle v2 kit (read1: 154 bp, read2: 154 bp, custom sequencing primers spiked into sequencing kit) at 10 pM loading concentration with 20% PhiX spike in. Resulting sequences were processed with USEARCH as above. The absolute bacterial density for a sample (A) was calculated by utilizing the weight of sample added (w) and proportion of reads mapping to spike in strain (ps) in the following formula: A = (1 – ps)/(ps * w). The absolute density of individual OTUs was calculated by rescaling the total sample absolute density by the relative abundance of sample OTUs.
16S FISH and imaging
Samples were fixed as with the MaP-seq protocol, embedded within paraffin blocks, 4 μm thick lumenal sections were cut and deparaffinized. 16S FISH was performed as previously described7,17. Briefly, previously validated FISH probes targeting abundant taxa present in the sample were obtained with conjugated fluorophores suitable for multiplex imaging: Erec482_a488 or Erec482_cy354 targeting Lachnospiraceae, Lab158_cy355 targeting Lactobacillaceae and Enterococcaceae, Ato291_cy556 targeting Coriobacteriaceae, Eub338_cy557 targeting Bacteria, and Non338_cy558 control probe (see Supplementary Table 4). Sections were incubated with probes at 10 ng/μL in FISH hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.5, 0.01% SDS, 10% formamide) at 47 °C for 4 hours. Sections were then incubated in preheated FISH wash buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.5) for 10 minutes, washed 3X times in PBS, incubated with 10 μg/mL DAPI in PBS for 10 minutes and washed 3X times in PBS. Sections were then mounted in mounting medium (Vector Laboratories H1000).
Images were acquired on a Nikon Eclipse Ti2 epifluorescence microscope with a SOLA-SE2 illuminator and Andor Zyla 4.2 plus camera controlled by Nikon Elements AR software. DAPI, FITC/GFP, RFP and CY5 filter cubes (Nikon 96359, 96362, 96364, 96366 respectively) were utilized. Large area four-color fluorescence scans with three 0.6 μm Z-stacks within the 4 μm section were performed with a Plan Apo λ 40X objective. The extended depth of focus (EDF) module was applied to resulting Z-stacks to obtain a focused image across the stack, and images across the entire section were stitched together.
Supplementary Material
Acknowledgements
We thank A. Kaufman for technical assistance, R. Rabadan and D. Vitkup for technical advice and discussions, and D. Pe’er and the CUMC Pathology Department for access to sequencing instruments. H.H.W. acknowledges specific funding from the NIH (1R01AI132403, 1R01DK118044), ONR (N00014-15-1-2704), and Burroughs Welcome Fund PATH (1016691) for this work. K.W.L. is partially supported by NIH R01GM110494. P.A.S. acknowledges support from NIH/NIBIB K01EB016071. R.U.S. is supported by a Fannie and John Hertz Foundation Fellowship and a NSF Graduate Research Fellowship (DGE-1644869).
Footnotes
Competing Interests
H.H.W. and R.U.S. are inventors on a provisional patent application filed by the Trustees of Columbia University in the City of New York regarding this work.
Data availability
All sequencing data is available at NCBI SRA under PRJNA541181.
Code availability
Code utilized in this study, microfluidic device designs and OTU tables can be accessed at http://github.com/ravisheth/mapseq.
References
- 1.Reichenbach T, Mobilia M & Frey E Mobility promotes and jeopardizes biodiversity in rock–paper–scissors games. Nature 448, 1046–1049 (2007). [DOI] [PubMed] [Google Scholar]
- 2.MacArthur RH & Wilson EO The theory of island biogeography. (1967).
- 3.Cordero OX & Datta MS Microbial interactions and community assembly at microscales. Current Opinion in Microbiology 31, 227–234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swidsinski A, Loening Baucke V, Verstraelen H, Osowska S & Doerffel Y Biostructure of Fecal Microbiota in Healthy Subjects and Patients With Chronic Idiopathic Diarrhea. Gastroenterology 135, 568–579.e2 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Yasuda K et al. Biogeography of the Intestinal Mucosal and Lumenal Microbiome in the Rhesus Macaque. Cell Host & Microbe 17, 385–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earle KA et al. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host & Microbe 18, 478–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE & Borisy GG Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences 113, E791–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI & Borisy GG Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U.S.A 21, 201711596–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson GP, Lee SM & Mazmanian SK Gut biogeography of the bacterial microbiota. 1–13 (2015). doi: 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SM et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 1–6 (2013). doi: 10.1038/nature12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagara Y, Takada T, Nagata Y, Kado S & Kushiro A Microscale spatial analysis provides evidence for adhesive monopolization of dietary nutrients by specific intestinal bacteria. PLoS ONE 12, e0175497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tropini C, Earle KA, Huang KC & Sonnenburg JL The Gut Microbiome: Connecting Spatial Organization to Function. Cell Host & Microbe 21, 433–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nava GM, Friedrichsen HJ & Stappenbeck TS Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J 5, 627–638 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedron T et al. A Crypt-Specific Core Microbiota Resides in the Mouse Colon. mBio 3, e00116–12–e00116–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valm AM, Welch JLM & Borisy GG CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Systematic and Applied Microbiology 35, 496–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geva-Zatorsky N et al. In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nature Medicine 21, 1091–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitaker WR, Shepherd ES & Sonnenburg JL Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell 169, 538–546.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira FC & Berry D Microbial nutrient niches in the gut. Environ Microbiol 19, 1366–1378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaldson GP et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wexler AG et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A 201525637–6 (2016). doi: 10.1073/pnas.1525637113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HJ, Boedicker JQ, Choi JW & Ismagilov RF Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proceedings of the National Academy of Sciences 105, 18188–18193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coyte KZ, Schluter J & Foster KR The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Amann R & Fuchs BM Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nature Reviews Microbiology 6, 339–348 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Coyne MJ & Comstock LE An Ecological Network of Polysaccharide Utilization among Human Intestinal Symbionts. Current Biology 24, 40–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji BW et al. Quantifying spatiotemporal dynamics and noise in absolute microbiota abundances using replicate sampling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ormerod KL et al. Genomic characterization of the uncultured Bacteroidales family S24–7 inhabiting the guts of homeothermic animals. Microbiome 1–17 (2016). doi: 10.1186/s40168-016-0181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakoff-Nahoum S, Foster KR & Comstock LE The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmody RN et al. Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host & Microbe 17, 72–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnenburg ED et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb CO, Ackerly DD, McPeek MA & Donoghue MJ Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst 33, 475–505 (2002). [Google Scholar]
- 32.Cavender-Bares J, Kozak KH, Fine PVA & Kembel SW The merging of community ecology and phylogenetic biology. Ecology Letters 12, 693–715 (2009). [DOI] [PubMed] [Google Scholar]
Online Methods References
- 33.Mazutis L et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc 8, 870–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parada AE, Needham DM & Fuhrman JA Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Walters W et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 1, e00009–15–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein AM et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bose S et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biology 1–16 (2015). doi: 10.1186/s13059-015-0684-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zilionis R et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Johansson MEV & Hansson GC Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol. Biol 842, 229–235 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apprill A, McNally S, Parsons R & Weber L Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol 75, 129–137 (2015). [Google Scholar]
- 43.Spencer SJ et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. 1–10 (2015). doi: 10.1038/ismej.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abate AR, Chen C-H, Agresti JJ & Weitz DA Beating Poisson encapsulation statistics using close-packed ordering. Lab Chip 9, 2628–2631 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Edgar RC & Flyvbjerg H Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Garrity GM, Tiedje JM & Cole JR Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao L et al. A catalog of the mouse gut metagenome. Nature Biotechnology 33, 1103–1108 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Maaten LVD & Hinton G Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008). [Google Scholar]
- 50.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb CO, Ackerly DD & Kembel SW Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Applied and Environmental Microbiology 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franks AH et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Applied and Environmental Microbiology 64, 3336–3345 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harmsen H, Elfferich P & Schut FA 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microbial Ecology in Health and Disease 11, 3–12 (1999). [Google Scholar]
- 56.Harmsen H et al. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Applied and Environmental Microbiology 66, 4523–4527 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amann RI et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and Environmental Microbiology 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallner G, Amann R & Beisker W Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.