Abstract
TP53 (p53) is the most frequently altered gene in human cancer. Identification of vulnerabilities imposed by TP53 alterations may enable effective therapeutic approaches. Through analyzing shRNA screening data, we identified TPRKB, a poorly characterized member of the tRNA-modifying EKC/KEOPS complex, as the most significant vulnerability in TP53-mutated cancer cell lines. In vitro and in vivo, across multiple benign-immortalized and cancer cell lines, we confirmed that TPRKB knockdown in TP53-deficient cells significantly inhibited proliferation, with minimal effect in TP53 wild-type cells. TP53 reintroduction into TP53-null cells resulted in loss of TPRKB sensitivity, confirming the importance of TP53 status in this context. Additionally, cell lines with mutant TP53 or amplified MDM2 (E3-ubiquitin ligase for TP53) also showed high sensitivity to TPRKB knockdown, consistent with TPRKB-dependence in a wide array of TP53-altered cancers. Depletion of other EKC/KEOPS complex members exhibited TP53-independent effects, supporting complex-independent functions of TPRKB. Lastly, we found that TP53 indirectly mediates TPRKB degradation, which was rescued by co-expression of PRPK, an interacting member of the EKC/KEOPS complex, or proteasome inhibition. Together, these results identify a unique and specific requirement of TPRKB in a variety of TP53-deficient cancers.
Keywords: TP53, TPRKB, synthetic lethality, non-oncogene addiction, EKC/KEOPS
INTRODUCTION
Tumor protein 53 (TP53 or p53) is a transcription factor that mediates the expression of genes involved in a myriad of cellular processes. In response to DNA damage or other genotoxic stressors, TP53 acts to regulate cell cycle, senescence, and apoptosis (1). Beyond these canonical functions, TP53 has also been implicated in cellular metabolism, autophagy, angiogenesis and migration (2). The importance of TP53 as a tumor suppressor is highlighted by the observation that approximately half of all cancers harbor inactivating TP53 mutations and these mutations are a driving force in cancer development and progression (3,4). Importantly, the majority of TP53 mutations involve the production of mutant TP53 that loses wild-type function while potentially gaining oncogenic capabilities, in addition to deleterious mutations or homozygous deletion.
Development of effective therapies for tumor suppressors, such as TP53, have been challenging in part due to the difficulty of developing therapeutic approaches that restore function. Several potential approaches for targeting TP53-deficient cancers have been described (5–8), including those exploiting non-oncogenic addiction to induce anti-tumorigenic cellular events, such as synthetic lethality(9). In this concept, genetic alterations render cancer cells dependent on genes that are not inherently oncogenic. The most advanced example of this is the use of PARP inhibitors in BRCA1/2 mutated cancers (10,11). Both BRCA1/2 and PARP1 play key roles in DNA damage repair, and when both pathways are simultaneously defective cells are unable to maintain sufficient DNA integrity and undergo mitotic catastrophe. Cancer cells harboring BRCA1/2 mutations are thus sensitive to PARP inhibitors, while similarly treated normal cells that maintain BRCA1/2 repair mechanisms remain largely viable.
Herein, we analyzed shRNA screening data from the Project Achilles cancer cell line compendium to identify TP53RK Binding Protein (TPRKB) as a specific vulnerability in TP53-altered cancers(12). TPRKB is a member of the evolutionarily conserved Endopeptidase-like Kinase Chromatin-associated protein complex/Kinase putative Endopeptidase and Other Proteins of Small size protein complex (EKC/KEOPS), along with TP53RK (PRPK), OSGEP, LAGE3, and C14ORF142 (13–17). EKC/KEOPS is responsible for the essential N6-threonylcarbamoyladenosine (t6A) modification of all ANN-codon recognizing tRNAs, and it is important for telomere length regulation in yeast(14,17–19). Interestingly, previous studies of the EKC/KEOPS complex have demonstrated that PRPK interacts with, phosphorylates, and activates TP53(13,20,21). Recently, germline mutations in EKC/KEOPS complex members have been linked to Galloway-Mowat syndrome, a rare condition characterized by early-onset nephrotic syndrome and microcephaly(22). Herein, we identify TPRKB as a vulnerability specifically in TP53-deficient cancers, with minimal effect in TP53 wild-type cells. Furthermore, we show that this reliance is independent of other EKC/KEOPS complex members, defining a novel function of TPRKB in human cancer.
MATERIALS AND METHODS
Cell culture and proliferation
H358, H196, Colo205, BxPC-3, HT-29, SJSA-1,93T449, CaOV3, HEK293T, MDA-MB-231, MDA-MB-468, and MCF10A were authenticated by and obtained from the American Type Culture Collection (ATCC). RKO and HCT116 cell lines were authenticated by and obtained from Horizon. HCC827 was a gift from Dr. David Beer’s lab. Hu09 and SAOS-2 were a gift from Dr. Arul Chinnaiyan’s lab. No further authentication of cells was performed. Upon receipt, cells were tested for Mycoplasma contamination using a commercially available kit and protocol (Sigma, LookOut Mycoplama PCR Detection Kit, MP0035). Negative cell lines were propagated and frozen until needed. Cell lines were typically used for experiments within 1.5 to 2 months post-thawing. H358, H196, HCC827, Colo205, BxPC-3, HT-29, and HCT116 were grown in RPMI containing 10% FBS. SJSA, 93T449, Hu09, SAOS-2, CaOV3, and HEK293T were grown in DMEM containing 10% FBS. MDA-MB-231 and MDA-MB-468 were grown in DMEM supplemented with non-essential amino acids and glutamax. MCF10A were grown in Mammary Epithelial Cell Growth Medium (Lonza). RKO cells were grown in EMEM media containing 10% FBS. Cell growth was monitored by either counting cell number using Beckman Z Coulter Counter or by measuring confluency using Incucyte (Essen Biosciences). For Coulter Counting experiments, typically 0.5×104 cells/well (SJSA-1, H196, 93944T, Hu09, SAOS-2) or 1×104 cells/well (for H358, MB-MDA-231, MB-MDA-468, MFC10A, HCT116, HT29, BxPC3, RKO and CaOV3) were plated on day 0 in a 24-well plate. Depending on the growth rate of cells, on either days 2, 4 and 6 after plating or days 2, 3, and 4 after plating cells were trypsinized for Coulter Counting analysis. All experiments utilized triplicate samples, with the average and standard error plotted. Two-sided t-test p-values <0.05 (*) and <0.001(**) for the last day of growth are indicated. For Incucyte experiments, either 1×103 cells/well or 4 ×103 cells/well were plated in 96-well plates in triplicates, and readings were taken every 4 hours. The 1-minute video reported is 50 frames taken over 7 days. All results were representative of at least two independent experiments.
RNA extraction and qPCR analyses
Cells were pelleted and lysed in lysis buffer (Purelink RNA Mini kit, Life technologies). RNA was extracted as per manufacturer’s instructions and quantified by NanoDrop 2000 spectrophotometer (Thermo Fisher). cDNA was prepared using High Capacity cDNA Reverse Transcription Kit, per manufacturer’s instruction (Applied Biosciences). SYBR green-based qPCR was performed in triplicate using various primers, as listed in Supplementary Table 1. HMBS was used as a normalization control for all experiments unless otherwise specified.
Tumor Xenograft Model
All procedures for mice experiments were approved by The University of Michigan University Committee on Use and Care of Animals (UCUCA). To evaluate the role of TPRKB in tumor formation, HCT116 and HCT116 TP53wt/R248W and SJSA-1 cells were infected with either control (scrambled sequence) or TPRKB specific shRNA (Supplementary Table 2) and propagated in medium containing 1ug/ml puromycin (Invivogen) for period of at least 10 days. 1×106 cells/side were injected subcutaneously in the flanks of athymic nude (Jackson labs). Each group consisted of 10 mice. Tumor was measured biweekly, and tumor volumes were calculated using following formula: pi/6(LxWxW), where L is length of the tumor and W is width of the tumor (23). Tumors were allowed to grow for 35–40 days at which point mice were sacrificed; tumors were collected and photographed.
Western Blot Analysis
Cell lysates were collected in NuPAGE LDS Sample Buffer and Reducing Agent (Life Technologies) at a 1x final concentration, sonicated, and denatured at 95°C for 5–15 minutes. NuPAGE 4–12% Bis-Tris gels (Life Technologies) were run in 1x NuPAGE MES SDS running buffer at 120V for 1.5–2 hours, followed by semi-dry transfer in 1x NuPAGE transfer buffer containing 20% methanol at 25V for 1 hour onto Immobilon-P PVDF membranes (Millipore). Membranes were blocked in either 5% milk for 5% Bovine Serum Albumin (based on manufacturer’s instructions) for 1 hour before probing with primary antibodies (Supplementary Table 3). Washes were completed with 1x TBS + 0.1% Tween-20. Signals were detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore). B-actin was used as a loading control unless otherwise specified.
Cell cycle analyses
H358 cells stably infected with vector control or TPRKB shRNA lentiviruses were plated in 6-well plates. Following 40 hour serum starvation, in order to synchronize the cell cycle, 10% serum containing RPMI was added to cells. Based on H358 doubling time of approximately 38 hours (ATCC), cells were collected 40 hours after serum reintroduction. Cells were then trypsinized, washed in Dulbecco’s Phosphate Buffered Saline (DPBS), and spun down. Pelleted cells were then resuspended in 0.5ml of 100% ethanol and stored at 4°C until further use. SJSA-1 cells that had been stably infected with vector control or TPRKB shRNAs were similarly processed, with the exception of collection occurring 24 hours post-serum-reintroduction with their respective media. Prior to staining, cells were re-pelleted, the ethanol was decanted, and cells were resuspended in DPBS containing 50ug/ml propidium iodide and 100ug/ml RNAse A. Cells were incubated in dark for 20 minutes before subjecting to flow-cytometry analyses. Data collected was further processed through ModFit software (Verity Software House). Data shown is a representative bar graph of two independent experiments.
DNA constructs and lentivirus production.
Lentiviral DNA vectors for TP53-V5 (22945), TP53 p.R175H-V5 (22936), TP53 p.R249S-V5 (22935), TP53 p.R273H-V5 (22934), and TP53 p.R280K-V5 (22933) were obtained from Bernard Futscher’s lab via Addgene. Additionally, RALB-HA (50989-from Anna Sablina’s lab), Rap2A-HA (19311- from David Sabatini’s lab), RagB-HA (19313- from David Sabatini’s lab), and RagD-HA (19316- from David Sabatini’s lab) were also obtained from Addgene. Lentiviral DNA vectors for PRPK-FLAG, TPRKB-HA, and TPRKB-Flag were created using cDNA generated from MCF10A cells, adding a tag through PCR, and cloned into a pLenti6 background with the pLenti6/V5 Directional TOPO Cloning Kit per the manufacturer’s instructions (ThermoFisher). Briefly, PCR was used to add our tag of interest (primer information in Supplementary Table 1) and create blunt end products. This was followed by TOPO cloning, gel purification, and transformation of STBL3 competent cells. 10–15 colonies were selected, and DNA was isolated with the use of PureLink Quick Plasmid Miniprep Kit per the manufacturer’s instructions (Invitrogen). DNA was then submitted for Sanger sequencing DNA Sequencing Core (University of Michigan Medical School) to verify the end products. All lentiviruses were synthesized either from the UMICH Vector Core (University of Michigan) or System Biosciences.
Transient DNA Transfections for Protein Stability Analysis
HEK293T cells were plated in a 6-well plate for 24 hours. FuGENE HD transfection reagent was then used to perform transfections per manufacturer’s protocol (Promega). In addition to plasmids of interest, pCDH-CMV-MCS-EF1a-copGFP (SBI Biosciences) construct was supplemented to normalize for total DNA transfection amounts between samples. Cell lysates were collected 24–48 hours later in NENT buffer: 100mM NaCl, 20mM Tris-HCl (ph = 8.0), 0.5 mM EDTA, and 0.5% (v/v) NP40. For analysis of protein degradation mechanism, cells were plated as described, but after 24 hours of transfection cells were treated with either DMSO or 500 nM bortezomib. After an additional 16–18 hours, cells were collected in NENT buffer for Western analysis.
Stable protein expressing or knockdown clones using recombinant viruses
Mammalian expression plasmids were generated or obtained from Addgene as described above. shRNA constructs were created using System Biosciences or purchased from Open Biosystems (Supplementary Table 2). DNA was made using Purelink Plasmid Midiprep kit (Life technologies) and submitted to the University of Michigan Vector Core for Lentiviral preparation. Active lentiviruses were infected to 50–60% confluent cells in either 6-well plate or 100-mm dish using polybrene (Millipore). 24 hours after infection, selection media was added. For exogenous expression plasmids, 5ug/ml Blasticidin containing medium was used (Invivogen). For knockdown clones, 1ug/ml puromycin containing medium was used. For clones that had over-expression of protein and knockdown of gene; media containing both 2.5ug blasticidin and 0.5ug puromycin were used. Subsequently to selection, clones were tested for over-expression and/or knockdown either by qPCR and/or Western analysis.
Co-immunoprecipitation experiments
For endogenous Co-IP experiments, cells were collected in NENT buffer, briefly sonicated, and spun down. ANTI-FLAG M2 Magnetic Beads (Sigma-Aldrich) were used to perform co-IP per the manufacturer’s instructions. Lysates were incubated with the beads overnight at 4°C, and eluted directly into 20 uL of 2x NuPAGE sample buffer (without reducing reagent). Samples were then used for Western blot analysis.
For exogenous Co-IP experiments, cells were transfected as described above. After 48 hours cells were collected in NENT buffer, briefly sonicated, and spun down. Antibodies (Supplementary Table 3) were used in conjunction with the Immunoprecipitation Kit-Dynabeads Protein G (Invitrogen) per the manufacturer’s instructions. After elution and denaturation, samples were used for Western blot analysis.
Genomic editing using CRISPR-Cas9
The CRISPR plasmid for knockout of TPRKB and PRPK was purchased from Sigma. The gRNA sequences are shown in Supplementary Table 1. The gRNA sequence for tagging of TPRKB was designed using the http://crispr.mit.edu/ website. The gRNA sequence (Supplementary Table 1) was cloned into CRISPR-Cas9 and gRNA expression vector plentiCRIPSV2 (Gift from Feng Zhang -Addgene plasmid #52961). To make TPRKB-Flag homologous recombination template, the forward oligo and reverse oligo containing the Flag sequence (underlined) were annealed to form double-stranded DNA following the protocol (http://www.origene.com/assets/documents/cDNA/pCas-Guide_System_Validation.pdf). The cells were transfected using lipofectamine 3000 following manufacturer’s protocol, and then were seeded into single cells following puromycin selection for 48 hrs. For the knockout experiments, 500ng of Cas9+sgRNA vector was used for transfection. For the knock-in experiments, 250 ng of Cas9+sgRNA vector and 250 ng of the annealed donor DNA were transfected in HEK293T cells in 24-well plate. Genomic DNA was extracted from the clonal lines using QuickExtract™ DNA Extraction Solution (QE09050, epicenter). Loci targeted by guide RNAs were amplified using the primers listed in Supplementary Table 1, and then sequenced using the forward primers.
RESULTS
Identification and validation of TPRKB dependency in TP53-mutant cell lines from Project Achilles
To identify potential vulnerabilities in cancers with specific genomic alterations, we mined data from the Broad Institute’s Project Achilles (12). Project Achilles contains information from genome-scale knockdown screens linked to observed cell survival in genomically characterized cancer cell lines. We analyzed the original shRNA data set (12), using the raw microarray log2 fold change in shRNA abundances for each cell line at the conclusion of the screening relative to the initial plasmid DNA reference pool fold change. Cell lines were annotated based on the presence or absence of hotspot (in oncogenes) or hotspot/deleterious (in tumor suppressors) mutation status and MDM2 status, and significant over-expression of mutations in highly growth inhibited cell lines was identified by a Fisher’s exact test. We confirmed several expected oncogenic vulnerabilities, such as BRAF in BRAF mutated cancer (data not shown). From this data, we also predicted ARID1B as a vulnerability specifically in ARID1A mutant cell lines (Fig S1). Importantly, requirement of ARID1B in ARID1A-deficient cancers was independently published, supporting the validity of our approach at identifying these relationships even in tumor suppressors (24).
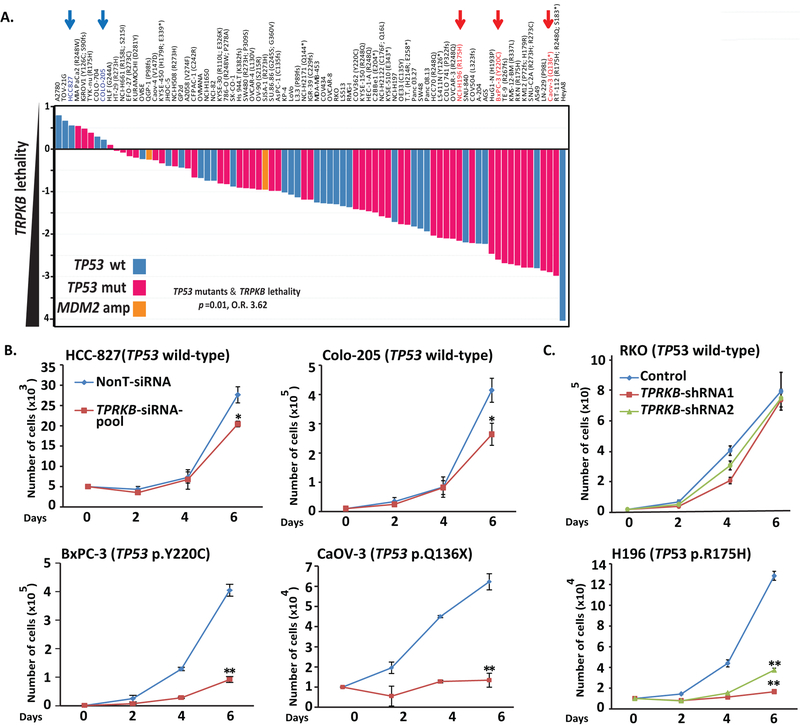
Hence, we were intrigued as only a single gene, TPRKB—a poorly characterized member of the EKC/KEOPS complex—was identified as a significant vulnerability in TP53 mutated (both hotspot and deleterious mutation) cancer cells (Fig 1A & S1). Importantly, analysis of the COSMIC (https://cancer.sanger.ac.uk/cosmic) (25) and MiPanda (http://mipanda.org) (26) databases demonstrated that TPRKB is both ubiquitously expressed across normal and cancer tissues/cells and infrequently genomically altered in cancer (Fig S1). To confirm the Project Achilles data, we used siRNA in select Project Achilles’ cancer cells lines, and confirmed marked decrease in proliferation in TP53-mutant vs. TP53 wild-type cells (Fig 1B & S1). Similar results were obtained with stable TPRKB knockdown using two independent shRNAs (Fig 1C & S1), validating the Project Achilles data identifying TPRKB-dependence in TP53 deficient cancers. Like ARID1A altered cell lines insensitive to ARID1B deficiency (reference (24) and Fig S1), we confirmed that some TP53 altered cell lines were insensitive to TPRKB depletion, such as the TP53-mutant HT-29 cell line, which was predicted to be non-responsive in the Project Achilles screen and confirmed in vitro (Fig S1), highlighting the need for further characterization of determinants of TPRKB sensitivity.
Figure 1. Identification and validation of TPRKB vulnerability in cancer cell lines with TP53 alterations.
A) TPRKB was the only gene identified in the Project Achilles genome-wide shRNA database as showing significant enrichment for dependency in TP53 altered cell lines (two-sided Fisher’s exact test odds ratio (O.R.) and p-value are shown for original (2015) TP53 annotation status; O.R. = 2.6 and p=0.06 for TP53 status and TPRKB dependency [> or < 1.4] for 2019 comprehensive cell line encyclopaedia [CCLE] TP53 annotation status). TPRKB dependency (fold-change in shRNA abundance versus control transfected cells) for cancer cell lines from Project Achilles is plotted, with cell lines ordered by increasing TPRKB dependency. The color of the bars indicates mutational status from 2019 CCLE annotation: blue bars indicate TP53 wild-type cells, red bars indicate TP53 hotspot/deleterious mutants, and orange bars indicate MDM2-amplifications. Blue and red arrows indicate cell lines with wild-type and mutant TP53, respectively, that were chosen for validation experiments. B) Differential effects of pooled siRNA against TPRKB (or scrambled control) on cell proliferation were confirmed in TP53 wt (Colo-205 and HCC-827) and TP53 mut cell lines (BxPC-3 and CaOV-3). C) As in B, but using independent shRNAs against TPRKB (or scrambled control) in RKO (TP53 wt) and H196 (TP53 mut). Confirmation of TPRKB knockdown is shown in Figures S1. All experiments utilized triplicate samples, with the average and standard error plotted.
* indicate p-values < 0.05 and ** indicate p-values <0.01.
Multiple types of TP53 alterations confer TPRKB sensitivity
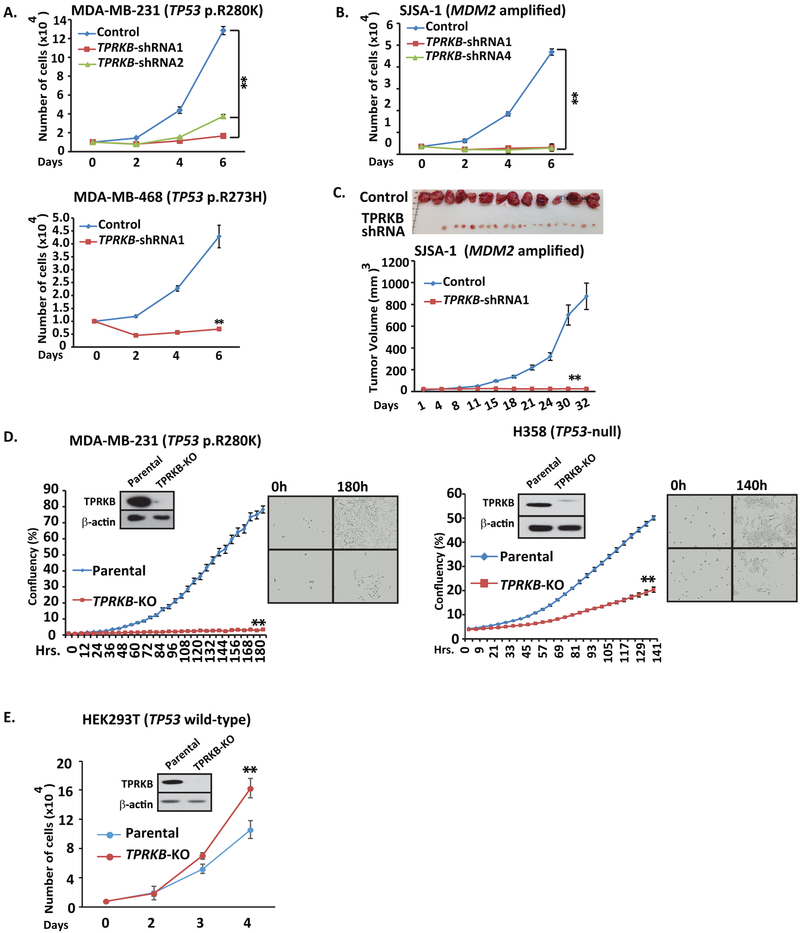
TP53 is genomically altered through multiple mechanisms, including hotspot mutations, deleterious mutations, and through activation of pathways that modulate TP53 protein, such as amplification of the E3-ubiquitin ligase MDM2. Hence, to determine if TPRKB dependency was conferred by a spectrum of TP53 perturbations, we tested for TPRKB sensitivity in TP53 mutant cell lines MDA-MB-231 (TP53 p.R280K) and MDA-MB-468 (TP53 p.R273H), as well as cell lines harboring MDM2 amplification including SJSA-1 & 93T449, Hu-09 (TP53 loss through fusion) and SAOS-2 (TP53 null). Importantly, all of these cell lines showed a striking decrease in cell proliferation upon TPRKB knockdown (Fig 2A, 2B, & S2). We confirmed these in vitro observations in vivo through SJSA xenografts in mice, which demonstrated a profound reduction of tumor size and burden in SJSA-1 cells with TPRKB knockdown (Fig 2C).
Figure 2. Various classes of TP53 perturbation result in marked TPRKB dependent proliferation.
Cancer cell lines with A) hotspot TP53 mutations (MDA-MB-231 and MDA-MB-469) or B) MDM2 amplification (SJSA-1; see Figure S2 for additional cell lines) were assessed for TPRKB dependent proliferation using shRNA. C) As in B, except using in vivo mouse xenografts with tumor volume plotted and tumors at sacrifice shown. D) CRISPR-Cas9 mediated TPRKB knockout in TP53 mut MDA-MB-231 and H358 (TP53 deep deletion) cells confirmed results from siRNA/shRNA. TPRKB knockout was confirmed by Western blotting, and % confluency was plotted with representative phase contrast photomicrographs at the indicated time points shown. E) Minimal proliferation inhibition was observed upon TPRKB knockout by CRISPR-Cas9 in benign immortalized TP53 wt cells (HEK293T). All experiments utilized triplicate samples, with the average and standard error plotted.
* indicate p-values < 0.05 and ** indicate p-values <0.01.
Confirmation of TPRKB sensitivity in TP53 mutant cancer cells through CRIPSR knockout
To unambiguously confirm TPRKB sensitivity in TP53 altered cancer cell lines, we used CRISPR-Cas9 to knockout either TPRKB or a reported interacting member of the EKC/KEOPS complex, PRPK, in various cell lines: HEK293T (TP53 wild-type), MDA-MB-231 (TP53 p.R280K) and H358 (TP53-null)(13). Sanger sequencing and Western analysis confirmed knockout of the genes and proteins, respectively (Fig 2D & 2E). Consistent with the siRNA and shRNA results described above, MDA-MB-231 and H358 TPRKB-knockout cells showed severely reduced cell proliferation, while HEK293T TPRKB-knockout cells exhibited slightly increased proliferation (Fig 2D & 2E). Interestingly, PRPK knockout in MDA-MB-231 cells only resulted in a marginal reduction in proliferation, while PRPK knockout in HEK293T cells resulted in increased proliferation, underscoring the greater dependence on TPRKB in TP53 perturbed cells (Fig S2). As PRPK stabilizes TPRKB (see below), we consider the modest proliferation defect observed upon PRPK-knockout as an indirect effect due to TPRKB reduction. Importantly, these results confirm those seen by siRNA and shRNA described above and demonstrate an essential role for TPRKB in a wide range of cancer cells with altered TP53 function.
TP53 reintroduction rescues proliferation upon TPRKB knockdown in TP53-null cells
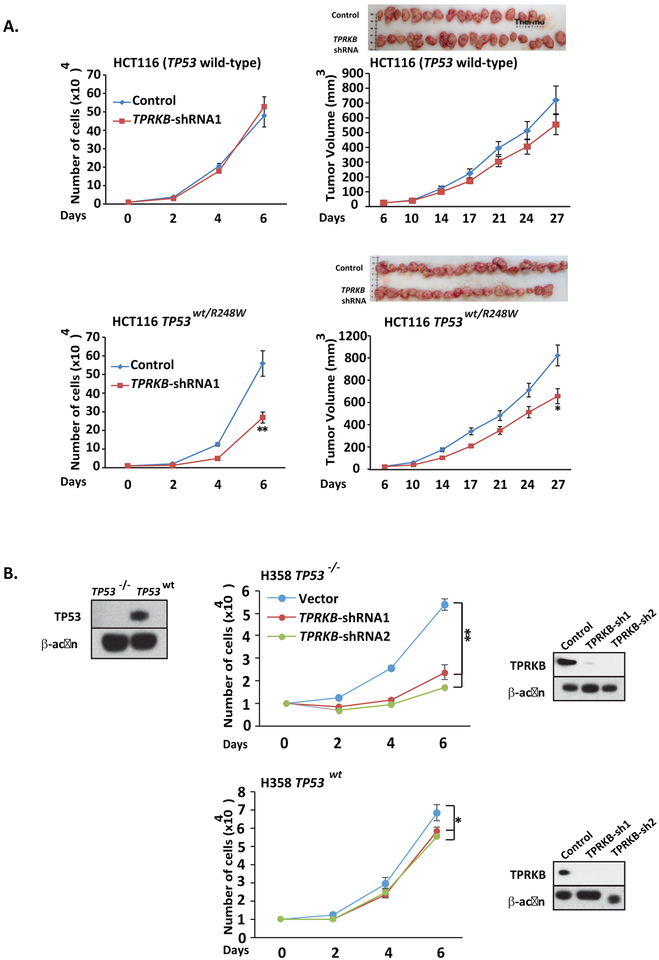
To confirm that the above effects of TPRKB knockdown/knockout in TP53 mutated cancer cells was directly TP53 dependent, we first utilized the isogenic colorectal cancer cell line HCT116 and HCT116 TP53wt/R248W. Notably, HCT116 is a TP53 wild-type cell line, and HCT116 TP53wt/R248W cell lines express TP53 p.R248W and wild-type TP53. As shown in Figure 3A, parental HCT116 cells with stable TPRKB shRNA knockdown did not show significant proliferative defects in vitro or in a mouse xenograft model compared to scrambled shRNA control cells. In contrast, stable TPRKB knockdown in HCT116 TP53wt/R248W resulted in modest, but significantly decreased proliferation in vitro and in vivo compared to scrambled shRNA control cells (Fig 3A).
Figure 3 – Isogenic cells show wild-type TP53 presence rescues proliferation defects from TPRKB knockdown.
A) TPRKB was stably knocked down in isogenic HCT116 (TP53 wild-type) and HCT116 TP53WT/R248W cells. The stable clones were assayed for cell proliferation and tumor formation in nude mice. B) TPRKB knockdown by shRNA was performed in H358 cells stably expressing LacZ control (left) or TP53 (right) and proliferation was monitored. Inset Western blot panels confirm TP53 over-expression from lentiviral transduction and TPRKB knockdown. All experiments utilized triplicate samples, with the average and standard error plotted.
* indicate p-values < 0.05 and ** indicate p-values <0.01.
As the above HCT116 isogenic system represents an exogenous TP53 deficient model, we sought to evaluate TPRKB dependency in an endogenous TP53 deficient model where TP53 could be reintroduced. Hence we created isogenic cell lines from TP53-null H358 lung cancer cells (H358 TP53−/−). We generated H358 cells through lentiviral infection that stably expressed wild-type TP53 (H358 TP53wt), recurrent TP53 mutations (H358 TP53R175H, H358 TP53R249S, H358 TP53R273H and H358 TP53R280K) or LacZ (H358 TP53−/− LacZ) as a control. As shown in Figure 3B, stable TPRKB knockdown reduced cell proliferation to a much greater extent in H358 TP53−/− LacZ compared to H358 TP53wt. In contrast, stable expression of the above TP53 mutations in H358 cells did not rescue the proliferation defect upon stable TPRKB knockdown (Fig S3). Taken together, these results confirm the TP53-dependent response to TPRKB depletion specifically in TP53 mutant or null cells.
TPRKB dependency in TP53 mutant cells is unique amongst EKC/KEOPS complex members
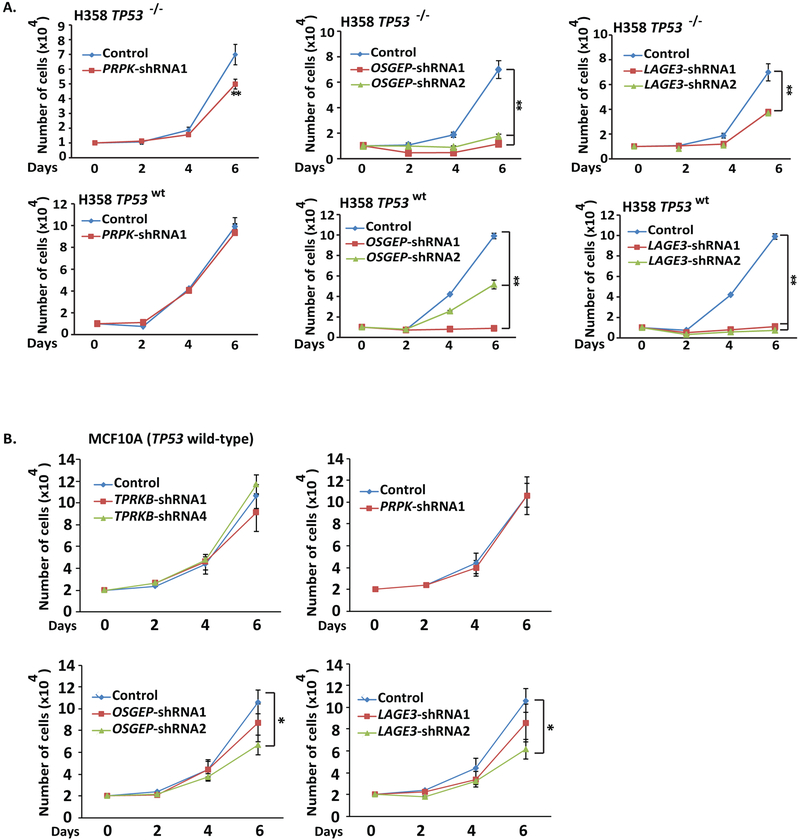
As described above, TPRKB is a member of EKC/KEOPS complex, which plays a major role in tRNA modification. Hence, we stably knocked down the other individual canonical complex members—PRPK, OSGEP and LAGE3—in H358 TP53−/− LacZ and H358 TP53wt cells (Fig S4). Consistent with the results of PRPK knockout in MDA-MB-231 cells described above, PRPK knockdown in H358 TP53−/− showed modestly reduced proliferation that was rescued by TP53 expression in H358 TP53wt cells (Fig 4A). In contrast, knockdown of OSGEP or LAGE3 significantly reduced cell proliferation independent of TP53 status in H358 cells. We then addressed the effect of individual EKC/KEOPS member knockdown in benign-immortalized TP53 wild-type MCF10A cells (Fig S4). Consistent with other TP53 wild-type models, TPRKB or PRPK knockdown in MCF10A had minimal impact on proliferation, while OSGEP and LAGE3 knockdown significantly affected cell proliferation (Fig 4B). In contrast, TP53 wild-type A-204 sarcoma cells did not exhibit reduced proliferation upon knockdown of any members of the EKC/KEOPS complex, demonstrating that OSGEP and LAGE3 phenotypes do not stratify by TP53 status (Fig S4). Our observations support general insensitivity to TPRKB or PRPK depletion in TP53 proficient cells, in contrast to non-TP53 related sensitivity to depletion of other EKC/KEOPS complex members.
Figure 4-. Knockdown of other EKC/KEOPS complex members does not produce the same TP53-dependent effects as TPRKB.
A) TPRKB is a member of the EKC/KEOPS complex, which includes the canonical proteins PRPK, OSGEP and LAGE3. To investigate the TP53 dependent effects of other EKC/KEOPS complex members on proliferation, PRPK, OSGEP or LAGE3 were knocked down using shRNA in H358 TP53−/− LacZ and H358 TP53WT cells as in Figure 3. B) Proliferation of benign immortalized MCF10A (TP53 wt) cells was similarly assessed after stable knockdown of the indicated EKC/KEOPS complex member by shRNA. All experiments utilized triplicate samples, with the average and standard error plotted.
* indicate p-values < 0.05 and ** indicate p-values <0.01.
Loss of TPRKB leads to cell cycle arrest and a reduction in the expression of anti-apoptotic proteins in TP53-deficient cells
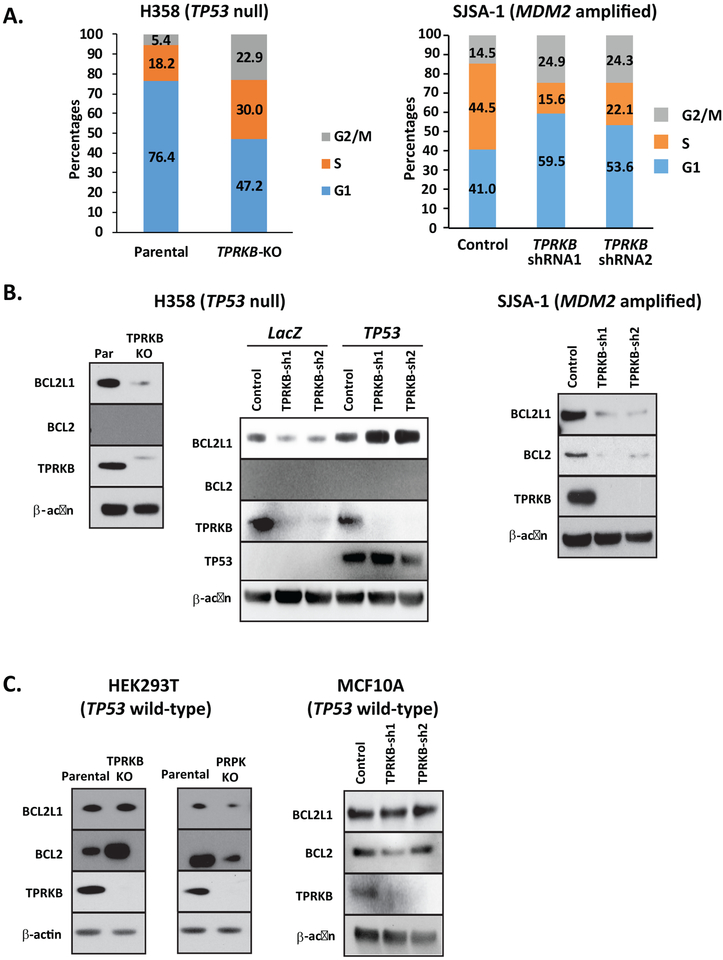
To investigate the mechanism of TPRKB dependency in TP53 altered cells, we first assessed the impact of TPRKB depletion on cell cycle progression in H358 TPRKB knockout cells and SJSA-1 TPRKB knockdown cells. Compared to H358 parental cells, H358-TPRKB knockout cells showed marked arrest in S and G2/M phase, while SJSA-1 TPRKB knockdown cells arrested in G1 and G2/M. (Fig 5A). As cell cycle arrest has been closely linked with modulation of the anti-apoptotic proteins B-cell lymphoma extra-large (Bcl-xL or BCL2L1) and B-cell lymphoma 2 (Bcl-2 or BCL2)(27), we assessed BCL2L1 and BCL2 in a panel of cell lines, including H358 parental, H358 TP53−/− LacZ, H358 TP53wt, HEK293T (TP53 wild-type), MCF10A (TP53 wild type), and SJSA-1 (MDM2-amplified) in the context of TPRKB depletion. Of note, compared to their respective parental lines, TP53-deficient H358, H358 TP53−/− LacZ, and SJSA-1 cells with TPRKB knockout or knockdown showed reduced BCL2L1 expression, while expression was largely unaltered in TP53 wild-type HEK293T, MCF10A, and H358 TP53wt cells with TPRKB loss (Fig 5B & 5C). Similarly, BCL2 expression was downregulated in SJSA-1 cells with TPRKB knockdown, while HEK293T and MCF10A showed increased and unchanged expression, respectively (Fig 5B & 5C). BCL2 was undetectable in H358 cells (Fig 5B). Taken together, these results suggest that the expression of anti-apoptotic factors may mediate TPRKB dependence in TP53-null cells.
Figure 5-. Loss of TPRKB leads to cell cycle arrest and reduced anti-apoptotic protein expression in TP53-deficient cells.
A) Serum stimulated synchronized H358 (parental) or CRISPR-Cas9 generated H358-TPRKB knockout (KO) cells and SJSA-1 control or TPRKB knockdown cells were assessed for cell cycle analysis by flow cytometry. The proportion of cells in G1, S, and G2/M is plotted. B) Anti-apoptotic protein (BCL2L1 and BCL2) expression was determined by Western blotting in H358 control, H358 TPRKB-KO, H358 TP53−/− LacZ, and H358 TP53WT and SJSA-1 control and TPRKB shRNA knockdown cells. C) Additional TP53 wild-type cell lines were assayed for BCL2 and BCL2L1 expression: HEK293T cells with TPRKB-KO or PRPK-KO and MCF10A control or TPRKB knockdown cells.
TP53 mediates TPRKB degradation, which can be partially rescued by either PRPK or inhibition of proteasomal machinery
As described above, while PRPK has been shown to interact with, phosphorylate, and activate TP53, exogenously over-expressed TPRKB and TP53 do not interact (13,20,21). However, TP53 and TPRKB interaction has not been assessed with endogenous proteins. Using CRISPR-Cas9, we endogenously tagged TPRKB with a Flag-epitope in HEK293T cells. We found that both TPRKB-Flag and endogenous PRPK are expressed exclusively in the cytosol (Fig S5), as opposed to both the nucleus and cytoplasm as previously reported (13). By IP-Western blotting, we observed the known interaction of PRPK and TPRKB, but TPRKB and TP53 did not interact (Fig 6A). Lastly, we confirmed that exogenously expressed TPRKB and TP53 did not interact (Fig S5). Our data is consistent with previous observations and supports an indirect relationship between TPRKB and TP53.
Figure 6. TP53 degrades TPRKB through the proteasome, while PRPK expression stabilizes TPRKB.
A) Parental HEK293T and HEK293T with CRISPR introduced FLAG epitope into the endogenous TPRKB locus (HEK293T-TPRKB-Flag) were used for co-immunoprecipitation. After Flag pulldown, samples were tested for endogenous TPRKB-Flag interaction with PRPK or TP53 B) Increasing amounts of exogenous TP53-V5 in HEK293T cells were used to observe effects of TP53 expression on TPRKB-HA protein levels by Western Blot. C) The impact of TP53 protein level on TPRKB expression was assessed through exogenous expression of TP53 in H358 cells and, conversely, knockdown of endogenous TP53 in HEK293T. D) Co-expression of PRPK-Flag and knockout of PRPK in HEK293T cells was used to determine PRPK effects on TPRKB protein levels. E) Using TPRKB knockout cells, we looked at PRPK expression by Western blot.
Given the interaction of PRPK with both TPRKB and TP53, we sought to determine whether TP53 may influence TPRKB stability through PRPK. Using exogenously expressed tagged proteins in HEK293T cells, we found that increasing amounts of TP53 led to a concentration-dependent reduction in TPRKB protein levels (Fig 6B). This observation was consistent in H358 cells, where stable exogenous expression of TP53 reduced TPRKB levels (Fig 6C). Likewise, even in HEK293T cells (insensitive to TPRKB knockdown), siRNA TP53 knockdown resulted in increased TPRKB levels (Fig 6C). We thus investigated the mechanism whereby TP53 mediates TPRKB expression by treating HEK293T cells co-expressing TPRKB and TP53 with the proteasome inhibitor bortezomib. As shown in Figure 6B, this led to a marked increase in TPRKB levels, even though a TP53-dependent reduction in TPRKB protein was still observable. Interestingly, TP53 only marginally reduced the expression of other unrelated proteins, but inhibition of the proteasome did not stabilize these levels (data not shown). Taken together, these data support the TP53 mediated degradation of TPRKB through the proteasome pathway.
In addition to interacting with, phosphorylating, and activating TP53, PRPK is the only component of the EKC/KEOPS complex that directly interacts with TPRKB (13,20,21). As we observed that PRPK was the only other member of the EKC/KEOPS complex that showed even modest differential response by TP53 status, we hypothesized that interaction with PRPK may mediate the TPRKB dependency of TP53-deficient cells. Through exogenous expression of tagged proteins, we found that PRPK is able to significantly stabilize TPRKB protein levels in both the absence (Fig 6D) or presence of exogenous TP53 expression (Fig S5). Importantly, however, co-expression of PRPK with other unrelated proteins did not prevent their TP53-mediated degradation, highlighting the specificity of PRPK-mediated TPRKB stabilization (Fig S5). Additionally, stable PRPK knockout in HEK293T cells (Fig 6D) or stable PRPK knockdown in various cell lines substantially reduced TPRKB levels (Fig S5). The markedly reduced TPRKB levels upon PRPK depletion suggests that the mild phenotypes observed with PRPK knockdown/knockout, as described above, are likely due to reduction in TPRKB levels. Conversely, we did not observe a significant change in PRPK expression with TPRKB depletion in in H358, MBA-MD-231, or HEK293T cells (Fig 6E & S5). Taken together, our data demonstrates that TP53-dependent degradation of TPRKB can be inhibited through stabilization by PRPK or through proteasomal pathway inhibition.
DISCUSSION
Herein, through in silico analysis coupled with in vitro and in vivo experimentation, we demonstrate that TPRKB, a member of the tRNA modifying EKC/KEOPS complex, is essential in cancer cells with TP53-alterations. Using isogenic cell lines, we found that TP53-null H358 lung carcinoma cells showed decreased proliferation upon TPRKB knockdown that was rescued by expression of wild-type, but not mutant, TP53. Likewise, HCT116 cells containing mutant TP53 were more sensitive to TPRKB knockdown than TP53 wild-type counterparts. Furthermore, TPRKB knockdown/knockout had minimal effect on proliferation in multiple benign immortalized or TP53 wild-type cancer cells.
TP53 can be deregulated in human cancers through multiple classes of genomic alterations, including missense, nonsense, and frameshift mutations, copy number loss, and degradation (e.g. by MDM2 amplification). Previous approaches to identifying synthetic lethal relationships in TP53 altered cancers have largely used single alteration classes, including TP53-null (28), specific hotspot TP53 mutation backgrounds (29–31) and TP53 deletion(32). Importantly, we found that TPRKB knockdown resulted in marked proliferative defects in TP53 null cancer cell lines (such as H358 described above), cell lines harboring TP53 hotspot missense mutations (e.g. H196 (TP53 p.R175H), MB-MDA 231, (TP53 p.R280K) and MBA-MB-468 (TP53 p.R273H)) as well as multiple cell lines harboring amplification of MDM2, a known E3-ubiquitin ligase responsible for degradation of TP53. Hence, our results suggest that TRPKB may represent a dependency across a larger spectrum of TP53 altered cancers than previous reports, and future efforts are aimed at determining the intricacies of response, particularly with regard to MDM2-mediated effects
Members of the EKC/KEOPS complex – TPRKB, PRPK, OSGEP, LAGE3, and recently identified C14ORF142 – are highly conserved from yeast to mammals. In yeast this complex has been shown to regulate telomere length maintenance, tRNA modification, and transcriptional processes(14,15,17–19). Further, the yeast ortholog of TPRKB, CGI-121, is non-essential for the tRNA modifying functions and instead acts as an allosteric regulator of the complex in this context(18). Importantly, yeast lack TP53, and there are relatively few studies examining the role of the EKC/KEOPS complex and its constituents in humans. Consistent with our data, a recent study in multiple myeloma demonstrated that knockdown of PRPK, an atypical kinase that can also interact with and phosphorylate TP53, inhibits cellular growth independent of TP53-status (33). PRPK expression has also been associated with invasion and metastasis potential of colorectal cancer (34). Little is known about the other EKC/KEOPS complex members in cancer, however, a recent study found that mutations in EKC/KEOPS complex members drive Galloway-Mowat syndrome, a rare condition characterized by early-onset nephrotic syndrome and microcephaly(22). Braun et al. found that mutation or knockdown of OSGEP, PRPK, or TPRKB led to reduced cellular proliferation in human podocytes, and knockout was embryonic lethal in zebra fish and mice. These results, coupled with our lack of effect in multiple TP53 wild-type benign immortalized cell lines, suggests that general dependency on the EKC/KEOPS complex may be related to TP53 status, cellular identity and development, which must be considered in any effort to therapeutically target TPRKB.
Although PRPK is the only member of the EKC/KEOPS complex to directly interact with TPRKB and TP53, we confirmed previous reports that TPRKB and TP53 do not directly interact(13). However, herein we demonstrate that TP53 induces TPRKB degradation in a concentration-dependent manner that can be alleviated either through PRPK expression or proteosomal inhibition (Fig 7A), providing a potential mechanism for TPRKB dependency only in the presence of TP53 alterations (Fig 7B). Our results further support the expression of anti-apoptotic proteins—including BCL2L1 and BCL2—as potential mediators of TPRKB dependency in H358 and SJSA-1 cells, consistent with the known convergence of TP53 and BCL proteins in cancer apoptosis (27). Ongoing studies are further exploring the mechanism driving TPRKB dependency across TP53 altered cancers.
Figure 7-. Model of TPRKB dependency in TP53-deficient cells.
A) Interactions between PRPK and TPRKB lead to TPRKB stabilization, while the presence of TP53 leads to reduced TPRKB levels. Additionally, TP53 is able to mediate TPRKB degradation through the proteasome, despite the fact that TPRKB and TP53 do not directly interact. Influences of these proteins may be key in phenotypes we witness. B) In cells that maintain wild-type TP53, loss of TPRKB does not have a profound effect on cell viability. In cells in which TP53 is perturbed, either through loss of wild-type protein, mutation of TP53, or disturbance of TP53 in some other manner (i.e. MDM2 amplification), TPRKB depletion leads to a reduction in cell proliferation.
In summary, we identified and validated TPRKB dependency across cancer cell lines harboring a range of TP53 alterations, including TP53 missense mutations, MDM2 amplifications, and TP53 loss, with minimal effect in benign or cancer cell lines with wild-type TP53. Hence, TPRKB may represent a therapeutic vulnerability that can be exploited for therapeutic targeting of TP53, the most frequently altered gene in human cancer.
Supplementary Material
IMPLICATIONS.
Cancer cells with genomic alterations in TP53 are dependent on TPRKB.
ACKNOWLEDGEMENTS
The authors thank the University of Michigan DNA Sequencing and Vector cores for Sanger sequencing and viral plasmids/production, respectively. This work was supported by the National Institutes of Health (R01 CA183857 to Scott. A. Tomlins). S.A.T. was supported by the A. Alfred Taubman Medical Research Institute.
Disclosures/Conflicts of Interest
S.A.T. has received travel support from ThermoFisher Scientific and had a separate sponsored research agreement with Compendia Bioscience/Life Technologies/ThermoFisher Scientific that was not used to support the research performed herein. S.A.T. is a co-founder of, consultant for and Laboratory Director of Strata Oncology. D.R.R. was an employee at Compendia Bioscience. The other authors have no competing interests to declare
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307–10 [DOI] [PubMed] [Google Scholar]
- 2.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nature reviews Cancer 2014;14:359–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell death and differentiation 2015;22:1239–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends in cell biology 2015;25:486–95 [DOI] [PubMed] [Google Scholar]
- 6.Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Current drug targets 2014;15:80–9 [DOI] [PubMed] [Google Scholar]
- 7.Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Frontiers in oncology 2015;5:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nature reviews Cancer 2018;18:89–102 [DOI] [PubMed] [Google Scholar]
- 9.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009;136:823–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21 [DOI] [PubMed] [Google Scholar]
- 11.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–7 [DOI] [PubMed] [Google Scholar]
- 12.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America 2011;108:12372–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyoshi A, Kito K, Aramoto T, Abe Y, Kobayashi N, Ueda N. Identification of CGI-121, a novel PRPK (p53-related protein kinase)-binding protein. Biochemical and biophysical research communications 2003;303:399–405 [DOI] [PubMed] [Google Scholar]
- 14.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 2006;124:1155–68 [DOI] [PubMed] [Google Scholar]
- 15.Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, et al. Yeast homolog of a cancer-testis antigen defines a new transcription complex. Embo J 2006;25:3576–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan LC, Maisonneuve P, Szilard RK, Lambert JP, Ng TF, Manczyk N, et al. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic acids research 2017;45:805–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, et al. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. Embo J 2011;30:873–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrochia L, Guetta D, Hecker A, Forterre P, Basta T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic acids research 2013;41:9484–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng J, He MH, Duan YM, Liu YT, Zhou JQ. Inhibition of telomere recombination by inactivation of KEOPS subunit Cgi121 promotes cell longevity. PLoS genetics 2015;11:e1005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe Y, Matsumoto S, Wei S, Nezu K, Miyoshi A, Kito K, et al. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. The Journal of biological chemistry 2001;276:44003–11 [DOI] [PubMed] [Google Scholar]
- 21.Facchin S, Lopreiato R, Ruzzene M, Marin O, Sartori G, Gotz C, et al. Functional homology between yeast piD261/Bud32 and human PRPK: both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS letters 2003;549:63–6 [DOI] [PubMed] [Google Scholar]
- 22.Braun DA, Rao J, Mollet G, Schapiro D, Daugeron MC, Tan W, et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nature genetics 2017;49:1529–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer chemotherapy and pharmacology 1989;24:148–54 [DOI] [PubMed] [Google Scholar]
- 24.Helming K, Wang XF, Wilson B, Vazquez F, Haswell J, Manchester H, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Cancer research 2014;74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. British journal of cancer 2004;91:355–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niknafs YS, Pandian B, Gajjar T, Gaudette Z, Wheelock K, Maz MP, et al. MiPanda: A Resource for Analyzing and Visualizing Next-Generation Sequencing Transcriptomics Data. Neoplasia 2018;20:1144–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janumyan YM, Sansam CG, Chattopadhyay A, Cheng N, Soucie EL, Penn LZ, et al. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. Embo J 2003;22:5459–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morandell S, Reinhardt HC, Cannell IG, Kim JS, Ruf DM, Mitra T, et al. A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell reports 2013;5:868–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada N, Watanabe Y, Yoshimura Y, Sakumoto H, Makishima F, Tsuchiya M, et al. Identification of a checkpoint modulator with synthetic lethality to p53 mutants. Anti-cancer drugs 2011;22:986–94 [DOI] [PubMed] [Google Scholar]
- 30.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 2013;155:844–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldwin A, Grueneberg DA, Hellner K, Sawyer J, Grace M, Li W, et al. Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. Proceedings of the National Academy of Sciences of the United States of America 2010;107:12463–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Zhang X, Han C, Wan G, Huang X, Ivan C, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature 2015;520:697–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hideshima T, Cottini F, Nozawa Y, Seo HS, Ohguchi H, Samur MK, et al. p53-related protein kinase confers poor prognosis and represents a novel therapeutic target in multiple myeloma. Blood 2017;129:1308–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zykova TA, Zhu F, Wang L, Li H, Bai R, Lim DY, et al. The T-LAK Cell-originated Protein Kinase Signal Pathway Promotes Colorectal Cancer Metastasis. EBioMedicine 2017;18:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.