Abstract
The sphingolipid ceramide regulates beta-oxidation of medium and long chain fatty acids in mitochondria. It is not known whether it also regulates oxidation of very long chain fatty acids (VLCFAs) in peroxisomes. Using affinity chromatography, co-immunoprecipitation, and proximity ligation assays we discovered that ceramide interacts with Hsd17b4, an enzyme critical for peroxisomal VLCFA oxidation and docosahexaenoic acid (DHA) generation. Immunocytochemistry showed that Hsd17b4 is distributed to ceramide-enriched mitochondria-associated membranes (CEMAMs). Molecular docking and in vitro mutagenesis experiments showed that ceramide binds to the sterol carrier protein 2-like domain in Hsd17b4 adjacent to peroxisome targeting signal 1 (PTS1), the C-terminal signal for interaction with peroxisomal biogenesis factor 5 (Pex5), a peroxin mediating transport of Hsd17b4 into peroxisomes. Inhibition of ceramide biosynthesis induced translocation of Hsd17b4 from CEMAMs to peroxisomes, interaction of Hsd17b4 with Pex5, and upregulation of DHA. This data indicates a novel role of ceramide as a molecular switch regulating interaction of Hsd17b4 with Pex5 and peroxisomal function.
1. Introduction
Ceramide has been implicated in various cellular and metabolic processes including cell signaling for proliferation and apoptosis, and function of mitochondria and other organelles and compartments [1, 2]. To date, analysis of metabolic function of ceramide has focused on shared lipid metabolites, while only a few studies investigated the interaction of ceramide with regulatory proteins of glucose or lipid metabolism. For many of the recently identified candidate proteins the function of ceramide interaction remains largely unknown. Using biotinylated ceramide immobilized on streptavidin-agarose beads we isolated ceramide binding proteins from human embryonic kidney (HEK) 293T cells, among which Hsd17b4 was identified by proteomics analysis. We determined the ceramide binding domain in Hsd17b4 and tested the function of ceramide binding for Hsd17b4 translocation and its effect on peroxisomal beta-oxidation of very long chain fatty acids (VLCFA) and generation of docosahexaenoic acid (DHA).
Hsd17b4 is one of the most important enzymes in VLCFA beta-oxidation. It is encoded by the hydroxysteroid (17-β) dehydrogenase 4 (HSD17B4) gene, which is also known as peroxisomal D-bifunctional protein (DBP) or multifunctional protein 2 (MFP-2) [3]. Hsd17b4 deficiency is typically associated with neonatal hypotonia, seizures, severely impaired psychomotor development, dysmorphism, loss of hearing and vision, and CNS abnormalities [4–6]. Hsd17b4 contains dehydrogenase, hydratase, and sterol carrier protein-2 like (Scp2-like) domains [7], among which the 2-enoyl-coenzyme A (CoA) hydratase domain and 3-hydroxyacyl-CoA dehydrogenase domain catalyze the second and third step of beta-oxidation, respectively. The Scp2-like domain is suggested to help bind sterols for oxidation by Hsd17b4, but it was also shown to bind to ceramide at high affinity (Kd = 5 nM) when expressed as part of Scp2 protein [8]. The Scp2-like domain contains a C-terminal targeting signal one (PTS1) signal peptide which binds to peroxisomal biogenesis factor 5 (Pex5), a peroxin that mediates transport of proteins into peroxisomes [7, 9].
We hypothesized that binding of ceramide to the Scp2-like domain may regulate interaction of Hsd17b5 with Pex5 and peroxisomal function. To test this hypothesis we performed molecular docking and in vitro mutagenesis of Hsd17b4, and tested binding of ectopically expressed deletion mutants to ceramide. We also depleted cells of ceramide to test the effect on interaction of Hsd17b4 with Pex5 and peroxisomal beta-oxidation of VLCFAs. Our data suggest that ceramide is a novel regulatory lipid for Hsd17b4 and peroxisomal function.
2. Materials and methods
2.1. Materials
Plasmids containing cDNA encoding pmTurquoise2-Peroxi (peroxisome marker described as CFP-Peroxisomes) was from Addgene, Watertown, MA (Cat#36203). Hsd17b4 mouse gene ORF cDNA clone was from Sino Biological company, Radnor, PA, USA (Cat#MG 52990-G), which was used as the template for generation of the GFP-Hsd17b4 truncations as described in the section on plasmid construction. Dulbecco Modified Eagle Medium (DMEM) was from Hyclone, Logan, Utah, USA, and Fetal Bovine Serum (FBS) was from Atlanta Biologicals, Flowery Branch, GA. Penicillin and streptomycin were purchased from GIBCO, Grand Island. N.Y (Cat#15146). HEK293T cells were received from Dr. Lin Mei (Augusta University, Augusta, GA) and Neuro2a (N2a) cells were obtained from ATCC, Manassas, VA (ATCC® CCL-131). Duolink PLA probe anti-rabbit plus (DUO92002), Duolink PLA probe anti-mouse minus (DUO92004), Duolink detection reagent red (DUO92008), Duolink detection reagent green (DUO92014), were purchased from Thermo Fisher Scientific (West Columbia, SC). Anti-ceramide rabbit IgG (1:100, our laboratory) was generated in our laboratory. Anti-Pex5 rabbit IgG was from Cell Signaling Technology, Danvers, MA (Cat#D7V4D) and Proteintech, Rosemont, IL (Cat#12545-1-AP). Anti-Hsd17b4-mouse antibody (NBP2-46005) and anti-Hsd17b4 rabbit antibody were from Novus, Centennial CO (NBP1-85296), anti-GFP mouse antibody was from Santa Cruz, Dallas, TX (Cat#K2713), and anti-GAPDH antibody was from Cell Signaling Technology (Cat# D16H11). Fumonisin B1 (FB1) was from Cayman, Michigan, USA (Cat#116355-83-0).
2.2. Construction of plasmids encoding cDNAs for marker proteins and Hsd17b4 protein domains
The fragment of Hsd17b4 cDNAs for the dehydratase domain, hydratase domain and Scp2-like domain were obtained by PCR amplification using the full-length mouse Hsd17b4 (ABclonal company, Cat#MG 52990-G) as the template. Then all of the fragments were subcloned into pEGFP-C1 vector, gift from Dr. Yanan Wang, Augusta University, Augusta, GA. Amplification of the dehydratase domain was achieved by PCR with a forward upstream primer 5’-CAG ATC TCG AGC TAT GGC TTC GCC GCT GAG GTT CGAC-3’ (XhoI site underlined) and a reverse primer5’-CAT GGA TCC GGC TGC AGG TGC CGC GTG ACT GGT-3’ (BamHI sites underlined). Amplification of the hydratase domain was achieved by PCR with a forward upstream primer 5’-CAG ATC TCG AGC TAT GTT CGT TGG TGC TGT TGG CCA T AAA-3’ (XhoI site underlined) and reverse primer 5’-CAT GGA TCC CTC TCC ACC CTC TGA AGG TGT CTG-3’ (BamHI site underlined). The primers used to amplify the Scp2-like domain were: forward primer: 5’-CAG ATC TCG AGC TAT GCT CCA GAG TGC CTC TGT GTT TGG GGAG-3’ (XhoI site underlined) and reverse primer 5’-CAT GGA TCC GAG CTT GGC ATA GTC TTT AAG A ATC (BamHI site underlined).
2.3. Cell culture and transfection with plasmids
All experiments involving mice were performed in compliance with the Animal Use Protocols approved by the Institutional Animal Care and Use Committee at the University of Kentucky. Primary astrocytes from nSMase2-deficient (fro/fro) C57B16 mice and their wild type littermates were prepared according to the protocol described previously [10, 11]. Astrocytes were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The treatment of HEK293T cells with 5 μM FB1 was performed for 48 h in DMEM/10% FBS as described in previous studies [12]. At this concentration of FBS, the contribution of ceramide from serum was <5% of endogenous ceramide and did not compromise ceramide depletion by FB1 [13–15]. The transfection HEK293T cells with different mutants of GFP-Hsd17b4 or Peroxisomes marker plasmid CFP-Peroxisomes via lipofectamine 3000 according to the manual. Forty-eight hours post transfection, cells were either harvest for ceramide-pull down experiments (see section on ceramide beads) or fixed for immunocytochemistry studies.
2.4. Immunocytochemistry and fluorescence microscopy
Cells grown on cover slips were fixed with 4% p-formaldehyde/0.5% glutaraldehyde/PBS for 20 min, followed by permeabilization with 0.2% Triton X-100 in PBS for 10 min at room temperature. Nonspecific binding sites were blocked with 3% ovalbumin/PBS for 1 h at 37 °C. Then the cells were incubated with primary antibodies at 4 °C overnight. The next day, cells on cover slips were washed by PBS and then incubated with secondary antibodies for 2 h at 37 °C. After washing, cover slips were mounted using Fluoroshield supplemented with DAPI (Sigma-Aldrich) to visualize the nuclei. Fluorescence microscopy was performed with a Nikon Ti2 Eclipse microscope equipped with NIS Elements software. Images were processed using a 3D deconvolution program as provided by the Elements software.
2.5. Image analysis of colocalization studies
Images (blinded identifiers) were randomly chosen and colocalization analyzed using the respective program in the NIS Elements software. The degree of colocalization was assessed by calculation of the Pearson’s correlation coefficient for two fluorescence channels in overlays as previously described [16]. PLA signals were also counted using the NIS Elements software. All of the data were collected from at least three independent cell cultures using at least five randomly selected areas/culture.
2.6. Proximity ligation assays (PLAs) and quantitative analysis
Cells grown on glass cover slips were fixed with 4% p-formaldehyde/0.5% glutaraldehyde/PBS for 20 min and then permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Nonspecific binding sites were blocked with Duolink PLA blocking solution (Sigma-Aldrich) for 1 h at 37 °C. Primary antibodies used were: anti-ceramide rabbit IgG (1:100, our laboratory), anti Pex5 rabbit IgG (1:200) and anti-Hsd17b4 mouse IgG (1:200). Secondary PLA probes: anti-rabbit PLUS affinity-purified donkey anti-rabbit IgG (H+L) and anti-mouse MINUS affinity-purified donkey anti-mouse IgG (H+L) were diluted 1:5 in antibody diluent buffer and samples incubated for 1 h at 37 °C. After washing, ligation and amplification step were performed following the manufacturer’s protocol (Sigma-Aldrich). Cover slips were mounted using Fluoroshield supplemented with DAPI (Sigma-Aldrich) to visualize the nuclei. Fluorescence microscopy and quantitation of PLA signals was performed as described in the previous sections. Images obtained with secondary antibody only were used as negative controls.
2.7. Preparation of binding proteins using ceramide beads
Biotinylated ceramide immobilized on streptavidin beads (P-BCer, Echelon Bioscience) and control beads (streptavidin beads saturated with biotin, P-B000, Echelon Bioscience) were incubated with equal amounts of cell lysates following a protocol as previously described [10]. In brief, primary cultured astrocytes or N2a cells or HEK293T cells were harvested in ceramide binding buffer (20 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA, pH 7.5 supplemented with protein inhibitor cocktail (Roche)). The cells were homogenized by a glass homogenizer on ice, and complete lysis of cells was achieved by sonication. After insoluble debris was removed by centrifugation at 20,800 x g for 30 min at 4°C, Triton X-100 was added to the supernatant at 0.25% of final concentration. 20 μ1 of a slurry of ceramide beads and control beads were added to the supernatant and incubated at 4°C overnight under rotary agitation. Beads were harvested by centrifugation at 4°C, 500 x g for 5 min and washed four times with ceramide binding buffer supplemented with 0.25% Triton X-100. The bound proteins were eluted by SDS-sample buffer containing 10% 2-mercaptoethanol and heating for 10 min at 60 °C, and then subjected to SDS-PAGE either for immunoblotting or Coomassie and /or silver staining. Stained bands were cut out and subjected to proteomics analysis.
2.8. Identification of ceramide binding proteins using mass spectrometry
Gel bands were excised from SDS-PAGE gels, cut into 1 mm3 pieces, de-stained using 50% acetonitrile in 25 mM ammonium bicarbonate, dried, reduced with dithiothreitol, alkylated using iodoacetamide, and digested overnight using trypsin. Peptides were extracted using 5% formic acid in 50% acetonitrile and then dried. Peptide digests were analyzed on an Orbitrap Fusion tribrid mass spectrometer (Thermo Fisher Scientific) coupled with an Ultimate 3000 nano-UPLC system (Thermo Fisher Scientific). Two microliters of reconstituted peptide was first trapped and washed on a Pepmap100 C18 trap (5 pm, 0.3 χ 5 mm) at 20 μl/min using 2% acetonitrile in water (with 0.1% formic acid) for 10 minutes and then separated on a Pepman 100 RSLC C18 column (2.0 μm, 75-μm χ 150-mm) using a gradient of 2 to 40% acetonitrile with 0.1% formic acid over 40 min at a flow rate of 300 nl/min and a column temperature of 40° C. Samples were analyzed by data-dependent acquisition in positive mode using an Orbitrap MS analyzer for precursor scan at 120,000 FWHM from 300 to 1500 m/z and ion-trap MS analyzer for MS/MS scans at top speed mode (3-second cycle time). Collision-induced dissociation (CID) was used as fragmentation method. Raw MS data were processed using Proteome Discoverer (vl.4, Thermo Fisher Scientific) and submitted for SequestHT search against the Uniprot database pertinent to the origin of the sample. Fixed value PSM validator algorithm was used for peptide spectrum matching validation. SequestHT search parameters were 10 ppm precursor and 0.6 Da product ion tolerance, with static carbamidomethylation (+57.021 Da).
2.9. Coimmunoprecipitation assays
HEK293T cells were seeded on the 100 mm dishes at 35-40% of density, and then incubated in the absence or presence of 5 μM FB1. Sixty hours post-addition of FB1, the untreated and treated cells were harvested and washed twice with cold PBS. The cell pellets were transferred into a Dounce homogenizer and disrupted and lysed with lipid binding buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5, supplemented with protein inhibitor cocktail (Roche)). Insoluble debris was removed by centrifugation at 20,800 x g for 30 min at 4°C and Triton X-100 was added to the supernatant at a final concentration of 0.25%. The protein concentrations in the supernatants from untreated cells and treated cells were adjusted to the same concentration and the same volume. Lysates were pre-cleared with Pierce protein A/G magnetic beads (Pierce Biotechnology, CAT#88802, Rockford, USA) and then incubated overnight with non-specific mouse IgG (control) or anti Hsd17b4 mouse IgG (Novus, NBP2-46005 ) at 4 °C under rotational movement. The next morning, 15 μL of Pierce protein A/G magnetic beads were added to the samples and incubated for 2 h at room temperature under rotational movement. The beads were harvested and washed three times by lipid binding buffer/0.25% Triton X-100. Protein bound to the beads was eluted by SDS-sample buffer containing 10% 2-mercaptoethanol for 30 min at room temperature and then subjected to SDS-PAGE for immunoblotting using anti-Pex 5 rabbit IgG. For immunoprecipitation of ceramide-enriched vesicles, HEK293T cells were lysed and incubated either with 5 μg/ml non-specific rabbit IgG (control) or with 5 μg/ml anti-ceramide rabbit IgG and vesicles prepared and protein analyzed following a previously published protocol [17].
2.10. Analysis of free fatty acids by mass spectrometry
Fatty acids were analyzed by chemical derivatization and electrospray ionization tandem mass spectrometry using adaptations of the methods of Bollinger et al. [18]. We used transesterification of free fatty acids to generate methylpyridinium derivatives that are readily detected by electrospray ionization mass spectrometry. This method has been extensively validated and widely used including other reports from our laboratory [18–23]. Cell pellets (~20 mg) or tissues (~20 mg) were placed in 0.5 mL of 0.1 M HCl in 1.5 mL microcentrifuge tubes. To each tube, three beads (stainlees steel beads, 3.2 mm diameter., Next Advance, USA) and 0.1 mL of internal standard (IS, 250 ng/mL of Octadecanoic-d35 acid) were added. The solutions were homogenized for 5 min with Bullet blender and transferred to a 12-mL borosilicate glass tube. After adding 1 mL of isooctane, the tubes were vortexed for 5 min, and phases separated by centrifugation at ~1000 x g for 10 min. The upper isooctane phase was removed via glass pipet and transferred to a 4-mL glass vial. The remaining aqueous phase was extracted once more with additional 1 mL of isooctane. The combined isooctane phases were evaporated to dryness under a stream of filtered nitrogen and derivatized with A-(4-aminomethylphenyl)pyridinium (AMPP).
Following the instruction of the AMP+ MaxSpec Kit (Cayman chemical, USA), 20 uL of cold acetonitrile/N, N-dimethylformamide (DMF) (4:1) solvent, 20 uL of the cold 1 M (l-(3-dimethylamino)propyl)-3-ethyl-carbodiimide hydrochloride (EDC) solution, 10 uL of the 1-hydroxybenzotriazole (HOBt) solution and 30 uL of the AMP+ solution were added to the dried material. The vial was mixed for 1 min on a vortex mixer, and the solution was transferred to a 200 uL vial insert using a 100 uL adjustable pipettor. Then the insert was placed in an autosampler vial and capped. The autosampler vial was placed in a 60°C incubator for 30 min. Samples were analyzed on the same day and kept in the auto-sampler rack at 4°C while queued for injection and LC-MS/MS analysis. Analysis was performed on an AB Sciex 4000 Q Trap coupled with an Exion LC system. The Analyst software package was used for data collection and analysis. Chromatography was carried out with a C8 reverse-phase column (Waters ACQUITY UPLC BEH C8, 2.1 × 50 mm, 1.7 μm) maintained at 40°C and the flow rate was set to 0.4 mL/min. Solvent A is 100 water/ 0.1 % formic acid, and solvent B is acetonitrile/ 0.1% formic acid. A gradient program was used as follow (T min/%A): 0/90, 0.5/90, 0.51/80, 10.0/30, 10.1/0, 12.0/0, 12.1/90, 15.0/90. The injection volume was 2.0 ul.
Mass spectrometer was equipped with an electrospray ionization (ESI) source and operated in positive mode under the following operating parameters: IonSpray Voltage 5.5 kV, Desolvation temperature 550°C, Ion Source Gas 1 40 psi, Ion Source Gas 2 50 psi, Curtain Gas 30 psi, Collision Gas Medium, Declustering Potential 160 V, Entrance Potential 10.0 V, and Collision Energy 55.0 V. Quantitative analysis was conducted by monitoring the precursor ion to production ion transitions of m/z 395.3/239.2 (Myristic acid), m/z 423.3/239.2 (Palmitic acid), m/z 451.4/239.2 (Stearic acid), m/z 449.3/239.2 [Oleic acid (n-9)], m/z 447.3/239.2 [Linoleic acid (ω-6)], m/z 479.4/239.2 (Arachidic acid), m/z 477.3/239.2 [Gadoleic acid (n-11)], m/z 475.3/239.2 [Dihomolinoleic acid(ω-6)], m/z 471.3/239.2 [Arachidonic acid (ω-6)], m/z 469.3/239.2 [EPA(o>3)], m/z 507.4/239.2 (Behenic acid), m/z 503.4/239.2 [Docosadienoic acid (ω-6)], m/z 495.3/239.2 [DHA (ω-3)], m/z 535.5/239.2 (Lignoceric acid), m/z 563.5/239.2 (Cerotic acid), and m/z 487.0/242.2 (Octadecanoic-d35 acid, IS) with a dwell time of 0.1 s.
3. Results
3.1. Hsd17b4 is a ceramide binding protein and colocalized with ceramide
To identify novel ceramide binding proteins, we performed affinity chromatography with biotin-linked ceramide immobilized on streptavidin agarose beads (Fig. 1 A). Biotin-saturated streptavidin beads were used as controls. Potential ceramide binding proteins were isolated from a lysate of human embryonic kidney (HEK293T) cells and mouse neuroblastoma Neuro2a (N2a) cells and then analyzed by SDS-gel electrophoresis. Figure 1B shows a unique silver-stained band at 80 kDa (arrow, N2a cells are shown) that was excised and subjected to proteomics analysis. We identified Hsd17b4 as a candidate for ceramide binding (proteomics results are shown in Supplemental Fig. 1). To confirm significance and specificity of ceramide binding to Hsd17b4 we compared the eluate of ceramide beads with those of other immobilized lipids and used lysates from different cell types for pull-down assays followed by immunoblots using anti-Hsd17b4 antibody. Fig. 1C shows that Hsd17b4 was pulled-down from HEK293T cells, N2a cells, and primary cultured astrocytes, indicating that Hsd17b4 from several cell types interacted with ceramide. Immobilized sphingomyelin did not pull down Hsd17b4, confirming its specificity for binding to ceramide (Fig. 1D). Sphingosine, also potentially bound by Hsd17b4 was not used for affinity chromatography because its biotinylated derivative is immobilized to streptavidin in the opposite direction of that of the ceramide derivative and its intracellular concentration is <5% of that of ceramide [13–15]. To confirm binding of Hsd17b4 to endogenous ceramide, we used protein A agarose and anti-ceramide rabbit IgG to pull down ceramide-containing vesicles from HEK293T cells, a method originally developed in our laboratory to identify proteins associated with ceramide-enriched membranes [17, 24], Figure 1E shows that only anti-ceramide IgG, but not control IgG, mediated isolation of Hsd17b4, indicating that Hsd17b4 was associated with ceramide-enriched membranes.
Figure 1. Hsd17b4 interacts and colocalizes with ceramide.
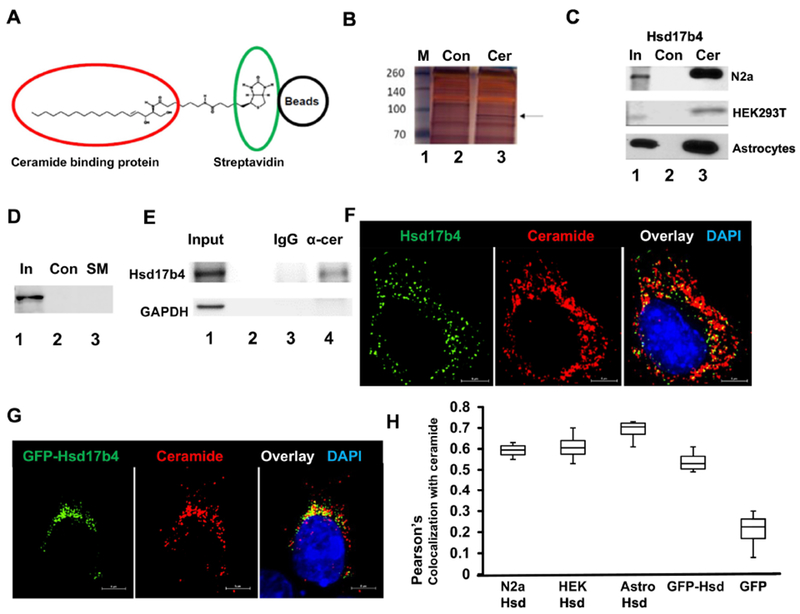
A. Principle of isolating ceramide binding proteins (red circle) by affinity chromatography using biotinylated ceramide immobilized on streptavidin (green circle) agarose beads (black circle). B. Protein interacting with ceramide was isolated using ceramide beads as shown in A. Protein was then separated by SDS-PAGE and silver-stained (Lane 3). Arrow indicates protein excised from the gel and further analyzed by mass spectrometry. Lane 2 shows control sample obtained with biotin-bound control beads. Lane 1, marker. C. Protein isolated from N2a cells, astrocytes, and HEK293T cells as in B, followed by SDS-PAGE and immunoblotting using antibody against Hsd17b4 (Lane 3). Lane 2 shows controls prepared as in A. Lane 1 shows input for control and ceramide beads. D. Affinity chromatography of protein from HEK293T cells using biotinylated sphingomyelin immobilized on streptavidin agarose beads. E. Coimmunoprecipitation of Hsd17b4 with α-ceramide IgG used to pull down ceramide-containing vesicles from HEK193T cells. Lane 1, input; lane 2, empty; lane 3, output control rabbit IgG; lane 4, output α-ceramide IgG. Immunoblot for GAPDH was used as negative control. F. Immunocytochemistry using N2a cells and antibodies against Hsd17b4 (green) and ceramide (red). G. HEK293T cells ectopically expressing EGFP-Hsd17b4 (green) were subjected to immunocytochemistry using antibody against ceramide. H. Colocalization analysis using Nikon NIS Elements program (Pearson’s coefficient) on immunocytochemistry for Hsd17b4 (or ectopically expressed EGFP-Hsd17b4) and ceramide in cell types as indicated in image legend. N=4. P<0.01
Next, we tested if Hsd17b4 was colocalized with ceramide in cells. Immunocytochemistry using antibodies against Hsd17b4 and ceramide showed that in N2a cells, Hsd17b4 was colocalized with ceramide in vesicular and membranous structures close to the nucleus and distributed throughout the cytosol (Fig. 1F). Colocalization with ceramide was also found for EGFP-Hsd17b4 ectopically expressed in HEK293T cells (Fig. 1G). A quantitative analysis (Pearson’s coefficient) showed that colocalization of Hsd17b4 or EGFP-Hsd17b4 with ceramide was similar in N2a cells, HEK293T cells, and primary cultured astrocytes, while there was no colocalization when expressing GFP as a negative control (Fig. 1H and Supplemental Fig. 2). These results suggested that Hsd17b4 and EGFP-Hsd17b4 interacted with ceramide in similar intracellular compartments in different cell types.
3.2. Scp2-like is the ceramide binding domain in Hsd1 7b4
Hsd17b4 is composed of three functional domains: dehydrogenase, hydratase, and the C-terminal Scp2-like domain, the latter harboring the PTS1 signaling sequence for binding of Hsd17b4 to Pex5, a peroxin mediating transport of Hsd17b4 into peroxisomes. Molecular docking (Swissdock) of the Scp2-like domain showed that there is a distinctive binding site occupied by the fatty acid or long chain base (sphingosyl) chain of ceramide (Fig. 2A). The sphingosyl chain binds adjacent to the PTS1 signal with a Gibbs energy of −10.3 kcal/mol corresponding to an apparent affinity constant of KD = 30 nM. In the extended or splayed chain conformation of ceramide with the sphingosyl residue protruding into the Scp2-like domain, the fatty acyl chain is available for anchoring Hsd17b4 to a lipid bilayer similar to proteins with covalent fatty acid modification (Fig. 2B). To test if the Scp2-like or other domains of Hsd17b4 bind to ceramide, we ectopically expressed N-terminal fusion proteins of these domains with EGFP (for domain structure of constructs see Fig. 3A). Forty-eight hours post-transfection, cells were lysed, insoluble material was removed by centrifugation, and the supernatant was subjected to affinity chromatography using ceramide-linked agarose beads. Figure 3B shows that EGFP-tagged dehydrogenase and hydratase domains were not pulled down using the beads, while EGFP-Scp2-like yielded a strong signal when using anti-GFP antibody on immunoblots of the pulled-down protein. We also transfected HEK293T cells with EGFP-Scp2-like plasmid to test for colocalization with ceramide. Figure 3C shows that EGFP-Scp2-like, but not EGFP, colocalized with ceramide similar to the distribution found with endogenous Hsd17b4 and EGFP-Hsd17b4. These results suggested that the SCP2-like domain mediated binding of Hsd17b4 to ceramide.
Figure 2: Modeling and construction of Hsd17b4 mutants.
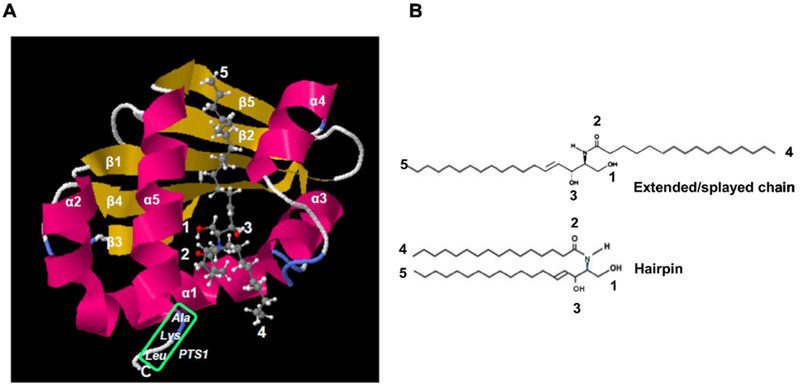
A. Molecular modeling of the Scp2-like domain using SwissDock with pdb file 1IKT (Scp2 like) and C16:0 ceramide (Zinc file 40164304.mol2) showing conformation with sphingosyl residue binding to protein (ΔG0’ = 10.3 kcal/mol, KD’ = 30 nM) and fatty acid residue accessible for membrane anchoring or immobilization on affinity gel as shown in Fig. 1A. Green box shows PTS1 sequence. Arrows point at residues indicated in legend at the upper left of image panel and B. B. Ext ended/splayed chain and hairpin conformation of C16:0 ceramide. The extended conformation is compatible with binding of ceramide shown in A.
Figure 3. Scp2-like domain in Hsd17b4 interacts with ceramide.
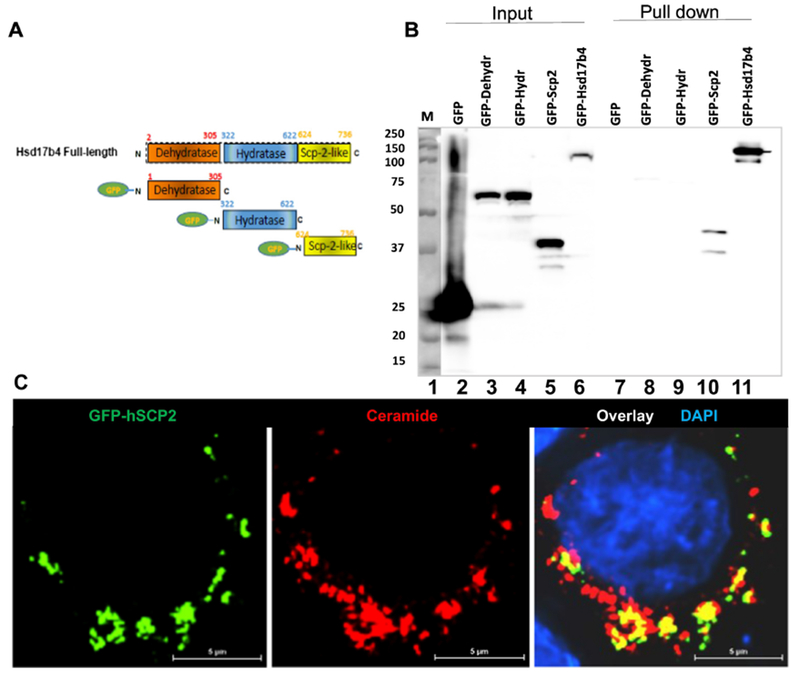
A. Domain structure of Hsd17b4 and EGFP-tagged constructs used for transfection. B. HEK293T cells were transfected with EGFP-tagged constructs encoding the three domains of Hsd17b4 or full length protein as depicted in A. Expression products were isolated from cell lysates using affinity chromatography of on ceramide beads, followed by SDS-PAGE and immunoblotting with anti-GFP antibody. Expression and affinity chromatography of EGFP was used as the negative control. Lane 1, marker; lanes 2-6, input; lane 7-11, output (pulled-down protein). C. Immunocytochemistry using anti-ceramide antibody and EGFP-tagged Scp2-like domain from Hsd17b4 (hsdSCP2) ectopically expressed in HEK293T cells.
3.3. Ceramide depletion induces translocation of Hsd17b4 to peroxisomes
While Hsd17b4 is a cytosolic protein distributed to peroxisomes, proteomics studies suggested that it is also present in MAMs [25, 26]. Most recently, we have reported that a portion of MAMs is comprised of a ceramide-enriched subcompartment we termed ceramide-enriched MAMs (CEMAMs) [27]. We tested if Hsd17b4 colocalizes with CEMAMs and peroxisomes by expressing MAM-specific and peroxisomal marker proteins and using anti-Hsd17b4 and anti-ceramide antibodies for immunocytochemistry in HEK293T cells. Figure 4A shows that a portion of Hsd17B4 was colocalized with MAMs as shown by SigmaR1-GFP expression (arrows). These MAMs were also labeled for ceramide (arrows in Fig. 4B) indicating that Hsd17B4 was distributed to CEMAMs. To test complex formation between ceramide and Hsd17b4 at CEMAMs we performed proximity ligation assays (PLAs), a technology using in situ PCR to amplify a fluorescence signal adjacent to two antigens if they are closer than 40 nm. We detected PLA signals for ceramide-Hsd17B4 complexes at SigmaR1-GFP containing MAMs suggesting that Hsd17B4 bound to ceramide in CEMAMs (Fig. 4C arrows). This data was consistent with the absence of PLA signals when cells were depleted of ceramide by incubation with FB1 (Fig. 4D). These results were also consistent with our data showing binding of Hsd17b4 to ceramide (Fig. 1B and C) and suggested that ceramide mediated association of cytosolic Hsd17b4 with CEMAMs.
Figure 4. Hsd17b4 is associated with ceramide at CEMAMs.
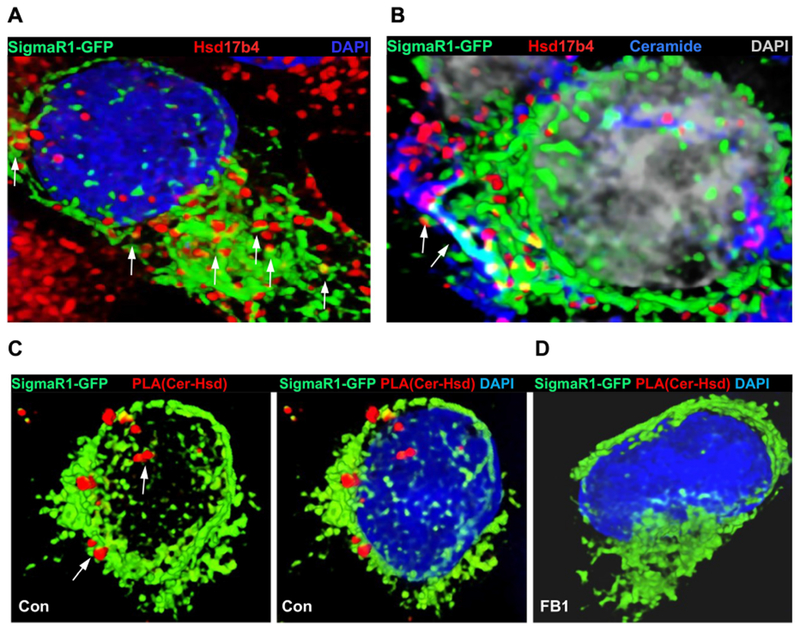
A, B. Immunocytochemistry using HEK293T cells expressing the MAM marker SigmaR1-GFP and antibodies against Hsd17b4 (red, A) and ceramide (blue, B). Arrows in B point at Hsd17B4 associated with MAMs, a portion of which are ceramide-enriched (CEMAMs). C, D. Proximity ligation assay (PLA, red) using anti-ceramide and Hsd17b4 antibodies in SigmR1-GFP (green) expressing HEK293T cells. DAPI staining is not shown in left panel to better visualize juxtaposition of PLA signals to MAMs. In D, ceramide depletion by incubation with FB1 obliterates PLA signals for the putative ceramide-Hsd17b4 complex.
3.4. Ceramide regulates interaction of Hsd17b4 with Pex5 at peroxisomes
Since Hsd17b4 is active in peroxisomes we tested if ceramide regulated translocation of Hsd17b4 to peroxisomes and its interaction with peroxisomal proteins. Figure 5A–C shows that FB1 treatment of HEK293T cells increased the level of colocalization between Hsd17b4 and peroxisomes as visualized by expression of the peroxisome marker Per-CFP (arrows). This result suggested that ceramide depletion led to translocation of Hsd17b4 from CEMAMs to peroxisomes.
Figure 5. Ceramide depletion induces translocation of Hsd17b4 to peroxisomes and interaction with Pex5.
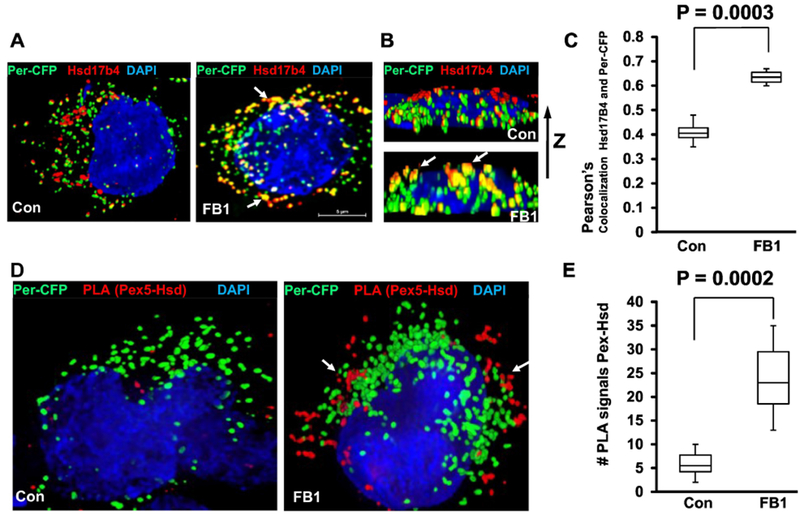
A-C. HEK293T cells expressing the peroxisome marker Per-CFP (green) were depleted of ceramide by FB1 treatment and subjected to immunocytochemistry using antibody against Hsd17b4 (red). A, top view; B, side view. Arrows point at peroxisomes co-localized with Hsd17b4. C, Quantitative analysis (Pearson’s coefficient) for colocalization of Per-CFP with Hsd17b4. N=5, P<0.01. D, E. PLAs using antibodies against Pex5 (rabbit IgG) and Hsd17b4 (mouse IgG) with HEK293T cells expressing Per-CFP (green). E is quantification of D for number of PLA signals. N=4.
Next, we tested if ceramide regulates the interaction between Hsd17b4 and Pex5 using PLA and co-immunoprecipitation analyses. PLA using antibodies against Hsd17b4 (mouse IgG) and Pex5 (rabbit IgG) showed that FB1 treatment increased the number of PLA signals for Hsd17b4-Pex5 complexes adjacent to ectopically expressed Per-CFP by 5-fold, indicating that complex formation due to ceramide depletion was upregulated at peroxisomes (Fig. 5D and E). Hsd17b4-Pex5 complex formation was confirmed by using lysates from cells treated with or without FB1 for co-immunoprecipitation assays with anti-Hsd17b4 IgG for the immunoprecipitation reaction and anti Pex5 rabbit IgG for SDS-PAGE/immunoblotting. Figure 6A shows that ceramide depletion increased the amount of co-immunoprecipitated Pex5 (lane 6 with FB1 vs. lane 4 without FB1). Interestingly, we also found that in ceramide-depleted cells, non-specific binding to control beads (lane 5) was visible, concurrent with 2-fold increased intensity of the Pex5 input band (lane 2 with FB1 vs. lane 1 without FB1). This data suggested that ceramide depletion led to increased Pex5 levels, which was confirmed by immunoblots of protein lysates from FB1-treated HEK293T cells (Fig. 6B and C).
Figure 6. Ceramide depletion by FB1 treatment upregulates Pex5 and generation DHA and EPA.
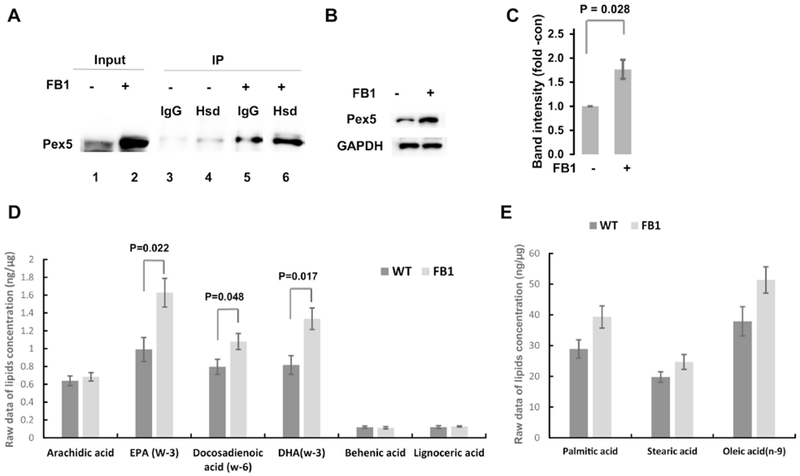
A. Coimmunoprecipitation analysis using cell lysates of HEK293T cells with or without prior treatment with 5 μM FB1 for 48 h. Antibody against Hsd17b4 was used for immunoprecipitation (IP) and anti-Pex5 antibody was used for the immunoblotting reaction (IB). Lanes 1, 2: input; lanes 3, 4: output without FB1; lanes 5, 6: output with FB1. FB1 treatment increases level of Pex 5 in input and output. IgG, non-specific mouse IgGused or IP. B, C. SDS-PAGE/immunoblot for protein levels of Pex5 in different cell types/tissue with or without FB1 treatment as in A. C is quantitation of B. N=5. P<0.05 as indicated in figure. D, E. Mass spectrometric analysis of free fatty acids in HEK293T cells with or without FB1 treatment as in A. D, 3-PUFAs and VLCFAs. E. Medium chain fatty acids. N=5. P<0.05 as indicated in figure.
3.5. Ceramide depletion increases generation of DHA and EPA
Our results showing that ceramide depletion induced Hsd17b4 translocation to peroxisomes and increased Pex5 levels suggested that peroxisomal function is also enhanced. We determined the level of DHA in FB1-treated HEK293T cells since Hsd17b4 is critical for its generation in peroxisomes. Figure 6D shows that the DHA level was increased by 60% in FB1-treated HEK293T cells, consistent with our observation that ceramide depletion enhanced translocation of Hsd17b4 to peroxisomes. Eicosapentaenoic acid (EPA) was also elevated, which could be attributed to retroconversion of DHA [28]. We tested the levels of free VLCFAs and MCFAs/LCFAs that are substrates for peroxisomal and mitochondrial beta-oxidation, respectively. The VLCFAs behenic acid (C22:0) and lignoceric acid (C24:0) were not altered in FB1-treated HEK293T cells, while levels of C14:0 to C18:0 fatty acids were increased (Fig. 6D and E). Increase of these fatty acid levels might have been due to inhibiting their incorporation into ceramide by FB1, while accumulation of VLCFAs was prevented by enhanced oxidation in peroxisomes.
4. Discussion
Beta-oxidation of fatty acids takes place in both, mitochondria and peroxisomes. While mitochondrial beta-oxidation of MCFAs and LCFAs is regulated by the respective ceramide species [29], it is unclear whether a similar regulation by ceramide also affects peroxisomal beta-oxidation of VLCFAs and generation of DHA. Hsd17b4 is central to peroxisomal generation of DHA and degradation of 17beta-estradiol, two lipids critical for neuronal survival and cancer suppression, respectively [30, 31]. Accordingly, downregulation of Hsd17b4 is associated with Alzheimer’s disease and neurodegeneration, while its upregulation is associated with liver cancer [30, 31]. To date, alteration of Hsd17b4 activity is attributed to mutations or different gene expression levels. It has not been investigated if other lipids, particularly those derived from incorporation of VLCFAs, may regulate Hsd17b4 activity. Ceramide is derived from covalent attachment of MCFAs, LCFAs, and VCLFAs to (di)hydrosphingosine, a reaction catalyzed by ceramide synthases (CerSs). Using the specific CerS inhibitor fumonisin B1 (FB1), we show for the first time that ceramide regulates translocation of Hsd17b4 and peroxisomal function.
Direct binding to proteins is a mechanism by which ceramide regulates lipid metabolism or cell signaling pathways. Hence, we hypothesized that ceramide may directly interact with Hsd17b4 to regulate peroxisomal function. This idea was corroborated by a recent study reporting that Hsd17b4 was isolated by binding to ceramide in a yeast surface immobilized ceramide for affinity chromatography of ceramide binding proteins, Hsd17b4 cDNA display assay to identify novel ceramide binding proteins [32]. When using was one of the prominent proteins we isolated from lysates of a variety of cell types, including HEK293T cells, N2a cells, and primary cultured astrocytes. Hsd17b4 was also found in ceramide-enriched vesicles isolated with anti-ceramide antibody. The identity of Hsd17b4 was confirmed by proteomics analysis and immunoblots of protein bound to ceramide. Further, failure of immobilized sphingomyelin to pull down Hsd17b4 from cell lysates indicated that sphingolipid binding of Hsd17b4 was specific for ceramide.
The observation that Hsd17b4 bound to ceramide immobilized on the surface of agarose beads suggested that binding occurred without ceramide being completely engulfed into the protein. A potential ceramide conformation simultaneously binding to the protein and a membrane could resemble that of extended or splayed-chain ceramide with the sphingosyl residue protruding into the protein and the fatty acid incorporated in the membrane (Fig. 2A and B). This conformation is related to the previously suggested “alkyl-protrusion model”, although in our model, membrane anchoring is mediated by the fatty acid similar to myristoylated or palmitoylated proteins [33–36]. The “splayed-chain conformation model” allows ceramide to stay in the membrane and therefore, it could serve as an anchor sequestering proteins such as atypical PKC (aPKC), glycogen synthase kinase 3β (GSK3), and tubulin to membranous compartments enriched with ceramide as found in our previous studies [27, 37, 38]. To date, there are only a few protein domains known to bind to ceramide [39], many of them proposing engulfment of the entire ceramide molecule into the binding site (hairpin conformation of ceramide). The proposed ceramide binding domains are the C1/CA3 domain in kinase suppressor of Ras [40], the START domain in ceramide transport protein (CERT) and StarD7 [41, 42], a hydrophobic (2 anti-parallel β-sheets + 1 a-helix) domain in the 12 inhibitor of protein phosphatase 2a (I2PP2a/SET) [43], a β-sandwich (2 anti-parallel β-sheets) motif in the core/DNA binding domain of p53 protein [44], and a putative ceramide interaction motif in the transmembrane α-helix of LAPTM4B protein [45]. None of these domains or motifs share considerable sequence similarity or are contained within Hsd17b4, although there are structurally similar alpha helix and antiparallel beta sheet motifs in the lipid binding pockets of the evolutionarily related START and Scp2-like domain in CERT and Hsd17b4, respectively [46]. Pull-down of Hsd17B4 by ceramide beads does not rule out the possibility that actual binding to ceramide is mediated by a different protein associated with Hsd17b4 in a complex. However, our in vitro mutagenesis experiments showed that the Scp2-like domain in Hsd17b4 mediates binding to ceramide, which is consistent with the results from a previous study reporting that the Scp2 protein binds directly to ceramide [8]. Scp2 protein transports sterols, and it is distributed to mitochondria and peroxisomes [47]. Neither the effect of ceramide binding to Scp2 protein nor the function of the respective Scp2-like domain in Hsd17b4 are known. However, it is known that the C-terminus of the Scp2-like domain in Hsd17b4 contains a PTS1 signal mediating association with peroxisomal proteins such as Pex5 [7, 9]. This data suggested that ceramide binding to the Scp2-like domain may regulate function of the PTS1 signal in translocation of Hsd17b4 to peroxisomes and its association with Pex5.
When using antibodies against ceramide and Hsd17b4, immunocytochemistry analysis showed that ceramide was significantly colocalized with Hsd17b4 in a variety of cell types (Pearson’s colocalization coefficient of 0.6-0.7), consistent with the results showing binding of the protein to ceramide. Colocalization with ceramide was confirmed by ectopic expression of EGFP-Hsd17b4 and EGFP-Scp2-like as well as PLAs for complex formation between ceramide and Hsd17b4. Initially, we assumed that ceramide in peroxisomes would assist with binding of Hsd17b4 to Pex5. This assumption was based on data from a previous study, suggesting that ceramide may facilitate fusion or biogenesis of peroxisomes [48], although direct visualization of ceramide in peroxisomes was not provided in this study. Using ceramide antibody we could not detect ceramide in peroxisomes, but in compartments associated with mitochondria. Most recently, our laboratory discovered that a subcompartment of mitochondria-associated membranes (MAMs) is specifically enriched with ceramide [27]. In the current study, we found that these ceramide-enriched MAMs (CEMAMs) colocalized with Hsd17b4, unless cells were depleted of ceramide by inhibition of CerSs with FB1. Colocalization of Hsd17b4 with CEMAMs was consistent with proteomics studies showing that MAMs contain Hsd17b4 [25, 26]. Therefore, our data suggested that cytosolic Hsd17b4 was distributed to CEMAMs by binding to ceramide, while another proportion of the enzyme was translocated to peroxisomes, probably by binding to Pex5.
Based on these observations, we revised our hypothesis in that ceramide does not assist with, but prevent peroxisomal translocation of Hsd17b4 and its interaction with Pex5. colocalization Colocalization studies as well as PLAs using antibodies against Hsd17b4 and Pex5 with FB1-treated HEK293T cells showed that ceramide depletion induced translocation of Hsd17b4 to peroxisomes and its complex formation with Pex5. Enhanced formation of Hsd17b4-Pex5 complexes by ceramide depletion was confirmed by coimmunoprecipitation studies. Figure 7 shows a model depicting regulation by ceramide of intracellular Hsd17b4 distribution and its interaction with Pex5. Our model implies that cytosolic Hsd17b4 may bind to either ceramide at CEMAMs or Pex5 at peroxisomes. Mutually exclusive binding to ceramide or Pex5 is mediated by the Scp2-like domain. While it is known that the C-terminal PTS1 sequence of the Scp2-like domain binds to Pex5 and initiates peroxisomal import of Hsd17b4 [9], our model explains for the first time how ceramide regulates this interaction and peroxisomal function. If the level of ceramide generation is low - as with FB1-mediated inhibition of CerSs - Hsd17b4 is translocated to peroxisomes and oxidizes VLCFAs that are not incorporate into ceramide or other lipids. If the ceramide level is high Hsd17b4 stays associated with CEMAMs and peroxisomal beta-oxidation of VLCFAs is downregulated. Our model does not exclude binding of other sphingolipids to regulate Hsd17b4 translocation and activity. Particularly, sphingosine or sphingosine-1-phosphate could occupy the binding site for the sphingosyl moiety in our “splayed-chain conformation” model of ceramide binding to the Scp2-like domain. However, the coimmunoprecipitation and studies using anti-ceramide antibody strongly suggest that ceramide is the main regulatory factor binding to Hsd17b4.
Figure 7. Model for ceramide regulation of Hsd17b4 translocation to peroxisomes, interaction with Pex5, and beta-oxidation of VLCFAs.
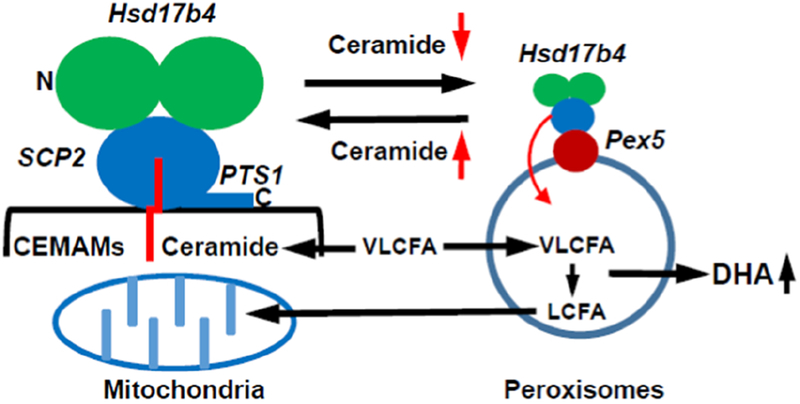
Hsd17b4 is anchored to CEMAMs by binding to ceramide, probably in the splayed-chain conformation with the sphingosyl residue (Sph) protruding into the Scp-2 like domain and the fatty acid (FA) incorporated into the membrane. Anchoring of Hsd17b4 to CEMAMs prevents the C-terminal PTS1 signal from binding to Pex5. Reduced incorporation of FAs into ceramide, e.g., experimentally induced by FB1-mediated inhibition of CerSs or genetic deficiency of nSMase2, leads to translocation of Hsd17b4 to peroxisomes and induces its interaction with Pex5 and peroxisomal import of the enzyme, which results in increased generation of DHA.
In addition to enhancing interaction of Hsd17b4 with Pex5, we found that the level of Pex5 was increased in HEK293T cells treated with FB1. This increase may arise from enhanced complex formation with Hsd17b4 thereby protecting Pex5 from proteolytic degradation as reported in previous studies [49–51]. Both, further characterization of the ceramide binding site in the Scp2-like domain of Hsd17b4 as well as the mechanism of increased Pex5 level due to ceramide depletion is currently investigated in our laboratory.
Increased Pex5 levels as well as enhanced complex formation with Hsd17b4 at peroxisomes suggested that ceramide depletion upregulates generation of DHA, a PUFA important for neuronal function. Mass spectrometric analysis showed that in HEK293T cells, ceramide depletion by FB1 treatment increased the concentration of free DHA by 60%. VLCFAs levels were not altered in FB1-treated cells, while those of of C14:0 to C18:0 fatty acids were increased. Increase of these fatty acid levels might have been due to inhibiting their incorporation into ceramide by FB1, while accumulation of VLCFAs was prevented by enhanced oxidation in peroxisomes.
The physiological significance of regulating peroxisomal function has been documented by many studies on peroxisomal dysfunction and disease. Dysfunction of peroxisomes is described for peroxisome biogenesis disorders arising from mutations of peroxins (e.g., Pex5) such as Zellweger syndrome, neonatal ALD, or infantile Refsum disease, or functional disorders such as D-bifunctional protein deficiency (mutation of Hsd17b4) or X-linked ALD (mutation of VLCFA transporter ABCD1) [5, 52]. The Pex5(−/−) mouse showed upregulation of a particular VLCFA containing ceramide species (C26:1/0 ceramide), but it is not known if this elevation is linked to the pathology of Zellweger disease [53]. Apart from genetic disorders, peroxisomal dysfunction is also observed in aging-related and neurodegenerative diseases such as AD [54–56].
To address potential correction of peroxisomal dysfunction, one will need to consider dietary supplementation with lipids generated in peroxisomes, namely DHA and plasmalogens. While supplementation with omega 3-fatty acids such as the DHA precursor linoleic acid is now a commonplace in dietary supplementation, the curative outcome in neurodegenerative disease is rather modest [57]. Recent studies suggest that neither exogenously added omega 3-fatty acids nor DHA fully compensate for lack of endogenously generated DHA. Our study shows that inhibiting ceramide generation thereby promoting Hsd17b4 translocation to peroxisomes and stimulating endogenous DHA generation is a promising approach to restore peroxisome function.
Supplementary Material
Highlights.
Hsd17b4, an enzyme critical for beta-oxidation of fatty acids in peroxisomes, is bound to ceramide at ceramide-enriched mitochondria-associated membranes (CEMAMs). Binding to ceramide prevents interaction of Hsd17b4 with the peroxisomal import protein Pex5.
Ceramide depletion induces translocation of Hsd17b4 from CEMAMs to peroxisomes, association of Hsd17b4 with Pex5, and upregulation of docosahexaenoic acid (DHA), a polyunsaturated fatty acid (PUFA) critical for brain function.
Our data indicates a novel role of ceramide as a molecular switch regulating peroxisomal function and generation of DHA.
Acknowledgements:
This work was the supported by the grants R01AG034389, R01NS095215, and NSF 1615874 to EB and a VA Merit Award I01CX001550 and P30 GM127211 to AJM. The authors thank the Department of Physiology (Chair Dr. Alan Daugherty). College of Medicine, University of Kentucky, Lexington, KY for institutional support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hannun YA, Luberto C, Ceramide in the eukaryotic stress response, Trends in cell biology, 10 (2000) 73–80. [DOI] [PubMed] [Google Scholar]
- [2].Hernandez-Corbacho MJ, Salama MF, Canals D, Senkal CE, Obeid LM, Sphingolipids in mitochondria, Biochimica et biophysica acta, 1862 (2017) 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de Launoit Y, Adamski J, Unique multifunctional HSD17B4 gene product: 17beta-hydroxysteroid dehydrogenase 4 and D-3-hydroxyacyl-coenzyme A dehydrogenase/hydratase involved in Zellweger syndrome, J Mol Endocrinol, 22 (1999) 227–240. [DOI] [PubMed] [Google Scholar]
- [4].Nascimento J, Mota C, Lacerda L, Pacheco S, Chorao R, Martins E, Garrido C, D-bifunctional protein deficiency: a cause of neonatal onset seizures and hypotonia, Pediatric neurology, 52 (2015) 539–543. [DOI] [PubMed] [Google Scholar]
- [5].Huyghe S, Mannaerts GP, Baes M, Van Veldhoven PP, Peroxisomal multifunctional protein-2: the enzyme, the patients and the knockout mouse model, Biochimica et biophysica acta, 1761 (2006) 973–994. [DOI] [PubMed] [Google Scholar]
- [6].Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, Fiumara A, Opitz JM, Levy-Lahad E, Klevit RE, King MC, Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome, American journal of human genetics, 87 (2010) 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Moller G, Leenders F, van Grunsven EG, Dolez V, Qualmann B, Kessels MM, Markus M, Krazeisen A, Husen B, Wanders RJ, de Launoit Y, Adamski J, Characterization of the HSD17B4 gene: D-specific multifunctional protein 2/17beta-hydroxysteroid dehydrogenase IV, The Journal of steroid biochemistry and molecular biology, 69 (1999) 441–446. [DOI] [PubMed] [Google Scholar]
- [8].Atshaves BP, Jefferson JR, McIntosh AL, Gallegos A, McCann BM, Landrock KK, Kier AB, Schroeder F, Effect of sterol carrier protein-2 expression on sphingolipid distribution in plasma membrane lipid rafts/caveolae, Lipids, 42 (2007) 871–884. [DOI] [PubMed] [Google Scholar]
- [9].Moller G, Luders J, Markus M, Husen B, Van Veldhoven PP, Adamski J, Peroxisome targeting of porcine 17beta-hydroxysteroid dehydrogenase type IV/D-specific multifunctional protein 2 is mediated by its C-terminal tripeptide AKI, Journal of cellular biochemistry, 73 (1999) 70–78. [DOI] [PubMed] [Google Scholar]
- [10].Kong JN, Zhu Z, Itokazu Y, Wang G, Dinkins MB, Zhong L, Lin HP, Elsherbini A, Leanhart S, Jiang X, Qin H, Zhi W, Spassieva SD, Bieberich E, Novel function of ceramide for regulation of mitochondrial ATP release in astrocytes, J Lipid Res, 59 (2018) 488–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dinkins MB, Enasko J, Hernandez C, Wang G, Kong J, Helwa I, Liu Y, Terry AV Jr., Bieberich E, Neutral Sphingomyelinase-2 Deficiency Ameliorates Alzheimer’s Disease Pathology and Improves Cognition in the 5XFAD Mouse, The Journal of neuroscience : the official journal of the Society for Neuroscience, 36 (2016) 8653–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, Bielawska A, Obeid LM, Hannun YA, Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line. Role for endogenous ceramide in mediating the action of exogenous ceramide, The Journal of biological chemistry, 277 (2002) 12960–12969. [DOI] [PubMed] [Google Scholar]
- [13].Hanamatsu H, Ohnishi S, Sakai S, Yuyama K, Mitsutake S, Takeda H, Hashino S, Igarashi Y, Altered levels of serum sphingomyelin and ceramide containing distinct acyl chains in young obese adults, Nutrition & diabetes, 4 (2014) e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Turner N, Lim XY, Toop HD, Osborne B, Brandon AE, Taylor EN, Fiveash CE, Govindaraju H, Teo JD, McEwen HP, Couttas TA, Butler SM, Das A, Kowalski GM, Bruce CR, Hoehn KL, Fath T, Schmitz-Peiffer C, Cooney GJ, Montgomery MK, Morris JC, Don AS, A selective inhibitor of ceramide synthase 1 reveals a novel role in fat metabolism, Nature communications, 9 (2018) 3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spassieva SD, Ji X, Liu Y, Gable K, Bielawski J, Dunn TM, Bieberich E, Zhao L, Ectopic expression of ceramide synthase 2 in neurons suppresses neurodegeneration induced by ceramide synthase 1 deficiency, Proceedings of the National Academy of Sciences of the United States of America, 113 (2016) 5928–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adler J, Parmryd I, Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient, Cytometry. Part A : the journal of the International Society for Analytical Cytology, 77 (2010) 733–742. [DOI] [PubMed] [Google Scholar]
- [17].He Q, Wang G, Dasgupta S, Dinkins M, Zhu G, Bieberich E, Characterization of an apical ceramide-enriched compartment regulating ciliogenesis, Molecular biology of the cell, 23 (2012) 3156–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bollinger JG, Naika GS, Sadilek M, Gelb MH, LC/ESI-MS/MS detection of FAs by charge reversal derivatization with more than four orders of magnitude improvement in sensitivity, Journal of lipid research, 54 (2013) 3523–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang WC, Adamec J, Regnier FE, Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding, Analytical chemistry, 79 (2007) 5150–5157. [DOI] [PubMed] [Google Scholar]
- [20].Wang M, Han RH, Han X, Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach, Analytical chemistry, 85 (2013) 9312–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Majkova Z, Layne J, Sunkara M, Morris AJ, Toborek M, Hennig B, Omega-3 fatty acid oxidation products prevent vascular endothelial cell activation by coplanar polychlorinated biphenyls, Toxicology and applied pharmacology, 251 (2011) 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Boyanovsky BB, Li X, Shridas P, Sunkara M, Morris AJ, Webb NR, Bioactive products generated by group V sPLA(2) hydrolysis of LDL activate macrophages to secrete pro-inflammatory cytokines, Cytokine, 50 (2010) 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Spencer M, Finlin BS, Unal R, Zhu B, Morris AJ, Shipp LR, Lee J, Walton RG, Adu A, Erfani R, Campbell M, McGehee RE Jr., Peterson CA, Kern PA, Omega-3 fatty acids reduce adipose tissue macrophages in human subjects with insulin resistance, Diabetes, 62 (2013) 1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krishnamurthy K, Dasgupta S, Bieberich E, Development and characterization of a novel anti-ceramide antibody, Journal of lipid research, 48 (2007) 968–975. [DOI] [PubMed] [Google Scholar]
- [25].Poston CN, Krishnan SC, Bazemore-Walker CR, In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM), Journal of proteomics, 79 (2013) 219–230. [DOI] [PubMed] [Google Scholar]
- [26].Calvo SE, Julien O, Clauser KR, Shen H, Kamer KJ, Wells JA, Mootha VK, Comparative Analysis of Mitochondrial N-Termini from Mouse, Human, and Yeast, Molecular & cellular proteomics : MCP, 16 (2017) 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kong JN, Zhu Z, Itokazu Y, Wang G, Dinkins MB, Zhong L, Lin HP, Elsherbini A, Leanhart S, Jiang X, Qin H, Zhi W, Spassieva SD, Bieberich E, Novel function of ceramide for regulation of mitochondrial ATP release in astrocytes, Journal of lipid research, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schuchardt JP, Ostermann AI, Stork L, Kutzner L, Kohrs H, Greupner T, Hahn A, Schebb NH, Effects of docosahexaenoic acid supplementation on PUFA levels in red blood cells and plasma, Prostaglandins, leukotrienes, and essential fatty acids, 115 (2016) 12–23. [DOI] [PubMed] [Google Scholar]
- [29].Fucho R, Casals N, Serra D, Herrero L, Ceramides and mitochondrial fatty acid oxidation in obesity, FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 31 (2017) 1263–1272. [DOI] [PubMed] [Google Scholar]
- [30].Pan LC, Xiao HY, Yin WJ, Lin Z, Correlation between HSD17B4 expression in rat liver cancer tissues and inflammation or proliferation, European review for medical and pharmacological sciences, 22 (2018) 3386–3393. [DOI] [PubMed] [Google Scholar]
- [31].Astarita G, Piomelli D, Towards a whole-body systems [multi-organ] lipidomics in Alzheimer’s disease, Prostaglandins, leukotrienes, and essential fatty acids, 85 (2011) 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bidlingmaier S, Ha K, Lee NK, Su Y, Liu B, Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing, Molecular & cellular proteomics : MCP, 15 (2016) 1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schutze S, Wickel M, Heinrich M, Winoto-Morbach S, Weber T, Brunner J, Kronke M, Use of affinity chromatography and TID-ceramide photoaffinity labeling for detection of ceramide-binding proteins, Methods in enzymology, 312 (2000) 429–438. [DOI] [PubMed] [Google Scholar]
- [34].Kronke M, The mode of ceramide action: the alkyl chain protrusion model, Cytokine & growth factor reviews, 8 (1997) 103–107. [DOI] [PubMed] [Google Scholar]
- [35].Skolova B, Hudska K, Pullmannova P, Kovacik A, Palat K, Roh J, Fleddermann J, Estrela-Lopis I, Vavrova K, Different phase behavior and packing of ceramides with long (C16) and very long (C24) acyls in model membranes: infrared spectroscopy using deuterated lipids, The journal of physical chemistry. B, 118 (2014) 10460–10470. [DOI] [PubMed] [Google Scholar]
- [36].Beddoes CM, Gooris GS, Bouwstra JA, Preferential arrangement of lipids in the long-periodicity phase of a stratum corneum matrix model, Journal of lipid research, 59 (2018) 2329–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kong JN, Hardin K, Dinkins M, Wang G, He Q, Mujadzic T, Zhu G, Bielawski J, Spassieva S, Bieberich E, Regulation of Chlamydomonas flagella and ependymal cell motile cilia by ceramide-mediated translocation of GSK3, Molecular biology of the cell, 26 (2015) 4451–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang G, Silva J, Krishnamurthy K, Tran E, Condie BG, Bieberich E, Direct binding to ceramide activates protein kinase Czeta before the formation of a pro-apoptotic complex with PAR-4 in differentiating stem cells, The Journal of biological chemistry, 280 (2005) 26415–26424. [DOI] [PubMed] [Google Scholar]
- [39].Canals D, Salamone S, Hannun YA, Visualizing bioactive ceramides, Chemistry and physics of lipids, 216 (2018) 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yin X, Zafrullah M, Lee H, Haimovitz-Friedman A, Fuks Z, Kolesnick R, A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation, Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology, 24 (2009) 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M, Molecular machinery for non-vesicular trafficking of ceramide, Nature, 426 (2003) 803–809. [DOI] [PubMed] [Google Scholar]
- [42].Bockelmann S, Mina JGM, Korneev S, Hassan DG, Muller D, Hilderink A, Vlieg HC, Raijmakers R, Heck AJR, Haberkant P, Holthuis JCM, A search for ceramide binding proteins using bifunctional lipid analogs yields CERT-related protein StarD7, Journal of lipid research, 59 (2018) 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Saddoughi SA, Gencer S, Peterson YK, Ward KE, Mukhopadhyay A, Oaks J, Bielawski J, Szulc ZM, Thomas RJ, Selvam SP, Senkal CE, Garrett-Mayer E, De Palma RM, Fedarovich D, Liu A, Habib AA, Stahelin RV, Perrotti D, Ogretmen B, Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis, EMBO Mol Med, 5 (2013) 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fekry B, Jeffries KA, Esmaeilniakooshkghazi A, Szulc ZM, Knagge KJ, Kirchner DR, Horita DA, Krupenko SA, Krupenko NI, C16-ceramide is a natural regulatory ligand of p53 in cellular stress response, Nature communications, 9 (2018) 4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhou K, Dichlberger A, Martinez-Seara H, Nyholm TKM, Li S, Kim YA, Vattulainen I, Ikonen E, Blom T, A Ceramide-Regulated Element in the Late Endosomal Protein LAPTM4B Controls Amino Acid Transporter Interaction, ACS central science, 4 (2018) 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chiapparino A, Maeda K, Turei D, Saez-Rodriguez J, Gavin AC, The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling, Progress in lipid research, 61 (2016) 30–39. [DOI] [PubMed] [Google Scholar]
- [47].Seedorf U, Ellinghaus P, Roch Nofer J, Sterol carrier protein-2, Biochimica et biophysica acta, 1486 (2000) 45–54. [DOI] [PubMed] [Google Scholar]
- [48].Boukh-Viner T, Guo T, Alexandrian A, Cerracchio A, Gregg C, Haile S, Kyskan R, Milijevic S, Oren D, Solomon J, Wong V, Nicaud JM, Rachubinski RA, English AM, Titorenko VI, Dynamic ergosterol- and ceramide-rich domains in the peroxisomal membrane serve as an organizing platform for peroxisome fusion, The Journal of cell biology, 168 (2005) 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kerssen D, Hambruch E, Klaas W, Platta HW, de Kruijff B, Erdmann R, Kunau WH, Schliebs W, Membrane association of the cycling peroxisome import receptor Pex5p, The Journal of biological chemistry, 281 (2006) 27003–27015. [DOI] [PubMed] [Google Scholar]
- [50].Platta HW, Girzalsky W, Erdmann R, Ubiquitination of the peroxisomal import receptor Pex5p, The Biochemical journal, 384 (2004) 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schwartzkopff B, Platta HW, Hasan S, Girzalsky W, Erdmann R, Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation, Bioscience reports, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Braverman NE, D’Agostino MD, Maclean GE, Peroxisome biogenesis disorders: Biological, clinical and pathophysiological perspectives, Developmental disabilities research reviews, 17 (2013) 187–196. [DOI] [PubMed] [Google Scholar]
- [53].Pettus BJ, Baes M, Busman M, Hannun YA, Van Veldhoven PP, Mass spectrometric analysis of ceramide perturbations in brain and fibroblasts of mice and human patients with peroxisomal disorders, Rapid Commun Mass Spectrom, 18 (2004) 1569–1574. [DOI] [PubMed] [Google Scholar]
- [54].Berger J, Dorninger F, Forss-Petter S, Kunze M, Peroxisomes in brain development and function, Biochimica et biophysica acta, 1863 (2016) 934–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fanelli F, Sepe S, D’Amelio M, Bernardi C, Cristiano L, Cimini A, Cecconi F, Ceru MP, Moreno S, Age-dependent roles of peroxisomes in the hippocampus of a transgenic mouse model of Alzheimer’s disease, Molecular neurodegeneration, 8 (2013) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lizard G, Rouaud O, Demarquoy J, Cherkaoui-Malki M, Iuliano L, Potential roles of peroxisomes in Alzheimer’s disease and in dementia of the Alzheimer’s type, Journal of Alzheimer’s disease : JAD, 29 (2012) 241–254. [DOI] [PubMed] [Google Scholar]
- [57].Ajith TA, A recent update on the effects of omega-3 fatty acids in Alzheimer’s disease, Current clinical pharmacology, (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


