Abstract
Purpose
To identify a profile of circulating tumor HPV DNA (ctHPVDNA) clearance kinetics that is associated with disease control after chemoradiotherapy (CRT) for HPV-associated oropharyngeal squamous cell carcinoma (OPSCC).
Patients and methods
A multi-institutional prospective biomarker trial was conducted in 103 patients with: 1) p16-positive OPSCC, 2) M0 disease 3) receipt of definitive CRT. Blood specimens were collected at baseline, weekly during CRT, and at follow-up visits. Optimized multianalyte digital PCR assays were used to quantify ctHPVDNA (types -16/18/31/33/35) in plasma. A control cohort of 55 healthy volunteers and 60 patients with non-HPV-associated malignancy, was also analyzed.
Results
Baseline plasma ctHPVDNA had high specificity (97%) and high sensitivity (89%) for detecting newly diagnosed HPV-associated OPSCC. Pre-treatment ctHPV16DNA copy number correlated with disease burden, tumor HPV copy number, and HPV integration status. We define a ctHPV16DNA favorable clearance profile as having high baseline copy number (>200 copies/mL) and >95% clearance of ctHPV16DNA by day 28 of CRT. Nineteen out of 67 evaluable patients had a ctHPV16DNA favorable clearance profile, and none had persistent or recurrent regional disease after CRT. In contrast, patients with adverse clinical risk factors (T4 or >10 pack years) and an unfavorable ctHPV16DNA clearance profile had a 35% actuarial rate of persistent or recurrent regional disease after CRT (p = 0.0049).
Conclusions and Relevance
A rapid clearance profile of ctHPVDNA may predict likelihood of disease control in patients with HPV-associated OPSCC patients treated with definitive CRT, and may be useful in selecting patients for de-intensified therapy.
Trial Registration
, ,
Keywords: HPV, digital PCR, Oropharyngeal Cancer, Radiation, Phase II
Introduction
The discovery of human papilloma virus (HPV) infection as a driver of oropharyngeal carcinogenesis has been a major advancement in head and neck oncology. Patients with HPV-associated oropharyngeal squamous cell carcinoma (OPSCC) have a significantly better prognosis than patients with HPV-negative OPSCC(1). The three-year local regional control and overall survival for HPV-associated vs. HPV negative are respectively: 86% vs. 65% and 82% vs. 46%(1). Further risk stratification of patients with HPV-associated OPSCC is done using tobacco smoking history: low risk (HPV-associated, ≤ 10 pack years, overall survival 93%) and intermediate risk (HPV-associated, >10 pack years, overall survival 71%)(1).
It is now standard clinical practice to use HPV, smoking status, and disease stage to risk stratify patients to facilitate prognostication, and therapeutic exploration of de-intensified treatment regimens is an active area of clinical trial research(2,3). Though the primary biomarker of HPV-status and secondary modifier of tobacco pack years (TPY) allow for significant prognostication, they are imprecise. For example, >10 TPY did not impact cancer control in HPV-associated OPSCC patients in a large retrospective study of 899 patients(4). Others have also reported lower tumor HPV copy number, presence of non-HPV16 viral strains, and HPV integration as being associated with adverse prognosis(5–12). Thus, there is a need to identify additional biomarkers of therapeutic sensitivity that may help to better risk stratify patients.
Circulating tumor DNA (ctDNA) is released by dying cancer cells and represents an accessible source for detecting tumor genetic biomarkers in many cancer types, including OPSCC(13). Plasma ctDNA can be evaluated serially over time to analyze the profile of molecular ctDNA responses during cancer therapy(14). The clinical utility of Epstein Barr Virus (EBV) ctDNA has been established for the early detection(15) of EBV-associated nasopharyngeal cancer and prognostication of patients after definitive chemoradiotherapy (CRT)(16). Similarly, plasma circulating tumor HPV16 DNA (ctHPV16DNA) is detectable in the majority of patients with HPV-associated OPSCC(12,17–20). Limited prior data has shown that ctHPV16DNA levels become largely undetectable post CRT in most patients, and that ctHPV16DNA levels may increase at the time of disease recurrence(12,19–21). These prior studies retrospectively analyzed limited subsets of patients using real-time PCR.
We have designed and validated a highly sensitive and specific digital PCR (dPCR) assay for absolute quantification of circulating tumor HPV16 DNA (ctHPV16DNA), as well as circulating tumor HPV DNA for the four most prevalent alternative high-risk HPV strains (18/31/33/35). We integrated these ctHPVDNA analyses into our ongoing multi-institutional prospective phase II clinical trials for patients with HPV-associated OPSCC, to investigate whether ctHPVDNA baseline levels and clearance kinetics improve risk stratification when combined with established clinical risk factors. We herein report our observations regarding a ctHPV16DNA rapid clearance profile that, in combination with established clinical risk factors, predicts local and regional disease control after definitive CRT.
Materials and Methods
Ethics Approval and Consent to Participate
All patients included in this study provided written informed consent to an IRB-approved prospective biomarker study at the University of North Carolina at Chapel Hill Cancer Hospital that was conducted in accordance with the U.S. Common Rule.
Study Design and Eligibility
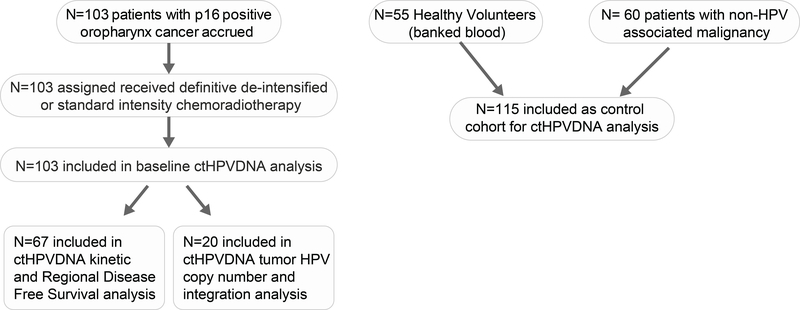
Patients with HPV-associated OPSCC were enrolled on an IRB approved (UNC IRB 11–1924) prospective biomarker study (NCT0316182). All patients provided written informed consent. The major eligibility criteria were: 1) age 18 years age or older, 2) biopsy proven squamous cell carcinoma of the oropharynx, 3) p16 positivity (defined as >70% of carcinoma cells showing nuclear reactivity), 4) T0 to T4, N0 to N3, M0 (AJCC 7th edition), and 5) receiving definitive CRT. Patients with T0 to T3, N0 to N2c (AJCC 7th edition) and with a tobacco smoking history of ≤ 10 TPY or >10 TPY and not actively smoking at time of diagnosis were co-enrolled on two institutional single arm phase II de-intensified CRT trial (NCT02281955 or NCT03077243). Fifty-five healthy volunteers and sixty patients with non-HPV-associated malignancy were also enrolled as a control cohort. A REMARK diagram of the patient cohorts analyzed in this study is provided in Figure 1.
Figure 1:
REMARK diagram of patient cohorts analyzed in this study.
Definitive Chemoradiotherapy
Patients not co-enrolled on the phase II de-intensification CRT trials received standard of care 70Gy intensity modulated radiotherapy (IMRT) with concurrent chemotherapy. All patients enrolled on the phase II de-intensification CRT trials received 60Gy IMRT with concurrent weekly intravenous cisplatin 30 mg/m2. Patients with T0-T2 N0–1 disease did not receive chemotherapy. All patients had a 3 month post-treatment PET/CT and clinical examination to determine need for neck dissection. Thereafter, patients underwent clinical examinations every 2–4 months for years 1–2, then every 6 months for years 3–5. Chest imaging was performed every 6 months.
Blood Collection and Extraction of Circulating DNA
De-identified blood specimens were collected pre-treatment, weekly during CRT, and with each post treatment follow-up visit in 10 mL cell-free DNA BCT® blood collection tubes (Streck 218962), and double-spun plasma (2000×g) was harvested within 3 days for storage at −80°C. for ctHPVDNA analysis. Plasma cell-free DNA (cfDNA) was extracted from 2–5 ml of plasma using the QIAamp circulating nucleic acid kit (Qiagen, Valencia, CA, USA; Catalog No. 55114). The amount of purified cfDNA was quantified with a Qubit® fluorometer and PicoGreen quantification reagents (Invitrogen, Carlsbad, CA, USA).
Chemicals and reagents
Droplet digital PCR reagents (2x ddPCR™ Supermix for Probes (No dUTP) Catalog # 186–3024; Pipet tips Catalog # 186–4121; Cartridge Catalog # 186–4109 and Sealing foil Catalog #181–4040) were purchased from Bio-Rad (Hercules, CA). Eppendorf™ 96-Well twin.tec™ PCR Plates (Catalog #E951020362), Falcon® 15ml Conical Centrifuge Tubes (Catalog # 352096), Falcon™ 50mL Conical Centrifuge Tubes (Corning #352098), Qubit™ dsDNA HS Assay Kit (Catalog # Q32851), and Qubit™ Assay Tubes (Catalog # Q32856) were purchased from Fisher scientific (Fair Hampton, NH). Bovine Serum Albumin was from Sigma (Catalog # A7906; Sigma–Aldrich, St. Louis, MO). Circulating DNA was quantified with Qubit (Invitrogen, Carlsbad, CA, USA). Target gene fragments for HPV subtype 16, 18, 31, 33, and 35 were synthesized as gBlocks from IDT for use as positive controls (Supplementary Table S1). These gBlocks were cloned into a pCR2.1-TOPO TA vector by Topo TA cloning kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The gene fragment constructs were verified by Sanger sequencing (Genewiz, New Jersey, USA). FAM-, TET- or HEX-conjugated Locked Nucleic Acids (LNA)-modified DNA oligonucleotide probes and FAM-ZEN dual quencher probes were synthesized by IDT (Integrated DNA Technology, San Jose, CA, see Supplementary Table S1).
ctHPV16DNA dPCR assay and detection of HPV variants
Primers and 5’ hydrolysis probes were designed to specifically detect a ~75bp amplicon within the E7 gene encoded by high-risk HPV strains 16, 18, 31, 33, and 35. Sequence details of the primers and probes used in this study are listed in Supplementary Table S1. Each reaction assay contained 10 μl of 2x dPCR™ Supermix for Probes (No dUTP), 0.9 μM of respective primers, 0.25 μM of respective probes, and 5–50 ng of cfDNA in a final volume of 20 μl. Digital droplet PCR assays were performed on the QX100 and/or QX200 platforms outfitted with an automated droplet generator (Bio-Rad). Each assay was validated against control DNA samples comprising either the complete genome for the HPV strain of interest, or the relevant portion of the E7 gene. All assays were specific for the HPV strain of interest, with no cross-reactivity to all other HPV strains tested. Standardized thresholds for positive and negative droplets were used across all samples. A dPCR assay for a control locus in the ESR1 gene was used to assess sample quality. All dPCR data was analyzed using QuantaSoft software version 1.7.4.0917 (Bio-Rad). Additional details of the dPCR assays used in this study are provided in the Supplementary Material.
Analysis of Tumor HPV copy number and integration status
Targeted exon sequencing was conducted after institutional review board (IRB) approval through the UNCseq (University of North Carolina, Chapel Hill, NC, http://www2.cscc.unc.edu/ unchreg/UNCseq, NCT01457196) pipeline, as previously described (22,23). In brief, sequencing data are routed through an automated pipeline. This workflow uses paired tumor and normal libraries to detect somatic mutations, large and small indels, structural variants, and copy number aberrations. Raw sequences are aligned to hg19_(hs37d5), supplemented with the HPV16 (NC_001526.2) and HPV 18 (NC_001357.1) genomes, using the Burrows-Wheeler Aligner (BWA) ‘mem’ algorithm. The alignments are then refined using our Assembly Based ReAlignment (ABRA(24)) procedure. ABRA generates a list of structural variants found in target locations, including HPV-human fusions, and permits alignment of both junction-containing and junction bridging reads. HPV-human chimeric reads were distinguished from non-chimeric HPV alignments, and these counts were normalized to the count of non-chimeric human alignments. Alignment details from ABRA were integrated with these normalized counts to generate circos plots for a subset of samples (25).
Statistical Analyses
Linear regression, t-tests, and survival analyses were performed in Prism 8 (Graphpad) software. Regional Disease-Free Survival (RDFS) was estimated using the Kaplan-Meier (KM) method. Regional Disease-Free Survival was measured from the time of the first dose of CRT and was defined as time to detection/development of persistent/recurrent disease in cervical lymph nodes. Two-sided log-rank test was applied to compare RDFS of different subgroups.
Results
Patient Characteristics
Between 2/2016 and 08/2018, 103 patients were enrolled. The clinical characteristics of the study population are shown in Table 1 and a REMARK diagram of cohorts for study analyses is presented in Figure 1. A total of 87 patients (84%) received de-intensified CRT on clinical trial (60 Gy). The majority of patients were never smokers or had ≤10 pack years of tobacco use (75%). HPV status was unknown in 48% (all tumors were p16 positive).
Table 1:
Patient Characteristics
| N=103 | % | |
|---|---|---|
| Age (mean) | 60 | NA |
| Gender | ||
| Male | 92 | 89% |
| Female | 11 | 11% |
| Tobacco use | ||
| Never smoker | 56 | 54% |
| ≤10 pack years | 21 | 21% |
| > 10 pack years | 26 | 25% |
| T Stage | ||
| T0 | 5 | 5% |
| T1 | 15 | 14% |
| T2 | 69 | 67% |
| T3 | 7 | 7% |
| T4 | 7 | 7% |
| N Stage | ||
| N0 | 5 | 4% |
| N1 | 16 | 16% |
| N2 | 82 | 80% |
| N3 | 0 | 0% |
| HPV Tissue Status | ||
| HPV+ | 44 | 43% |
| HPV − | 10 | 9% |
| HPV unknown | 49 | 48% |
| Radiation Dose | ||
| 60 Gy | 87 | 84% |
| 70 Gy | 16 | 16% |
| Chemotherapy | ||
| Yes | 87 | 84% |
| No | 16 | 16% |
Cancer Control Outcomes
The median follow-up (f/u) for 67 patient subset for whom weekly blood samples were collected and a post-treatment PET scan was available was 16.5 months (range 3.9 – 32.9). Ten patients had a neck dissection because of incomplete radiographic response on the 3 month post-treatment PET/CT. Five of 10 patients had a positive neck dissection specimen. Three patients had recurrent disease: 0 local, 2 regional and distant, and 1 distant only. The actuarial 1-year local control, regional control, and distant metastasis free survival, and OS were 100%, 91%, 94%, and 98%, respectively. Actuarial 1-year estimate of RDFS (5 positive neck dissection specimens and 2 regional recurrences) was 91%.
Plasma circulating tumor HPVDNA is detectable in a majority of oropharyngeal cancer patients
A digital PCR (dPCR) assay was developed to amplify and quantify a 73bp region of the HPV16 E7 gene. The assay does not cross-detect other HPV subtypes including HPV-6/11/18/31/33/35 (Supplementary Figure S1A). A titration analysis reveals exceptional linearity (R2 = 0.99) and low variance over 5 orders of magnitude (Supplementary Figure S1B).
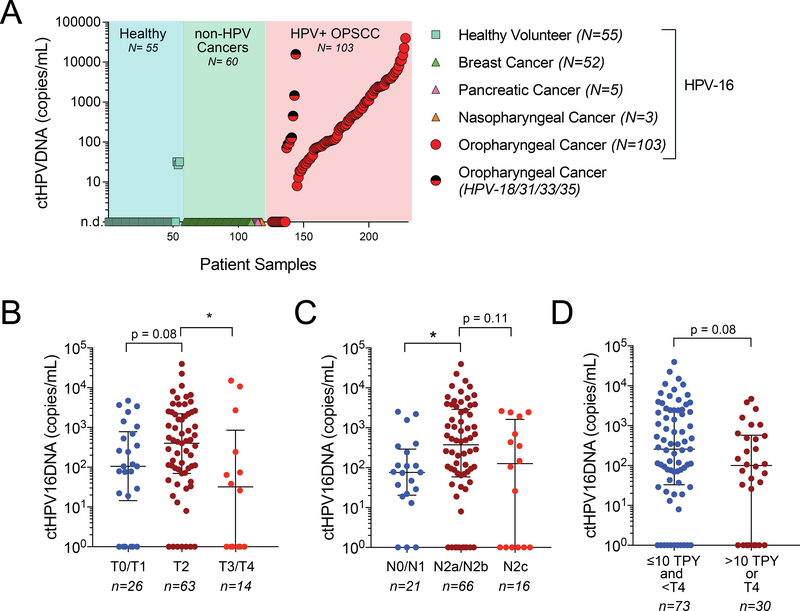
ctHPV16DNA was undetectable in 52 out of 55 healthy volunteers (Figure 2A). Three female volunteers, ages 20, 21, and 31, had low but detectable levels of ctHPV16DNA. We were unable to access the clinical history of these healthy volunteers to establish cervical HPV status. ctHPV16DNA was not detected in plasma DNA extracted from 60 patients with non-HPV associated malignancy (Figure 2A). Eighty four out of 103 (82%) patients with HPV-associated OPSCC had detectable pre-treatment ctHPV16DNA (Figure 2B), and median copy number 419 copies/mL plasma (range 8 – 22,579). The 19 samples with undetectable ctHPV16DNA were also analyzed by specific dPCR assays for alternative high-risk HPV strains (HPV 18/31/33/35, Supplementary Figure S2). Eight out of 19 patients with undetectable baseline ctHPV16DNA were positive for ctHPVDNA from an alternative high-risk HPV strain (one HPV31, three HPV33, and four HPV35), with median copy number of 124 copies/mL (range 71 – 15,829). Four of the remaining 11 patients who were negative for ctHPVDNA for all high-risk strains tested were analyzed by tumor genomic sequencing, and two were found to have a TP53 mutation. Based on the analysis of 103 cases and 115 controls, we estimate that ctHPVDNA testing using an optimized multianalyte dPCR assay has 97% specificity and 89% sensitivity to identify patients with newly diagnosed and non-metastatic HPV-associated OPSCC.
Figure 2. Detection of plasma ctHPVDNA in HPV-associated oropharyngeal cancer patients.
(A) Measurement of ctHPVDNA copies/mL plasma in 55 healthy volunteers, 60 patients with non-HPV associated cancers, and 103 patients with HPV-associated OPSCC. Two-toned (red-black) circles denote patients who were negative for ctHPV16DNA but positive for ctHPVDNA from an alternative high-risk HPV strain (-18/31/33/35). (B-D) Baseline levels of ctHPV16DNA stratified by (B) tumor stage, (C) nodal stage, and (D) clinical risk factors. Median and inter-quartile range is shown for each graph. *, p<0.05; p-values based on a two-sided Mann-Whitney test.
Baseline ctHPV16DNA levels correlate with tumor burden and clinical risk factors
There was a trend towards higher baseline plasma ctHPV16DNA levels in patients with T2 tumors compared to T0/T1 tumors (Figure 2B). Despite having a relatively low number of T3/T4 tumors in the cohort, these patients had significantly lower baseline levels of ctHPV16DNA relative to patients with T2 tumors (Figure 2B), suggesting that larger tumor size may be associated with lower rates of ctHPVDNA release. Consistent with a prior study (12), we observed significantly higher ctHPV16DNA levels in patients with N2a/N2b versus N0/N1 disease (AJCC 7th edition) (Figure 2C). Again, there was a trend towards lower baseline ctHPV16DNA in patients presenting with N2c disease, relative to the N2a/N2b patient subgroup (Figure 2C). Similar trends were seen when AJCC 8th edition N stage classifications were used (Supplementary Figure S3). These findings suggest that tumor burden alone may not explain the variable levels of pre-treatment ctHPV16DNA in OPSCC patients. Notably, patients with adverse clinical risk factors (> 10 TPY or T4) had lower baseline ctHPV16DNA levels than patients with low clinical risk (≤10 TPY and <T4) (Figure 2D). Thus, although ctHPV16DNA is detectable in a majority of patients with newly diagnosed HPV-associated OPSCC, low baseline and undetectable ctHPV16DNA levels may be associated with clinically higher-risk disease.
Rapid clearance kinetics of ctHPV16DNA during CRT in a subset of patients
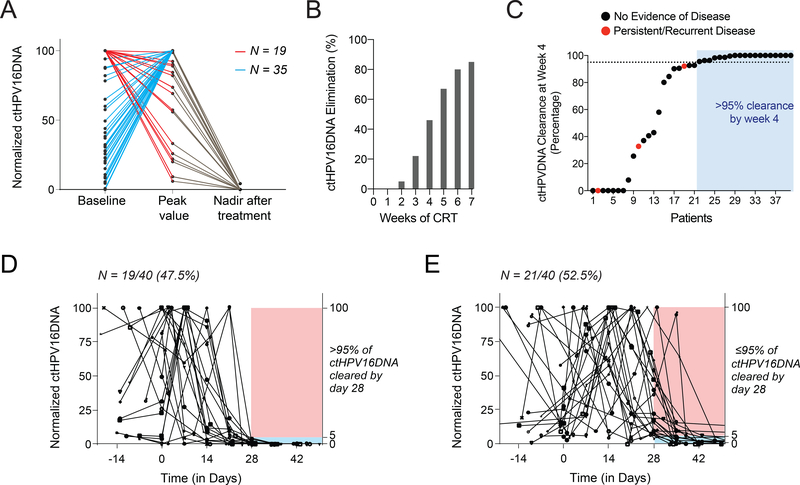
We next investigated kinetics of ctHPV16DNA clearance during CRT in a subset of 67 patients for whom weekly blood samples were collected and a post-treatment PET scan result was available to assess treatment response (median follow-up 16.5 months). Fifty-four out of 67 patients had detectable baseline ctHPV16DNA. In 35 out of 54 patients (65%), ctHPV16DNA levels increased after initiating CRT before diminishing later in the course of therapy (Figure 3A). Patients with ≤200 copies/mL of baseline ctHPV16DNA had labile clearance kinetics (Supplementary Figure S4), possibly due to a lower reliability of signal detection or non-uniform responses to treatment. Thus, we analyzed weekly ctHPV16DNA clearance kinetics in the 40 patient subset (out of 67) who had >200 copies/mL baseline ctHPV16DNA. Levels of ctHPV16DNA diminished during CRT, and approximately 80% of patients had eliminated all ctHPV16DNA by the end of CRT (Figure 3B). Furthermore, ctHPV16DNA had cleared in 92%, 94%, and 100% of patients by 6 months, 1 year, and 2 years post-CRT, respectively. Patients with detectable ctHPV16DNA at the end of week 6 of CRT did not have a higher incidence of residual or recurrent disease (Supplementary Figure S5). Alternatively, we measured percent clearance of ctHPV16DNA at week 4 relative to pre-treatment levels as a potential indicator of CRT sensitivity, as has previously been investigated for EBV-associated nasopharyngeal cancer(26). Using a median cutoff of >95% clearance at week 4 (day 25–31), we identified 19 patients with a ctHPV16DNA rapid clearance profile (Figure 3C). These patients had a consistent drop in ctHPV16DNA during CRT, and all patients had a clinical complete response to CRT. In contrast, the remaining 21 patients had delayed clearance kinetics (</= 95% clearance at week 4), exhibited more unpredictable ctHPV16DNA profiles (Figure 3D), and 3 of these patients had persistence/recurrent disease after CRT (2 positive neck dissections, 1 distant metastasis). Thus, ctHPV16DNA rapid clearance kinetics, defined as >95% clearance by week 4, may be a biomarker of CRT sensitivity and greater likelihood of disease control.
Figure 3. Variable kinetics of ctHPV16DNA clearance during chemoradiotherapy.
(A) Normalized ctHPV16DNA abundance (relative to the highest measured value for each patient) at baseline, peak value between week 1–3 of CRT, and nadir after treatment (within 6 months of completing therapy) for 54 evaluable patients with detectable ctHPV16DNA. In one subgroup (red lines, N = 19) ctHPV16DNA levels decrease immediately after initiating CRT, whereas in the second subgroup (blue lines, N = 35) ctHPV16DNA levels initially spike followed by a subsequent decrease. (B) Percentage of patients with ctHPV16DNA elimination at different time points after initiating CRT. (C) Percentage clearance of ctHPV16DNA at week 4 in 40 patients with baseline ctHPV16DNA levels >200 copies/mL. Red dots indicate patients that later developed residual or recurrent disease. (D) Rapid ctHPV16DNA clearance kinetics in patients with >95% clearance of baseline ctHPV16DNA levels by week 4 of CRT. (E) Delayed ctHPV16DNA clearance kinetics in patients with 95% or less clearance of baseline ctHPV16DNA levels by week 4 of CRT.
Low baseline ctHPV16DNA levels correlates with low tumor HPV copy number and HPV integration
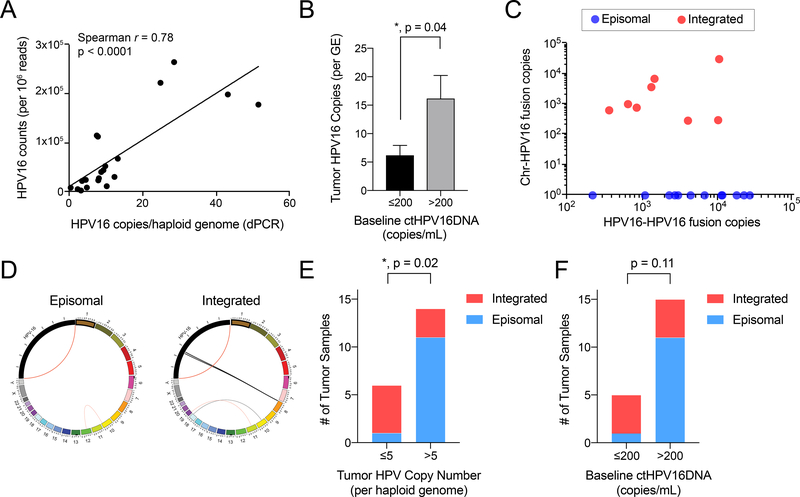
Twenty HPV16 positive patients in our cohort were co-enrolled on an institutional cancer genomic registry study (NCT01457196) and underwent next-generation sequencing (NGS) of their diagnostic tissue biopsy using the UNCSeq platform (27), as well as quantification of HPVDNA using dPCR. We observed excellent correlation between HPV16 copies per haploid genome measured by dPCR and HPV16 read counts per 106 total aligned reads by NGS (Figure 4A). Patients with low baseline ctHPV16DNA (</= 200 copies/mL) had a significantly lower tumor HPV copy number than patients with high baseline ctHPV16DNA (>200 copies/mL) (Figure 4B). We next evaluated HPV structure in tumors using a validated insertion-deletion detection algorithm(24) to quantify HPV-chromosomal fusion reads that are indicative of HPV integration into the somatic genome (Figure 4C). As a positive control, we detected end-to-start fusions of the HPV genome (“HPV-HPV fusion”) in all samples, which is expected because the HPV reference genome is deposited as a linear sequence yet originates as a circular episome in nature. There was evidence for HPV integration in 8 out of 20 patients (40%, Figure 4C). Circos plots are shown for two representative tumors without and with HPV integration (Figure 4D). A significantly higher proportion of patients with low tumor HPV copy number (≤5 copies/haploid genome) had HPV integration (Figure 4E, p=0.02). Similarly, patients with low baseline ctHPV16DNA (≤200 copies/mL) were more likely to have HPV integration (Figure 4F, p=0.11). Thus, low baseline levels of ctHPV16DNA are indicative of lower tumor HPV copy number and a greater likelihood of HPV integration, both of which represent adverse tumor genomic features.
Figure 4. Correlation between ctHPV16DNA and tumor HPV genomic features.
(A) Spearman correlation between copies of HPV16 detected in tumor biopsy samples by dPCR and NGS (B) Tumor HPV16 copies per genome equivalent (GE) in patients grouped by baseline ctHPV16DNA levels. Mean ± SEM is shown. P value calculated using a two-tailed, unpaired t-test with unequal variances. (C) Normalized NGS counts for hybrid HPV-chromosomal reads (indicative of HPV integration) relative to HPV-HPV fusion reads (positive control). (D) Circos plots illustrating the detecting HPV and chromosomal rearrangements in a tumor with purely episomal HPV (left panel) and HPV integration into a locus on chromosome 8 (right panel). (E-F) Number of tumor samples with integrated versus episomal HPV after grouping patients by (E) tumor HPV copy number (per GE) or (F) baseline ctHPV16DNA (copies/mL). P values calculated by a two-tailed Fisher’s exact test.
ctHPV16DNA rapid clearance profile and correlation to local and regional disease control
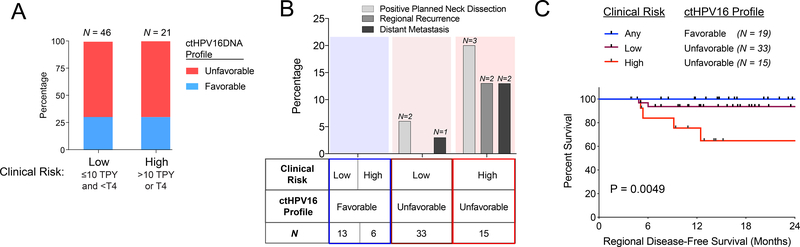
We next analyzed correlation between ctHPV16DNA clearance profile and disease control in the same 67 patient subset with available weekly blood samples, post-treatment PET scans, and subsequent clinical follow-up (median 16.5 months). We defined a favorable ctHPV16DNA clearance profile as high baseline level (>200 copies/mL) with rapid clearance (>95% clearance by week 4). Twenty-eight percent of the patient cohort (19/67) had a favorable ctHPV16DNA clearance profile, which did not differ based on established clinical risk factors (Figure 5A). The remaining patients had an unfavorable ctHPV16DNA profile (undetectable or ≤200 copies/mL at baseline, or ≤ 95% clearance by week 4). Patients with an unfavorable ctHPV16DNA clearance profile had a higher frequency of regional disease persistence/recurrence and distant metastasis, particularly amongst the clinical high-risk patient subset (Figure 5B). We next quantified actuarial persistent/recurrent regional disease-free survival (RDFS) after CRT. Patients with a favorable ctHPV16DNA clearance profile had 100% RDFS (Figure 5C). Among patients with an unfavorable ctHPV16DNA clearance profile, there was an interaction with established clinical risk factors. Patients with clinical low-risk disease (≤10 TPY and <T4) had ~90% RDFS, whereas patients with clinical high-risk disease (>10 TPY or T4) had ~65% RDFS at 18 months post-CRT (Figure 5C, p = 0.0049). These findings suggest that favorable ctHPV16DNA clearance profile identifies a low-risk subset of HPV-associated OPSCC patients (possibly regardless of clinical risk factors) that are highly responsive to definitive CRT.
Figure 5. A favorable ctHPV16DNA clearance profile correlates with disease control in OPSCC patients treated with CRT.
(A) Percentages of clinical low-risk and clinical high-risk patients with a favorable ctHPV16DNA profile, which is defined as baseline ctHPV16DNA >200 copies/mL and >95% clearance by week 4. (B) Percentage of patients within each subgroup who experience a positive neck dissection, regional recurrence, or distant metastasis. (C) Kaplan-Meier analysis of regional disease-free survival stratified by clinical risk and ctHPV16DNA profile. P value calculated using a two-tailed log-rank test for trend.
Discussion
We have prospectively analyzed ctHPVDNA baseline levels and clearance kinetics in a cohort of newly diagnosed HPV-associated OPSCC patients treated with definitive CRT, 84% of whom were treated with de-intensified CRT on a multi-institutional phase II study. Our findings add to a growing body of evidence that ctHPVDNA is a promising biomarker for early detection of HPV-associated OPSCC (sensitivity 89% and specificity 97%), similar to the utility of ctEBVDNA in screening programs for EBV-associated nasopharyngeal cancer(15). Significantly, 11% of our cohort had undetectable ctHPVDNA from 5 of the most common HPV strains that have been linked to OPSCC, suggesting that some of these cases may represent false positives due to p16-based identification, consistent with another recent report(28).
Although baseline ctHPVDNA copy number correlated with disease burden, we observed paradoxically lower plasma ctHPVDNA levels in patients with adverse clinical risk factors (Figure 2D, >10 TPY or T4 disease). By analyzing matched tumor genomics, we discovered that patients with low pre-treatment ctHPV16DNA levels (≤ 200 copies/mL) had lower tumor HPV copy number and a higher likelihood of HPV integration, which are associated with worse outcomes in OPSCC(5,6,8–12,29). In contrast, patients with abundant pre-treatment ctHPV16DNA levels (>200 copies/mL plasma) were most likely to have tumors with high copy and episomal HPV, which correlates with favorable prognosis in both cervical cancer and OPSCC(8,9,11,14). Thus, beyond simply a measure of tumor burden, high baseline plasma ctHPV16DNA levels (>200 copies/mL) are indicative of favorable tumor genomic biomarkers.
We also found that the rate of ctHPV16DNA clearance correlates with CRT sensitivity. Using week 4 of CRT as a benchmark timepoint, we defined a favorable ctHPV16DNA clearance profile as elevated baseline levels (>200 copies/mL) that are rapidly cleared (>95% clearance of pre-treatment levels by week 4). Patients with a favorable ctHPV16DNA clearance profile had excellent RDFS (~100% at 18 months). Amongst patients with an unfavorable ctHPV16DNA clearance profile, those with adverse clinical risk factors (>10 TPY or T4) experienced a high rate of residual or recurrent regional nodal disease (~35% at 18 months), which resembles disease control rates after CRT for HPV negative OPSCC.
Although there are parallels between ctHPVDNA in OPSCC and ctEBVDNA in NPC with regards to early detection, our findings highlight a few clinically significant differences as well. First, whereas high pre-treatment levels of ctEBVDNA are associated with worse prognosis in NPC(16), we find that the opposite is true for ctHPVDNA in OPSCC (i.e., low pre-treatment ctHPVDNA levels are associated with worse clinical outcomes). Second, a recent study demonstrated the prognostic significance of ctEBVDNA at the end of CRT as predictive of NPC recurrence(30). We did not observe a similar correlation with ctHPVDNA, as patients with detectable signal at the end of CRT were not enriched for those that later developed residual or recurrent disease. However, a high ctHPVDNA clearance percentage at week 4 (i.e., >95%) was associated with a lower rate of regional disease persistence/recurrence, similar to what has been shown in EBV-associated NPC(26). A possible explanation for these discrepant findings may be due to the highly variable HPV copy numbers per tumor genome that are observed in HPV-associated OPSCC (see Figure 4A–B). Tumors with high-copy, episomal HPV may have as many as 50–100 copies per cell, which may make it possible to detect small amounts of residual ctHPVDNA even when CRT has effectively treated all of the active disease. Thus, the specific ctDNA profiles associated with clinical outcomes may differ according to the distinct clinical and biological features of EBV-associated NPC and HPV-associated OPSCC, respectively.
The potential clinical utility of ctHPV16DNA monitoring in OPSCC is broad, especially in light of recent evidence that some forms of de-intensified therapy are not effective for all patients with HPV-associated OPSCC(31,32). Future studies should prospectively investigate whether ctHPVDNA kinetics can dynamically guide in “real-time” the intensity of CRT that may be required. Can patients with ≤ 10 TPY and a favorable ctHPV16DNA profile be further de-escalated (reduce radiation to 50 or 54Gy)? Is de-intensified CRT efficacious in patients with >10 TPY and a favorable ctHPV16DNA profile? Our data suggests that de-intensified CRT should not be offered to patients with an unfavorable ctHPVDNA profile and >10 TPY or T4 disease. Our findings illustrate that dynamic monitoring of ctHPVDNA during CRT may facilitate personalized treatment decisions to maximize tumor control while also minimizing treatment-associated morbidity.
Another potential clinical utility of dynamic ctHPVDNA monitoring may be in the post-treatment, surveillance setting. Our findings indicate that the sensitivity of ctHPVDNA appears to be quite high, and its use to detect cancer recurrence after definitive treatment may be advantageous. ctHPVDNA based surveillance may detect recurrences earlier than standard clinical visits and occasional radiographic surveillance. Early detection may result in a greater occurrence of the oligometastatic state and thus allow for more aggressive salvage treatment (e.g. metastasectomy, radiosurgery) that may have greater efficacy. Furthermore, ctHPVDNA surveillance may be more cost effective than current surveillance approaches.
Limitations
Although our trial included over 100 patients, it is underpowered due to low occurrence of disease persistence/recurrence and limited follow-up. Despite these limitations, we observed an early trend in worse RDFS (Figure 5C) in patients with >10 TPY and unfavorable ctHPVDNA kinetics. Validation of our findings using a clinical-grade test in independent and larger patient cohorts will be necessary to confirm our findings.
Conclusions and Future Directions
Circulating tumor HPVDNA is present in most patients with newly diagnosed HPV-associated OPSCC. Our findings suggest that a rapid clearance profile of ctHPV16DNA clearance during CRT may have clinical utility in stratifying patients with HPV-associated OPSCC based on likelihood of disease control. Future studies assessing ctHPVDNA as an integral biomarker to guide treatment de-intensification are warranted, and may facilitate personalized treatment decisions based on tumor biology in addition to clinical risk factors. Finally, prospective evaluation of ctHPVDNA as a biomarker in other HPV-associated malignancies (e.g. cervical and anal cancers) should be evaluated.
Supplementary Material
Translational relevance.
Plasma circulating tumor HPV DNA (ctHPVDNA) is a promising biomarker for monitoring treatment responses in patients with HPV-associated oropharyngeal squamous cell carcinoma (OPSCC). This study characterizes baseline levels and clearance kinetics of ctHPVDNA in a cohort of patients treated with definitive chemoradiotherapy (CRT). A favorable clearance profile was defined as high baseline ctHPVDNA levels (>200 copies/mL) that are rapidly eliminated during CRT (>95% clearance by day 28). Patients with a favorable clearance profile, regardless of tobacco pack years, had excellent cancer control (Regional DFS = 100%), while those with unfavorable clearance profile and adverse clinical factors (T4, > 10 tobacco pack years) had significantly worse cancer control (Regional DFS = 65%). Future studies assessing ctHPVDNA as an integral biomarker to guide treatment de-intensification are warranted, and may facilitate personalized treatment decisions based on tumor biology in addition to clinical risk factors.
Acknowledgments
We are grateful to the patients who agreed to participate in our research study. We thank L. Marks, J. Tepper, S. Earp, C. Moody, A. Amelio, M. Gulley, Gupta lab members, and colleagues in the UNC Department of Radiation Oncology for critical feedback and guidance. The Department of Radiation Oncology at the UNC School of Medicine, UNC Lineberger Cancer Center, the University Cancer Research Fund, and the University of Florida School of Medicine Department of Radiation Oncology supported this research study. G.P.G. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. Additional funding support was provided through NCI grants U10CA181009 (D.N.H.) and P30 CA016086 (J.S.P., UNC Lineberger).
Funding/Support: UCRF, BWF, University of North Carolina School of Medicine, Department of Radiation Oncology, Lineberger Comprehensive Cancer Center, and University of Florida School of Medicine, Department of Radiation Oncology
Role of the Funder/Sponsor: The funders had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Funding Source: Burroughs Wellcome Fund, University Cancer Research Fund, National Cancer Institute
Footnotes
Conflict of Interest: B.S.C, S.K., and G.P.G. have filed a U.S. patent application on aspects of the technology described in this manuscript (Application No. 62/732,117). B.S.C. and G.P.G. serve on the advisory board of Naveris, Inc, and hold equity interest. All other authors have no pertinent conflicts of interest to declare.
References
- 1.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363(1):24–35 doi 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chera BS, Amdur RJ, Tepper J, Qaqish B, Green R, Aumer SL, et al. Phase 2 Trial of De-intensified Chemoradiation Therapy for Favorable-Risk Human Papillomavirus-Associated Oropharyngeal Squamous Cell Carcinoma. International journal of radiation oncology, biology, physics 2015;93(5):976–85 doi 10.1016/j.ijrobp.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 3.Chera BS, Amdur RJ, Tepper JE, Tan X, Weiss J, Grilley-Olson JE, et al. Mature results of a prospective study of deintensified chemoradiotherapy for low-risk human papillomavirus-associated oropharyngeal squamous cell carcinoma. Cancer 2018;124(11):2347–54 doi 10.1002/cncr.31338. [DOI] [PubMed] [Google Scholar]
- 4.O’Sullivan B, Huang SH, Siu LL, Waldron J, Zhao H, Perez-Ordonez B, et al. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2013;31(5):543–50 doi 10.1200/JCO.2012.44.0164. [DOI] [PubMed] [Google Scholar]
- 5.Bratman SV, Waldron JN, Liu FF. Human Papillomavirus Genotypes Conferring Poor Prognosis in Head and Neck Squamous Cell Carcinoma-Reply. JAMA Oncol 2017;3(1):125–6 doi 10.1001/jamaoncol.2016.3397. [DOI] [PubMed] [Google Scholar]
- 6.Mazul AL, Rodriguez-Ormaza N, Taylor JM, Desai DD, Brennan P, Anantharaman D, et al. Prognostic significance of non-HPV16 genotypes in oropharyngeal squamous cell carcinoma. Oral Oncol 2016;61:98–103 doi 10.1016/j.oraloncology.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin HJ, Joo J, Yoon JH, Yoo CW, Kim JY. Physical status of human papillomavirus integration in cervical cancer is associated with treatment outcome of the patients treated with radiotherapy. PLoS One 2014;9(1):e78995 doi 10.1371/journal.pone.0078995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walline HM, Goudsmit CM, McHugh JB, Tang AL, Owen JH, Teh BT, et al. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head Neck 2017;39(5):840–52 doi 10.1002/hed.24729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koneva LA, Zhang Y, Virani S, Hall PB, McHugh JB, Chepeha DB, et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol Cancer Res 2018;16(1):90–102 doi 10.1158/1541-7786.MCR-17-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan IM, DiNardo LJ, Windle B. Integration of Human Papillomavirus Genomes in Head and Neck Cancer: Is It Time to Consider a Paradigm Shift? Viruses 2017;9(8) doi 10.3390/v9080208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nulton TJ, Kim NK, DiNardo LJ, Morgan IM, Windle B. Patients with integrated HPV16 in head and neck cancer show poor survival. Oral Oncol 2018;80:52–5 doi 10.1016/j.oraloncology.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao H, Banh A, Kwok S, Shi X, Wu S, Krakow T, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. International journal of radiation oncology, biology, physics 2012;82(3):e351–8 doi 10.1016/j.ijrobp.2011.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulz P, Heitzer E, Geigl JB, Speicher MR. Patient monitoring through liquid biopsies using circulating tumor DNA. Int J Cancer 2017;141(5):887–96 doi 10.1002/ijc.30759. [DOI] [PubMed] [Google Scholar]
- 14.Rostami A, Bratman SV. Utilizing circulating tumour DNA in radiation oncology. Radiother Oncol 2017;124(3):357–64 doi 10.1016/j.radonc.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2017;377(6):513–22 doi 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 16.Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350(24):2461–70 doi 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 17.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6(224):224ra24 doi 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 2015;7(293):293ra104 doi 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higginson DS, Yarusi B, Chan S, Mitrani L, Thompson C, McBride SM, Riaz N, and Lee N. Use of Human Papillomavirus 16 (HPV16) Cell Free DNA for Assessment of Response to Chemoradiation in HPV-Associated Oropharyngeal Cancer. International Journal of Radiation Oncology Biology Physics 2015;93(3S). [Google Scholar]
- 20.Ahn SM, Chan JY, Zhang Z, Wang H, Khan Z, Bishop JA, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg 2014;140(9):846–54 doi 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahlstrom KR, Li G, Hussey CS, Vo JT, Wei Q, Zhao C, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer 2015;121(19):3455–64 doi 10.1002/cncr.29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeck WR, Parker J, Carson CC, Shields JM, Sambade MJ, Peters EC, et al. Targeted next generation sequencing identifies clinically actionable mutations in patients with melanoma. Pigment Cell Melanoma Res 2014;27(4):653–63 doi 10.1111/pcmr.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Wang A, Walter V, Patel NM, Eberhard DA, Hayward MC, et al. Combined Targeted DNA Sequencing in Non-Small Cell Lung Cancer (NSCLC) Using UNCseq and NGScopy, and RNA Sequencing Using UNCqeR for the Detection of Genetic Aberrations in NSCLC. PLoS One 2015;10(6):e0129280 doi 10.1371/journal.pone.0129280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mose LE, Wilkerson MD, Hayes DN, Perou CM, Parker JS. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 2014;30(19):2813–5 doi 10.1093/bioinformatics/btu376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19(9):1639–45 doi 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung SF, Chan KC, Ma BB, Hui EP, Mo F, Chow KC, et al. Plasma Epstein-Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol 2014;25(6):1204–8 doi 10.1093/annonc/mdu117. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery ND, Parker JS, Eberhard DA, Patel NM, Weck KE, Sharpless NE, et al. Identification of Human Papillomavirus Infection in Cancer Tissue by Targeted Next-generation Sequencing. Appl Immunohistochem Mol Morphol 2016;24(7):490–5 doi 10.1097/PAI.0000000000000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nauta IH, Rietbergen MM, van Bokhoven A, Bloemena E, Lissenberg-Witte BI, Heideman DAM, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol 2018;29(5):1273–9 doi 10.1093/annonc/mdy060. [DOI] [PubMed] [Google Scholar]
- 29.Groves IJ, Coleman N. Human papillomavirus genome integration in squamous carcinogenesis: what have next-generation sequencing studies taught us? J Pathol 2018;245(1):9–18 doi 10.1002/path.5058. [DOI] [PubMed] [Google Scholar]
- 30.Chan ATC, Hui EP, Ngan RKC, Tung SY, Cheng ACK, Ng WT, et al. Analysis of Plasma Epstein-Barr Virus DNA in Nasopharyngeal Cancer After Chemoradiation to Identify High-Risk Patients for Adjuvant Chemotherapy: A Randomized Controlled Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018:JCO2018777847 doi 10.1200/JCO.2018.77.7847. [DOI] [PubMed] [Google Scholar]
- 31.Gillison ML, Trotti AM, Harris J, Eisbruch A, Harari PM, Adelstein DJ, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet 2018. doi 10.1016/S0140-6736(18)32779-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet 2018. doi 10.1016/S0140-6736(18)32752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.