Abstract
The dependence of capillary zone electrophoresis separations on the charge state of analyte is useful for the analysis of many post-translational modifications in proteins. In this work, we coupled CZE to an Orbitrap Fusion Lumos Tribrid platform with advanced peak determination algorithm for phosphoproteomics analysis. A linear polyacrylamide coated capillary with very low electroosmotic flow was used for the separation. The optimal injection volume is between 100 nL and 150 nL of phosphopeptides dissolved in 30 mM ammonium bicarbonate (pH 8.2) buffer, which produces a dynamic pH junction sample injection. Larger injection volumes resulted in serious peak broadening and decreased numbers of phosphopeptide identifications. The optimized system identified 4405 phosphopeptides from 220 ng of enriched phosphopeptides from mouse brain, which represents the state-of-the-art result for single shot CZE-ESI-MS/MS based phosphoproteome analysis. We found that the migration time for phosphopeptides are much longer than nonphosphopeptides and increased along with the number of the phosphorylation sites on the peptides, as expected for the additional negative charges associated with the phosphate groups. We also investigated the phosphorylation site motifs; a number of motifs appeared in CZE-ESI-MS/MS data but not in LC-ESI-MS/MS data, which suggested the complementary performance of the techniques. Data are available via ProteomeXchange with identifier PXD012888.
Keywords: Phosphoproteomics, CZE, Orbitrap Fusion Lumos Tribrid, Advanced Peak Determination Algorithm
Introduction
Proteins are converted to their mature form through post-translational modification (PTM) processing.1 Protein phosphorylation,2 which affects metabolism, growth, division, differentiation, apoptosis, membrane transport, and immunity, is the most common PTM in signal transduction and is a central mechanism in health and disease.3,4 Identification of low abundant phosphoproteins remains challenging.5–7
Mass spectrometry (MS) has emerged as an indispensable technology for global PTM analysis due to its capacity to localize modifications to a single residue.8–13 Advances in MS-based phosphoproteomics have made possible the identification of tens of thousands of phosphopeptides,14–16 which have provided valuable data on the complex networks and functions coordinated by phosphorylation.
Currently, reversed phase liquid chromatography-electrospray ionization-tandem mass spectrometry (RPLC-ESI-MS/MS) dominates MS-based phosphoproteomics due to its excellent separation performance.17–21 Nevertheless, there are limitations to this approach. Very short and polar peptides (e.g., peptides with several phosphorylation sites) tend to flush out of the chromatographic column during sample loading, whereas high hydrophobic peptides are difficult to elute from the column.
Capillary zone electrophoresis (CZE) is an intriguing alternative technology for phosphopeptide separation before mass spectrometry. CZE separations are based on the size-to-charge ratio of the peptide, and these separations are exquisitely sensitive to changes in the charge of a peptide. Moreover, the migration time of phosphopeptides in CZE can be accurately predicted which will be helpful for estimating the confidence of phosphopeptide identifications and overcoming the problem of underestimating the false discovery rates for phosphopeptides identifications by the traditional target-decoy approach.22,23
Following the development of stable and sensitive CZE-MS interfaces,24,25 CZE- ESI-MS/MS has attracted recent interest for shotgun proteomic analysis.26,27 Much effort has been made to overcome its poor concentration detection limit,28,29 to increase the separation window, and to improve throughput.30–32
CZE has also been used for study of the phosphoproteome. Cao et al. developed on-line Fe(III) immobilized metal-ion affinity chromatography (IMAC)-CZE-ESI-MS and applied it to sub-pmol analysis of phosphopeptides.33 This platform was applied to the analysis of phosphopeptide mixtures and a phosphoprotein digest. This work has been expanded by use of multiple stage tandem mass spectrometry (MSn, n = 2,3) to isolate and fragment target ions to provide more reliable assignments of phosphorylated residues34. Figeys et al. described a solid-phase extraction (SPE)-CZE-ESI-MS/MS system for the automated, sensitive analysis of minor peptide species in complex samples such as phosphopetides generated by the proteolytic digestion of endothelial nitric oxide synthase.35 More recently, Ludwig et al. systematically compared UPLC-ESI-MS/MS and CZE-ESI-MS/MS for phosphorylated peptide identifications (IDs) using an enriched phosphoproteome from the MCF-10A cell line.36 UPLC generated more phosphorylated peptide IDs than CZE from 2 μg sample loadings. However, CZEMS/MS consistently and significantly outperformed UPLC-MS/MS in terms of phosphorylated peptide and total peptide IDs identified from 2–200 ng sample. By using a dynamic pH junction method, 2,313 phosphorylated peptides were identified from 1 μg sample with single-shot CZE-ESI-MS/MS in a 100 min analysis. To improve the identification performance of CZE-ESI-MS/MS, the phosphopeptides enriched from PC-12 pheochromocytoma cells were first fractionated by using RPLC then analyzed by CZE-MS.22,37 However, the performance of single-shot CZE-ESI-MS/MS for phosphoproteomics analysis has so far fallen far below that of the state-of-art RPLC-MS/MS system, which has identifies over 16,000 phosphopeptides from 1 μg sample in 3 h.16
In this work, we coupled CZE to an Orbitrap Fusion Lumos Tribrid mass spectrometer with advanced peak determination (APD) algorithm for phosphoproteomics analysis. A linear polyacrylamide (LPA) coated capillary was prepared by surface-confined aqueous reversible addition-fragmentation chain transfer (SCARAFT) polymerization method and had very low electroosmotic flow (EOF), which produced a long separation window.32 The dynamic pH junction technique was used for sample injection that filled ~7.5 % of the capillary volume without excessive band broadening to improve concentration detection limits. More recently, we identified over 27000 peptides with single shot CZE-ESI-MS/MS analysis on this platform.38
METHODS AND MATERIALS
Reagents and Chemicals.
Formic acid (FA), acetic acid (HAc), trifluoroacetic acid (TFA), acrylamide, 4,4 -azobis(4-cyanovaleric acid), cyanomethyl [3-(trimethoxysilyl)propyl] trithiocarbonate, ammonium acetate, bovine pancreas TPCK-treated trypsin, tris(2-carboxyethyl)phosphine (TCEP), and iodoacetamide (IAA) were purchased from Sigma-Aldrich (St. Louis, USA). Methanol was purchased from Honeywell Burdick & Jackson (Wicklow, Ireland). Uncoated fused silica capillary (50 μm i.d. × 350 μm o.d.) was purchased from Polymicro Technologies (Phoenix, AZ). Water was deionized by a Nano Pure system from Thermo Scientific (Marietta, OH).
Preparation of the coated capillaries by surface-confined aqueous reversible-addition fragmentation transfer (SCARAFT) polymerization method.
Preparation of the linear polyacrylamide(LPA) coated capillary by that SCARAFT method is similar to our original report.32 The detailed process is described in the Supporting information.
Phosphopeptides enrichment from mouse brain sample.
The detailed protocol for preparation of mouse brain tryptic digest is presented in the Supporting information. All animal procedures were performed according to protocols approved by the University of Wisconsin Institutional Animal Care and Use Committee. For phosphopeptide enrichment, the dried whole mouse brain tryptic peptides were dissolved in 80% acetonitrile/6% TFA and enriched for phosphopeptides using titanium(IV) immobilized metal ion affinity chromatography. In brief, 200 μL of magnetic titanium(IV) beads (ReSyn Biocsciences) were washed twice with 80% acetonitrile/6% TFA (all washes are 1 mL), incubated with the sample on a shaker for 30 min, washed again three times with 80% acetonitrile/6% TFA, once with 80% acetonitrile, once with 0.5 M glycolic acid/80% acetonitrile, and once again with 80% acetonitrile. Phosphopeptides were eluted with 1% ammonium hydroxide/50% acetonitrile. Phospho-depleted peptides, for protein analyses, were saved from the unbound portion following the incubation step.
CZE-ESI-MS/MS analysis.
The CZE system consists of two high-voltage power supplies (Spellman CZE 1000R) and an electrokinetically pumped electrospray interface. The interface was used to couple the CZE separation capillary to a mass spectrometer (see below). The electrospray emitter was made from borosilicate glass capillary (1.0 mm o.d. × 0.75 mm i.d., 10 cm long) pulled with a Sutter instrument P-1000 flaming/brown micropipette puller; the size of the emitter opening was 15–20 μm. The electrospray sheath flow buffer was 10% (v/v) methanol with 0.5%FA. A LPA coated capillary (50 μm i.d.×350 μm o.d., 100 cm long) prepared by using SCARAFT method was used for the CZE separation. The dynamic pH junction is commonly used for improving the concentration sensitivity of CZE.39,40 In this work, titanium(IV) IMAC enriched phosphopeptides were dissolved in 30 mM ammonium bicarbonate (pH 8.2). The background electrolyte was 1 M HAc. This combination produces a dynamic pH junction following injection. The sample was injected by pressure. 23.8 kV was appliedat the injection end of the capillary for separation and 1.8 kV on the sheath flow reservoir for electrospray in positive polarity. Voltage programming was controlled by LabView software.
Mass Spectrometer Operating Parameters.
Two different mass spectrometers were used in these experiments. Q Exactive HF mass spectrometer (Thermo Fisher Scientific) was operated in positive mode. A data-dependent acquisition with top 10 method was used. The S-lens RF level was set at 60, and the heated capillary was at 300 °C. Full scan resolution was set to 60,000 at m/z 200. Full scan target was 3.00E+06 with a maximum fill time of 15 ms. Full MS scans were acquired over the 300−1350 m/z range. Target value for fragment scans was set at 1.00E+05 with a maximum fill time of 110 ms. and intensity threshold was kept at 1.00E+05. Isolation width was set at 1.4 Th. A fixed first mass of 100 was used. Normalized collision energy was set at 28.
For experiments using an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) with instrument control software version 3.0, typical analyses used a 240,000 resolving power survey scan with an AGC of 106 in positive mode, followed by MS/MS of the most intense precursors for 1 s. The scan range was 300–1350 m/z. The MS/MS analyses were performed by 0.7 m/z isolation with the quadrupole, normalized HCD collision energy of 30%, and analysis of fragment ions in the ion trap using the “Rapid” scan speed scanning from 200 to 1200 m/z. Dynamic exclusion was set to 20 s. Monoisotopic precursor selection (MIPS) was set to Peptide. For MS/MS analyses, the maximum injection time was set to 22 ms, with an AGC target of 30,000, and charge states unknown, 1, or >5 were excluded. The advanced peak determination was either off or on for SPD and APD analyses, respectively.
Database search and data analysis.
Database searching of the raw files was performed in Proteome Discoverer 2.2 (Thermo) with Sequest HT and MS Amanda 2.0 database search engines.41,42 Databases with taxonomy as mouse were downloaded from Uniprot. Searching the reversed database was performed to evaluate the false discovery rate. For the data obtained on a Q-Exactive HF mass spectrometer, precursor tolerance was set to 10 ppm and fragment mass tolerance was 0.05 Da. For data obtained on a Fusion Lumos mass spectrometer, precursor tolerance was set to 10 ppm and fragment mass tolerance was 0.4 Da. Full tryptic digestion was selected and allowed up to two missed cleavages. Carbamidomethylation (C) was set as fixed modifications. Oxidation (M), deamidated (NQ), acetylation (K), and phosphorylation (S, T, Y) were set as variable modifications. Database search results were evaluated with Percolator (version 2.04) software. On the peptide level, peptide confidence value as high was used to filter the peptide identification, and the corresponding false discovery rate on peptide level was less than 1%. On the protein level, protein grouping was enabled. PhosphoRS was used for determining phosphorylation site probabilities.43
Raw MS files were also analyzed by MaxQuant (version 1.6.1.0.).44 MS/MS spectra were searched by the Andromeda search engine against the Uniprot mouse protein database. The database also included common contaminants. MaxQuant analysis included an initial search with a precursor mass tolerance of 20 ppm, main search precursor mass tolerance of 6 ppm and fragment mass tolerance of 20 ppm. The search included: trypsin as enzyme, variable modifications of oxidation (M), deamidation (NQ) and phospho (STY), and fixed modification of carbamidomethyl cysteine. Minimal peptide length was set to six amino acids and the maximum number of missed cleavages was set to two. The false discovery rate (FDR) was set to 0.01 for both peptide and protein identifications.
Motifs analysis was performed in Motif-X (http://motif-x.med.harvard.edu).45,46 The parameters were set as: occurrence of 20 and a significance of 0.000001, the mouse fasta database was used as background.
RESULTS AND DISCUSSION
The effect of the injection volume on the identification numbers.
We investigated the relationship between the injection volumes and the number of identified phosphopeptides, Figure 1. The numbers of phosphopeptide IDs increased by ~25% when the sample injection volume was increased from 49 nL (97 ng) to 104 nL (208 ng). However, the identification numbers decreasing slightly when the injection volumes were further increased to 208 nL (416 ng) and 402 nL (804) ng. These results were unexpected because identification numbers usually reached a plateau when the loading amount increased to the maximum column capacity. The number of identified phosphopeptides was increased from 2168 (N=2, RSD=3.4%) to 3005 (N=2, RSD=0.5%) when the separation time was extended from 100 min to 140 min for the run with 402 nL injection. However, the number still less than 3207(N=2, 0.7%) obtained under the optimal injection volume of 104 nL with 100 min separation time. We analyzed the peak widths distributions for different sample loading amounts, the results are shown in Figure S1 in the Supporting information. We found significant peak broadening when the injection volume was greater than ~150 nL (corresponding to 300 ng). It is known that the optimal injection volume for CZE is usually less than 1% of the total capillary volume to maintain its high separation efficiency. Though the injection volume could be increased by using the dynamic pH junction without compromise the CZE separation performance, we concluded that the optimal injection volume is between 100 nL and 150 nL under the dynamic pH junction conditions used in this work.
Figure 1.
The relationship between the injection volume and the number of phosphopeptide identifications. Experimental conditions: Q Exactive HF mass spectrometer; 50 μm i.d. × 350 μm o.d. × 100 cm long LPA coated capillary; 2 mg/mL of enriched phosphopeptides from mouse brain digest; 1 M HAc separation buffer; 23.8 kV separation voltage; and 1.8 kV spray voltage in positive polarity. Errorbars are standard deviations of the measurements.
Coupling to an Orbitrap Fusion Lumos Tribrid platform with APD algorithm.
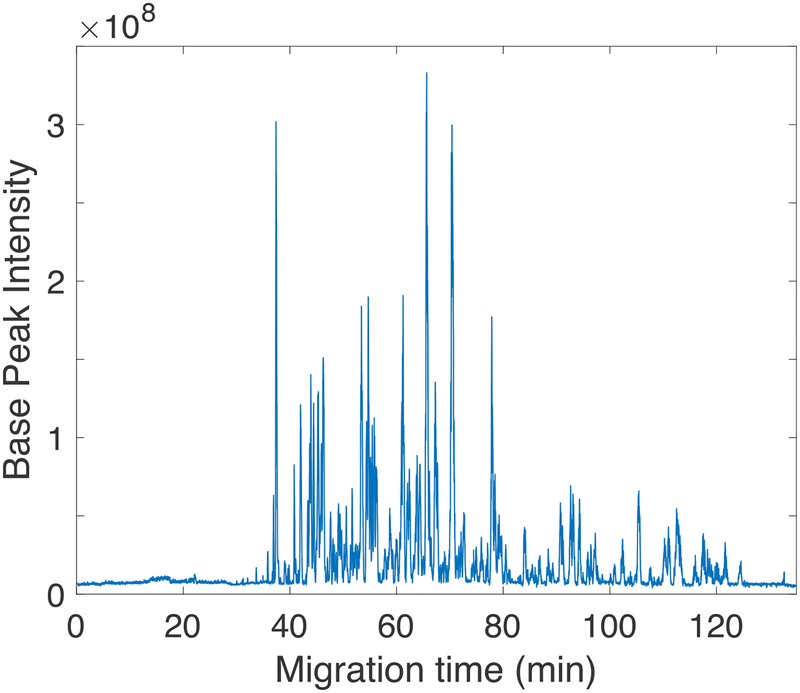
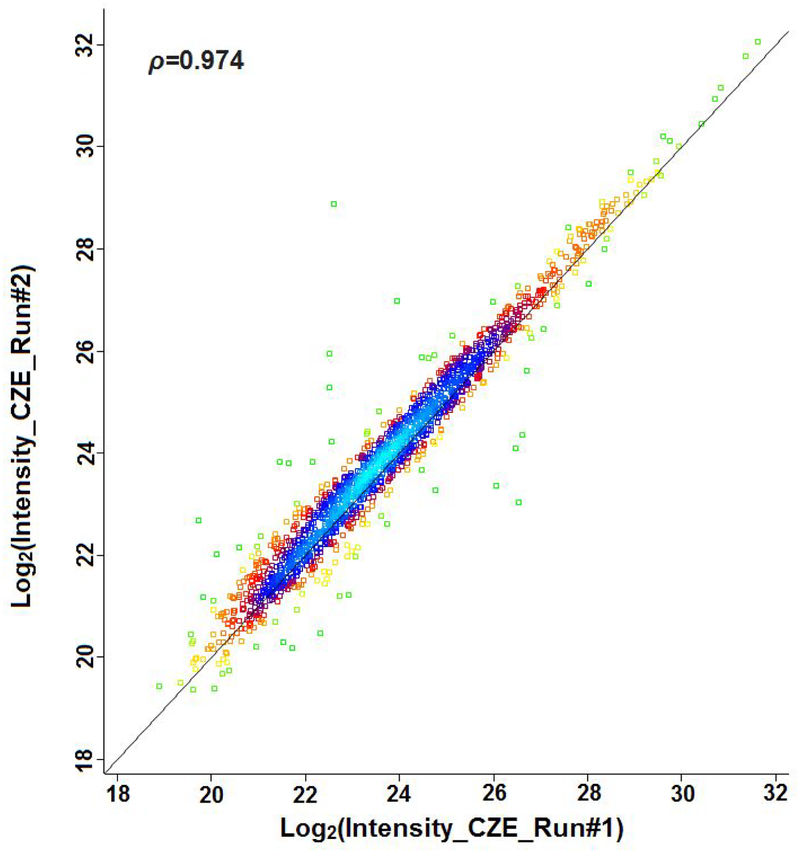
To further improve the identification performance, we coupled CZE to an Orbitrap Fusion Lumos Tribrid platform that employed an APD algorithm. Coon’s group recently described the APD algorithm that used an iterative approach to improve the annotation of peptides’ charge states at the densely populated m/z regions in LC-MS based shotgun proteomics.16 This feature is especially useful for CZE-MS based proteomics due to the not uniform distribution of peptides throughout the separation space.47 We identified 4,405 phosphopeptides and 1,998 protein groups with better than 75% phosphosite localization probability from 220 ng of enriched phosphopeptides from mouse brain digest by using single shot CZE-ESI-MS/MS analysis, Figure 2. Scatter plot between replicates was shown in Figure 3, pearson correlation coefficient of 0.974 is obtained, demonstrating the good reproducibility and high label-free quantification precision of the CZE-MS method.
Figure 2.
Single shot CZE-ESI-MS/MS analysis of phosphopeptides. Experimental conditions: Orbitrap Fusion Lumos Tribrid mass spectrometer; 220 ng enriched phosphopeptides from mouse brain digest; other conditions are same as in Figure 1.
Figure 3.
Scatter plot between the replicates of CZE-MS. Experimental conditions are same as in Figure 2. Data are plotted in a green-yellow-red-blue color map corresponding to the density of peptides observed for the two runs.
Migration time and pKa distribution of the identified phosphopeptides.
We analyzed the migration time distribution of peptides and phosphopeptides with different numbers of phosphorylation sites, Figure 4. Peptides without phosphorylation sites tend to migrate faster than phosphorylated peptides, Figure 4(A). Moreover, the migration time of the phosphorylated peptides increased with the increase of the number of the phosphorylation sites, Figure 4(B). The results suggest adjustment of EOF to decrease the migration time of peptides with multiple phosphorylation sites without degrading the identification performance would be a fruitful strategy to increase the throughput of CZE-MS for phosphoproteomics analysis.
Figure 4.
Migration time distributions. (A) Identified peptides. (B) Phosphopeptides with 2 and 3 phosphorylation sites.
We also investigated the pKa distribution of the identified peptides and phosphopeptides, Figure 5. As expected, most of the phosphopeptides have a low pI values. A large proportion of the Ti(IV) IMAC enriched phosphopeptides originate from acidic kinase substrates.48,49
Figure 5.
pKa distribution of (A) all the identified peptides and (B) the phosphopeptides with 2 and 3 phosphorylation sites.
Enrichment analysis of identified phosphorylated proteins.
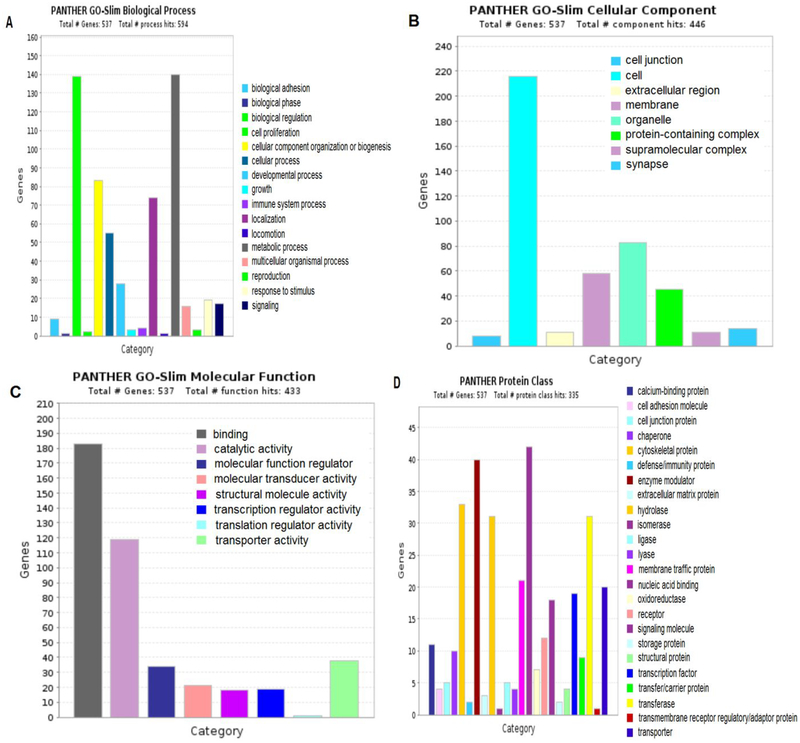
The phosphorylated proteins identified by CZE-MS were used for the Gene Ontology (GO) biological process, cellular component, molecular function and protein class enrichment analysis through PANTHER classification system.50 The identified phosphorylated proteins are mainly involved in metabolic process, biological regulation and cellular component organization or biogenesis et al., Figure 6(A). For cellular component, we also identified many phosphorylated proteins from organelle and membrane, Figure 6(B). For molecular function, most of the identified phosphorylated proteins have binding, catalytic and transporter activity, Figure 6(C). Many kinases were detected in the phosphorylated proteins identified by CZE-MS. For example, RAF proto-oncogene serine/threonine-protein kinase(Q99N57) which acts as a regulatory link between the membrane-associated Ras GTPases and the MAPK/ERK cascade, and this critical regulatory link functions as a switch determining cell fate decisions including proliferation, differentiation, apoptosis, survival and oncogenic transformation. Ren et al had demonstrated that the PI3K/AKT signaling pathway inhibits ERK signaling via AKT1-dependent phosphorylation of RAF1 on Ser259 in endothelial cells.51
Figure 6.
Enrichment analysis of identified phosphorylated proteins by CZE-MS.(A) Biological process. (B) Cellular component. (C) Molecular function. (D) Protein class.
Motifs analysis of the identified phosphorylation sites and comparison with LC-MS.
In this study, Motif-X was used to extract the overrepresented motifs from the identified phosphorylation sites using the mouse fasta database as background, Figure S2 in the Supporting information. We found many proline-directed kinase substrates (with a proline shown on the +1 position) that are very common in the phosphoproteome.52 Many motifs with D/E were also found for the identified phosphorylation sites, as expected for phosphopeptides originating from acidic kinase substrates. Moreover, we found some motifs from the phosphorylation sites identified by using CZE-MS method didn’t appear in the LC-MS data. For example, 16 motifs were found with threonine as central foreground residue in our data, 10 of which were also found in the data from LC-MS method. In addition, six motifs only appeared in the CZE-MS data (Motif 7, 9, 12, 13, 14, 16 of motifs found with a phosphorylated threonine in Figure S2). We also compared the identified phosphorylated proteins by CZE-MS and LC-MS. 961 protein groups were identified by both methods, while 559 protein groups were only identified by CZE-MS and 4104 protein groups were only identified by LC-MS. These results suggested that CZE-MS and LC-MS have complementary performance for phosphoproteome analysis, and data from both methods appears necessary for deep and more complete phosphoproteome mapping.
Conclusions
The advanced peak determination algorithm for Orbitrap Fusion Lumos Tribrid platform improves charge state determination, increasing the number of precursors available for data-dependent analysis. We coupled CZE with this system for phosphoproteomics analysis. CZE separates peptides based on their charge to size ratio, which forms the basis of an accurate algorithm for prediction of electrophoretic mobility. However, the distribution of peptides is not uniform in CZE and there are typically dense regions where large numbers of peptides appear. The advanced peak algorithm is particularly useful in this case because it is able to annotate peptides’ charge states in the densely populated m/z regions.
A LPA coated capillary with very low EOF coupled with the dynamic pH junction technique further improved the CZE separation performance. 4,405 phosphopeptides were identified from 220 ng of enriched phosphopeptides from mouse brain, which is the state-of-the-art result for single shot CZE-ESI-MS/MS based phosphoproteome analysis. We analyzed the migration time distribution of the identified peptides and found that the migration times for phosphopeptides were much longer than for unphosphorylated peptides, and increased along with the number of the phosphorylation sites on the peptides due to the increased negative charge on the peptide. Moreover, the number of identified phosphopeptides could be further improved by systematically optimizing the MS parameters for CZE-MS.53 We compared the phosphorylation site motifs and identified phosphorylated proteins for LC-MS and CZE-MS, the results demonstrated the complementary performance of CZE-MS and LC-MS techniques.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. William Boggess and Mathew Champion of the University of Notre Dame Mass Spectrometry and Proteomic Facility for their assistance. This work was supported by grants from the National Institute of Health (R01GM096767, R01HD084399, R35 GM 118110, and P41GM108538).
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
Supporting Information
Preparation of coated capillary
Capillary tip etching
Mouse-brain phosphopeptide enrichment
Figure S1 – peak width distributions
Figure S2 – phosphorylation site motifs
Figure S3 – Motifs found with a phosphorylated threonine in LC-ESI-MS/MS data
Table S1 – Excel spread sheet with phosphopeptide identifications.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository,54 with the dataset identifier PXD012888. The authors declare no competing financial interest.
REFERENCES
- (1).Walsh CT; Garneau-Tsodikova S; Gatto GJ Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed 2005, 44, 7342–7372. [DOI] [PubMed] [Google Scholar]
- (2).Ubersax JA; Ferrell JE Jr. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 2007, 8, 530–541. [DOI] [PubMed] [Google Scholar]
- (3).Cozzone AJ Protein-phosphorylation in bacteria. Trends Biochem. Sci 1984, 9, 400–403. [Google Scholar]
- (4).Olsen JV; Blagoev B; Gnad F; Macek B; Kumar C; Mortensen P; Mann M Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [DOI] [PubMed] [Google Scholar]
- (5).Ahn NG; Resing KA Toward the phosphoproteome. Nat. Biotechnol 2001, 19, 317–318. [DOI] [PubMed] [Google Scholar]
- (6).Oda Y; Nagasu T; Chait BT Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol 2001, 19, 379–382. [DOI] [PubMed] [Google Scholar]
- (7).Pinkse MWH; Uitto PM; Hilhorst MJ; Ooms B; Heck AJR Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem 2004, 76, 3935–3943. [DOI] [PubMed] [Google Scholar]
- (8).Mann M; Jensen ON Proteomic analysis of post-translational modifications. Nat. Biotechnol 2003, 21, 255–261. [DOI] [PubMed] [Google Scholar]
- (9).Merrill AE; Coon JJ Quantifying proteomes and their post-translational modifications by stable isotope label-based mass spectrometry. Curr. Opin. Chem. Biol 2013, 17, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhou H; Watts JD; Aebersold R A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol 2001, 19, 375–378. [DOI] [PubMed] [Google Scholar]
- (11).Ficarro SB; McCleland ML; Stukenberg PT; Burke DJ; Ross MM; Shabanowitz J; Hunt DF; White FM Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol 2002, 20, 301–305. [DOI] [PubMed] [Google Scholar]
- (12).Claverol S; Burlet-Schiltz O; Gairin JE; Monsarrat B Characterization of protein variants and post-translational modifications: ESI-MSn analyses of intact proteins eluted from polyacrylamide gels. Mol. Cell. Proteomics 2003, 2, 483–493. [DOI] [PubMed] [Google Scholar]
- (13).Gerber SA; Rush J; Stemman O; Kirschner MW; Gygi SP Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci U S A 2003, 100, 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ferries S; Perkins S; Brownridge PJ; Campbell A; Eyers PA; Jones AR; Eyers CE Evaluation of Parameters for Confident Phosphorylation Site Localization Using an Orbitrap Fusion Tribrid Mass Spectrometer. J. Proteome Res 2017, 16, 3448–3459. [DOI] [PubMed] [Google Scholar]
- (15).Hogrebe A; von Stechow L; Bekker-Jensen DB; Weinert BT; Kelstrup CD; Olsen JV Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun 2018, 9, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hebert AS; Thöing C; Riley NM; Kwiecien NW; Shiskova E; Huguet R; Cardasis HL; Kuehn A; Eliuk S; Zabrouskov V; Westphall MS; McAlister GC; Coon JJ Improved Precursor Characterization for Data-Dependent Mass Spectrometry. Anal. Chem 2018, 90, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lim KB; Kassel DB Phosphopeptides enrichment using on-line two-dimensional strong cation exchange followed by reversed-phase liquid chromatography/mass spectrometry. Anal. Biochem 2006, 354, 213–219. [DOI] [PubMed] [Google Scholar]
- (18).Feng S; Pan C; Jiang X; Xu S; Zhou H; Ye M; Zou H Fe3+ immobilized metal affinity chromatography with silica monolithic capillary column for phosphoproteome analysis. Proteomics 2007, 7, 351–360. [DOI] [PubMed] [Google Scholar]
- (19).Villén J; Beausoleil SA; Gerber SA; Gygi SP Large-scale phosphorylation analysis of mouse liver. Proc. Nat. Acad. Sci 2007, 104, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Macek B; Gnad F; Soufi B; Kumar C; Olsen JV; Mijakovic I; Mann M Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 2008, 7, 299–307. [DOI] [PubMed] [Google Scholar]
- (21).Yao Y; Dong J; Dong M; Liu F; Wang Y; Mao J; Ye M; Zou H An immobilized titanium (IV) ion affinity chromatography adsorbent for solid phase extraction of phosphopeptides for phosphoproteome analysis. J. Chromatogr. A 2017, 1498, 22–28. [DOI] [PubMed] [Google Scholar]
- (22).Chen D; Ludwig KR; Krokhin OV; Spicer V; Yang Z; Shen X; Hummon AB; Sun L Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Large-Scale Phosphoproteomics with the Production of over 11,000 Phosphopeptides from the Colon Carcinoma HCT116 Cell Line. Anal. Chem 2019, 91, 2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fu Y; Qian X Transferred subgroup false discovery rate for rare post-translational modifications detected by mass spectrometry. Mol. Cell. Proteomics 2014, 13, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wojcik R; Dada OO; Sadilek M; Dovichi NJ Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun. Mass Spectrom 2010, 24, 2554–2560. [DOI] [PubMed] [Google Scholar]
- (25).Moini M Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem 2007, 79, 4241–4246. [DOI] [PubMed] [Google Scholar]
- (26).Li Y; Champion MM; Sun L; Champion PAD; Wojcik R; Dovichi NJ Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry as an alternative proteomics platform to ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry for samples of intermediate complexity. Anal. Chem 2012, 84, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wojcik R; Zhu G; Zhang Z; Yan X; Zhao Y; Sun L; Champion MM; Dovichi NJ Capillary zone electrophoresis as a tool for bottom-up protein analysis. Bioanalysis 2016, 8, 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang Y; Fonslow BR; Wong CCL; Nakorchevsky A; Yates JR Improving the comprehensiveness and sensitivity of sheathless capillary electrophoresis-tandem mass spectrometry for proteomic analysis. Anal. Chem 2012, 84, 8505–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zhang Z; Sun L; Zhu G; Yan X; Dovichi NJ Integrated strong cation-exchange hybrid monolith coupled with capillary zone electrophoresis and simultaneous dynamic pH junction for large-volume proteomic analysis by mass spectrometry. Talanta 2015, 138, 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhang Z; Yan X; Sun L; Zhu G; Dovichi NJ Integrated strong cation-exchange hybrid monolith coupled with capillary zone electrophoresis and simultaneous dynamic pH junction for large-volume proteomic analysis by mass spectrometry. Anal. Chem 2015, 87, 4572–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Boley DA; Zhang Z; Dovichi NJ Multisegment injections improve peptide identification rates in capillary zone electrophoresis-based bottom-up proteomics. J. Chromatogr. A 2017, 1523, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhang Z; Peuchen EH; Dovichi NJ Surface-Confined Aqueous Reversible Addition-Fragmentation Chain Transfer (SCARAFT) Polymerization Method for Preparation of Coated Capillary Leads to over 10 000 Peptides Identified from 25 ng HeLa Digest by Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry. Anal. Chem 2017, 89, 6774–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cao P; Stults JT Phosphopeptide analysis by on-line immobilized metal-ion affinity chromatography-capillary electrophoresis-electrospray ionization mass spectrometry. J. Chromatogr. A 1999, 853, 225–235. [DOI] [PubMed] [Google Scholar]
- (34).Cao P; Stults JT Mapping the phosphorylation sites of proteins using on-line immobilized metal affinity chromatography/capillary electrophoresis/electrospray ionization multiple stage tandem mass spectrometry. Rapid Commun. Mass Spectrom 2000, 14, 1600–1606. [DOI] [PubMed] [Google Scholar]
- (35).Figeys D; Corthals GL; Gallis B; Goodlett DR; Ducret A; Corson MA; Aebersold R Data-dependent modulation of solid-phase extraction capillary electrophoresis for the analysis of complex peptide and phosphopeptide mixtures by tandem mass spectrometry: application to endothelial nitric oxide synthase. Anal. Chem 1999, 71, 2279–2287. [DOI] [PubMed] [Google Scholar]
- (36).Ludwig KR; Sun L; Zhu G; Dovichi NJ; Hummon AB Over 2300 phosphorylated peptide identifications with single-shot capillary zone electrophoresis-tandem mass spectrometry in a 100 min separation. Anal. Chem 2015, 87, 9532–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Faserl K; Sarg B; Gruber P; Lindner HH Investigating capillary electrophoresis-mass spectrometry for the analysis of common post-translational modifications. Electrophoresis 2018, 39, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zhang Z; Hebert AS; Westphall MS; Qu Y; Coon JJ; Dovichi NJ Production of Over 27 000 Peptide and Nearly 4400 Protein Identifications by Single-Shot Capillary-Zone Electrophoresis-Mass Spectrometry via Combination of a Very-Low-Electroosmosis Coated Capillary, a Third-Generation Electrokinetically-Pumped Sheath-Flow Nanospray Interface, an Orbitrap Fusion Lumos Tribrid Mass Spectrometer, and an Advanced-Peak-Determination Algorithm. Anal. Chem 2018, 90, 12090–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Aebersold R; Morrison HD Analysis of dilute peptide samples by capillary zone electrophoresis. J. Chromatogr. A 1990, 516, 79–88. [DOI] [PubMed] [Google Scholar]
- (40).Britz-McKibbin P; Chen DDY Selective focusing of catecholamines and weakly acidic compounds by capillary electrophoresis using a dynamic pH junction. Anal. Chem 2000, 72, 1242–1252. [DOI] [PubMed] [Google Scholar]
- (41).Eng JK; McCormack AL; Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom 1994, 5, 976–989. [DOI] [PubMed] [Google Scholar]
- (42).Dorfer V; Pichler P; Stranzl T; Stadlmann J; Taus T; Winkler S; Mechtler K MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res 2014, 13, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Taus T; Köcher T; Pichler P; Paschke C; Schmidt A; Henrich C; Mechtler K Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res 2011, 10, 5354–5362. [DOI] [PubMed] [Google Scholar]
- (44).Cox J; Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- (45).Schwartz D; Gygi SP An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol 2005, 23, 1391–1398. [DOI] [PubMed] [Google Scholar]
- (46).Chou MF; Schwartz D Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics 2011, 35, 13.15.1–13.15.24. [DOI] [PubMed] [Google Scholar]
- (47).Krokhin OV; Anderson G; Spicer V; Sun L; Dovichi NJ Predicting Electrophoretic Mobility of Tryptic Peptides for High-Throughput CZE-MS Analysis. Anal. Chem 2017, 89, 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zhou H; Ye M; Dong J; Corradini E; Cristobal A; Heck AJ; Zou H; Mohammed S Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc 2013, 8, 461–480. [DOI] [PubMed] [Google Scholar]
- (49).Post H; Penning R; Fitzpatrick MA; Garrigues LB; Wu W; MacGillavry HD; Hoogenraad CC; Heck AJR; Altelaar AFM Robust, Sensitive, and Automated Phosphopeptide Enrichment Optimized for Low Sample Amounts Applied to Primary Hippocampal Neurons. J. Proteome Res 2016, 16, 726–737. [DOI] [PubMed] [Google Scholar]
- (50).Mi H; Muruganujan A; Huang X; Ebert D; Mills C; Guo X; Thomas PD Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc 2019, 14, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ren B; Deng Y; Mukhopadhyay A; Lanahan AA; Zhuang ZW; Moodie KL; Mulligan-Kehoe MJ; Byzova TV; Peterson RT; Simons M ERK1/2-Akt1 crosstalk regulates arteriogenesis in mice and zebrafish. J. Clin. Invest 2010, 120, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dong M; Bian Y; Dong J; Wang K; Liu Z; Qin H; Ye M; Zou H Selective Enrichment of Cysteine-Containing Phosphopeptides for Subphosphoproteome Analysis. J. Proteome Res 2015, 14, 5341–5347. [DOI] [PubMed] [Google Scholar]
- (53).Zhang Z; Dovichi NJ Optimization of mass spectrometric parameters improve the identification performance of capillary zone electrophoresis for single-shot bottom-up proteomics analysis. Anal. Chim. Acta 2018, 1001, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Brazma A; Jarnuczak AF; Csordas A; Inuganti A; Kundu DJ; Pérez E; Bai J; Bernal-Llinares M; Walzer M; Hewapathirana S; Ternent T; Vizcaíno JA; Perez-Riverol Y; Griss J; Mayer G; Uszkoreit J; Eisenacher M; Pfeuffer J; Sachsenberg T; Cox J; Tiwary S; Yılmaz Ş; Audain E The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2018, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.