Abstract
A large body of genetic data from schizophrenia-related research has identified an assortment of genes and disturbed pathways supporting involvement of complex genetic components for schizophrenia spectrum and other psychotic disorders. Advances in genetic technology and expanding studies with searchable genomic databases have led to multiple published reports, allowing us to compile a master list of known, clinically relevant, or susceptibility genes contributing to schizophrenia. We searched key words related to schizophrenia and genetics from peer-reviewed medical literature sources, authoritative public access psychiatric websites and genomic databases dedicated to gene discovery and characterization of schizophrenia. Our list of 560 genes were arranged in alphabetical order in tabular form with gene symbols placed on high-resolution human chromosome ideograms. Genome wide pathway analysis using GeneAnalytics was carried out on the resulting list of genes to assess the underlying genetic architecture for schizophrenia. Recognized genes of clinical relevance, susceptibility or causation impact a broad range of biological pathways and mechanisms including ion channels (e.g., CACNA1B, CACNA1C, CACNA1H), metabolism (e.g., CYP1A2, CYP2C19, CYP2D6), multiple targets of neurotransmitter pathways impacting dopamine, GABA, glutamate, and serotonin function, brain development (e.g., NRG1, RELN), signaling peptides (e.g., PIK3CA, PIK4CA) and immune function (e.g., HLA-DRB1, HLA-DQA1) and interleukins (e.g., IL1A, IL10, IL6). This summary will enable clinical and laboratory geneticists, genetic counselors, and other clinicians to access convenient pictorial images of the distribution and location of contributing genes to inform diagnosis and gene-based treatment as well as provide risk estimates for genetic counseling of families with affected relatives.
Keywords: schizophrenia spectrum, genetic biomarkers, gene distribution and location, autism spectrum disorder
INTRODUCTION
Schizophrenia is a chronic debilitating psychiatric disorder that affects approximately 1% of the general population; occurring equally across gender and ethnicity [National Institute of Mental Health Web site, http://www.nimh.nih.gov/health/topics/schizophrenia/index.shtml]. The disorder is characterized by delusions, hallucinations, disorganized speech and behavior, and other symptoms that hinder daily functioning [American Psychiatric Association, 2013]. These symptoms and other factors associated with schizophrenia have serious negative effects on individuals and society as a whole including high risk of suicide, unemployment, and substance abuse [Andreasen and Black, 2006; Andrew et al., 2012]. Despite the relatively low frequency of illness, the economic burden for both the direct and indirect costs of schizophrenia is estimated to be in the tens of billions of dollars [Chong et al., 2014]. Schizophrenia is recognized as one of the most burdening dis-abilities globally by the World Health Organization [WHO, 2008]. Given the seriousness and devastating effects of this disorder, both for the diagnosed individual and society, it is important for researchers to understand factors that contribute to schizophrenia including genetics, the focus of this review.
Several decades of research support the role of genetics in the development, diagnosis, and treatment of schizophrenia [Riley and Kendler, 2006; Kavanagh et al., 2015]. An approximate 10-fold increased frequency of schizophrenia is reported among relatives with affected family members compared with relatives of normal controls (i.e., 4.8% to 0.5%, respectively) [Kendler and Diehl, 1993]. Twin studies in schizophrenia have reported concordance rates as high as 85% for monozygotic twins, and 25% for dizygotic twins [Franzek and Beckmann, 1998]. Additionally, schizophrenia prevalence in a study of over two million Swedish families showed a significantly increased frequency and risk of schizophrenia in adopted children from biological parents with schizophrenia when compared to those children adopted away from parents without schizophrenia [Lichtenstein et al., 2009]. Combined twin and family studies and meta-analysis approaches have estimated that the heritability of schizophrenia is as high as 80% [Cardno and Gottesman, 2000].
The importance of genes in the etiology of schizophrenia is reinforced by advances in genetic technology and consortium studies with combined patient databases and genetic analytical approaches (e.g., linkage, genome wide association studies [GWAS], chromosomal variants, functional assessments, next generation sequencing and high-resolution microarrays utilizing single nucleotide polymorphism, and copy number probes). No clear relationship has been found to explain observed genetic risks and specific DNA variants or pathways with biological processes or protein alterations, but over-lapping data from regions of the human genome does support linkage (e.g., DTNBP1, NRG1, TAAR6) or identification of candidate genes (e.g., COMT, AKT1, RGS4) in schizophrenia [Riley and Kendler, 2006]. The emergence of a number of replicated genetic linkage studies have identified targeted chromosomal regions of interest and candidate gene locations. For example, there is growing evidence for linkage replication across several chromosomal regions including 1q, 5q, 6p, 6q, 8p, 10p, 13q, 15q, and 22q [Riley and Kendler, 2006]. Recently, the Schizophrenia Working Group of the Psychiatric Genomics Consortium [2014] identified 108 potential schizophrenia-associated genetic loci. Focused attention on molecular genetics and these chromosome regions have identified several potential candidate genes for schizophrenia.
Schizophrenia spectrum and other related disorders, which include schizoaffective, schizophreniform, delusional, and schizotypal personality disorders, are also increased in family members of those with schizophrenia and can be components of single-gene defects, such as COMT and PRODH mutations or copy number variants as a component of syndromic cytogenetic disorders including the 22q11.2 deletion, but most individuals with schizophrenia are non-syndromic [Riley and Kendler, 2006]. The high-heritability estimates for schizophrenia further emphasize the need to explore genetic causation for diagnosis and to inform and guide gene-based treatment options depending on specific genetic disturbances or lesions. A large number of validated genes are now recognized as playing a pivotal role in schizophrenia [Riley and Kendler, 2006; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Kavanagh et al., 2015]. Hence, a current list is needed of recognized clinically relevant, susceptible, or known genes as genetic biomarkers for schizophrenia. These genetic biomarkers can then be assessed to identify the role in schizophrenia and for new diagnostic approaches in the clinical evaluation of those presenting with schizophrenia as well as supplying more accurate and informative genetic counseling for family members.
The goal of our study was to utilize high-resolution chromosome ideograms (850 band level) and plot the location of genes identified by searching the literature, genetic databases, and consortium and authoritative federally sponsored websites for genes playing a documented role in schizophrenia. In tabular form, the individual gene symbols will be listed in alphabetical order along with their expanded names or description and chromosome location. The reader will then be able to quickly identify the gene of interest and access conveniently the visual image of the location and distribution of specific genes on individual high-resolution chromosome ideograms. To further assess the underlying genetic architecture of schizophrenia, we used the GeneAnalytics (http://geneanalytics.genecards.org/) and VarElect (http://varelect.genecards.org/) data analysis computer tools to map the identified genes to tissues and cells, diseases, phenotypes, molecular pathways, and biological processes with the greatest overlap and probable relevance to schizophrenia.
METHODS
For our study, we used computer-based authoritative internet websites, schizophrenia genomics consortiums databases and peer-reviewed medical literature reports to search for genes associated with schizophrenia. We initially examined the literature and databases utilizing pertinent key words (i.e., human genes, gene variants, genetics, mutations, schizophrenia) searching for sources with involvement of genetics in the etiology of schizophrenia. Source materials included genetic linkage and functional associations from peer-reviewed research articles and authoritative computer website genomic databases on this topic in humans. Disequilibrium data, single nucleotide polymorphisms, or copy number variation supporting a relationship between the individual gene and schizophrenia were reviewed. We found data from whole-genome sequencing of families with relatives having schizophrenia and large collaborative research consortiums on schizophrenia and psychosis gene expression or transcriptome profiles, genetic linkage, copy number variation, and genome wide association studies (GWAS). Hundreds of published research articles were reviewed from PubMed (https://www.ncbi.nlm.nih.gov/pubmed) and a list of genes were compiled having more than one source contributing to schizophrenia. Our report compiled a current list of recognized genes with proposed associations with schizophrenia causation, pathology, or course with possible impact on treatment response (e.g., drug metabolism). The number of clinically relevant genes impacting schizophrenia is expected to expand as additional causal genes and gene/phenotype associations will be reported in the future using expanding genetic tools such as next generation sequencing, copy number variant analysis and data from large collaborative research consortiums [International Schizophrenia Consortium, 2009; Stefansson et al., 2009; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011; Ripke et al., 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014].
Our approach also focused on integrated online gene disease catalogs particularly Mendelian Inheritance in Man (OMIM) (www.OMIM.org) and other respected and searchable sources, such as the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/), PubMed (https://www.ncbi.nlm.nih.gov/pubmed), GeneCards (https://www.genecards.org), and Phenopedia (http://www.hugenavigator.net/HuGENavigator/startPagePhenoPedia.do). The literature sources that were searched consisted of both primary research articles and meta-analyses with reviews summarizing genetic evidence from several sources. If possible, specific sources for genes associated with schizophrenia embedded in meta-analyses and reviews were extracted and used in development of the master gene summary table. Genes located within potential loci regions were not included, unless a specific gene was located or documented in another source.
For each gene fitting our criteria, the gene symbol, expanded name, chromosome band location, and a reference source for each gene was compiled in a master summary table (Table I). The master table was organized alphabetically by gene symbol. The most current gene information, such as location and symbol, was obtained from OMIM and GeneCards with each gene plotted on high-resolution chromosome ideograms (850 band level) (Fig. 1).
TABLE I.
Currently Recognized Genes for Schizophrenia and Their Chromosome Locations
| Gene symbol | Gene name | Location | References |
|---|---|---|---|
| ABATa | 4-Aminobutyrate aminotransferase | 16p13.2 | Tabarés-Seisdedos et al. [2011] |
| ABCA13 | ATP (adenosine triphosphate)-binding cassette, subfamily A, member 13 | 7p12.3 | Knight et al. [2009] |
| ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 7q21.12 | Tabarés-Seisdedos et al. [2011] |
| ACADVL | Acyl-CoA dehydrogenase, very long chain | 17p13.1 | Sanders et al. [2013] |
| ACE | Angiotensin I converting enzyme | 17q23.3 | Tabarés-Seisdedos et al. [2011] |
| ACHE | Acetylcholinesterase | 7q221 | Tabarés-Seisdedos et al. [2011] |
| ACSL6 | Acyl-CoA synthetase long-chain family member 6 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| ACSM1 | Acyl-CoA synthetase medium-chain family member 1 | 16p12.3 | Athanasiu et al. [2010] |
| ADCY1 | Adenylate cyclase 1 | 7p12.3 | Wright et al. [2013] |
| ADCY9 | Adenylate cyclase 9 | 16p13,3 | Wright et al. [2013] |
| ADCYAP1 | Adenylate cyclase activating polypeptide 1 | 18p11.32 | Tabarés-Seisdedos et al. [2011] |
| ADIPOQ | Adiponectin, C1Q, and collagen domain containing | 3q27.3 | Tabarés-Seisdedos et al. [2011] |
| ADORA2Aa | Adenosine A2a receptor | 22q11.23 | Tabarés-Seisdedos et al. [2011] |
| ADRA1A | Adrenoceptor alpha 1A | 8p21.2 | Tabarés-Seisdedos et al. [2011] |
| ADRA2A | Adrenoceptor alpha 2A | 10q25.2 | Tabarés-Seisdedos et al. [2011] |
| ADRA2C | Adrenoceptor alpha 2C | 4p16.3 | Tabarés-Seisdedos et al. [2011] |
| ADRB3 | Adrenoceptor beta 3 | 8p11.23 | Tabarés-Seisdedos et al. [2011] |
| ADRBK2 | Adrenergic, beta, receptor kinase 2 | 22q12.1 | Tabarés-Seisdedos et al. [2011] |
| ADSS | Adenylosuccinate synthase | 1q44 | Tabarés-Seisdedos et al. [2011] |
| AHCYL2 | Adenosylhomocysteinase-like 2 | 7q32.1 | Wright et al. [2013] |
| AHI1a | Abelson helper integration site 1 | 6q23.3 | Ingason et al. [2010] |
| AKT1a | V-Akt murine thymoma viral oncogene homolog 1 | 14q32.33 | Shi et al. [2008] |
| AKT3 | V-Akt murine thymoma viral oncogene homolog 3 | 1q44 | Ripke et al. [2013] |
| ALDH3B1 | Aldehyde dehydrogenase 3 family, member B1 | 11q13.2 | Tabarés-Seisdedos et al. [2011] |
| ALDH5A1a | Aldehyde dehydrogenase 5 family, member A1 | 6p22.31 | Tabarés-Seisdedos et al. [2011] |
| ANK3a | Ankyrin 3 | 10q21.2 | Athanasiu et al. [2010] |
| ANKK1 | Ankyrin repeat and kinase domain containing 1 | 11q23.2 | Tabarés-Seisdedos et al. [2011] |
| APBA2a | Amyloid beta (A4) precursor protein-binding, family A, member 2 | 15q13.1 | Tabarés-Seisdedos et al. [2011] |
| APOD | Apolipoprotein D | 3q29 | Tabarés-Seisdedos et al. [2011] |
| APOE | Apolipoprotein E | 19q13.32 | Allen et al. [2008] |
| APOL1 | Apolipoprotein L-I | 22q12.3 | Mimmack et al. [2002] |
| APOL2 | Apolipoprotein L-II | 22q12.3 | Mimmack et al. [2002] |
| APOL4 | Apolipoprotein L-IV | 22q12.3 | Mimmack et al. [2002] |
| ARa | Androgen receptor | Xq12 | Tabarés-Seisdedos et al. [2011] |
| ARHGAP18 | Rho GTPase activating protein 18 | 6p22.33 | Potkin et al. [2009] |
| ARHGAP44 | Rho GTPase activating protein 44 | 17p12 | Wright et al. [2013] |
| ARNTL | Aryl hydrocarbon receptor nuclear translocator-like | 11p15.2 | Tabarés-Seisdedos et al. [2011] |
| ARRB2 | Arrestin, beta 2 | 17p13.2 | Tabarés-Seisdedos et al. [2011] |
| ARVCF | Armadillo repeat gene deleted in velocardiofacial syndrome | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| AS3MT | Arsenic (+3 oxidation state) methyltransferase | 10q24.32 | Aberg et al. [2006] |
| ASCL1 | Achaete-scute family bHLH transcription factor 1 | 12q23.2 | Tabarés-Seisdedos et al. [2011] |
| ASTN2a | Astrotactin 2 | 9q33.1 | Wang et al. [1996] |
| ATF2 | Activating transcription factor 2 | 2q31.1 | Tabarés-Seisdedos et al. [2011] |
| ATF4 | Activating transcription factor 4 | 22q131 | Tabarés-Seisdedos et al. [2011] |
| ATF5 | Activating transcription factor 5 | 19q13.33 | Tabarés-Seisdedos et al. [2011] |
| ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 | 18q21.1 | Lauriat et al. [2006] |
| ATXN1 | Ataxin 1 | 6p22.33 | Wang et al. [1996] |
| ATXN8OS | Ataxin 8 opposite strand | 13q21.33 | Tabarés-Seisdedos et al. [2011] |
| AUTS2a | Autism susceptibility candidate 2 | 7q11.22 | Tabarés-Seisdedos et al. [2011] |
| AXIN1 | Axis inhibitor 1 | 16p13.3 | Wright et al. [2013] |
| B3GNT2 | Beta-1,3-n-acetylglucosaminyltransferase 2 | 2p15 | Sanders et al. [2013] |
| BARD1 | BRCA1 (breast cancer 1 gene) associated RING domain 1 | 2q35 | van Schijndel et al. [2009] |
| BAI3 | Brain-specific angiogenesis inhibitor 3 | 6q12 | Wright et al. [2013] |
| BCL2L13 | BCL2 (B-cell CLL/lymphoma 2)-like 13 (apoptosis facilitator) | 22q11.21 | Wright et al. [2013] |
| BCL2L2 | BCL2-like 2 | 14q11.2 | Sanders et al. [2013] |
| BCL9 | B-cell CLL (chronic lymphocytic leukemia)/lymphoma 9 | 1q21.2 | Xu and He [2010] |
| BDNFa | Brain-derived neurotrophic factor | 11p14.1 | Wockner et al. [2014] |
| BIK | BCL2-interacting killer | 22q13.31 | Sanders et al. [2013] |
| BRD1 | Bromodomain containing 1 | 22q13.33 | Kushima et al. [2010] |
| BTN2A2 | Butyrophilin, subfamily 2, member A2 | 6p22.2 | Shi et al. [2009] |
| BTN3A1 | Butyrophilin, subfamily 3, member A1 | 6p22.2 | Shi et al. [2009] |
| BTN3A2 | Butyrophilin, subfamily 3, member A2 | 6p22.2 | Shi et al. [2009] |
| C12orf65 | Chromosome 12 open reading frame 65 | 12q24.31 | Ripke et al. [2013] |
| C2orf82 | Chromosome 2 open reading frame 82 | 2q37.1 | Ripke et al. [2013] |
| CACNA1Ba | Calcium channel, voltage-dependent, N-type, alpha 1B subunit | 9q34.3 | Tabarés-Seisdedos et al. [2011] |
| CACNA1Ca | Calcium channel, voltage-dependent, L-type, alpha 1C subunit | 12p13.33 | Neale and Sklar [2015] |
| CACNA1Ha | Calcium channel, voltage-dependent, T-type, alpha 1H subunit | 16p13.3 | Wright et al. [2013] |
| CACNB2a | Calcium channel, voltage-dependent, beta 2 subunit | 10p12.33 | Neale and Sklar [2015] |
| CACNG2 | Calcium channel, voltage-dependent, gamma subunit 2 | 22q12.3 | Tabarés-Seisdedos et al. [2011] |
| CAMK2B | Calcium/calmodulin-dependent protein kinase II beta | 7p13 | Novak et al. [2000] |
| CARTPT | CART (cocaine and amphetamine regulated transcript) prepropeptide | 5q13.2 | Tabarés-Seisdedos et al. [2011] |
| CBSa | Cystathionine-beta-synthase | 21q22.3 | Tabarés-Seisdedos et al. [2011] |
| CCDC68 | Coiled-coil domain containing 68 | 18q21.2 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| CCK | Cholecystokinin | 3p22.1 | Tabarés-Seisdedos et al. [2011] |
| CCKAR | Cholecystokinin A receptor | 4p15.2 | Ocklenburg et al. [2013] |
| CCL2 | Chemokine (C-C motif) ligand 2 | 17q12 | Tabarés-Seisdedos et al. [2011] |
| CD28 | Antigen CD28 | 2q33.2 | Frydecka et al. [2015] |
| CDC42 | Cell division cycle 42 | 1p36.12 | Wright et al. [2013] |
| CDC42SE2 | CDC42 (cell division cycle 42) small effector 2 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| CDK6 | Cyclin-dependent kinase 6 | 7q21.2 | Wright et al. [2013] |
| CEACAM21 | Carcinoembryonic antigen-related cell adhesion molecule 21 | 19q13.2 | Alkelai et al. [2012] |
| CERK | Ceramide kinase | 22q13.31 | Wright et al. [2013] |
| CHAT | Choline acetyltransferase | 10q11.23 | Tabarés-Seisdedos et al. [2011] |
| CHD1a | Chromodomain helicase DNA (deoxyribonucleic acid) binding protein 1 | 5q21.1 | Wright et al. [2013] |
| CHGA | Chromogranin A | 14q32.12 | Sun et al. [2008] |
| CHGB | Chromogranin B | 20p12.3 | Tabarés-Seisdedos et al. [2011] |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | 1q32.1 | Ohi et al. [2010] |
| CHL1 | Cell adhesion molecule L1-like | 3p26.3 | Tabarés-Seisdedos et al. [2011] |
| CHRM1 | Cholinergic receptor, muscarinic 1 | 11q12.3 | Tabarés-Seisdedos et al. [2011] |
| CHRM2 | Cholinergic receptor, muscarinic 2 | 7q33 | Tabarés-Seisdedos et al. [2011] |
| CHRM5 | Cholinergic receptor, muscarinic 5 | 15q14 | Tabarés-Seisdedos et al. [2011] |
| CHRNA3 | Cholinergic receptor, nicotinic, alpha 3 (neuronal) | 15q25.1 | Tabarés-Seisdedos et al. [2011] |
| CHRNA4 | Cholinergic receptor, nicotinic, alpha 4 (neuronal) | 20q13.33 | Tabarés-Seisdedos et al. [2011] |
| CHRNA5 | Cholinergic receptor, nicotinic, alpha 5 (neuronal) | 15q25.1 | Jackson et al. [2013] |
| CHRNA7a | Cholinergic receptor, nicotinic, alpha 7 (neuronal) | 15q13.3 | Harrison and Weinberger [2005] |
| CHRNB2 | cholinergic receptor, nicotinic, beta 2 (neuronal) | 1q21.3 | Tabarés-Seisdedos et al. [2011] |
| CLDN5 | Claudin 5 | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| CLINT1 | Clathrin interactor 1 | 5q33.3 | Tabarés-Seisdedos et al. [2011] |
| CLOCK | Clock circadian regulator | 4q12 | Zhang et al. [2011] |
| CMYA5 | Cardiomyopathy-associated protein 5 | 5q14.1 | Chen et al. [2011b] |
| CNNM2 | Cyclin M2 | 10q24.32 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| CNP | Cyclic nucleotide phosphodiesterase | 17q21.2 | Tabarés-Seisdedos et al. [2011] |
| CNR1a | Cannabinoid receptor 1 | 6q15 | Tabarés-Seisdedos et al. [2011] |
| CNTF | Ciliary neurotrophic factor | 11q12.1 | Tabarés-Seisdedos et al. [2011] |
| CNTNAP2a | Contactin-associated protein-like 2 | 7q35 | Wang et al. [1996] |
| CNTNAP5a | Contactin-associated protein-like 5 | 2q14.3 | Tabarés-Seisdedos et al. [2011] |
| COMT | Catechol-O-methyltransferase | 22q11.21 | Allen et al. [2008] |
| COMTD1 | Catechol-O-methyltransferase domain containing 1 | 10q22.1 | Wockner et al. [2014] |
| CPLX2 | Complexin 2 | 5q35.2 | Tabarés-Seisdedos et al. [2011] |
| CRKL | V-crk avian sarcoma virus CT10 oncogene homolog-like | 22q11.21 | Wright et al. [2013] |
| CSF2RA | Colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | Xp22.33 | Loe-Mie et al. [2010] |
| CSF2RB | Granulocyte-macrophage colony-stimulating factor receptor, beta | 22q12.3 | Tabarés-Seisdedos et al. [2011] |
| CSMD1a | CUB (complement C1r/C1s, Uegf, Bmp1) and Sushi multiple domains 1 | 8p23.2 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4 | 2q33.2 | Frydecka et al. [2015] |
| CTNNA3a | Catenin, alpha 3 | 10q21.3 | Xu and He [2010] |
| CYBB | Cytochrome b-245, beta polypeptide | Xp11 4 | Sanders et al. [2013] |
| CYP17A1 | Cytochrome P450, family 17, subfamily A, polypeptide 1 | 10q24.32 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 | 15q24.1 | Tabarés-Seisdedos et al. [2011] |
| CYP2C19 | Cytochrome P450, family 2, subfamily C, polypeptide 19 | 10q23.33 | Tabarés-Seisdedos et al. [2011] |
| CYP2D6 | Cytochrome P450, family 2, subfamily D, polypeptide 6 | 22q13.2 | Allen et al. [2008] |
| CYP3A4 | Cytochrome P450, family 3, subfamily A, polypeptide 4 | 7q22.1 | Tabarés-Seisdedos et al. [2011] |
| CYP3A5 | cytochrome P450, family 3, subfamily A, polypeptide 5 | 7q22.1 | Tabarés-Seisdedos et al. [2011] |
| DAO | D-amino-acid oxidase | 12q24.11 | Harrison and Weinberger [2005] |
| DAOA | D-amino acid oxidase activator | 13q33.2 | Shi et al. [2008] |
| DAOAAS | DAOA (D-amino acid oxidase activator) antisense RNA (ribonucleic acid) 1 | 13q33.2 | Chumakov et al. [2002] |
| DBH | Dopamine beta-hydroxylase | 9q34.2 | Tabarés-Seisdedos et al. [2011] |
| DBNDD2 | Dysbindin domain containing 2 | 20q13.12 | Sanders et al. [2013] |
| DBP | D site of albumin promoter binding protein | 19q13.33 | Sanders et al. [2013] |
| DCDC2 | Doublecortin domain containing 2 | 6p22.31 | Tabarés-Seisdedos et al. [2011] |
| DDCa | Dopa decarboxylase | 7p12.1 | Tabarés-Seisdedos et al. [2011] |
| DGCR2 | DiGeorge syndrome critical region gene 2 | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| DGCR8 | Digeorge syndrome critical region gene 8 | 22q11.21 | Chun et al. [2014] |
| DICER1 | Dicer, Drosophila, homolog of, 1 | 14q32.13 | Sanders et al. [2013] |
| DISC1a | Disrupted in schizophrenia 1 | 1q42.2 | Harrison and Weinberger [2005] |
| DISC2 | Disrupted in schizophrenia 2 | 1q42.2 | Millar et al. [2004] |
| DKK2 | Dickkopf, Xenopus, homolog of, 2 | 4q25 | van Schijndel et al. [2009] |
| DLG1 | Discs, large homolog 1 (Drosophila) | 3q29 | Uezato et al. [2012] |
| DLG2 | Discs, large homolog 2 (Drosophila) | 11q14 1 | Tabarés-Seisdedos et al. [2011] |
| DLG4a | Discs, large homolog 4 (Drosophila) | 17p11.2 | Tabarés-Seisdedos et al. [2011] |
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 | 19p13.2 | Guidotti et al. [2000] |
| DOC2A | Double C2-like domains, alpha | 16p11.2 | Tabarés-Seisdedos et al. [2011] |
| DOCK4a | Dedicator of cytokinesis 4 | 7q31.1 | Alkelai et al. [2012] |
| DPP10a | Dipeptidyl-peptidase X | 2q14.1 | Tabarés-Seisdedos et al. [2011] |
| DPYD | Dihydropyrimidine dehydrogenase | 1p21.3 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| DPYSL2 | Dihydropyrimidinase-like 2 | 8p21.2 | Tabarés-Seisdedos et al. [2011] |
| DR1 | Down-regulator of transcription 1, TBP (TATA box-binding protein)-binding | 1p221 | Wright et al. [2013] |
| DRD1a | Dopamine receptor D1 | 5q35,2 | Allen et al. [2008] |
| DRD2a | Dopamine receptor D2 | 11q23.2 | Shi et al. [2008] |
| DRD3a | Dopamine receptor D3 | 3q13.31 | Williams et al. [1998] |
| DRD4 | Dopamine receptor D4 | 11p15.5 | Wockner et al. [2014] |
| DRD5 | Dopamine receptor D5 | 4p16.1 | Tabarés-Seisdedos et al. [2011] |
| DRP2 | Dystrophin-related protein 2 | Xq22.1 | Tabarés-Seisdedos et al. [2011] |
| DTNBP1 | Dystrobrevin-binding protein1 | 6p22.33 | Shi et al. [2008] |
| EGF | Epidermal growth factor | 4q25 | Tabarés-Seisdedos et al. [2011] |
| EGR2a | Early growth response 2 | 10q21.3 | Tabarés-Seisdedos et al. [2011] |
| EGR3 | Early growth response 3 | 8p21.3 | Tabarés-Seisdedos et al. [2011] |
| ELK1 | ELK1, member of ETS oncogene family | Xp11.23 | Sanders et al. [2013] |
| EML5 | Echinoderm microtubule-associated protein like 5 | 14q31.3 | Chen et al. [2011a] |
| EN2a | Engrailed 2 | 7q36.3 | Wright et al. [2013] |
| ERBB3 | V-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 | 12q13.2 | Tabarés-Seisdedos et al. [2011] |
| ERBB4a | V-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 | 2q34 | Sun et al. [2008] |
| ESR1a | Estrogen receptor 1 | 6q25.1 | Tabarés-Seisdedos et al. [2011] |
| FAAH | Fatty acid amide hydrolase | 1p33 | Tabarés-Seisdedos et al. [2011] |
| FABP3a | Fatty acid binding protein 3 | 1p35.2 | Tabarés-Seisdedos et al. [2011] |
| FABP5a | Fatty acid binding protein 5 | 8q21.13 | Tabarés-Seisdedos et al. [2011] |
| FABP7a | Fatty acid binding protein 7 | 6q22.31 | Tabarés-Seisdedos et al. [2011] |
| FAM7A | Family with sequence similarity 7A | 15q13.2 | Riley et al. [2002] |
| FAM69A | Family with sequence similarity 69, member A | 1p22.1 | Sanders et al. [2013] |
| FAM84A | Family with sequence similarity 84, member A | 2p24.3 | Wright et al. [2013] |
| FAM135Ba | Family with sequence similarity 135, member B | 8q24.23 | Wright et al. [2013] |
| FEZ1 | Fasciculation and elongation protein zeta 1 | 11q24.2 | Tabarés-Seisdedos et al. [2011] |
| FGF1 | Fibroblast growth factor 1 | 5q31.3 | Tabarés-Seisdedos et al. [2011] |
| FGR | Feline Gardner-Rasheed sarcoma viral oncogene homolog | 6q21 | Tabarés-Seisdedos et al. [2011] |
| FMNL2 | Formin-like 2 | 2q23.3 | Wright et al. [2013] |
| FNBP1L | Formin binding protein 1-like | 1p22.1 | Wright et al. [2013] |
| FOXP2a | Forkhead box P2 | 7q31.1 | Tabarés-Seisdedos et al. [2011] |
| FTCDNL1 | Formiminotransferase cyclodeaminase n-terminal like | 2q33.1 | Ripke et al. [2013] |
| FXYD2 | FXYD domain containing ion transport regulator 2 | 11q23.3 | Sun et al. [2008] |
| FXYD6 | FXYD domain containing ion transport regulator 6 | 11q23.3 | Choudhury et al. [2007] |
| FYN | FYN oncogene related to SRC (v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog) | 6q21 | Tabarés-Seisdedos et al. [2011] |
| FZD3 | Frizzled class receptor 3 | 8p21.1 | Tabarés-Seisdedos et al. [2011] |
| GABBR1 | GABA (gamma-aminobutyric acid) B receptor, 1 | 6p22.1 | Tabarés-Seisdedos et al. [2011] |
| GABRA1a | GABA A receptor, alpha 1 | 5q34 | Petryshen et al. [2005] |
| GABRA2 | GABA A receptor, alpha 2 | 4p12 | Tabarés-Seisdedos et al. [2011] |
| GABRA4a | GABA A receptor, alpha 4 | 4p12 | Tabarés-Seisdedos et al. [2011] |
| GABRA6 | GABA A receptor, alpha 6 | 5q34 | Petryshen et al. [2005] |
| GABRB2 | GABA A receptor, beta 2 | 5q34 | Allen et al. [2008] |
| GABRB3a | GABA A receptor, beta 3 | 15q12 | Tabarés-Seisdedos et al. [2011] |
| GABRG1 | GABA A receptor, gamma-1 | 4p12 | Tabarés-Seisdedos et al. [2011] |
| GABRG2 | GABA A receptor, gamma-2 | 5q34 | Tabarés-Seisdedos et al. [2011] |
| GABRG3 | GABA A receptor, gamma-3 | 15q12 | Tabarés-Seisdedos et al. [2011] |
| GABRR1 | GABA A receptor, rho 1 | 6q15 | Wang et al. [1996] |
| GABRR2 | GABA A receptor, rho 2 | 6q15 | Wang et al. [1996] |
| GABRP | GABA A receptor, pi | 5q35.1 | Petryshen et al. [2005] |
| GAD1a | Glutamate decarboxylase 1 | 2q31.1 | Tabarés-Seisdedos et al. [2011] |
| GAD2 | Glutamate decarboxylase 2 | 10p12.1 | Tabarés-Seisdedos et al. [2011] |
| GBP2 | Guanylate binding protein 2, interferon-inducible | 1p22.2 | Sanders et al. [2013] |
| GBP4 | Guanylate binding protein 4 | 1p22.2 | Sanders et al. [2013] |
| GCLC | Glutamate-cysteine ligase, catalytic subunit | 6p12,1 | Gysin et al. [2007] |
| GCLM | Glutamate-cysteine ligase, subunit | 1p22.1 | Tosic et al. [2006] |
| GDNF | Glial cell derived neurotrophic factor | 5p13.2 | Tabarés-Seisdedos et al. [2011] |
| GFRA2 | GDNF family receptor alpha 2 | 8p21.3 | Tabarés-Seisdedos et al. [2011] |
| GLIS2 | GLIS (GLI-similar) family zinc finger 2 | 16p13.3 | Wright et al. [2013] |
| GLO1a | Glyoxalase I | 6p21.2 | Sanders et al. [2013] |
| GLS | Glutaminase | 2q32.2 | Tabarés-Seisdedos et al. [2011] |
| GLUD1 | Glutamate dehydrogenase 1 | 10q23.2 | Tabarés-Seisdedos et al. [2011] |
| GLUL | Glutamate-ammonia ligase | 1q25.3 | Tabarés-Seisdedos et al. [2011] |
| GNB1La | Guanine nucleotide binding protein, beta 1-like | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| GNB3 | Guanine nucleotide binding protein, beta-3 | 12p13.31 | Tabarés-Seisdedos et al. [2011] |
| GPHNa | Gephyrin | 14q23.3 | Lionel et al. [2013] |
| GPR50 | G protein-coupled receptor 50 | Xq28 | Thomson et al. [2005] |
| GPR85 | G protein-coupled receptor 85 | 7q31.1 | Matsumoto et al. [2008] |
| GPX1a | Glutathione peroxidase 1 | 3p21.31 | Tabarés-Seisdedos et al. [2011] |
| GRAMD4 | GRAM domain containing 4 | 22q13.31 | Wright et al. [2013] |
| GRIA1 | Glutamate receptor, ionotropic, AMPA (alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate) 1 | 5q33.2 | Tabarés-Seisdedos et al. [2011] |
| GRIA2 | Glutamate receptor, ionotropic, AMPA 2 | 4q32.1 | Tabarés-Seisdedos et al. [2011] |
| GRIA3 | Glutamate receptor, ionotropic, AMPA 3 | Xq25 | Tabarés-Seisdedos et al. [2011] |
| GRIA4 | Glutamate receptor, ionotropic, AMPA 4 | 11q22.3 | Tabarés-Seisdedos et al. [2011] |
| GRID1a | Glutamate receptor, ionotropic, delta 1 | 10q23.2 | Tabarés-Seisdedos et al. [2011] |
| GRIK3 | Glutamate receptor, ionotropic, kainate 3 | 1p34.3 | Dai et al. [2014] |
| GRIK4 | Glutamate receptor, ionotropic, kainate 4 | 11q23.3 | Tabarés-Seisdedos et al. [2011] |
| GRIK5 | Glutamate receptor, ionotropic, kainate 5 | 19q13.2 | Tabarés-Seisdedos et al. [2011] |
| GRIN1a | Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | 9q34.3 | Begni et al. [2003] |
| GRIN2Aa | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 16p13.2 | Tabarés-Seisdedos et al. [2011] |
| GRIN2Ba | Glutamate receptor, ionotropic, N-methyl D-aspartate 2B | 12p13.1 | Allen et al. [2008] |
| GRIN2C | Glutamate receptor, ionotropic, N-methyl D-aspartate 2C | 17q25.1 | Tabarés-Seisdedos et al. [2011] |
| GRIN2D | Glutamate receptor, ionotropic, N-methyl D-aspartate 2D | 19q13.33 | Tabarés-Seisdedos et al. [2011] |
| GRIN3A | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 9q31.1 | Takata et al. [2013] |
| GRIN3B | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3B | 19p13.3 | Tabarés-Seisdedos et al. [2011] |
| GRIP1a | Glutamate receptor interacting protein 1 | 12q14.3 | Tabarés-Seisdedos et al. [2011] |
| GRM2 | Glutamate receptor, metabotropic 2 | 3p21.2 | Tabarés-Seisdedos et al. [2011] |
| GRM3 | Glutamate receptor, metabotropic 3 | 7q21.11 | Harrison and Weinberger [2005] |
| GRM4a | Glutamate receptor, metabotropic 4 | 6p21.31 | Tabarés-Seisdedos et al. [2011] |
| GRM5a | Glutamate receptor, metabotropic 5 | 11q14.3 | Wright et al. [2013] |
| GRM7 | Glutamate receptor, metabotropic 7 | 3p26.1 | Tabarés-Seisdedos et al. [2011] |
| GRM8a | Glutamate receptor, metabotropic 8 | 7q31.33 | Tabarés-Seisdedos et al. [2011] |
| GSK3A | Glycogen synthase kinase 3 alpha | 19q13.2 | Tabarés-Seisdedos et al. [2011] |
| GSK3Ba | Glycogen synthase kinase 3 beta | 3q13.33 | Luo et al. [2014] |
| GSTM1a | Glutathione S-transferase mu 1 | 1p13.3 | Tabarés-Seisdedos et al. [2011] |
| GSTP1 | Glutathione S-transferase pi 1 | 11q13.2 | Tabarés-Seisdedos et al. [2011] |
| GSTT1 | Glutathione S-transferase theta 1 | 22q11.23 | Tabarés-Seisdedos et al. [2011] |
| GSTT2 | Glutathione S-transferase theta 2 | 22q11.23 | Tabarés-Seisdedos et al. [2011] |
| HCRTR1 | Hypocretin receptor 1 | 1p35.2 | Tabarés-Seisdedos et al. [2011] |
| HERC2a | HECT domain and RCC1-like domain 2 | 15q13.1 | Sanders et al. [2013] |
| HINT1 | Histidine triad nucleotide-binding protein 1 | 5q23.3 | Tabarés-Seisdedos et al. [2011] |
| HIST1H2AG | Histone cluster 1, H2AG | 6p22.1 | Shi et al. [2009] |
| HIST1H2BC | Histone cluster 1, H2BC | 6p22.2 | Sanders et al. [2013] |
| HIST1H2BD | Histone cluster 1, H2BD | 6p22.2 | Sanders et al. [2013] |
| HIST1H2BG | Histone cluster 1, H2BG | 6p22.2 | Sanders et al. [2013] |
| HIST1H2BH | Histone cluster 1, H2BH | 6p22.2 | Sanders et al. [2013] |
| HIST1H2BI | Histone cluster 1, H2BI | 6p22.2 | Stefansson et al. [2009] |
| HIST1H2BJ | Histone gene cluster 1, H2B histone family, member J | 6p22.1 | Loe-Mie et al. [2010] |
| HIST1H2BK | Histone cluster 1, H2BK | 6p22.1 | Sanders et al. [2013] |
| HLA-Aa | Major histocompatibility complex, class I, A | 6p22.1 | Tabarés-Seisdedos et al. [2011] |
| HLA-B | Major histocompatibility complex, class I, B | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| HLA-DQA1 | Major histocompatibility complex, class II, DQ alpha 1 | 6p21.32 | Jia et al. [2012] |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 6p21.32 | Tabarés-Seisdedos et al. [2011] |
| HLA-DRB1a | Major histocompatibility complex, class II, DR beta 1 | 6p21.32 | Tabarés-Seisdedos et al. [2011] |
| HLA-DRB3 | Major histocompatibility complex, class II, DR beta 3 | 6p21.3 | de Jong et al. [2012] |
| HLA-DRB9 | Major histocompatibility complex, class II, DR beta 9 (pseudogene) | 6p21.32 | Ripke et al. [2013] |
| HNMT | Histamine N-methyltransferase | 2q22.1 | Sanders et al. [2013] |
| HOMER1a | Homer, Drosophila, homolog of, 1 | 5q14.1 | Tabarés-Seisdedos et al. [2011] |
| HOMER2 | Homer, Drosophila, homolog of, 2 | 15q25.2 | Tabarés-Seisdedos et al. [2011] |
| HP | Haptoglobin | 16q22.2 | Allen et al. [2008] |
| HRH1 | Histamine receptor H1 | 3p25.3 | Tabarés-Seisdedos et al. [2011] |
| HRH2 | Histamine receptor H2 | 5q35.2 | Tabarés-Seisdedos et al. [2011] |
| HRH3 | Histamine receptor H3 | 20q13.33 | Tabarés-Seisdedos et al. [2011] |
| HSPA1A | Heat shock 70 kDa protein 1A | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| HSPA1B | Heat shock 70 kDa protein 1B | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| HSPA1L | Heat shock 70 kDa protein-like 1 | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| HSPA6 | Heat-shock 70 kDa protein 6 | 1q23.3 | Brzustowicz et al. [2002]] |
| HSPA7 | Heat-shock 70 kDa protein7 | 1q23.3 | Brzustowicz et al. [2002] |
| HTR1A | 5-Hydroxytryptamine receptor 1A | 5q12.3 | Tabarés-Seisdedos et al. [2011] |
| HTR1Ba | 5-Hydroxytryptamine receptor 1B | 6q14.1 | Tabarés-Seisdedos et al. [2011] |
| HTR1D | 5-Hydroxytryptamine receptor 1D | 1p36.12 | Tabarés-Seisdedos et al. [2011] |
| HTR2Aa | 5-Hydroxytryptamine receptor 2A | 13q14.2 | Williams et al. [1997] |
| HTR2C | 5-Hydroxytryptamine receptor 2C | Xq23 | Wright et al. [2013] |
| HTR3Aa | 5-Hydroxytryptamine receptor 3A | 11q23.2 | Tabarés-Seisdedos et al. [2011] |
| HTR3B | 5-Hydroxytryptamine receptor 3B | 11q23.2 | Tabarés-Seisdedos et al. [2011] |
| HTR3Ca | 5-Hydroxytryptamine receptor 3C | 3q27.1 | Tabarés-Seisdedos et al. [2011] |
| HTR3D | 5-Hydroxytryptamine receptor 3D | 3q27.1 | Tabarés-Seisdedos et al. [2011] |
| HTR3E | 5-Hydroxytryptamine receptor 3E | 3q27.1 | Tabarés-Seisdedos et al. [2011] |
| HTR4 | 5-Hydroxytryptamine receptor 4 | 5q32 | Tabarés-Seisdedos et al. [2011] |
| HTR5A | 5-Hydroxytryptamine receptor 5A | 7q36.2 | Tabarés-Seisdedos et al. [2011] |
| HTR6 | 5-Hydroxytryptamine receptor 6 | 1p36.13 | Tabarés-Seisdedos et al., 2011] |
| HTR7a | 5-Hydroxytryptamine receptor 7 | 10q23.31 | Tabarés-Seisdedos et al. [2011] |
| ICAM1 | Intercellular adhesion molecule 1 | 19p13.2 | Tabarés-Seisdedos et al. [2011] |
| IFITM3 | Interferon-induced transmembrane protein 3 | 11p15.5 | Sanders et al. [2013] |
| IL10 | Interleukin 10 | 1q32.1 | Tabarés-Seisdedos et al. [2011] |
| IL12B | Interleukin 12B | 5q33.3 | Tabarés-Seisdedos et al. [2011] |
| IL1A | Interleukin 1, alpha | 2q13 | Tabarés-Seisdedos et al. [2011] |
| IL1B | Interleukin 1, beta | 2q13 | Xu and He [2010] |
| IL1RN | Interleukin 1 receptor antagonist | 2q14.2 | Tabarés-Seisdedos et al. [2011] |
| IL3 | Interleukin 3 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| IL3RA | Interleukin 3 receptor, alpha | Xp22.33 | Tabarés-Seisdedos et al. [2011] |
| IL4 | Interleukin 4 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| IL6 | Interleukin 6 | 7p15.3 | Tabarés-Seisdedos et al. [2011] |
| IMMP2La | Inner mitochondrial membrane peptidase, subunit 2, S. cerevisiae, homolog of | 7q31.1 | Tabarés-Seisdedos et al. [2011] |
| IMPA2 | Myo-inositol monophosphatase 2 | 18p11.21 | Tabarés-Seisdedos et al. [2011] |
| INSIG2 | Insulin-induced gene 2 | 2q14.2 | Tabarés-Seisdedos et al. [2011] |
| IPO5 | Importin 5 | 13q32.2 | Tabarés-Seisdedos et al. [2011] |
| ITIH3 | Inter-alpha-trypsin inhibitor heavy chain 3 | 3p21.1 | Ripke et al. [2013] |
| JAG2 | Jagged 2 | 14q32.33 | Tabarés-Seisdedos et al. [2011] |
| JARID2a | Jumonji, AT-rich interactive domain 2 | 6p22.33 | Tabarés-Seisdedos et al. [2011] |
| KCNH2 | Potassium voltage-gated channel, subfamily H, member 2 | 7q36.1 | Hashimoto et al. [2013] |
| KCNH7 | Potassium voltage-gated channel, subfamily H (eag-related), member 7 | 2q24.2 | Zhang et al. [2012] |
| KCNN3 | Potassium channel, calcium-activated, intermediate/small conductance, subfamily N, member 3 | 1q21.3 | Chandy et al. [1998] |
| KIAA0513 | KIAA0513 designated gene | 16q24.1 | Lauriat et al. [2006] |
| KIAA1644 | KIAA1644 designated gene | 22q13 | Wright et al. [2013] |
| L1RE1 | LINE1 (long interspersed nuclear element) retrotransposable element 1 | 22q11.1-q11.2 | Bundo et al. [2014] |
| LAMA5 | Laminin, alpha 5 | 20q13.33 | van Schijndel et al. [2009] |
| LEPa | Leptin | 7q32.1 | Tabarés-Seisdedos et al. [2011] |
| LEPR | Leptin receptor | 1p31.3 | Tabarés-Seisdedos et al. [2011] |
| LPL | Lipoprotein lipase | 8p21.3 | Tabarés-Seisdedos et al. [2011] |
| LRRC4a | Leucine rich repeat containing 4 | 7q32.1 | Wright et al. [2013] |
| LSM1 | LSM1 protein | 8p11.23 | Shi et al. [2011] |
| LTA | Lymphotoxin alpha | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| M1AP | Meiosis 1 associated protein | 2p13.1 | Ripke et al. [2013] |
| MAD1L1 | Mitotic arrest-deficient 1, yeast, homolog-like 1 | 7p22.3 | Ripke et al. [2013] |
| MAG | Myelin-associated glycoprotein | 19q13.12 | Bahn [2002] |
| MAGI1 | Membrane-associated guanylate kinase, WW, and PDZ domain containing 1 | 3p14.1 | Sun et al. [2008] |
| MAGI2 | Membrane-associated guanylate kinase, WW, and PDZ domain containing 2 | 7q21.11 | Sun et al. [2008] |
| MAGI3 | Membrane-associated guanylate kinase, WW, and PDZ domain containing 3 | 1p13.2 | Sun et al. [2008] |
| MAOAa | Monoamine oxidase A | Xp11.3 | Tabarés-Seisdedos et al. [2011] |
| MAOBa | Monoamine oxidase B | Xp11.3 | Rice et al. [1984] |
| MAP2K4 | Mitogen-activated protein kinase kinase 4 | 17p12 | Wright et al. [2013] |
| MAPK14 | Mitogen-activated protein kinase 14 | 6p21.31 | Tabarés-Seisdedos et al. [2011] |
| MAPK3a | Mitogen-activated protein kinase 3 | 16p11.2 | Wright et al. [2013] |
| MAPT | Microtubule-associated protein tau | 17q21.31 | Tabarés-Seisdedos et al. [2011] |
| MAU2 | MAU2 (maternally affected uncoordination 2) sister chromatid cohesion factor | 19p13.11 | Ripke et al. [2013] |
| MCHR1 | Melanin-concentrating hormone receptor 1 | 22q13.2 | Tabarés-Seisdedos et al. [2011] |
| MDGA1 | MAM (meprin, A5 protein, and protein tyrosine phosphatase Mu) domain containing glycosylphosphatidylinositol anchor 1 | 6p21.2 | Ocklenburg et al. [2013] |
| MECP2a | Methyl CpG binding protein 2 | Xq28 | Tabarés-Seisdedos et al. [2011] |
| MED1 | Mediator complex subunit 1 | 17q12 | Wright et al. [2013] |
| MED12a | Mediator complex subunit 12 | Xq13.1 | Tabarés-Seisdedos et al. [2011] |
| MED15 | Mediator complex subunit 15 | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| MEGF10 | Multiple epidermal growth factor-like domains 10 | 5q23.2 | Tabarés-Seisdedos et al. [2011] |
| MGRN1 | Mahogunin ring finger 1 | 16p13.3 | Wright et al. [2013] |
| MGST1 | Microsomal glutathione S-transferase 1 | 12p12.3 | Wockner et al. [2014] |
| MICB | Major histocompatibility complex class I chain-related gene B | 6p21.33 | Tabarés-Seisdedos et al. [2011] |
| MIR137 | MicroRNA 137 | 1p21.3 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| MIR30E | MicroRNA 30e | 1p34.2 | Tabarés-Seisdedos et al. [2011] |
| MKL1 | Megakaryoblastic leukemia 1 | 22q13.2 | Luo et al. [2014] |
| MLC1 | Megalencephalic leukoencephalopathy with subcortical cysts 1 | 22q13.33 | Tabarés-Seisdedos et al. [2011] |
| MMP16 | Matrix metallopeptidase 16 | 8q21.3 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| MMP9 | Matrix metallopeptidase 9 | 20q13.12 | Tabarés-Seisdedos et al. [2011] |
| MOXD1 | Monooxygenase, DBH-like 1 | 6q23.2 | Sanders et al. [2013] |
| MPO | Myeloperoxidase | 17q22 | Tabarés-Seisdedos et al. [2011] |
| MTHFRa | Methylenetetrahydrofolate reductase | 1p36.22 | Zintzaras [2006] |
| MYH9 | Myosin, heavy chain 9, non-muscle | 22q12.3 | Tabarés-Seisdedos et al. [2011] |
| MYO16a | Myosin XVI | 13q33.3 | Rodriguez-Murillo et al. [2014] |
| NALCN | Sodium leak channel, non-selective | 13q33.1 | Wang et al. [1996] |
| NCAM1 | Neural cell adhesion molecule 1 | 11q23.2 | Tabarés-Seisdedos et al. [2011] |
| NDE1 | NudE neurodevelopment protein 1 | 16p13.11 | Tabarés-Seisdedos et al. [2011] |
| NDEL1 | NudE neurodevelopment protein 1-like 1 | 17p13.1 | Tabarés-Seisdedos et al. [2011] |
| NETO1 | Neuropilin and tolloid-like 1 | 18q22.3 | Banno et al. [2011] |
| NEUROD2 | Neuronal differentiation 2 | 17q12 | Wright et al. [2013] |
| NEUROG1 | Neurogenin 1 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| NLGN1a | Neuroligin 1 | 3q26.31 | Tabarés-Seisdedos et al. [2011] |
| NLRP1 | NLR (NOD-like microbial receptor) family, pyrin domain containing 1 | 17p13.2 | Sanders et al. [2013] |
| NOD2 | Nucleotide-binding oligomerization domain containing 2 | 16q12.1 | van Schijndel et al. [2009] |
| NOS1 | Nitric oxide synthase 1 | 12q24.22 | Weber et al. [2014] |
| NOS1APa | Nitric oxide synthase 1 adaptor protein | 1q23.3 | Weber et al. [2014] |
| NOS3 | Nitric oxide synthase 3 | 7q36,1 | Tabarés-Seisdedos et al. [2011] |
| NOTCH2 | Notch, Drosophila, homolog of, 2 | 1p11.2 | Tabarés-Seisdedos et al. [2011] |
| NOTCH3a | Notch, Drosophila, homolog of, 3 | 19p13.12 | Tabarés-Seisdedos et al. [2011] |
| NOTCH4 | Notch, Drosophila, homolog of, 4 | 6p21.32 | Ikeda et al. [2011] |
| NPAS3 | Neuronal PAS domain protein 3 | 14q13.1 | Macintyre et al. [2010] |
| NPPC | Natriuretic peptide precursor C | 2q37.1 | Roussos and Haroutunian [2014] |
| NPY | Neuropeptide Y | 7p15.3 | Tabarés-Seisdedos et al. [2011] |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 16q22.1 | Tabarés-Seisdedos et al. [2011] |
| NR4A1a | Nuclear receptor subfamily 4, group A, member 1 | 12q13.13 | Tabarés-Seisdedos et al. [2011] |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2q24.1 | Tabarés-Seisdedos et al. [2011] |
| NRG1a | Neuregulin 1 | 8p12 | Wockner et al. [2014] |
| NRG2 | Neuregulin 2 | 5q31.2 | Wright et al. [2013] |
| NRG3 | Neuregulin 3 | 10q23.1 | Kao et al. [2010] |
| NRGN | Neurogranin | 11q24.2 | Wockner et al. [2014] |
| NRXN1a | Neurexin 1 | 2p16.3 | Kirov et al. [2008] |
| NRXN3a | Neurexin 3 | 14q24.3 | Tabarés-Seisdedos et al. [2011] |
| NSF | N-ethylmaleimide-sensitive factor | 17q21.31 | Mirnics et al. [2000] |
| NT5C2 | 5-prime-nucleotidase, cytosolic II | 10q24.33 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| NTF3 | Neurotrophin 3 | 12p13.31 | Tabarés-Seisdedos et al. [2011] |
| NTNG1a | Netrin G1 | 1p13.3 | Tabarés-Seisdedos et al. [2011] |
| NTRK1a | Neurotrophic tyrosine kinase, receptor, type 1 | 1q23.1 | van Schijndel et al. [2009] |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 | 9q21.33 | Tabarés-Seisdedos et al. [2011] |
| NTRK3a | Neurotrophic tyrosine kinase, receptor, type 3 | 15q25.3 | Tabarés-Seisdedos et al. [2011] |
| NUDT9P1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 9 pseudogene 1 | 10q23.32 | Tabarés-Seisdedos et al. [2011] |
| NUMBL | Numb, Drosophila, homolog-like | 19q13.2 | Sun et al. [2008] |
| OLIG2 | Oligodendrocytes lineage transcription factor 2 | 21q22.11 | Georgieva et al. [2006] |
| OXT | Oxytocin | 20p13 | Tabarés-Seisdedos et al. [2011] |
| OXTRa | Oxytocin receptor | 3p25.3 | Tabarés-Seisdedos et al. [2011] |
| PAFAH1B1a | Platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 | 17p13.3 | Tabarés-Seisdedos et al. [2011] |
| PAHa | Phenylalanine hydroxylase | 12q23.2 | Tabarés-Seisdedos et al. [2011] |
| PAWR | PRKC (protein kinase 3), apoptosis, WT1 (Wilms tumor 1), regulator | 12q21.2 | Tabarés-Seisdedos et al. [2011] |
| PBRM1a | Polybromo 1 | 3p21.1 | Kondo et al. [2013] |
| PCGEM1 | Prostate-specific gene PCGEM1 | 2q32.3 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| PCM1 | Pericentriolar material 1 | 8p22 | Moens et al. [2010] |
| PCNT | Pericentrin | 21q22.3 | Tabarés-Seisdedos et al. [2011] |
| PDE4Ba | Phosphodiesterase 4B, cAMP-specific | 1p31.3 | Millar et al. [2005] |
| PDLIM5 | PDZ (pSD95, Dlg1, and zo-1) and LIM Lin11, Isl-1, and Mec-3) domain 5 | 4q22.3 | Tabarés-Seisdedos et al. [2011] |
| PDYN | Prodynorphin | 20p13 | Tabarés-Seisdedos et al. [2011] |
| PEMT | Phosphatidylethanolamine N-methyltransferase | 17p11.2 | Tabarés-Seisdedos et al. [2011] |
| PER3 | Period circadian clock 3 | 1p36.23 | Tabarés-Seisdedos et al. [2011] |
| PGBD1 | PiggyBac transposable element derived 1 | 6p22.1 | Alkelai et al. [2012] |
| PGP | Phosphoglycolate phosphatase | 19p13.11 | Wright et al. [2013] |
| PHF3 | PHD (plant homeodomain) finger protein 3 | 6q12 | Wright et al. [2013] |
| PHOX2B | Paired-like homeobox 2B | 4p13 | Toyota et al. [2004] |
| PI4KA | Phosphatidylinositol 4-kinase, catalytic, alpha | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| PICK1 | Protein interacting with C kinase 1 | 22q13.1 | Tabarés-Seisdedos et al. [2011] |
| PIK3C3 | Phosphatidylinositol 3-kinase, class 3 | 18q12.3 | Tabarés-Seisdedos et al. [2011] |
| PIK4CA | Phosphatidylinositol 4-kinase, catalytic, alpha | 22q11.21 | Sullivan [2005] |
| PIP4K2A | Phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | 10p12.2 | Tabarés-Seisdedos et al. [2011] |
| PLA2G1B | Phospholipase A2, group IB | 12q24.31 | Tabarés-Seisdedos et al. [2011] |
| PLA2G4A | Phospholipase A2, group IVA | 1q31.1 | Tabarés-Seisdedos et al. [2011] |
| PLA2G4C | Phospholipase A2, group IVC | 19q13.33 | Tabarés-Seisdedos et al. [2011] |
| PLA2G4D | Phospholipase A2, group IVD | 15q15.1 | Tabarés-Seisdedos et al. [2011] |
| PLA2G6 | Phospholipase A2, group VI | 22q13.1 | Tabarés-Seisdedos et al. [2011] |
| PLA2G7 | Phospholipase A2, group VII | 6p12.3 | Tabarés-Seisdedos et al. [2011] |
| PLAA | Phospholipase A2-activating protein | 9p21.2 | Athanasiu et al. [2010] |
| PLP1 | Proteolipid protein 1 | Xq22.2 | Aberg et al. [2006] |
| PLXNA2 | Plexin A2 | 1q32.2 | Allen et al. [2008] |
| PNOC | Prepronociceptin | 8p21.1 | Tabarés-Seisdedos et al. [2011] |
| POM121L2 | POM121 (POM121 transmembrane nucleoporin) transmembrane nucleoporin-like 2 | 6p22.1 | Aberg et al. [2006] |
| PON1a | Paraoxonase 1 | 7q21.3 | Tabarés-Seisdedos et al. [2011] |
| PPARG | Peroxisome proliferator-activated receptor gamma | 3p25.2 | Tabarés-Seisdedos et al. [2011] |
| PPP1R1Ba | Protein phosphatase 1, regulatory (inhibitor) subunit 1B | 17q12 | Meyer-Lindenberg et al. [2007] |
| PPP2R2B | Protein phosphatase 2, regulatory subunit B, beta | 5q32 | Tabarés-Seisdedos et al. [2011] |
| PPP3CC | Protein phosphatase 3, catalytic subunit, gamma isoform | 8p21.3 | Wockner et al. [2014] |
| PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | 1q21.1 | Wright et al. [2013] |
| PRKCA | Protein kinase C, alpha | 17q24.2 | Tabarés-Seisdedos et al. [2011] |
| PRKCD | Protein kinase C, delta | 3p21.1 | Sanders et al. [2013] |
| PRODHa | Proline dehydrogenase | 22q11.21 | Sullivan [2005] |
| PRODH2 | Proline dehydrogenase 2 | 19q13.12 | Harrison and Weinberger [2005] |
| PRNP | Prion protein | 20p13 | Tabarés-Seisdedos et al. [2011] |
| PRRT4 | Proline-rich transmembrane protein 4 | 7q32.1 | Wright et al. [2013] |
| PRSS16 | Protease, serine, 16 | 6p22.1 | Stefansson et al. [2009] |
| PTBP3 | Polypyrimidine tract-binding protein 3 | 9q32 | Sanders et al. [2013] |
| PTGS2a | Prostaglandin-endoperoxide synthase 2 | 1q31.1 | Tabarés-Seisdedos et al. [2011] |
| PTPN21 | Protein tyrosine phosphatase, non-receptor type 21 | 14q31.3 | Chen et al. [2011a] |
| PTPRZ1 | Protein tyrosine phosphatase, receptor-type, zeta 1 | 7q31.32 | Tabarés-Seisdedos et al. [2011] |
| QKI | QKI, KH (K homology) domain containing, RNA binding | 6q26 | Aberg et al. [2006] |
| QPCT | Glutaminyl-peptide cyclotransferase | 2p22.2 | Ripke et al. [2013] |
| RAI1a | Retinoic acid induced 1 | 17p11.2 | Toulouse et al. [2003] |
| RANBP2 | Ran binding protein 2 | 2q12.3 | Wright et al. [2013] |
| RAPGEF6 | Rap guanine nucleotide exchange factor 6 | 5q31.1 | Tabarés-Seisdedos et al. [2011] |
| RELNa | Reelin | 7q22.1 | van Schijndel et al. [2009] |
| REST | RE1-silencing transcription factor | 4q12 | Loe-Mie et al. [2010] |
| RGS2 | Regulator of G-protein signaling 2 | 1q31.2 | Tabarés-Seisdedos et al. [2011] |
| RGS4 | Regulator of G-protein signaling 4 | 1q23.3 | Harrison and Weinberger [2005] |
| RGS9 | Regulator of G-protein signaling 9 | 17q24.1 | Tabarés-Seisdedos et al. [2011] |
| RNF144 | Ring finger protein 144A | 2p25.2 | Xu and He [2010] |
| RNF5 | Ring finger protein 5 | 6p21.32 | de Jong et al. [2012] |
| RSRC1 | Arginine/serine-rich coiled-coil 1 | 3q25.32 | Potkin et al. [2009] |
| RTN4 | Reticulon 4 | 2p16.1 | Tabarés-Seisdedos et al. [2011] |
| RTN4R | Reticulon 4 receptor | 22q11.21 | Sinibaldi et al. [2004] |
| RXRB | Retinoid X receptor, beta | 6p21.32 | Tabarés-Seisdedos et al. [2011] |
| S100A10 | S100 calcium-binding protein A10 | 1q21.3 | Sanders et al. [2013] |
| S100B | S100 calcium-binding protein beta | 21q22.3 | Tabarés-Seisdedos et al. [2011] |
| SBK1 | SH3 (SRC homology 3) domain-binding kinase 1 | 16p11.2 | Wright et al. [2013] |
| SDCCAG8 | Serologically defined colon cancer antigen 8 | 1q43 | Ripke et al. [2013] |
| SELENBP1 | Selenium-binding protein 1 | 1q21.3 | Tabarés-Seisdedos et al. [2011] |
| SEMA3D | Semaphorin 3D | 7q21.11 | Tabarés-Seisdedos et al. [2011] |
| SEZ6L2a | Seizure-related 6 homolog (mouse)-like 2 | 16p11.2 | Wright et al. [2013] |
| SGK1 | Serum/glucocorticoid-regulated kinase 1 | 6q23.2 | Sanders et al. [2013] |
| SHANK3a | SH3 and multiple ankyrin repeat domains 3 | 22q13.33 | Gauthier et al. [2010] |
| SHMT1 | Serine hydroxymethyltransferase, cytosolic | 17p11.2 | Tabarés-Seisdedos et al. [2011] |
| SHOX | Short stature homeobox | Xp22.33 | Loe-Mie et al. [2010] |
| SIGMAR1 | Sigma non-opioid intracellular receptor 1 | 9p13.3 | Ohi et al. [2011] |
| SLC17A1 | Solute carrier family 17 (organic anion transporter), member 1 | 6p22.2 | Shi et al. [2009] |
| SLC17A3 | Solute carrier family 17 (organic anion transporter), member 3 | 6p22.2 | Shi et al. [2009] |
| SLC17A6 | Solute carrier family 17 (vesicular glutamate transporter), member 6 | 11p14.3 | Tabarés-Seisdedos et al. [2011] |
| SLC17A7 | Solute carrier family 17 (vesicular glutamate transporter), member 7 | 19q13.33 | Tabarés-Seisdedos et al. [2011] |
| SLC18A1 | Solute carrier family 18 (vesicular glutamate transporter), member 1 | 8p21.3 | Tabarés-Seisdedos et al. [2011] |
| SLC18A2 | Solute carrier family 18 (vesicular glutamate transporter), member 2 | 10q25.3 | Tabarés-Seisdedos et al. [2011] |
| SLC1A1a | Solute carrier family 1, member 1 | 9p24.2 | Allen et al. [2008] |
| SLC1A2 | Solute carrier family 1 (glial high-affinity glutamate transporter), member 2 | 11p13 | Tabarés-Seisdedos et al. [2011] |
| SLC1A3 | Solute carrier family 1 (glial high-affinity glutamate transporter), member 3 | 5p13.2 | Tabarés-Seisdedos et al. [2011] |
| SLC1A4 | Solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 2p14 | Tabarés-Seisdedos et al. [2011] |
| SLC1A6 | Solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | 19p13.12 | Tabarés-Seisdedos et al. [2011] |
| SLC24A5 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 | 15q21.1 | Tabarés-Seisdedos et al. [2011] |
| SLC39A8 | Solute carrier family 39 (zinc transporter), member 8 | 4q24 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| SLC5A7 | Solute carrier family 5 (sodium/choline cotransporter), member 7 | 2q12.3 | Wright et al. [2013] |
| SLC6A2 | Solute carrier family 6 (neurotransmitter transporter, noradrenaline), member 2 | 16q12.2 | Tabarés-Seisdedos et al. [2011] |
| SLC6A3a | Solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | 5p15.33 | Tabarés-Seisdedos et al. [2011] |
| SLC6A4a | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 17q11.2 | Allen et al. [2008] |
| SLC6A9 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 1p34.1 | Tabarés-Seisdedos et al. [2011] |
| SLC9A3R2 | Solute carrier family 9, member 3, regulator 2 | 16p13.3 | Wright et al. [2013] |
| SLCO6A1 | Solute carrier organic anion transporter family, member 6A1 | 5q21.1 | Ripke et al. [2013] |
| SLX4 | SLX4, S. cerevisiae, homolog of | 16p13.3 | Wright et al. [2013] |
| SMARCA2 | SW1/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 2 | 9p24.3 | Loe-Mie et al. [2010] |
| SNAP25 | Synaptosomal-associated protein, 25kDa | 20p12.2 | Dai et al. [2014] |
| SNAP29 | Synaptosomal-associated protein, 29-kDa | 22q11.21 | Sun et al. [2008] |
| SNX19a | Sorting nexin 19 | 11q24.3 | Ripke et al. [2013] |
| SOD2 | Superoxide dismutase 2 | 6q25.3 | Tabarés-Seisdedos et al. [2011] |
| SOX10 | SRY (sex determining region Y)-box 10 | 22q13.1 | Yuan et al. [2012] |
| SP4 | Sp4 transcription factor | 7p15.3 | Tabarés-Seisdedos et al. [2011] |
| SP8 | Sp8 transcription factor | 7p21.1 | Kondo et al. [2013] |
| SRC | V-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | 20q11.23 | Pitcher et al. [2011] |
| SRD5A1 | Steroid-5-alpha-reductase 1 | 5p15.31 | Tabarés-Seisdedos et al. [2011] |
| SREBF1 | Sterol regulatory element binding transcription factor 1 | 17p11.2 | Tabarés-Seisdedos et al. [2011] |
| SREBF2 | Sterol regulatory element binding transcription factor 2 | 22q13.2 | Tabarés-Seisdedos et al. [2011] |
| SRR | Serine racemase | 17p13.3 | Tabarés-Seisdedos et al. [2011] |
| ST3GAL1 | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 | 8q24.22 | Xu and He [2010] |
| ST6GAL2 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | 2q12.3 | Wright et al. [2013] |
| ST8SIA2a | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 15q26.1 | Tabarés-Seisdedos et al. [2011] |
| STAC2 | SH3 and cysteine-rich domain 2 | 17q12 | Wright et al. [2013] |
| STT3A | Oligosaccharyltransferase complex, catalytic subunit STT3A | 11q24.2 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| STX1B | Syntaxin 1B | 16p11.2 | Wright et al. [2013] |
| STX6a | Syntaxin 6 | 1q25.3 | Sanders et al. [2013] |
| SULT4A1 | Sulfotransferase family 4A, member 1 | 22q13.31 | Tabarés-Seisdedos et al. [2011] |
| SYN2a | Synapsin II | 3p25.2 | Saviouk et al. [2007] |
| SYN3a | Synapsin lll | 22q12.3 | Kao et al. [1998] |
| SYNGR1 | Synaptogyrin 1 | 22q13.1 | Tabarés-Seisdedos et al. [2011] |
| SYT11 | Synaptotagmin 11 | 1q22 | Sanders et al. [2013] |
| TAAR6 | Trace amine-associated receptor 6 | 6q23.2 | Duan et al. [2004] |
| TACR1 | Tachykinin receptor 1 | 2p12 | Tabarés-Seisdedos et al. [2011] |
| TAF15 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa | 17q12 | Wright et al. [2013] |
| TAP2 | Transporter, ATP-binding cassette, major histocompatibility complex, 2 | 6p21.32 | Tabarés-Seisdedos et al. [2011] |
| TBX1a | T-box 1 | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| TCF4a | Transcription factor 4 | 18q21.2 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| TF | Transferrin | 3q22.1 | Aberg et al. [2006] |
| THa | Tyrosine hydroxylase | 11p15.5 | Tabarés-Seisdedos et al. [2011] |
| TIMELESS | Timeless, Drosophila, homolog of | 12q13.3 | Tabarés-Seisdedos et al. [2011] |
| TMED5 | Transmembrane emp24 protein transport domain containing 5 | 1p22.1 | Wright et al. [2013] |
| TMEM245 | Transmembrane protein 245 | 9q31.3 | Xu and He [2010] |
| TNF | Tumor necrosis factor | 6p21.33 | Sacchetti et al. [2007] |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 1p36.22 | Tabarés-Seisdedos et al. [2011] |
| TNR | Tenascin R | 1q25.1 | Tabarés-Seisdedos et al. [2011] |
| TP53 | Tumor protein p53 | 17p13.1 | Allen et al. [2008] |
| TPH1 | Tryptophan hydroxylase 1 | 11p15.1 | Allen et al. [2008] |
| TPH2a | Tryptophan hydroxylase 2 | 12q21.1 | Tabarés-Seisdedos et al. [2011] |
| TRAX | Translin-associated factor X | 1q42.2 | Cannon et al. [2005] |
| TRIM26 | Tripartite motif containing 26 | 6p22.1 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
| TSNARE1 | T-SNARE (soluble NSF attachment protein receptor) domain containing 1 | 8q24.3 | Ripke et al. [2013] |
| TSNAX | Translin-associated factor X | 1q42.2 | Tabarés-Seisdedos et al. [2011] |
| TSPAN33 | Tetraspanin 33 | 7q32.1 | Wright et al. [2013] |
| TXNDC5 | Thioredoxin domain containing 5 | 6p24.3 | Tabarés-Seisdedos et al. [2011] |
| UBE3Ca | Ubiquitin protein ligase E3C | 7q36.3 | Wright et al. [2013] |
| UFD1L | Ubiquitin fusion degradation 1 like | 22q11.21 | Tabarés-Seisdedos et al. [2011] |
| UGT1A4 | UDP (uridine 5’-diphospho-) glucuronosyltransferase 1 family, polypeptide A4 | 2q37.1 | Tabarés-Seisdedos et al. [2011] |
| UHMK1 | U2AF (U2 auxiliary factor) homology motif kinase 1 | 1q23.3 | Dumaine et al. [2011] |
| VAMP4 | Vesicle-associated membrane protein 4 | 1q24.3 | Sanders et al. [2013] |
| VDR | Vitamin D receptor | 12q13.11 | Tabarés-Seisdedos et al. [2011] |
| VIPR2 | Vasoactive intestinal peptide receptor 2 | 7q36,3 | Vacic et al. [2011] |
| VKORC1 | Vitamin K epoxide reductase complex, subunit 1 | 16p11.2 | Wright et al. [2013] |
| VRK2 | Vaccinia related kinase 2 | 2p16.1 | Stefansson et al. [2009] |
| WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1-like 1 | 8p11.23 | Shi et al. [2011] |
| WWC1 | WW, C2, and coiled-coil domain-containing 1 | 5q34 | Tabarés-Seisdedos et al. [2011] |
| XBP1 | X-box binding protein 1 | 22q12.1 | Tabarés-Seisdedos et al. [2011] |
| XKR4 | XK, Kell blood group complex subunit-related family, member 4 | 8q12.1 | Tabarés-Seisdedos et al. [2011] |
| YES | V-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 6q21 | Tabarés-Seisdedos et al. [2011] |
| YWHAEa | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon isoform | 17p13.3 | Ikeda et al. [2008] |
| YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, Eta | 22q12.3 | Tabarés-Seisdedos et al. [2011] |
| ZBTB42 | Zinc finger and BTB (BR-C, ttk and bab) domain containing 42 | 14q32.33 | Ocklenburg et al. [2013] |
| ZDHHC8 | Zinc finger, DHHC (Asp-His-His-Cys)-type containing 8 | 22q11.21 | Sullivan [2005] |
| ZEB2 | Zinc finger E-box binding homeobox 2 | 2q22.3 | Ripke et al. [2013] |
| ZNF184 | Zinc finger protein 184 | 6p22.1 | Shi et al. [2009] |
| ZNF385D | Zinc finger protein 385D | 3p24.3 | Xu and He [2010] |
| ZNF804Aa | Zinc finger protein 804A | 2q32.1 | Williams et al. [2011] |
| ZSWIM6 | Zinc finger, SWIM (SWI2/SNF2 and MuDR)-type containing 6 | 5q12.1 | Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium [2011] |
Overlap with recognized Autism Spectrum Disorders (ASD) genes.
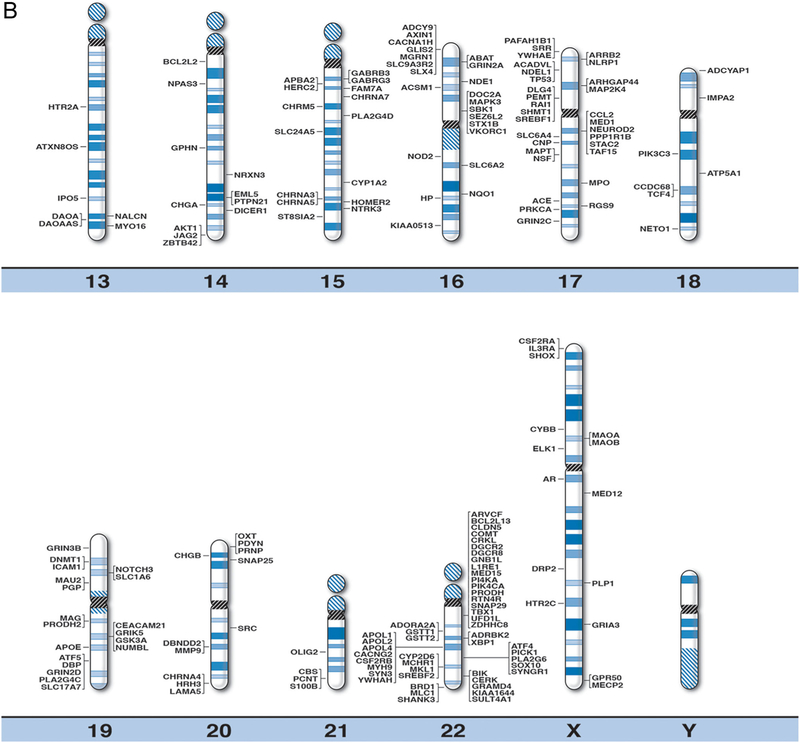
FIG. 1.
High resolution human chromosome ideograms (850 band level) with gene symbols representing currently recognized genes for schizophrenia plotted on chromosome bands for each of the 560 genes. The centromere area is highlighted in black which separates the upper short ‘p’ arm from the lower long ‘q’ arm for each chromosome. The gene symbols are arranged in alphabetical order with the expanded name and precise chromosome band location listed in Table 1. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmgb].
Genome wide pathway analysis was carried out for the derived gene master list and all mapped genomic variants associated with schizophrenia using a commercially available software program and data analysis application, GeneAnalytics (http://geneanalytics.genecards.org/) which is part of the GeneCards Suite developed by LifeMap Sciences (http://www.lifemapsc.com/products/genecards-suite-premium-tools/). GeneCards Suite provides enhanced gene variant prioritization, gene feature, and pathway analysis through three premium analytical tools: VarElect, the NGS phenotyper, GeneAnalytics™, a novel gene set analysis tool, and GeneALaCart, the GeneCards batch querying application. These applications are integrated with GeneCards® human gene database, MalaCards human disease database, PathCards, human biological pathways database, and LifeMap Discovery® tissues and cells database to provide an extensive universe of data from human genes, proteins, cells, biological pathways, diseases and their relationships.
The GeneAnalytics program was used to organize mammalian gene sets into functional categories based upon tissues and cells, diseases, pathways, GO-biological processes, GO- molecular function, phenotypes, and compounds (endogenous or exogenous) with functional links to the queried schizophrenia gene set. Gene ontology (GO) terms (e.g., superpathways and GO-biological function) were scored based upon transformation of the binomial P-value which is equivalent to a corrected P-value with significance defined at P < 0.0001. Disease-matching scores were derived based upon the number of overlapping genes and the nature of the gene-disease association. Tissues and cells were scored using a matching algorithm that weighs tissue specificity, abundance, and function of the gene. Related pathways were then grouped into superpathways to improve inferences, pathway enrichment, reduce redundancy, and rank genes within a biological mechanism via the multiplicity of constituent pathways.
RESULTS AND DISCUSSION
Based on existing literature in peer-reviewed journal articles and computer genomic-based websites for schizophrenia, we identified a total of 560 recognized or proposed, clinically relevant or susceptibility genes for schizophrenia. VarElect NGS phenotyping of the derived list of genes identified a direct relationship between 465 of those genes and schizophrenia phenotype; 86 genes were indirectly associated with schizophrenia phenotype; two genes were found to be unrelated to schizophrenia phenotype (COMTD1, L1RE1) and seven genes were not recognized by the VarElect application. Our analysis considered the spatial relationship of the identified genes to ascertain any patterning or potential for hot spot regions relevant to causation or course. This analysis was necessarily adjusted for the total length of the chromosome which is related to the total number of genes per chromosome. Our master list of genes or genetic markers provides additional support for previous reports of targeted chromosomal regions associated with schizophrenia. For example, chromosomes 1, 6, and 22 contained genetic regions previously identified as areas of interest for schizophrenia [Riley and Kendler, 2006]. These three chromosomes also possessed over 30% of the total number of genes identified on our master gene list but reflect only 15% of the chromosomal material by size in humans [Shaffer et al., 2012] (Table II). When examining the distribution of genes by chromosome arm for these top three chromosomes (i.e., 1, 6, 22), we found that chromosome 1, considered the largest human chromosome with nearly equal length short (p) and long (q) arms, contained a similar number of genes on each chromosome arm. In contrast, chromosome 22 is the second smallest chromosome but had the third highest number of genes recorded for schizophrenia. Chromosome 22 is an acrocentric chromosome as are chromosomes 13, 14, 15, and 21 and contain only ribosomal genes on their p arms. Chromosome 6 is approximately two-thirds the length of chromosome 1, but showed an unexpectedly high number of schizophrenia genes in relationship to its size. In addition, the short arm of chromosome 6 is about one-half the size of the long arm but contained three times the number of genes for schizophrenia than the long arm. Interestingly, the major histocompatibility immune function gene complex is located on the short arm of chromosome 6.
TABLE II.
The Number of Schizophrenia Genes With Distribution Among Chromosomes
| Chromosome | Schizophrenia genes | Percentage of total genes | Genes on p arm | Genes on q arm |
|---|---|---|---|---|
| 6 | 66 | 11.8 | 49 | 17 |
| 1 | 57 | 10.2 | 27 | 30 |
| 22 | 48 | 8.6 | 0 | 48 |
| 7 | 39 | 7.0 | 9 | 30 |
| 2 | 37 | 6.6 | 10 | 27 |
| 5 | 35 | 6.2 | 4 | 31 |
| 17 | 30 | 5.3 | 15 | 15 |
| 11 | 27 | 4.8 | 8 | 19 |
| 3 | 25 | 4.5 | 14 | 11 |
| 16 | 22 | 3.9 | 17 | 5 |
| 8 | 21 | 3.7 | 15 | 6 |
| 10 | 20 | 3.7 | 3 | 17 |
| 19 | 19 | 3.4 | 7 | 12 |
| 12 | 18 | 3.2 | 5 | 13 |
| 15 | 15 | 2.7 | 0 | 15 |
| X | 15 | 2.7 | 7 | 8 |
| 4 | 14 | 2.5 | 7 | 7 |
| 9 | 12 | 2.1 | 4 | 8 |
| 14 | 11 | 2.0 | 0 | 11 |
| 20 | 11 | 2.0 | 5 | 6 |
| 18 | 7 | 1.2 | 2 | 5 |
| 13 | 7 | 1.2 | 0 | 7 |
| 21 | 4 | 0.7 | 0 | 4 |
| Y | 0 | 0.0 | 0 | 0 |
| Total | 560 | 100 | 208 | 352 |
p, short arm; q, long arm.
Chromosomes are arranged by the highest to lowest number of contained schizophrenia genes.
Genome wide pathway analysis found the identified genes with a proposed role in schizophrenia and of potential clinical relevance for susceptibility, causation, treatment response, or course of illness impacted a broad range of biological pathways, mechanisms, tissues, and cell types including significant association with the cerebellum, cerebral cortex, medulla, thalamus, hypothalamus, pons, and amygdala. These genes were significantly associated with 10 recognized disease states with strongest overlap to schizophrenia (score = 126.6, 141 of 239 genes) supporting the validity of our compiled list to predict phenotype. Additionally, disease states, such as obesity (score = 32.3, 44 of 576 genes), breast cancer (score = 32.2, 47 of 781 genes), malaria (score = 28.8, 35 of 335 genes), rheumatoid arthritis (score = 24.2, 37 of 502 genes), bipolar disorder (score = 23.0, 20 of 31 genes), lung cancer (score = 22.4, 34 of 622 genes), colorectal cancer (score = 22.2,37 of 807), and obsessive-compulsive disorder (score = 211, 15 of 17) significantly overlapped with our compiled gene list and may suggest genetic pleiotropic relationships with clinical relevance to co-morbidity and outcome. Not surprisingly, 20 different GeneAnalytics molecular pathways associated with our gene list impacted ionotropic glutamate and GABA activity, protein/amino acid binding, transporter activity and receptor binding/function for dopamine and serotonin were broadly represented by significant associations with 69 biological processes in 95 superpathways. Pathways and mechanisms included ion channels (e.g., CACNA1B, CACNA1C, CACNA1H), metabolic enzymes (e.g., CYP1A2, CYP2C19, CYP2D6), brain development (e.g., NRG1, RELN), signaling peptides (e.g., PIK3CA, PIK4CA), and immune function (e.g., HLA-A, HLA-DRB1) and interleukins [e.g., IL1A, IL10, IL6]). Multiple genes involved with neurotransmitter function were identified impacting dopamine (e.g., DRD1, DRD2), GABA (e.g., GABBR1, GABRA1), and serotonin (e.g., HTR1A, HTR2C) pathways with a large number directly tied to glutamate processing (e.g., GAD1, GLUL) and signaling (e.g., GRIK3, GRM5).
To further assess the role of genetics in the neurodevelopment and function, we compared our list of proposed schizophrenia genes with a separate list of genes recently recognized in Autism Spectrum Disorders (ASD) [Butler et al., 2015]. Researchers have sought to uncover the similarities and differences between these two common neurobehavioral disorders [Cheung et al., 2010; Meyer et al., 2011; Hommer and Swedo, 2015]. We found overlap with a shared group of 116 genes between the two disorders (Table I), and the list of genes, their names, expression, and function may begin to suggest possible mechanisms including epigenetics playing a role in schizophrenia as well as a multifactorial basis. VarElect NGS phenotyping similarly identified a direct relationship between 191 genes from our schizophrenia gene list and autism phenotype; 352 genes were indirectly associated with autism phenotype; 10 genes were found to be unrelated to autism phenotype (seven genes were not recognized). The master list of recognized genes represented several molecular mechanisms further supporting that schizophrenia is not due to a single condition but results from a combination of neurochemical disturbances and manifest themselves differently across affected people. The changes that occur over time in those with schizophrenia may represent endophenotypes of the disorder that are genetically based and include interactions with environmental factors (epigenetics). More studies are required in schizophrenia to address these observations and patterns in affected individuals. Research involving the common genes should further advance our knowledge in the causation of brain disorders, psychiatry, neurological function, and development.
Better understanding of factors contributing to schizophrenia can be advanced through the genetic testing and analysis of several types of tissue sources (e.g., peripheral blood and saliva) and not solely brain specimens. Progress in the field of genetics has made tissue sources, such as the ones previously mentioned, readily available for testing; helping to accelerate the potential for discovery of novel genes associated with schizophrenia. For example, a growing number of biological specimens exist at tissue bank repositories for obtaining RNA to study transcription or coding with non-coding expression pattern profiles in individuals with schizophrenia, along with other brain disorders.
The authors encourage the use of this collection of recognized or proposed clinically relevant candidate and susceptibility genes and biomarkers for schizophrenia in their evaluation of patients and families presenting for genetic services requiring accurate information for diagnosis, genetic counseling risk estimates, and natural history and prognostic concerns. Early detection of schizophrenia, possibly through the use of genetic involvement, may help reduce the potentially devastating long-term effects of the disorder for an individual [Abbas and Lieberman, 2015]. A comprehensive approach should include a clinical genetics evaluation to rule out dysmorphic features or recognizable syndromes in which schizophrenia is a feature, cytogenetic or metabolic testing, identification of single gene disorders, or the use of high-resolution microarrays to identify subtle structural abnormalities supported by next generation DNA sequencing analysis for discovery of new gene mutations or variants. Interviews with parents for collection of family history and individual medical records are needed for support from health caregivers and their record overviews of medical history information. Additionally, a three-generation family pedigree should be obtained to include recording of developmental milestones and description with recording of onset of atypical behaviors and triggering events, if any.
A current list of medications and ongoing treatments with list of previous medications and outcomes, along with diagnostic or surgical procedures, should be obtained and may be helpful, given the role genetics play in medication metabolism and effects as several key cytochrome p450 hepatic enzymes involved with medication/drug metabolism were identified and included in our master list [Smith et al., 2015]. Pharmacogenetics has also been shown to contribute in weight gain as side effects associated with antipsychotic medications [Kao and Müller, 2013; Smith et al., 2015]. Also, much of the time a single type of antipsychotic medication is not sufficient to manage symptoms and these medications are often taken in conjunction with other psychoactive medications, such as mood stabilizers and antidepressants [Abbas and Lieberman, 2015]. Genetic information regarding medication metabolism and selectivity could potentially aid health care providers to optimize safety and efficacy of treatment plans for their patients. Furthermore, brain imaging and electroencephalogram patterns should be reviewed, if available, along with analysis of routine laboratory test results to assess drug screens, liver, vision, kidney and cardiac function, metabolic and immune status, complete blood counts, and neurological system findings.
In summary, we illustrated the master list of recognized or proposed clinically relevant, susceptible genes for schizophrenia as genetic biomarkers by plotting on individual high-resolution chromosome ideograms. We generated a table to increase awareness required for diagnosis, genetic testing, and counseling purposes for family members presenting for neuropsychiatric and genetic services. Creating a master list of genes related to schizophrenia is a complicated process; new genes are continually identified, but not all genes are equally important as biomarkers or certain to be causative. The authors encourage the undertaking of additional research to further investigate the causal relationships between specific genes and schizophrenia and understanding the molecular pathways that lead to this debilitating neuropsychiatric disorder.
ACKNOWLEDGMENTS
We would like to acknowledge the support of the Headley Family Scholarship and the National Institutes of Health (NICHD grant number HD02528). We also thank Lorie Gavulic for excellent artistic design and preparation of chromosome ideograms.
Grant sponsor: Headley Family Scholarship; Grant sponsor: National Institutes of Health; Grant number: HD02528.
REFERENCES
- Abbas AI, Lieberman JA. 2015. Pharmacological treatments for schizophrenia In: Nathan PE, Gorman JM, editors. A guide to treatments that work. New York, NY: Oxford University Press; pp 175–215. [Google Scholar]
- Aberg K, Saetre P, Jareborg N, Jazin E. 2006. Human QKI, a potential regulator of mRNA expression of hum an oligodendrocyte-related genes involved in schizophrenia. Proc Nat Acad Sci 103:7482–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, Liu Y, Bukszár J, McClay JL, Khachane AN, Andreassen OA, Blackwood D, Corvin A, Djurovic S, Gurling H, Ophoff R, Pato CN, Pato MT, Riley B, W ebb T, Kendler K, O’Donovan M, Craddock N, Kirov G, Owen M, Rujescu D, St Clair D, Werge T, Hultman CM, Delisi LE, Sullivan P, van den Oord EJ. 2013. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiat 70:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkelai A, Lupoli S, Greenbaum L, Kohn Y, Kanyas-Sarner K, Ben-Asher E, Lancet D, Macciardi F, Lerer B. 2012. DOCK4 and CEACAM21 as novel schizophrenia candidate genes in the Jewish population. Int J Neuro-psychopharmacol 15(4):459–469. [DOI] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen B, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. 2008. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: The SzGene database. Nat Genet 40:827–834. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. 2013. Diagnostic and Statistical Manual of Mental Disorders, 5th edition Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- Andreasen NC, Black DW. 2006. Introductory Textbook of Psychiatry, 4th edition Washington, DC: American Psychiatric Publishing, Inc. [Google Scholar]
- Andrew A, Knapp M, McCrone P, Parsonage M, Trachtenberg M. 2012. Effective interventions in schizophrenia: The economic case A report prepared for the Schizophrenia Commmission. London: Rethink Mental Illness. [Google Scholar]
- Athanasiu L, Mattingsdal M, Kähler AK, Brown A, Gustafsson O, Agartz I, Giegling I, Muglia P, Cichon S, Rietschel M, Pietiläinen OP, Peltonen L, Bramon E, Collier D, Clair DS, Sigurdsson E, Petursson H, Rujescu D, Melle I, Steen VM, Djurovic S, Andreassen OA. 2010. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res 44(12):748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S 2002. Gene expression in bipolar disorder and schizophrenia: New approaches to old problems. Bipolar Disord 4(Suppl. 1):70–72. [DOI] [PubMed] [Google Scholar]
- Banno M, Koide T, Aleksic B, Yamada K, Kikuchi T, Kohmura K, Adachi Y, Kawano N, Kushima I, Ikeda M, Inada T, Yoshikawa T, Iwata N, Ozaki N. 2011. A case control association study and cognitive function analysis of Neuropilin and Tolloid-Like 1 gene and schizophrenia in the Japanese population. PLoS ONE 6:e28929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begni S, Moraschi S, Bignotti S, Fumagalli F, Rillosi L, Perez J, Gennarelli M. 2003. Association between the G1001C polymorphism in the GRIN1 gene promoter region and schizophrenia. Biol Psychiat 53:617–619. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hayter JE, Hodgkinson KA, Chow EWC, Bassett AS. 2002. Fine mapping of the schizophrenia susceptibility locus on chromosome 1q22. Hum Hered 54:199–209. [DOI] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, Sunaga F, Toritsuka M, Ikawa D, Kakita A, Kato M, Kasai K, Kishimoto T, Nawa H, Okano H, Yoshikawa T, Kato T, Iwamoto K. 2014. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81:306–313. [DOI] [PubMed] [Google Scholar]
- Butler MG, Rafi SK, Manzardo AM. 2015. High-resolution chromosome ideogram representation of currently recognized genes for autism spectrum disorders. Int J Mol Sci 16(3):6464–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TGM, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. 2005. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiat 62:1205–1213. [DOI] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. 2000. Twin studies of schizophrenia: From bow-and-arrow concordances to Star Wars Mx and functional genomics. Am J Med Genet 97:12–17. [PubMed] [Google Scholar]
- Chandy KG, Fantino E, Wittekindt O, Kalman K, Tong LL, Ho TH, Gutman GA, Crocq MA, Ganguli R, Nimgaonkar V, Morris-Rosendahl DJ, Gargus JJ. 1998. Isolation of a novel potassium channel gene hSKCa3 containing a polymorphic CAG repeat: A candidate for schizophrenia and bipolar disorder? Molec Psychiat 3:32–37. [DOI] [PubMed] [Google Scholar]
- Chen J, Lee G, Fanous AH, Zhao Z, Jia P, O’Neill A, Walsh D, Kendler KS, Chen X, International Schizophrenia Consortium. 2011a. Two non-synonymous markers in PTPN21, identified by genome-wide association study data-mining and replication, are associated with schizophrenia. Schizophr Res 131:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z, Guo A, van den Oord E, Sullivan PF, Shi J, Levinson DF, Gejman PV, Sanders A, Duan J, Owen MJ, Craddock NJ, O ‘Donovan MC, Blackman J, Lewis D, Kirov GK, Qin W, Schwab S, Wildenauer D, Chowdari K, Nimgaonkar V, Straub RE, Weinberger DR, O’Neill FA, Walsh D, Bronstein M, Darvasi A, Lencz T, Malhotra AK, Rujescu D, Giegling I, Werge T, Hansen T, Ingason A, Nöethen MM, Rietschel M, Cichon S, Djurovic S, Andreassen OA, Cantor RM, Ophoff R, Corvin A, Morris DW, Gill M, Pato CN, Pato MT, Macedo A, Gurling HM, McQuillin A, Pimm J, Hultman C, Lichtenstein P, Sklar P, Purcell SM, Scolnick E, St Clair D, Blackwood DH, Kendler KS, GROUP Investigators, International Schizophrenia Consortium. 2011b. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry 16:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Yu K, Fung G, Leung M, Wong C, Li Q, Sham P, Chua S, McAlonan G. 2010. Autistic disorders and schizophrenia: Related or remote? An anatomical likelihood estimation. PloS One 5(8):e12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HY, Chaiyakunapruk N, Wu DBC, Lee KKC, Chiou CF. 2014. Global economic burden of schizophrenia: A systematic review. Value Health 7(17):A767. [DOI] [PubMed] [Google Scholar]
- Choudhury K, McQuillin A, Puri V, Pimm J, Datta S, Thirumalai S, Krasucki R, Lawrence J, Bass NJ, Quested D, Crombie C, Fraser G, Walker N, Nadeem H, Johnson S, Curtis D, St Clair D, Gurling HM. 2007. A genetic association study of chromosome 11q 22–24 in two different samples implicates the FXYD6 gene, encoding phosphohippolin, in susceptibility to schizophrenia. Am J Hum Genet 80:664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, Debailleul V, Simon AM, Caterina D, Dufaure I, Malekzadeh K, Belova M, Luan JJ, Bouillot M, Sambucy JL, Primas G, Saumier M, Boubkiri N, Martin-Saumier S, Nasroune M, Peixoto H, Delaye A, Pinchot V, Bastucci M, Guillou S, Chevillon M, Sainz-Fuertes R, Meguenni S, Aurich-Costa J, Cherif D, Gimalac A, Van Duijn C, Gauvreau D, Ouellette G, Fortier I, Raelson J, Sherbatich T, Riazanskaia N, Rogaev E, Raeymaekers P, Aerssens J, Konings F, Luyten W, Macciardi F, Sham PC, Straub RE, Weinberger DR, Cohen N, Cohen D. 2002. Genetic and physiological data implicating the new hum an gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Nat Acad Sci 99:13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Westmoreland JJ, Bayazitov IT, Eddins D, Pani AK, Smeyne RJ, Yu J, Blundon JA, Zakharenko SS. 2014. Specific disruption of thalamic inputs to the auditory cortex in schizophrenia models. Science 344:1178–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Wang Y, Yuan J, Zhou X, Jiang D, Li J, Zhang Y, Yin H, Duan S. 2014. Meta-analyses of 10 polymorphisms associated with the risk of schizophrenia. Biomed Rep 2:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong S, van Eijk KR, Zeegers DW, Strengman E, Janson E, Veldink JH, van den Berg LH, Cahn W, Kahn RS, Boks MP. Ophoff RA; PGC Schizophrenia (GWAS) Consortium. 2012. Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur J Hum Genet 20(9):1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Martinez M, Sanders AR, Hou C, Saitou N, Kitano T, Mowry BJ, Crowe RR, Silverman JM, Levinson DF, Gejman PV. 2004. Polymorphisms in the trace amine receptor 4 (TRAR4) gene on chromosome 6q23.2 are associated with susceptibility to schizophrenia. Am J Hum Genet 75:624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaine J, Maucuer A, Barbet A, Manceau V, Deshommes J, Meary A, Szöke A, Schürhoff F, Llorca PM, Lancon C, Leboyer M, Jamain S. 2011. Genetic and moleculate exploration of UHMK1 in schizophrenic patients. Psychiatr Genet 21:315–318. [DOI] [PubMed] [Google Scholar]
- Franzek E, Beckmann H. 1998. Different genetic background of schizophrenia spectrum psychoses: A twin study. Am J Psychiatry 155(1): 76–83. [DOI] [PubMed] [Google Scholar]
- Frydecka D, Beszłej JA, Pawlak-Adamska E, Misiak B, Karabon L, Tomkiewicz A, Partyka A, Jonkisz A, Szewczuk-Bogusławska M, Zawadzki M, Kiejna A. 2015. CTLA4 and CD28 gene polymorphisms with respect to affective symptom domain in schizophrenia. Neuro-psychobiology 71:158–167. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Côté M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Néri C, Dubé MP, Samuels ME, Marineau C, Stone EA, Awadalla P, Barker PA, Carbonetto S, Drapeau P, Rouleau GA. S2D Team. 2010. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Nat Acad Sci 107:7863–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva L, Moskvina V, Peirce T, Norton N, Bray NJ, Jones L, Holmans P, Macgregor S, Zammit S, Wilkinson J, Williams H, Nikolov I, Williams N, Ivanov D, Davis KL, Haroutunian V, Buxbaum JD, Craddock N, Kirov G, Owen MJ, O’Donovan MC. 2006. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Nat Acad Sci 103:12469–12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, Uzunov D, Costa E. 2000. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: A postmortem brain study. Arch Gen Psychiat 57:1061–1069. [DOI] [PubMed] [Google Scholar]
- Gysin R, Kraftsik R, Sandell J, Bovet P, Chappuis C, Conus P, Deppen P, Preisig M, Ruiz V, Steullet P, Tosic M, Werge T, Cuénod M, Do KQ. 2007. Impaired glutathione synthesis in schizophrenia: Convergent genetic and functional evidence. Proc Nat Acad Sci 104(42): 16621–16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. 2005. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry 2005(10):40–68. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Yamamori H, Kamino K, Morihara T, Iwase M, Kazui H, Takeda M. 2013. The KCNH2 gene is associated with neurocognition and the risk of schizophrenia. World J Biol Psychiatry 14(2):114–120. [DOI] [PubMed] [Google Scholar]
- Hommer RE, Swedo SE. 2015. Schizophrenia and autism—Related disorders. Schizophr Bull 41(2):313–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Hikita T, Taya S, Uraguchi-Asaki J, Toyo-oka K, Wynshaw-Boris A, Ujike H, Inada T, Takao K, Miyakawa T, Ozaki N, Kaibuchi K, Iwata N. 2008. Identification of YWHAE, a gene encoding 14–3-3epsilon, as a possible susceptibility gene for schizophrenia. Hum Molec Genet 17:3212–3222. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kinoshita Y, Okochi T, Kawashima K, Kushima I, Ito Y, Nakamura Y, Kishi T, Okumura T, Fukuo Y, Williams HJ, Hamshere ML, Ivanov D, Inada T, Suzuki M, Hashimoto R, Ujike H, Takeda M, Craddock N, Kaibuchi K, Owen MJ, Ozaki N, O’Donovan MC, Iwata N. 2011. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry 69(15):472–478. [DOI] [PubMed] [Google Scholar]
- Ingason A, Giegling I, Cichon S, Hansen T, Rasmussen HB, Nielsen J, Jürgens G, Muglia P, Hartmann AM, Strengman E, Vasilescu C, Mühleisen TW, Djurovic S, Melle I, Lerer B, Möller HJ, Francks C, Pietiläinen OP, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Walshe M, Vassos E, Di Forti M, Murray R, Bonetto C, Tosato S, Investigators GROUP, Cantor RM, Rietschel M, Craddock N, Owen MJ, Peltonen L, Andreassen OA, Nöthen MM, St Clair D, Ophoff RA, O ‘Donovan MC, Collier DA, Werge T, Rujescu D. 2010. A large replication study and meta-analysis in European samples provides further support for associaiton of AHI1 markers with schizophrenia. Hum Molec Genet 19:1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460(7256):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KJ, Fanous AH, Chen J, Kendler KS, Chen X. 2013. Variants in the 15q25 gene cluster are associated with risk for schizophrenia and bipolar disorder. Psychiatr Genet 23(1):20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, International Schizophrenia Consortium, Zhao Z. 2012. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput Biol 8(7):e1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao A, Müller D. 2013. Genetics of antipsychotic-induced weight gain: Update and current perspectives. Pharmacogenomics 14(16):2067–2083. [DOI] [PubMed] [Google Scholar]
- Kao HT, Porton B, Czernik AJ, Feng J, Yiu G, Haring M, Benfenati F, Greengard P. 1998. A third member of the synapsin gene family. Proc Nat Acad Sci 95:4667–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, Law AJ. 2010. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci USA 107(35):15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh DH, Tansey KE, O’Donovan MC, Owen MJ. 2015. Schizophrenia genetics: Emerging themes for a complex disorder. Mol Psychiatry 20(1):72–76. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Diehl SR. 1993. The genetics of schizophrenia: A current, genetic-epidemiologic perspective. Schizophr Bull 19:261–285. [DOI] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O’Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. 2008. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet 17(3):458–465. [DOI] [PubMed] [Google Scholar]
- Knight HM, Pickard BS, Maclean A, Malloy MP, Soares DC, McRae AF, Condie A, White A, Hawkins W, McGhee K, van Beck M, MacIntyre DJ, Starr JM, Deary IJ, Visscher PM, Porteous DJ, Cannon RE, St Clair D, Muir WJ, Blackwood DH. 2009. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am J Hum Genet 85(6):833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Ikeda M, Kajio Y, Saito T, Iwayama Y, Aleksic B, Yamada K, Toyota T, Hattori E, Ujike H, Inada T, Kunugi H, Kato T, Yoshikawa T, Ozaki N, Iwata N. 2013. Genetic variants on 3q21 and in the Sp8 transcription factor gene (SP8) as susceptibility loci for psychotic disorders: A genetic association study. PLoS One 8(8):e70964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushima I, Aleksic B, Ikeda M, Yamanouchi Y, Kinoshita Y, Ito Y, Nakamura Y, Inada T, Iwata N, Ozaki N. 2010. Association study of bromodomain-containing 1 gene with schizophrenia in Japanese population. Am J Med Genet B Neuropsychiatr Genet 153B:786–791. [DOI] [PubMed] [Google Scholar]
- Lauriat TL, Dracheva S, Kremerskothen J, Duning K, Haroutunian V, Buxbaum JD, Hyde TM, Kleinman JE, McInnes LA. 2006. Characterization of KIAA0513, a novel signaling molecule that interacts with modulators of neuroplasticity, apoptosis, and the cytoskeleton. Brain Res 1121:1–11. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. 2009. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet 373(9659):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel AC, Vaags AK, Sato D, Gazzellone MJ, Mitchell EB, Chen HY, Costain G, Walker S, Egger G, Thiruvahindrapuram B, Merico D, Prasad A, Anagnostou E, Fombonne E, Zwaigenbaum L, Roberts W, Szatmari P, Fernandez BA, Georgieva L, Brzustowicz LM, Roetzer K, Kaschnitz W, Vincent JB, Windpassinger C, Marshall CR, Trifiletti RR, Kirmani S, Kirov G, Petek E, Hodge JC, Bassett AS, Scherer SW. 2013. Rare exonic deletions implicate the synaptic organizer gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum Molec Genet 22:2055–2066. [DOI] [PubMed] [Google Scholar]
- Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, Aggerbeck L, Pupko T, Gorwood P, Simonneau M, Moalic JM. 2010. SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum Molec Genet 19:2841–2857. [DOI] [PubMed] [Google Scholar]
- Luo XJ, Huang L, Oord EJ, Aberg KA, Gan L, Zhao Z, Yao YG. 2014. Common variants in the MKL1 gene confer risk of schizophrenia. Schizophr Bull 41(3):715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G, Alford T, Xiong L, Rouleau GA, Tibbo PG, Cox DW. 2010. Association of NPAS3 exonic variation with schizophrenia. Schizophr Res 120(1–3)):143–149. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Straub RE, Marenco S, Nicodemus KK, Matsumoto S, Fujikawa A, Miyoshi S, Shobo M, Takahashi S, Yarimizu J, Yuri M, Hiramoto M, Morita S, Yokota H, Sasayama T, Terai K, Yoshino M, Miyake A, Callicott JH, Egan MF, Meyer-Lindenberg A, Kempf L, Honea R, Vakkalanka RK, Takasaki J, Kamohara M, Soga T, Hiyama H, Ishii H, Matsuo A, Nishimura S, Matsuoka N, Kobori M, Matsushime H, Katoh M, Furuichi K, Weinberger DR. 2008. The evolutionarily conserved Gprotein-coupled receptor SREB2/GPR85 influences brain size, behavior, and vulnerability to schizophrenia. Proc Nat Acad Sci 105:6133–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Dammann O. 2011. Schizophrenia and autism: Both shared and disorder-specific pathogenesis via perinatal inflammation?. Pediatr Res 69:26R–33R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH, Egan MF, Huffaker SS, Mattay VS, Kolachana B, Kleinman JE, Weinberger DR. 2007. Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J Clin Invest 117:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thom son PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. 2005. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 310:1187–1191. [DOI] [PubMed] [Google Scholar]
- Millar JK, James R, Brandon NJ, Thomson PA. 2004. DISC1 and DIS C2: Discovering and dissecting molecular mechanisms underlying psychiatric illness. Ann Med 36(5):367–378. [DOI] [PubMed] [Google Scholar]
- Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, McKenna PJ, Jones PB, Arai H, Starkey M, Emson PC, Bahn S. 2002. Gene expression analysis in schizophrenia: Reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Nat Acad Sci 99:4680–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. 2000. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 28:53–67. [DOI] [PubMed] [Google Scholar]
- Moens LN, Ceulemans S, Alaerts M, Van Den Bossche MJ, Lenaerts AS, De Zutter S, Norrback KF, Adolfsson R, Del-Favero J. 2010. PCM1 and schizophrenia: A replication study in the northern Swedish population. Am J Med Genet B Neuropsychiatr Genet 153B:1240–1243. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sklar P. 2015. Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Curr Opin Neurobiol 30:131–138. [DOI] [PubMed] [Google Scholar]
- Novak G, Seeman P, Tallerico T. 2000. Schizophrenia: Elevated mRNA for calcium-calmodulin-dependent protein kinase II-beta in frontal cortex. Molec Brain Res 82:95–100. [DOI] [PubMed] [Google Scholar]
- Ocklenburg S, Beste C, Güntürkün O. 2013. Handedness: A neurogenetic shift of perspective. Neurosci Biobehav Rev 37(10):2788–2793. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yasuda Y, Yoshida T, Takahashi H, Iike N, Iwase M, Kamino K, Ishii R, Kazui H, Fukumoto M, Takamura H, Yamamori H, Azechi M, Ikezawa K, Tanimukai H, Tagami S, Morihara T, Okochi M, Yamada K, Numata S, Ikeda M, Tanaka T, Kudo T, Ueno S, Yoshikawa T, Ohmori T, Iwata N, Ozaki N, Takeda M. 2010. The chitinase 3-like 1 gene and schizophrenia: Evidence from a multi-center case-control study and meta-analysis. Schizophr Res 116(2–3):126–132. [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Middleton FA, Tahl AR, Rockwell GN, Purcell S, Aldinger KA, Kirby A, Morley CP, McGann L, Gentile KL, Waggoner SG, Medeiros HM, Carvalho C, Macedo A, Albus M, Maier W, Trixler M, Eichhammer P, Schwab SG, Wildenauer DB, Azevedo MH, Pato MT, Pato CN, Daly MJ, Sklar P. 2005. Genetic investigation of chromosome 5q GABAA receptor subunit genes in schizophrenia. Mol Psychiatry 10(12):1074–1088. [DOI] [PubMed] [Google Scholar]
- Pitcher GM, Kalia LV, Ng D, Goodfellow NM, Yee KT, Lambe EK, Salter MW. 2011. Schizophrenia susceptibility pathway neuregulin 1-ErbB4 suppresses Src upregulation of NMDA receptors. Nature Med 17:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G, Macciardi F. 2009. Gene discovery through imaging genetics: Identification of two novel genes associated with schizophrenia. Mol Psychiatry 14(4):416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J, McGuffin P, Goldin LR, Shaskan EG, Gershon ES. 1984. Platelet monoamine oxidase (MAO) activity: Evidence for a single major locus. Am J Hum Genet 36:36–43. [PMC free article] [PubMed] [Google Scholar]
- Riley B, Kendler K. 2006. Molecular genetic studies of schizophrenia. Europ J Hum Genet 14(6):669–680. [DOI] [PubMed] [Google Scholar]
- Riley B, Williamson M, Collier D, Wilkie H, Makoff A. 2002. A 3-Mb map of a large segmental duplication overlapping the alpha-7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13-q14. Genomics 79:197–209. [DOI] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Hol-mans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O’Neill FA, Owen MJ, Milovancevic MP, Posthum a D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Multicenter Genetic Studies of Schizophrenia Consortium, Levinson DF, Gejman PV, Kendler KS, Laurent C, Mowry BJ, O’Donovan MC, Owen MJ, Pulver AE, Riley BP, Schwab SG, Wildenauer DB, Dudbridge F, Holmans P, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, O’Neill FA, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Psychosis Endophenotypes International Consortium, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Lin K, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Powell J, Rujescu D, Van Os J, Walshe M, Weisbrod M, Wiersma D, Wellcome Trust Case Control Consortium, Donnelly P, Barroso I, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, Barroso I, Deloukas P, Mathew CG, Blackwell JM, Brown MA, Corvin AP, McCarthy MI, Spencer CC, Bramon E, Corvin AP, O’Donovan MC, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. 2013. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45(8): 1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Xu B, Roos JL, Abecasis GR, Gogos JA, Karayiorgou M. 2014. Fine mapping on chromosome 13q 32–34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropscyhopharmacology 39(4):934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Haroutunian V. 2014. Schizophrenia: Susceptibility genes and oligodendroglial and myelin related abnormalities. Front Cell Neurosci 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti E, Bocchio-Chiavetto L, Valsecchi Pl, Scassellati C, Pasqualetti P, Bonvicini C, Corsini P, Rossi G, Cesana BM, Barlati S, Gennarelli M. 2007. G308A tum or necrosis factor alpha functional polymorphism and schizophrenia risk: Meta-analysis plus association study. Brain Behav Immun 21:450–457. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Göring HH, Duan J, Drigalenko EI, Moy W, Freda J, He D, Shi J, MGS, Gejman PV. 2013. Transcriptome study of differential expression in schizophrenia. Hum Molec Genet 22(24):5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviouk V, Moreau MP, Tereshchenko IV, Brzustowicz LM. 2007. Association of synapsin 2 with schizophrenia in families of Northern European ancestry. Schizophr Res 96(1–3):100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. 2011. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 43(10):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, McGowan-Jordan J, Schmid M. 2012. ISCN 2013: An international system for human cytogenetic nomenclature (2013). Unionville, CT: Karger Medical and Scientific Publishers. [Google Scholar]
- Shi J, Badner JA, Gershon ES, Liu C. 2008. Genetic associations with schizophrenia: Meta-analyses of 12 candidate genes. Schizophr Res 104:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. 2009. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460(7256):753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J, Zhang F, Chen J, Zhou G, Ji W, Li B, Xu Y, Liu D, Wang P, Yang P, Liu B, Sun W, Wan C, Qin S, He G, Steinberg S, Cichon S, Werge T, Sigurdsson E, Tosato S, Palotie A, Nöthen MM, Rietschel M, Ophoff RA, Collier DA, Rujescu D, Clair DS, Stefansson H, Stefansson K, Ji J, Wang Q, Li W, Zheng L, Zhang H, Feng G, He L. 2011. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet 43(12):1224–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi L, De Luca A, Bellacchio E, Conti E, Pasini A, Paloscia C, Spalletta G, Caltagirone C, Pizzuti A, Dallapiccola B. 2004. Mutations of the Nogo-66 receptor (RTN4R) gene in schizophrenia. Hum Mutat 24(6):534–535. [DOI] [PubMed] [Google Scholar]
- Smith T, Sharp S, Manzardo AM, Butler MG. 2015. Pharmacogenetics informed decision making in adolescent psychiatric treatment: A clinical case report. Int J Mol Sci 16(3):4416–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Böttcher Y, Olesen J, Breuer R, Möller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Réthelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Genetic Risk and Outcome in Psychosis (GROUP), Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jönsson EG, Terenius L, Agartz I, Petursson H, Nöthen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. 2009. Common variants conferring risk of schizophrenia. Nature 460:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. 2005. The genetics of schizophrenia. PLoS Med 2(7):e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. 2008. Candidate genes for schizophrenia: A survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet 147B(7):1173–1181. [DOI] [PubMed] [Google Scholar]
- Tabarés-Seisdedos R, Dumont N, Baudot A, Valderas JM, Climent J, Valencia A, Crespo-Facorro B, Vieta E, Gómez-Beneyto M, Martínez S, Rubenstein JL. 2011. No paradox, no progress: Inverse cancer comorbidity in people with other complex diseases. Lancet Oncol 12:604–608. [DOI] [PubMed] [Google Scholar]
- Takata A, Iwayama Y, Fukuo Y, Ikeda M, Okochi T, Maekawa M, Toyota T, Yamada K, H attori E, Ohnishi T, Toyoshima M, Ujike H, Inada T, Kunugi H, Ozaki N, Nanko S, Nakamura K, Mori N, Kanba S, Iwata N, Kato T, Yoshikawa T. 2013. A population-specific uncommon variant in GRIN3A associated with schizophrenia. Biol Psychiatry 73(6):532–539. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Thom son AM, Dunbar DR, Grassie MA, Condie A, Walker MT, Smith DJ, Pulford DJ, Muir W, Blackwood DH, Porteous DJ. 2005. Sex-specific association between bipolar affective disorder in women and GPR50, and X-linked orphan G protein-coupled receptor. Molec Psychiat 10:470–478. [DOI] [PubMed] [Google Scholar]
- Tosic M, Ott J, Barral S, Bovet P, Deppen P, Gheorghita F, Matthey ML, Parnas J, Preisig M, Saraga M, Solida A, Timm S, Wang AG, Werge T, Cuénod M, Do KQ. 2006. Schizophrenia and oxidative stress: Glutamate cysteine ligase modifier as a susceptibility gene. Am J Hum Genet 79:586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulouse A, Rochefort D, Roussel J, Joober R, Rouleau GA. 2003. Molecular cloning and characterization of human RAI1, a gene associated with schizophrenia. Genomics 82:162–171. [DOI] [PubMed] [Google Scholar]
- Toyota T, Yoshitsugu K, Ebihara M, Yamada K, Ohba H, Fukasawa M, Minabe Y, Nakamura K, Sekine Y, Takei N, Suzuki K, Itokawa M, Meerabux JM, Iwayama-Shigeno Y, Tomaru Y, Shimizu H, Hattori E, Mori N, Yoshikawa T. 2004. Association between schizophrenia with ocular misalignment and polyalanine length variation in PMX2B. Hum Mol Genet 13(5):551–561. [DOI] [PubMed] [Google Scholar]
- Uezato A, Kimura-Sato J, Yamamoto N, Iijima Y, Kunugi H, Nishikawa T. 2012. Further evidence for a male-selective genetic association of synapse-associated protein 97 (SAP97) genet with schizophrenia. Behav Brain Funct 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, Iakoucheva LM, Krastoshevsky O, Krause V, Larach-Walters V, Welsh DK, Craig D, Kelsoe JR, Gershon ES, Leal SM, Dell Aquila M, Morris DW, Gill M, Corvin A, Insel PA, McClellan J, King MC, Karayiorgou M, Levy DL, DeLisi LE, Sebat J. 2011. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schijndel JE, van Loo KMJ, van Zweeden M, Djurovic S, Andreassen OA, Hansen T, Werge T, Kallunki P, Pedersen JT, Martens GJ. 2009. Three-cohort targeted gene screening reveals a non-synonymous TRKA polymorphism associated with schizophrenia. J Psychiatr Res 43:1195–1199. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu XF, Aragam N. 2010. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res 124:192–199. [DOI] [PubMed] [Google Scholar]
- Wang S, Detera-Wadleigh SD, Coon H, Sun CE, Goldin LR, Duffy DL, Byerley WF, Gershon ES, Diehl SR. 1996. Evidence of linkage disequilibrium between schizophrenia and the SCA1 CAG repeat on chromosome 6p23. Am J Hum Genet 59:731–736. [PMC free article] [PubMed] [Google Scholar]
- Weber H, Klamer D, Freudenberg F, Kittel-Schneider S, Rivero O, Scholz CJ, Volkert J, Kopf J, Heupel J, Herterich S, Adolfsson R, Alttoa A, Post A, Grußendorf H, Kramer A, Gessner A, Schmidt B, Hempel S, Jacob CP, Sanjuán J, Moltö MD, Lesch KP, Freitag CM, Kent L, Reif A. 2014. The genetic contribution of the NO system at the glutamatergic post-synapse to schizophrenia: Further evidence and meta-analysis. Eur Neuropsychopharmacol 24(1):65–85. [DOI] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, M orris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Molecular Genetics of Schizophrenia Collaboration (MGS) International Schizophrenia Consortium (ISC), SGENE-plus GROUP, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O’Donovan MC. 2011. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry 16:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, McGuffin P, Nothen M, Owen MJ. 1997. Meta-analysis of association betwen the 5-HT2a receptor T102C polymorphism and schizophrenia. Lancet 349:1221. [DOI] [PubMed] [Google Scholar]
- Williams J, Spurlock G, Holmans P, Mant R, Murphy K, Jones L, Cardno A, Asherson P, Blackwood D, Muir W, Meszaros K, Aschauer H, Mallet J, Laurent C, Pekkarinen P, Seppala J, Stefanis CN, Papadimitriou GN, Macciardi F, Verga M, Pato C, Azevedo H, Crocq MA, Gurling H, Owen MJ. 1998. A meta-analysis and transmission disequilibrium study of association between the dopamine D3 receptor gene and schizophrenia. Mol Psychiatry 3:141–149. [DOI] [PubMed] [Google Scholar]
- Wockner LF, Noble EP, Lawford BR, Young RM, Morris CP, Whitehall VL, Voisey J. 2014. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry 4(1):e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2008. The global burden of disease: 2004 update. Geneva, Switzerland: World Health Organization. [Google Scholar]
- Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. 2013. Potential impact of miR-137 and its targets in schizophrenia. Front Genet 4:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Aragam N, Li X, Villla EC, Wang L, Briones D, Petty L, Posada Y, Arana TB, Cruz G, Mao C, Camarillo C, Su BB, Escamilla MA, Wang K. 2013. BCL9 and C9orf5 are associated with negative symptoms in schizophrenia: Meta-analysis of two genome-wide association studies. PLos One 8(1):e51674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, He L. 2010. Covergent evidence shows a positive association of interleukin-1 gene complex locus with susceptibility to schizophrenia in the Caucasian population. Schizophr Res 120:131–142. [DOI] [PubMed] [Google Scholar]
- Yuan A, Yi Z, Sun J, Du Y, Yu T, Zhang C, Liu Y, Zhou Y, Liu D, Li H, Xu Y, Cheng Z, Li W, Yu S. 2012. Effect of SOX10 gene polymorphism on early onset schizophrenia in Chinese Han population. Neurosci Lett 521:93–97. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liao G, Liu C, Sun L, Liu Y, Wang Y, Jiang Z, Wang Z. 2011. The association of CLOCK gene T3111C polymorphism and hPER3 gene 54-nucleotide repeat polymorphism with Chinese Han people schizophrenics. Mol Biol Rep 38(1):349–354. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lu S, Meng L, Min Z, Tian J, Valenzuela RK, Guo T, Tian L, Zhao W, Ma J. 2012. Genetic evidence for the association between the early growth response 3 (ERG3) gene and schizophrenia. PLoS ONE 7:e30237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras E 2006. C677T and A1298C methylenetetrahydrofolate reductase gene polymorphisms in schizophrenia, bipolar disorder and depression: A meta-analysis of genetic association studies. Psychiatr Genet 16:105–115. [DOI] [PubMed] [Google Scholar]