Abstract
The aim of this study was to correlate the status of the KLF6 tumor suppressor gene including loss of heterozygozity (LOH), mutation and alternative splicing in human pancreatic cancer with tumor grade and survival.
Whereas neither KLF6 loss nor mutation was identified, expression of the KLF6 alternative splice forms was significantly increased in pancreatic tumor samples and cell lines. These cancers demonstrated marked cytoplasmic KLF6 expression, consistent with over-expression and accumulation of KLF6 splice form(s), which lack a nuclear localization signal. In addition, KLF6 splicing correlated significantly with tumor stage and survival.
In summary, pancreatic cancer displays a novel pattern of KLF6 dysregulation through selectively increased expression of KLF6 splice variants. Therefore, determination of KLF6 mRNA splicing levels may represent a novel biomarker predicting prognosis.
Keywords: Pancreatic cancer, krüppel-like factor 6, tumor suppressor gene
INTRODUCTION
Krüppel-like factor 6 (KLF6) is a member of the Krüppel-like factor family of transcription factors; this family contains at least twenty members, which are defined by their common 81 amino acid C-terminal DNA-binding domain (1–3). Krüppel-like factors exhibit a remarkable range of activities regulating cell growth and differentiation in virtually all tissues (4, 5). They may function either as transcriptional activators or repressors, depending on the cell type and promoter context (4, 5).
The KLF6 gene was originally cloned from human placenta (6) and activated rat hepatic stellate cells (7). Chromosomal deletion of the region containing the KLF6 locus (10p15) in prostate cancer (8), combined with its characterization as a growth suppressor, led to the identification of KLF6 as a tumor suppressor gene frequently inactivated in prostate cancer (8). Growth suppressive mechanisms of KLF6 include transcriptional induction of p21 in a p53-independent manner (8), upregulation of TGFβ1 and its receptors (9), inactivation of c-jun (10), inhibition of other proto-oncogene signaling pathways (11), and sequestration of cyclin D1 (12). In addition to prostate cancer (8, 13), inactivation of KLF6 by loss and/or mutation has now been identified in several other cancers, including gastric (14), colorectal (15), hepatocellular (16), and ovarian carcinoma (17). In addition, downregulation of KLF6 mRNA has been identified in primary non small cell lung carcinoma (18), and reduced expression of KLF6 mRNA is associated with worse prognosis in prostate cancer (19).
We recently described an additional mechanism of KLF6 inactivation through the generation of alternative splice products of the KLF6 gene that are over-expressed in prostate and ovarian cancers (17, 20). These splice forms inhibit the function of the wild-type, full length protein, even in the absence of KLF6 loss or inactivating mutation (20). Mechanisms underlying generation of these splice forms in cancer have not been fully clarified, however a pathway in somatic cells has been identified in which presence of a G to A polymorphism in the first intron of the KLF6 gene leads to increased splicing that is associated with enhanced risk of prostate cancer (20). Moreover, splicing is correlated with Ras oncogene activation in hepatocellular carcinoma, which directly increases KLF6 splicing in cultured cells (21). Abrogation of splice form expression through use of specific siRNAs reduces cell growth in culture and prostate tumor xenografts, thereby confirming their growth-promoting, tumorigenic activity (17, 22). The net activity of KLF6 is thus represented by the relative expression of full length to splice form mRNA, which can be expressed simply as the ratio of KLF6wt/KLF6 splice form mRNA, as determined by both quantitative real time PCR and western blotting.
Carcinoma of the pancreas is among the most lethal of solid organ tumors. The disease typically presents late, when curative resection is not possible (23). Moreover, no specific biomarkers have been identified to enable early diagnosis (24). In addition, advanced pancreatic cancer is generally resistant to chemotherapy (25), and a response rate of only one-quarter or less can be expected with standard agents like gemcitabine (26). The molecular mechanisms underlying the development of chemotherapy resistance in pancreatic cancer are not clear.
In the present study we have examined KLF6 gene expression in pancreatic cancer. Specifically, we have characterized the frequency of loss of heterozygosity (LOH), mutation and alternative splicing in 24 well-characterized primary pancreatic adenocarcinoma samples and correlated these findings with clinically relevant disease endpoints.
MATERIALS AND METHODS
Clinical data
Pancreatic cancer tissues were obtained from 24 patients (13 female, 11 male) undergoing a pylorus-preserving Whipple resection due to pancreatic ductal adenocarcinoma. In all patients an R0 resection was performed. Normal pancreatic tissues were obtained from 8 individuals (3 female, 5 male) through an organ donor program. Immediately following surgical removal, all tissue samples were either fixed in formaldehyde or frozen in liquid nitrogen. All cancer tissue samples were graded independently by a pathologist, and classified histologically as ductal adenocarcinoma of the pancreas. All clinical data in Berne and Heidelberg were registered in a prospective database between January 1995 and December 2003 (Table 1).
Table 1: Clinical data.
Clinical data and mRNA KLF6wt/sp in patients with pancreatic carcinoma (n=24) and normal pancreas* (n=8). G = grading; pT, pN, pM according to the TNM system; R = classification of residual tumor.
| Patient | Gender | Age | G | UICC | pT | pN | pM | R | Survival [months] | KLF6wt/sp mRNA |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 68 | 3 | II | 3 | 0 | 0 | 0 | 10 | 1.85 |
| 2 | M | 69 | 3 | II | 3 | 0 | 0 | 0 | 7 | 1.75 |
| 3 | M | 66 | 3 | III | 4 | 1 | 0 | 0 | 10 | 1.85 |
| 4 | M | 53 | 1 | I | 1 | 0 | 0 | 0 | 28 | 2.33 |
| 5 | W | 66 | 2 | III | 3 | 1 | 0 | 0 | 69 | 2.63 |
| 6 | M | 70 | 2 | III | 3 | 1 | 0 | 0 | 6 | 2.04 |
| 7 | W | 77 | 2 | III | 4 | 1 | 0 | 0 | 4 | 1.32 |
| 8 | M | 75 | 1 | I | 2 | 0 | 0 | 0 | 54 | 3.57 |
| 9 | M | 61 | 3 | II | 4 | 0 | 0 | 0 | 18 | 2.38 |
| 10 | W | 71 | 2 | IV | 4 | 1 | 1 | 0 | 11 | 2.27 |
| 11 | W | 60 | 2 | III | 3 | 1 | 0 | 0 | 9 | 2.27 |
| 12 | W | 52 | 3 | III | 4 | 1 | 0 | 0 | 5 | 1.75 |
| 13 | M | 72 | 1 | III | 2 | 1 | 0 | 0 | 30 | 2.63 |
| 14 | M | 75 | 1 | II | 3 | 0 | 0 | 0 | - | 2.17 |
| 15 | W | 52 | 1 | I | 2 | 0 | 0 | 0 | 19 | 2.13 |
| 16 | M | 76 | 2 | III | 2 | 1 | 0 | 0 | 17 | 2.08 |
| 17 | W | 83 | 1 | I | 2 | 0 | 0 | 0 | 27 | 3.57 |
| 18 | W | 73 | 2 | III | 4 | 1 | 0 | 0 | 27 | 2.50 |
| 19 | W | 52 | 3 | III | 2 | 1 | 0 | 0 | 8 | 0.48 |
| 20 | W | 56 | 2 | III | 2 | 1 | 0 | 0 | 11 | 1.52 |
| 21 | W | 64 | 1 | II | 3 | 0 | 0 | 0 | 21 | 2.08 |
| 22 | M | 57 | 3 | III | 3 | 1 | 0 | 0 | 11 | 1.59 |
| 23 | W | 69 | 3 | III | 3 | 1 | 0 | 0 | 9 | 1.82 |
| 24 | W | 81 | 3 | III | 3 | 1 | 0 | 0 | 6 | 1.39 |
| 25* | W | 50 | 5.50 | |||||||
| 26* | M | 38 | 1.61 | |||||||
| 27* | M | 43 | 3.45 | |||||||
| 28* | M | 25 | 4.00 | |||||||
| 29* | F | 45 | 2.94 | |||||||
| 30* | F | 50 | 3.45 | |||||||
| 31* | M | 39 | 5.26 | |||||||
| 32* | M | 41 | 2.63 |
The median age of the patients undergoing a pancreaticoduodenectomy for pancreatic cancer was 68 years (range: 52–83 years). The median age of the normal pancreatic organ donors was 42 years (range: 25–50 years). According to the Classification of the UICC (International Union Against Cancer), there were four patients with stage I tumors, five with stage II, 14 with stage III, and one with stage IV tumors. Tumor grading (27) was well differentiated in 7 cases, moderately differentiated in 8 cases, and undifferentiated in 9 cases. The median survival in the group of patients with pancreatic carcinoma was 11 months (range: 8–27); (Table 2). The study was approved by the Ethics Committee of the University of Berne, Switzerland, and the University of Heidelberg, Germany.
Table 2: Correlation of clinical data with mRNA KLF6wt/sp.
Clinical data and ratio of mRNA KLFwt/sp expression were correlated and statistical analysis was performed using the Spearman test. Additionally, Mann Whitney Test was used to analyze the differences between KLF6 ratio of normal patients and cancer patients.
| Characteristics | Normal pancreas KLF6wt/sp (avg) | Pancreas cancer KLF6wt/sp (avg) | P-value |
|---|---|---|---|
| Patients (n) | 3.45(8) | 2.08 (24) | 0.002 |
| Grading | 0.001 | ||
| 1 (n) | - | 2.33 (7) | |
| 2 (n) | - | 2.18 (8) | |
| 3 (n) | - | 1.75 (9) | |
| Tumor stage | 0.076 | ||
| I (n) | - | 2.95 (4) | |
| II (n) | - | 2.08 (5) | |
| III (n) | - | 1.84 (14) | |
| IV (n) | 1.5 (1) | ||
| Survival | 0.0001 |
Pancreatic cancer cell lines
Seven human pancreatic carcinoma cell lines were used: The moderately differentiated human pancreatic adenocarcinoma cell lines T3M4, Capan-1, BxPC-3, and the less differentiated human pancreatic carcinoma cell lines AsPC-1, Colo-357, PANC-1, and Mia PaCa-2 were obtained from the American Tissue Type Culture Collection (Rockville, MD). Cell lines were cultured in Dulbecco Modified Eagle Medium (DMEM; Life Technology, Rockville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Life Technology), penicillin G (100 units/mL), and streptomycin (100 μg/mL). Cells were grown as a monolayer culture at 37 °C in humidified air with 5% (Capan-1, Colo-357, AsPC-1, T3M4, BxPC-3) or 10% CO2 (Mia PaCa-2 and PANC-1). Unless otherwise indicated, all chemicals were purchased from Sigma Chemicals (St. Louis, MO).
Immunohistochemistry
Paraffin-embedded tissue sections (2–4 μm in thickness) were subjected to immunostaining using the Dako Envision + System (Dako Diagnostics AG, Zürich, Switzerland). Tissue sections for each tissue sample were deparaffinized with xylene and rehydrated through graded alcohol into distilled water. Endogenous peroxidase activity was quenched by incubating the slides in 0.03 % hydrogen peroxide and sodium azide, followed by washing in Tris-buffered saline. The sections were then incubated overnight at 4°C with rabbit polyclonal antibody KLF6 (sc7158, Santa Cruz Biotechnology, Santa Cruz, CA) diluted in 0.05 mol/L Tris-HCL buffer containing 1% bovine serum albumin. KLF6 antibodies recognizing the amino terminus region (amino acids 28–201) conserved in all KLF6 splice forms were raised in rabbits. Bound antibody was detected with a streptavidin-biotin-horseradish peroxidase (HRP) system (DAKO Diagnostics AG) in which slides were successively incubated with biotinylated antirabbit IgG, streptavidin-HRP, and 3–3′diaminobenzidine (DAB). To ensure antibody specificity, control slides were incubated either in the absence of primary antibody or with a nonspecific IgG antibody; immunostaining was not detected in either case. All slides were analyzed by two independent observers blinded to patient status. Any differences in the findings were resolved by joint review and consultation with a third observer.
Quantitative PCR for KLF6wt and KLF6sp in human pancreatic cancer tissue and cultered pancreas tumor cells
For quantifying target gene expression, RNA isolation from cultured cells and patient samples was performed using RNeasy Mini and Midi kits (Qiagen). All RNA was treated with DNase (Qiagen). RNA (1 μg) was reverse transcribed for each reaction using first-strand cDNA synthesis with random primers (Promega, Madison, WI). Real time PCR reactions were optimized to amplify either KLF6 wild type alone (KLF6wt), or total KLF6 mRNA (a primer designed and validated as previously described (20, 22) to detect both wtKLF6 and all KLF6 splice forms). Thus KLF6 splice form mRNA (KLF6sp) expression was calculated by determining the difference in absolute amount of these two PCR products. Thus, expression of KLF6wt (i.e., full length) mRNA and KLF6total mRNA (= KLF6wt + KLF6sp) were determined by quantitative real-time PCR using the following PCR primers on an ABI PRISM 7900HT Sequence Detection System (APPLIED Biosystems, Foster City, CA): KLF6wt forward 5’-CGG ACG CAC ACA GGA GAA AA-3’ and KLF6wt reverse 5’-CGG TGT GCT TTC GGA AGT G-3’; KLF6total forward 5’-CTG CCG TCT CTG GAG GAG T-3’ and KLF6total reverse 5’- TCC ACA GAT CTT CCT GGC TGT C-3’. All experiments were performed in triplicate and normalized to GAPDH mRNA expression. To calculate the fold change in KLF6sp, the fold change in total KLF6 was divided by the fold change in KLF6wt alone. The clinical data of the patients were correlated with the ratio of KLF6wt/KLF6sp mRNA expression. All quantitative real time PCR data shown in Results represent three independent real time PCR reactions that have each been performed in triplicate.
Loss of KLF6 heterozygosity (LOH) analysis in human pancreatic cancer
Fluorescent LOH analysis was performed using genomic microdissected DNA from matched normal/pancreatic cancer as previously described (8). Fluorescently labeled microsatellite markers flanking KLF6 and ordered according to the Marshfield map were generated. PCR was performed according to manufacturer’s suggestions (Perkin Elmer, Boston, MA); markers D10S591 and D10S594 that flank the KLF6 gene, as well as three KLF6 specific markers, KLF6M1, KLF6M2, and KLF6M4. Primer sequences for KLF6 specific markers were as follows: KLF6M1 F: 5’ GAG GGA GTG AGG CTT TCT GTT 3’; KLF6M1 R: 5’ TTT CCA GCC CAC TGT CTT CTT GAC 3’; KLF6M2 F: 5’ ATG GCC CTG ACT TCT 3’; KLF6M2 R: 5’TAC TTG CGG AGC GTG AGC C 3’; KLF6M4 F: GCA TTA AGA ATA GTG AAG GC 3’; KLF6M4 R: 5’ GAT GTG TTT GGC TCA GGG A 3’. The exponential range of the PCR was determined for each sample, and was between 30–38 cycles. The data were analyzed using the ABI Genescan and Genotyper software packages (Perkin Elmer) and allelic loss was scored by two independent observers as described before (16). In our system, a relative allele ratio of less than 0.7 was defined as loss of heterozygosity. The XLOH was confirmed at least twice for each marker. LOH analysis of the TP53 gene locus was performed as described above using three microsatellite markers flanking its locus: D17S796, D17S578 and D17S786.
Western Blot
Pancreatic cancer cell lines (n=7), human pancreatic cancer tissues (n=7) and human normal pancreas tissue (n=5) were homogenized in ice-cold suspension buffer (10 mM Tris-HCL, pH 7.6, 100 mM NaCl) containing a complete protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany). The homogenized material was collected and centrifuged at 4° C for 30 minutes at 14000 × g to remove the insoluble material. The protein concentration of the supernatant was measured by spectrophotometry using the BCA protein assay method (Pierce, Rockford, IL, USA). A total of 40 μg protein/lane was separated by SDS-polyacrylamide gel electrophoresis. After transfer to nitrocellulose membranes, blots were incubated with a polyclonal antibody against KLF6. After washing, blots were incubated with anti-rabbit IgG (Amersham International, Amersham, Bucks, UK) conjugated with horseradish peroxidase. Visualization was performed by the enhanced chemoluminescent method (Amersham International, Freiburg, Germany).
Statistical Analysis
The age and survival of all patients are reported as median and range. Furthermore, we analyzed the median KLF6wt/sp ratio according to the four tumor stages (UICC) and tumor grades (I, II, III) (27). Differences between the groups were analyzed using Kruskal-Wallis and Mann-Whitney-U tests for non-parametric data with p< 0.05 considered statistically significant. For the relationship between survival analysis and KLF6wt/sp, log-rank test was used. Additionally, in a Cox-regression analysis we tested KLF6wt/sp ratio together with stage, and grade as continuous covariates for an independent prognostic factor versus survival. Correlation of KLF6wt/sp with tumor grading, tumor stage and survival was analyzed by the Spearmans test. Bivariate analysis was done by Mann-Whitney test.
RESULTS
Enhanced cytoplasmic KLF6 expression in pancreatic carcinoma
Normal pancreas tissue
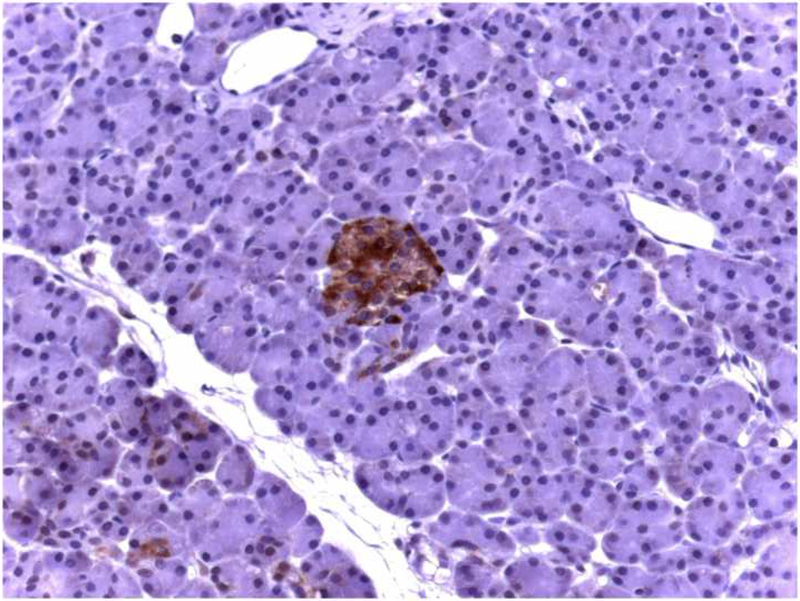
We first assessed the pattern of KLF6 expression in normal pancreatic tissue by immunostaining of paraffin sections using a KLF6 polyclonal antibody (n=15). KLF6-immunoreactivity was present in scattered islet cells, and weak staining was also apparent in ductal cells. No KLF6 immunostaining was apparent in acinar cells or pancreatic micro-vessels (Figure 1A).
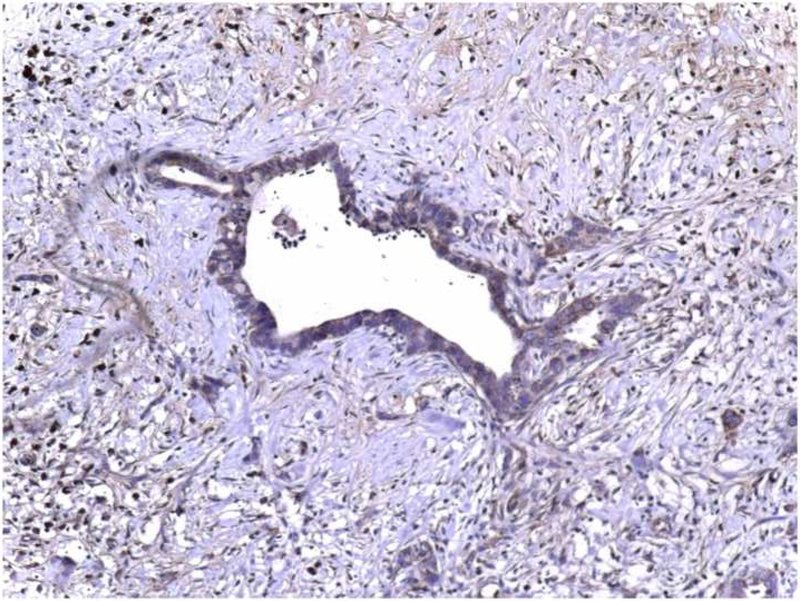
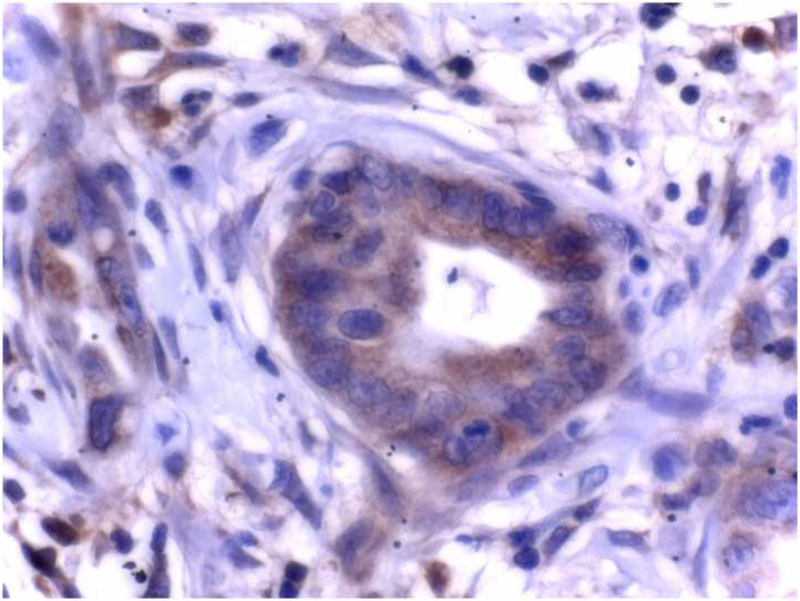
Figure 1: Localization of KLF6 in normal human pancreatic tissue and pancreatic cancer samples.
(A) Normal pancreas, displays KLF6 immunostaining in scattered islet cells, faint staining in ductal cells, but no staining in acinus cells (× 50). (B) Ductal adenocarcinoma of the pancreas, demonstrating scattered cytoplasmatic KLF6 immunoreactivity, with some stromal staining. (× 50). (C) Higher power immunostaining for KLF6 in a separate pancreatic adenocarcinoma demonstrating the cytoplasmic and stromal staining (× 400). Insert showing no immunoreactivity of ductal cancer cells in control tissue.
Ductal adenocarcinoma of the pancreas
In contrast to normal pancreas, ductal carcinoma cells displayed marked KLF6 immunoreactivity within the cytoplasm of tumor cells. Acinar cells and normal ducts within the cancer tissue were negative, and only some micro-vessels surrounding the tumor exhibited KLF6 immunoreactivity. Islet cells in the normal pancreas tissue within the cancer samples had similar staining as islets within normal controls. These findings were consistent among 15 samples analyzed, and a representative photomicrograph is shown (Figures 1B and 1C).
KLF6 protein isoforms in normal pancreas and pancreatic carcinoma
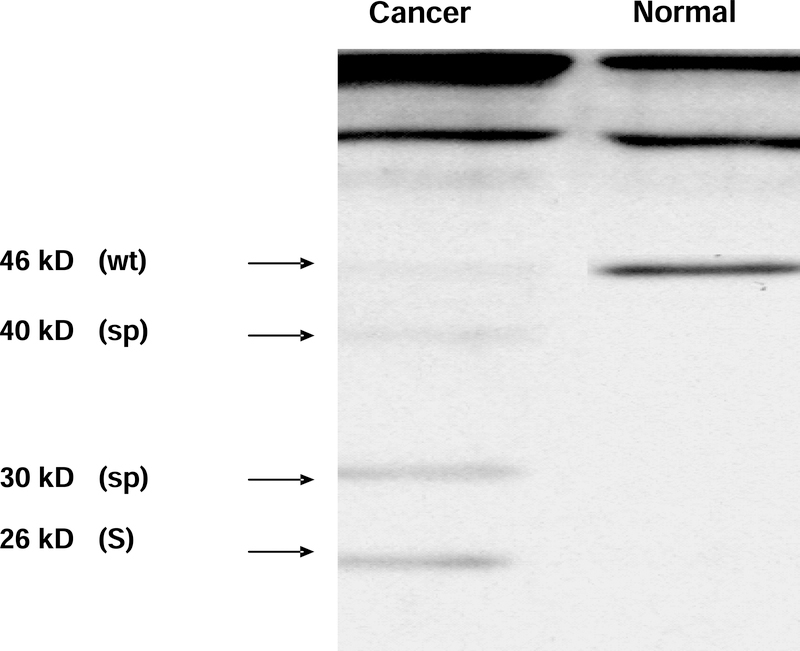
Given the marked over-expression of cytoplasmic KLF6 in pancreatic tumor samples we sought to specifically define which KLF6 isoforms were present in both normal and cancerous pancreatic tissue. Full length wild-type KLF6 (wtKLF6) typically migrates on Western blot as a single or double band at ~46 kD, whereas alternative splice products have lower molecular weights, with the predominant splice form, SV1, detectable as a 26 kD protein (20). Accordingly, we examined both normal pancreas and carcinoma for the presence and relative expression of the various KLF6 isoforms present in these tissues. In normal pancreas only KLF6 wild type (46 kD) was detected. In contrast, in human pancreatic cancer tissue, bands of 40, 30, and 26 kD were identified in addition to KLF6. These lower MW bands are consistent with the previously described alternatively spliced products of KLF6 (20) (Figure 2).
Figure 2: Expression of KLF6wt protein and KLF6 splice forms in normal human pancreatic tissue and pancreatic cancer samples.
Representative Western blots are shown with pancreatic cancer at the left column and normal pancreas at the right column. Pancreatic cancer samples revealing a faint protein band at 46 kD representing KLFwt and bands at 26 kD, 30 kD, and 40 kD representing known forms of KLF6. In contrast, KLF6 protein expression in normal pancreas showing only one strong band at 46 kD representing KLF6wt.
Loss of heterozygosity (LOH) analysis of KLF6
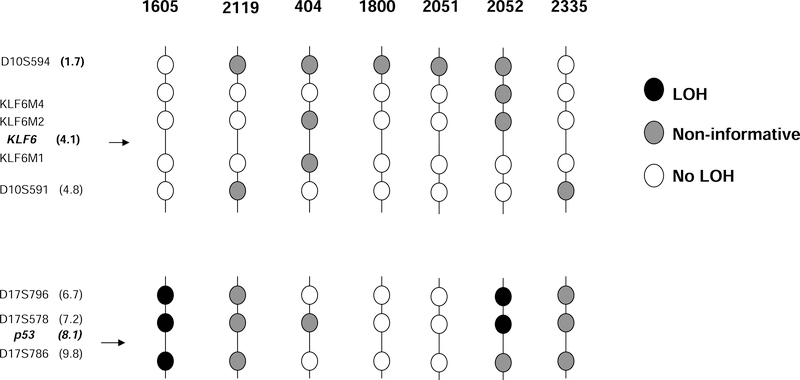
Our previous studies have demonstrated a widely variable frequency of LOH of the KLF6 locus in human cancers, depending on the tumor type. For example, in prostate cancer, ~70% LOH was detected (8), whereas in hepatocellular carcinoma there was 39% LOH (16). Interestingly none of the pancreatic carcinomas analyzed displayed LOH of the KLF6 locus. To validate the methodology, findings for KLF6 were compared to LOH of the p53 locus, which was present in two of the seven samples (Figure 3). This frequency of LOH at the p53 locus is considerably lower than that reported recently (28), although p53 LOH is typically more common in invasive, non-resectable pancreatic cancers (29). Moreover, LOH detection is greatly increased by microdissection, which was not performed in our study (30). However, genetic divergence of pancreatic cancer with respect to tumor suppressor gene alterations is common (31).
Figure 3: Loss of heterozygosity (LOH) of the KLF6 and p53 genes in human pancreatic cancer samples.
LOH of the KLF6 and p53 locus was analyzed in tumor tissue from seven patients using microsatellite markers (vertical axis, with patient # above each axis) from the 10p15 region and KLF6-specific markers KLF6M1, M2 and M4. These markers flank the KLF6 gene by approximately 40 Kb centromerically, 10 Kb and 20 Kb telomerically. The lower half displays microsatellite markers flanking the p53 gene. Black filled circle - LOH; gray - non-informative (NI); white circle - no evidence of loss.
Increased KLF6 alternative mRNA splicing in pancreatic cancers and cell lines
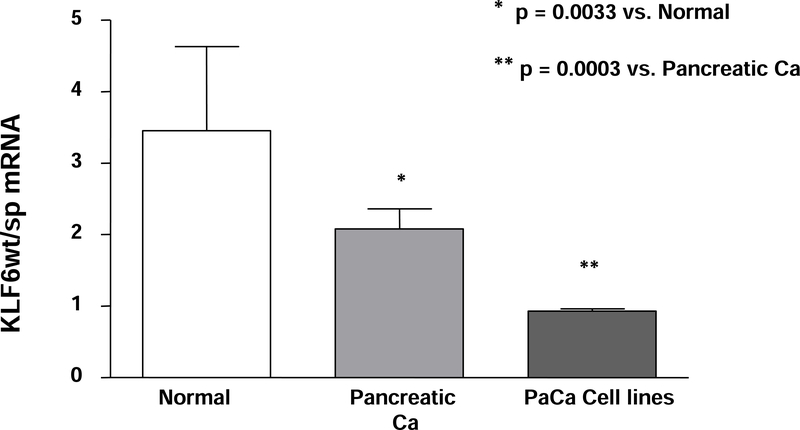
We used quantitative real time PCR method to compare KLF6 splicing in normal and malignant pancreatic tissues. The expression of KLF6wt and total mRNA splice forms were analyzed in 24 pancreatic cancers and in 8 normal pancreatic tissues. The real time PCR data, together with clinical characteristics of the patients are presented in Table 1. As shown in Table 2, in normal pancreas the median KLF6wt/sp ratio was 3.45 (range: 2.71–4.95) compared to 2.08 in pancreatic tumors (range: 1.75–2.37) (Table 2) (p=0.03). The reduced ratio of KLF6wt/sp was almost entirely due to higher expression of KLF6sp in the cancer samples.
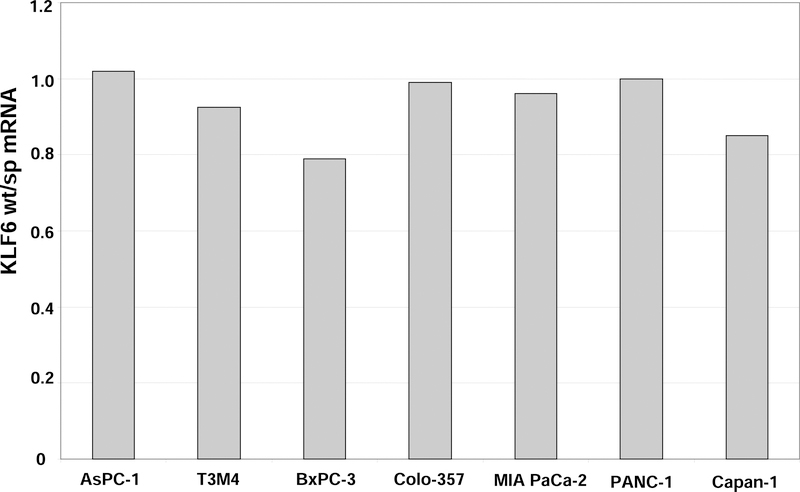
We also analyzed KLF6 alternative splicing in pancreatic cancer cell lines (AsPC-1, T3M4, BxPC-3, MIA PaCa-2, PANC-1, Capan-1). In all cell lines examined, the ratio of KLF6wt to KLF6sp was consistently reduced (Figure 4): BxPC-3: 0.79, Capan-1: 0.85, PANC-1: 1, and AsPC-1: 1.02). KLF6wt/sp in pancreatic cancer cell lines was significantly lower than in primary human pancreatic cancer or normal pancreatic tissue (p=0.003 and p=0.0003, respectively; Figure 4). These data indicate that KLF6 alternative splicing is increased in pancreatic cancer cell lines even more than in primary pancreatic cancers.
Figure 4:
(A) Increased KLF6wt/sp mRNA ratio in human pancreatic cancer cell lines
Quantitative real-time PCR of extracted total RNA from the seven human pancreatic cancer cell lines was performed as described in Materials and Methods. KLF6sp expression was calculated by determining the difference between KLF6 total mRNA and KLF6wt mRNA alone. All analyses were performed in triplicate and normalized to GAPDH mRNA expression, and values are markedly reduced compared to normal pancreatic tissue and primary pancreatic cancers (see Figure 4B).
(B): Decreasing ratio of KLF6wt/sp mRNA in pancreatic cancer and pancreatic cancer cell lines.
The mean ratio of KLF6wt/sp mRNA of 24 human pancreatic cancer tissues compared to specimens of normal human pancreas from 8 individuals. These data are compared to the median ratio of KLF6wt/sp mRNA from the 7 human pancreatic cancer cell lines in panel A. Error bars represent the SEM of three different experiments. Statistical analysis revealed a significantly higher ratio in normal pancreatic tissues versus pancreatic cancer tissues (p=0.0033) and pancreatic cancer tissues versus pancreatic cancer cell lines (p=0.0003), respectively.
Correlation between KLF6wt/sp ratio and clinical data
We examined the relationships between the ratio of wild type to splice form mRNA (KLF6wt/sp) and both tumor grade and clinical data in patients with pancreatic cancer. In well-differentiated cancer samples (G1), the median ratio of KLF6wt/sp was 2.33 (range: 2.13–3.57), compared to 2.18 (range: 1.65–2.44) in moderately differentiated tumors, (G2), and 1.75 (range: 1.49–1.83) in poorly differentiated cancer samples (G3). KLF6wt/sp was significantly related to the tumor grade (G1-G3), with more poorly differentiated tumors having a lower KLF6wt/sp ratio than well-differentiated tumors (p=0.001; Table 2). Additionally, bivariate analysis demonstrated a significant difference between KLF6wt/sp >/< 2 and grading (p<0.001).
We also examined the relationship between the relative ratio of KLF6wt/sp as a function of disease stage according to UICC criteria. In UICC stage I, the median KLF6 wt/sp ratio was 2.95 (range: 2.18–3.57); (median survival: 28 months), in UICC II, 2.08 (range: 1.8–2.3); (median survival: median 14 months), and in UICC III, 1.84 (range: 1.48/2.3); (median survival: 10 months). There was only one patient with UICC IV who survived 11 months after surgery; this patient had a KLF6wt/sp ratio of 1.55. There was a trend towards correlation between KLF6wt/sp and tumor stage (UICC), but did not reach statistical significance (p=0.076) (Table 2).
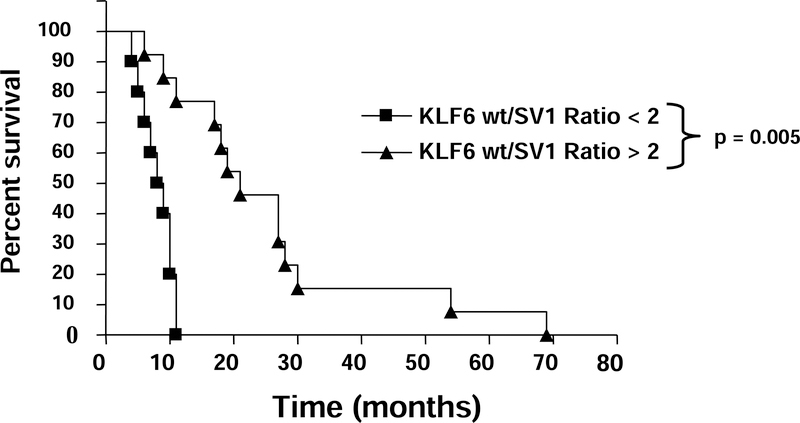
However, the KLF6wt/sp ratio at the time of resection was highly correlated with patient survival (p<0.001). (Table 2) The bivariate anaylsis and the log-rank test revealed significantly longer survival in those patients with KLF6wt/sp ratio > 2 (median: 21 months; range: 14–19) than the survival of patients with a KLF6wt/sp ratio < 2 (median: 9 months; range: 6–10); (p=0.005) (Figure 5). Furthermore, the Cox-regression revealed KLF6 ratio as an independent marker for survival (p=0.006).
Figure 5: The ratio of KLF6wt/sp mRNA expression ratio correlates with survival in patients with pancreatic cancer.
The KLF6wt/sp ratio was determined from mRNA that was extracted from tumors harvested immediately following resection in patients with proven ductal adenocarcinoma of the pancreas. The ratio was significantly correlated with overall survival in these patients. The median survival of patients with KLF6wt/sp ratio >2 (n=14) was 21 months, which was significantly longer than the median survival of 9 month in patients (n=10) with KLF6wt/sp ratio ≤ 2 (p=0.005).
DISCUSSION
In the present study we have characterized the KLF6 allele status and the expression of KLF6 mRNA in human pancreatic tumors and cancer cell lines. With the recent discovery that KLF6 mRNA is alternatively spliced in human prostate cancer (20), we focused on pancreatic cancer because of its highly lethal nature, our limited understanding of underlying mechanisms, and because of the availability of a very well characterized set of tumors associated with detailed clinical data, including survival. The findings build upon a substantial body of data implicating inactivation of KLF6 in the pathogenesis of a number of human cancers (8, 13, 15, 16), but provide new information regarding the prognostic value of KLF6 alternative splicing.
These data reinforce the potential role of enhanced alternative splicing of KLF6 as a growth-promoting mechanism in human cancer. Moreover, these are the first data to correlate the ratio of KLF6 wt/sp mRNA at the time of surgery for pancreatic cancer with reduced survival.
KLF6 immunostaining of pancreatic tissue identified specific cytoplasmic accumulation within tumor cells, similar to colorectal cancer (15). Although not clearly understood at the time of the earlier report (15), a likely mechanism appears to be the specific accumulation of KLF6 splice forms, which can accumulate in the cytoplasm because it lacks a nuclear localization signal. In pancreatic cancer, the tumor cells and microvessels exhibited KLF6 immunoreactivity, whereas no staining was observed in ductal cells of normal pancreas tissue. Western blot analysis of pancreatic cancer tissue using the same antibody used for immunohistochemistry confirmed the presence of lower MW KLF6 isoforms of ~40, ~30, and 26 kD. It is possible, but less likely, that the cytoplasmic accumulation represented full length KLF6, as this antibody does not distinguish between full length KLF6 and its splice forms. However, full length KLF6 typically appears in the nucleus in normal tissues but not in the cytoplasm (32).
While our study did not examine the biologic activity of KLF6 splice forms in normal pancreas and pancreatic cancer tissues, previous studies in prostate (20, 22) and ovarian (17) cancers clearly indicate a growth-promoting activity of the SV1 isoform. The mechanism of SV1’s proliferative activity is not fully clarified, but likely reflects in part the sequestration of wild type, full length KLF6 protein in the cytoplasm (data not shown). Alternatively, a mechanism independent of direct KLF6 antagonism cannot be excluded. Regardless, the KLF6 SV1 isoform functionally antagonizes the ability of KLF6wt to suppress cell proliferation and tumorgenicity in vivo (20, 22). Increased alternative KLF6 splicing has an inhibitory effect on p21 and possibly other transcriptional targets (20, 22).
Two recent studies, one in primary lung cancer samples and the other in esophageal cancer cell lines have documented reduced expression of KLF6 mRNA as a result of gene silencing due to hypermethylation (33, 34). Furthermore, de novo KLF6 methylation may contribute to gene inactivation in astrocytic glioma, where mutations and LOH are shown to play only a minor role in KLF6 inactivation (35). Regardless of the mechanism, decreased expression of wild type KLF6 has been identified in other tumors by microarray analysis (36). However, the mechanisms underlying this reduction have not been elucidated, and the results were not validated by real time PCR.
In contrast, in the present study the reduced ratio of KLF6wt/sp mRNA in pancreatic tumor samples was primarily due to enhanced splice form expression rather than reduced KLF6 full length mRNA. It appears that net KLF6 activity is regulated in part by a critical balance between KLF6wt and alternatively KLF6 spliced forms (ratio mRNA KLF6wt/sp). However, it is not clear whether the biologic effects of this ratio are the same regardless of whether the ratio is altered as a result of an increase in splice form expression, or a reduction in full-length mRNA expression. Of great significance, however, an increased KLF6wt/sp ratio is associated with increased tumor differentiation. Moreover, the association of increased KLF6wt/sp ratio with survival in patients with pancreas cancer raises the possibility of using this ratio as an independent predictor of prognosis. However, larger, prospective studies are required to establish such a role.
In conclusion, pancreatic cancer is associated with enhanced alternative splicing of the KLF6 tumor suppressor gene without associated LOH or gene mutation. Based on its close correlation with survival, the ratio of KLF6wt/sp mRNA is a potential prognostic marker whose value should be further validated in animal models and prospective human studies.
ACKNOWLEDGMENTS
Funding has been obtained from the NIH (DK37340), to S.L.F., the U.S. Department of Defense, DAMD17-03-1-0100 to S.L.F., and the Hella-Bühler Fund (2004-01) to H.F. The study sponsors had no involvement in study design, in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision making to submit the manuscript for publication.
Footnotes
CONFLICT OF INTEREST
There are no conflict of interest to be declared by the authors of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem 2001;276:34355–34358. [DOI] [PubMed] [Google Scholar]
- 2.Schuh R, Aicher W, Gaul U, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell 1986;47:1025–1032. [DOI] [PubMed] [Google Scholar]
- 3.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol 2000;32:1103–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol 2003;4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci 1999;24:236–240. [DOI] [PubMed] [Google Scholar]
- 6.Koritschoner NP, Bocco JL, Panzetta-Dutari GM, Dumur CI, Flury A, Patrito LC. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J Biol Chem 1997;272:9573–9580. [DOI] [PubMed] [Google Scholar]
- 7.Ratziu V, Lalazar A, Wong L, et al. Zf9, a Kruppel-like transcription factor upregulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci USA 1998;95:9500–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narla G, Heath KE, Reeves HL, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001;294:2563–2566. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y, Ratziu V, Choi SG, et al. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem 1998;273:33750–33758. [DOI] [PubMed] [Google Scholar]
- 10.Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, Chatton B, Bocco JL. A new role for the Kruppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene 2004;23:8196–8205. [DOI] [PubMed] [Google Scholar]
- 11.Kimmelman AC, Qiao RF, Narla G, et al. Suppression of glioblastoma tumorigenicity by the Kruppel-like transcription factor KLF6. Oncogene 2004;23:5077–5083. [DOI] [PubMed] [Google Scholar]
- 12.Benzeno S, Narla G, Allina J, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res 2004;64:3885–3891. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Hyytinen ER, Sun X, et al. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol 2003;162:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho YG, Kim CJ, Park CH, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene 2005;24:4588–4590. [DOI] [PubMed] [Google Scholar]
- 15.Reeves HL, Narla G, Ogunbiyi O, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology 2004;126:1090–1103. [DOI] [PubMed] [Google Scholar]
- 16.Kremer-Tal S, Reeves HL, Narla G, et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology 2004;40:1047–1052. [DOI] [PubMed] [Google Scholar]
- 17.Difeo A, Narla G, Hirshfeld J, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res 2006;12:3730–3739. [DOI] [PubMed] [Google Scholar]
- 18.Ito G, Uchiyama M, Kondo M, et al. Kruppel-like factor 6 is frequently downregulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res 2004;64:3838–3843. [DOI] [PubMed] [Google Scholar]
- 19.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer 2. J Clin Invest 2004;113:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narla G, Difeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res 2005;65:1213–1222. [DOI] [PubMed] [Google Scholar]
- 21.Yea S, Narla G, Zhao X, et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008; 134: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narla G, Difeo A, Yao S, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread 2. Cancer Res 2005;65:5761–5768. [DOI] [PubMed] [Google Scholar]
- 23.Schneider G, Siveke JT, Eckel F, Schmid RM. Pancreatic cancer: basic and clinical aspects. Gastroenterology 2005; 128:1606–1625. [DOI] [PubMed] [Google Scholar]
- 24.Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer 2005;41:2213–2236. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart AC, Rothenberg ML, Berlin JD. Treatment for pancreatic cancer: current therapy and continued progress. Gastroenterology 2005;128:1642–1654. [DOI] [PubMed] [Google Scholar]
- 26.Burris HA III, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol 1997;15:2403–2413. [DOI] [PubMed] [Google Scholar]
- 27.Luttges J, Schemm S, Vogel I, Hedderich J, Kremer B, Kloppel G. The grade of pancreatic ductal carcinoma is an independent prognostic factor and is superior to the immunohistochemical assessment of proliferation. J Pathol 2000;191:154–161. [DOI] [PubMed] [Google Scholar]
- 28.Heinmoller E, Dietmaier W, Zirngibl H, et al. Molecular analysis of microdissected tumors and preneoplastic intraductal lesions in pancreatic carcinoma. Am J Pathol 2000;157:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada K p16 and p53 gene alterations and accumulations in the malignant evolution of intraductal papillary-mucinous tumors of the pancreas. J Hepatobiliary Pancreat Surg 2002;9:76–85. [DOI] [PubMed] [Google Scholar]
- 30.Heinmoller E, Bockholt A, Werther M, et al. Laser microdissection of small tissue samples--application to chronic pancreatitis tissues. Pathol Res Pract 2003;199:363–371. [DOI] [PubMed] [Google Scholar]
- 31.Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T. Genetic progression and divergence in pancreatic carcinoma. Am J Pathol 2000;156:2123–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojima S, Hayashi S, Shimokado K, et al. Transcriptional activation of urokinase by the Kruppel-like factor Zf9/COPEB activates latent TGF-beta1 in vascular endothelial cells. Blood 2000;95:1309–1316. [PubMed] [Google Scholar]
- 33.Wikman H, Kettunen E, Seppanen JK, et al. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene 2002;21:5804–5813. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell 2002;2:485–495. [DOI] [PubMed] [Google Scholar]
- 35.Jeng YM, Hsu HC. KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer 2003;105:625–629. [DOI] [PubMed] [Google Scholar]
- 36.Glinsky GV, Bersezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]