Abstract
DNA damage triggers diverse cancers, particularly hepatocellular carcinoma (HCC), but the intrinsic link between DNA damage and tumorigenesis remains unclear. Due to its role as an epigenetic and transcriptional regulator, histone deacetylase 3 (HDAC3) is essential for DNA damage control and is often aberrantly expressed in human HCC. In this study, we used individual class I HDAC member-deficient mice to demonstrate that K9 in histone H3 (H3K9), which is the critical site for the assembly of DNA damage response complexes, is exclusively targeted by HDAC3. Ablation of HDAC3 disrupted the deacetylation and consequent trimethylation of H3K9 (H3K9me3), the first step in double-strand break (DSB) repair, and led to the accumulation of damaged DNA. Simultaneously, hyperacetylated H3K9 (H3K9ac) served as a transcriptional activator and enhanced multiple signaling pathways to promote tumorigenesis. Together these results show that HDAC3 targets the H3K9ac/H3K9me3 transition to serve as a critical regulator that controls both DNA damage repair and the transcription of many tumor-related genes. Moreover, these findings provide novel insights into the link between DNA damage and transcriptional reprogramming in tumorigenesis.
Keywords: HDAC3, H3K9, liver cancer, DNA damage repair, transcriptional reprogramming
Introduction
Hepatocellular carcinoma (HCC) often occurs when hepatocytes suffer continuous chronic injuries, particularly those that induce chromatin or DNA damage (1). DNA is vulnerable to various genotoxic stresses derived from endogenous metabolites, environmental and dietary carcinogens, and certain anti-inflammatory drugs, etc. (2, 3). Each human cell is estimated to be subjected to approximately 70,000 lesions daily (4), and these damages can accumulate and result in genetic changes, particularly in the presence of insufficient DNA damage repair mechanisms, which play a pivotal role in triggering tumorigenesis (1, 5).
Efficient DNA damage repair is accurately controlled by dynamic histone modulations. Once a DNA double-stranded break (DSB) occurs, histone H2A.X is phosphorylated into γH2A.X (6, 7), and this step is followed by recruitment of the MRE11–RAD50–NBS1 (MRN) complex and the phosphorylation of CK2. CK2 phosphorylation releases HP1β from chromatin to expose H3K9me3 (trimethylation of histone H3 at K9), and this exposure allows the binding of Tip60, a histone acetyltransferase (HAT) that acts as a core factor for assembly of the repair complex (7, 8). Activation of Tip60 by DSBs requires the interaction of Tip60 with H3K9me3, and the transition of H3K9ac (acetylation of H3K9) to H3K9me3 is therefore one of the key histone modifications that contributes to the DNA damage response (DDR) and acts as the first step in DSB repair either by homologous recombination (HR) or nonhomologous end joining (NHEJ).
The activation of proto-oncogenes, which is often accompanied by the inactivation of tumor-suppressor genes, represents a typical example of transcriptional reprogramming (9) and is a distinctive characteristic of malignancies. Although triggered by various toxic stresses, transcriptional reprogramming in diverse malignancies often leads to the widespread deregulation of gene expression profiles and the disruption of signaling networks that control proliferation and cellular functions, and these effects result in malignant characteristics (10, 11). Both DNA damage and transcriptional reprogramming are considered the critical triggers that drive malignant transformation, but the intrinsic association between them remains obscure.
Because histone modulation plays unique roles in both DNA damage control and gene transcription, we speculate that histone modulation defects play a central role in mutation accumulation and transcriptional reprogramming. Histone acetylation and deacetylation, which are catalyzed by a pair of antagonistic enzyme families, HATs and histone deacetylases (HDACs), respectively, are reportedly the most important histone modulators. Class I HDAC members, including HDACs 1, 2, 3 and 8 (12), are widely distributed in the nucleus, where they exert overlapping but distinct functions in regulating diverse pathophysiological processes, including metabolism, mitosis, apoptosis, and tumorigenesis (13, 14). Among HDACs, HDAC3 plays an extraordinary role in DNA damage control and genomic stability. The germ-line deletion of HDAC3 results in early embryonic lethality (15), and the absence of HDAC3 in mouse embryonic fibroblasts (MEFs) leads to cell cycle-dependent DNA damage and apoptosis (16). Moreover, hepatic HDAC3 loss in mice significantly impairs genomic stability, activates c-Myc and Ras signaling and robustly disrupts p53 and Wnt signaling, which results in the early onset of spontaneous HCC (17, 18).
HDAC3 is crucial in cell cycle regulation, although the molecular mechanism remains controversial. The downregulation of HDAC3 increases p21WAF1/cip1 expression and thereby induces G1-phase arrest in liver cancer cells (19). HDAC3 facilitates the phosphorylation of H3S10 by Aurora B, which is crucial for the G2/M transition in mitosis (20). HDAC3 also controls the G2/M transition by regulating the CDK1 levels (21), and the inactivation of HDAC3 impairs S-phase progression by either disturbing replication fork progression or modulating cyclin A acetylation (22). We recently reported that HDAC3 plays a unique role in the cytoplasm, where it promotes IL6/STAT3 signaling at the early stage of liver regeneration (23).
Distinct from HDAC1 and HDAC2, which are extensively elevated in HCC (24), HDAC3 has seldom been found to be significantly increased, and its levels are reduced in approximately 13% of cases (17). Moreover, nuclear receptor corepressor (NCOR1), the cosuppressor of HDAC3 that is essential for HDAC3 activation, is inactive or reduced by at least 2-fold in nearly 1/3 of all HCC tissues (17). Thus, decreased HDAC3/NCOR1 signaling is frequently found in HCC. The liver-specific deletion of HDAC3 in mice leads to accumulated DNA damage, chronic liver injury, metabolic disorder, transcriptional reprogramming, and, eventually, spontaneous HCC onset and thus represents an appropriate model for simulating the histopathological course of clinical HCC, particularly in patients with decreased HDAC3/NCORI signaling.
Although the functions of HDAC3 in liver damage and cancers in mice have been well established, the signaling pathways through which HDAC3 regulates DNA damage repair and transcriptional reprogramming remain obscure. Understanding the cellular and molecular basis of HDAC3 in tumorigenesis is important for gaining insight into the relationship among DNA damage, transcriptional reprogramming and malignant transformation, which will be helpful for developing novel strategies for HCC prevention and treatment.
Materials and Methods
Human HCC samples
Specimens of HBV-associated HCC were collected from patients who underwent curative resection at West China Hospital, Sichuan University, with written informed consent. The procedures used for human sample collection and use were approved by the ethics committee of West China Hospital, Sichuan University (Chengdu, China). Twenty paired human liver cancer and adjacent nontumor tissues were used for the analysis of gene expression, and the detail clinical information for each patient was listed in Supplementary Table 1.
Animals
HDAC3loxP/loxP frozen embryos were purchased from the European Mouse Mutant Archive (EMMA). Alb-Cre and Alb-CreERT transgenic mice were purchased from Shanghai Biomodel Organism Science & Technology Development Co., Ltd, China. HDAC3loxP/loxP mice were intercrossed with Albumin-Cre or Albumin-CreERT transgenic mice to obtain constitutive (Alb-Cre:Hdac3−/−) or inducible (Alb-CreERT:Hdac3−/−) liver-specific HDAC3-deficient mice, respectively (23). HDAC1, HDAC2 or HDAC8 conditional-knockout mice are generated using the same strategy (25, 26). Littermates without Cre were used as wild-type controls (WT). Fumarylacetoacetate hydrolase-deficient (Fah−/−) mice were used (27) to assess the liver repopulation capacity of hepatic progenitor cells (HPCs). All the mice used were fed a normal chow diet and housed with corncob bedding under SPF conditions. The animal procedures and care were conducted in accordance with national and international laws and policies and approved by the Animal Care and Use Committee of Sichuan University.
Histology, immunohistochemistry, Oil red O and immunofluorescence staining
Mouse livers harvested at defined points were fixed with 10% buffered formalin for 48 h and then processed for sectioning by embedding in paraffin. The paraffin sections (4 μm) were prepared for H&E, Periodic Acid-Schiff (PAS) and immunohistochemistry staining. Fresh liver tissues were collected, embedded in Tissue Tek OCT compound (Sakura Finetek) and maintained frozen. The tissue blocks were further cut into 5-μm sections for Oil red O staining. Apart from special requirements, the excised livers were further fixed overnight in 10% buffered formalin, washed with PBS, dehydrated in 30% sucrose at 4 °C overnight, and then embedded in OCT. Sections (4 μm) were prepared for immunofluorescence staining using a previously described procedure (23). The antibodies used in this study are listed in Supplementary Table 2.
HPC isolation and transplantation
Liver single cells were isolated by two-step collagenase digestion and gradient centrifugation. To capture CD133+ progenitor cells, the cells were labeled with primary CD133 antibody (mouse IgG1, Miltenyi Biotec) and subsequently magnetically isolated using the EasySep PE Selection Kit (Stemcell Technologies) according to the manufacturer’s instructions. A total of 5 × 105 CD133+ HPCs were injected into the spleen of each Fah−/− mouse as previously described (27).
Western blotting
Liver tissue was lysed in RIPA buffer and centrifuged at 13,000 rpm and 4 °C for 15 min. Western blotting was performed using standard protocols. The antibodies used in this study are listed in Supplementary Table 2.
Real-time RT-PCR analysis
RNA from tissues was extracted using the TRIzol reagent (Invitrogen) and further purified using the RNeasy kit (Qiagen). cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad), and qRT-PCR was performed with the Bio-Rad CFX96 System. The sequences of the indicated primers are presented in Supplementary Table 3.
Statistical analysis
The statistical analyses were performed using Microsoft Excel software or Prism GraphPad 7. The results are expressed as the means ± s.e.m. The significance of the differences between groups was tested using unpaired, two-tailed Student’s t test with Welch correction. A P-value < 0.05 was considered significant.
For more details, see the Supplementary Material.
Results
HDAC3 reduction leads to similar gene expression profiles in human and mouse liver cancers
The gene expression profile analysis of 20 pairs of human HCC tissues and their corresponding adjacent tissues showed that the levels of HDAC1 and HDAC2 were increased to various levels in HCC tissues, whereas the HDAC8 levels were substantially decreased in these tissues. Notably, although HDAC3 was mildly to moderately (≥1.25-fold) decreased in five cases (25%), NCOR1, the cosuppressor of HDAC3, was reduced by more than 1.5-fold in 13 cases (65%) and reduced by at least 2-fold in seven cases (35%) (Fig. 1A), which is consistent with the GEO (17) and TCGA datasets.
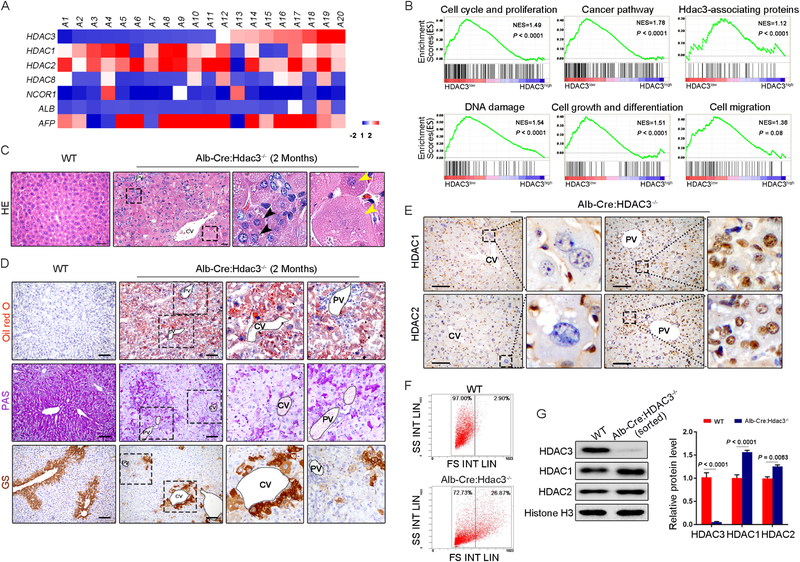
Fig. 1. HDAC3 reduction leads to gradual hepatocyte dysfunction, increasing DNA damage and compensatory elevation of HDAC1 and HDAC2.
(A) Heatmap representing the mRNA levels in human HCC samples (n = 20). (B) GSEA enrichment plots showing the comparison of the gene expression profiles in HDAC3low-HCC and HDAC3high-HCC with the gene sets in spontaneous liver cancer in an Alb-Cre:Hdac3−/− mouse as indicated. NES, normalized enrichment score; the P value indicates the significance of the enrichment score. (C) H&E-stained liver sections. The black arrowheads indicate the periportal hepatocytes, and the yellow arrowheads indicate the degenerative hepatocytes in the central lobule. Note the disappearance of the nucleus. Scale bar, 50 μm. (D) Oil Red O staining, PAS staining and GS immunohistochemistry staining of liver sections. Scale bar, 100 μm. (E) HDAC1 and HDAC2 staining in sections of Alb-Cre:Hdac3−/− livers. Scale bar, 50 μm. (F) Flow cytometric sorting of hepatocytes from WT and Alb-Cre:Hdac3−/− livers. (G) Immunoblotting was performed using total protein lysates from WT and Alb-Cre:Hdac3−/− hepatocytes sorted by flow cytometry. The levels of HDAC1, HDAC2 and HDAC3 in each genotype are expressed as ratios to histone H3 (n = 3 samples per group). WT, wild type; CV, central vein; PV, portal vein. The data are presented as the means ± s.e.m.; the P value was determined by two-tailed Student’s t test.
We established liver-specific HDAC3-ablation mice as described previously (28), and the mice developed spontaneous HCC within 6 months after (Supplementary Fig. S1A–E). To explore whether hepatic HDAC3-inactive mice could be used to at least partially mimic the HCC tumorigenic process in humans, the expression patterns of HDAC3low-HCC and HDAC3high-HCC were compared with those in datasets from spontaneous liver cancers in Alb-Cre:Hdac3−/− mice through gene set enrichment analysis (GSEA). Based on the GSEA results, the transcriptome profile of HDAC3low-HCC was a significant match to the signatures of HDAC3-inactive mouse tumors, including genes related to cell cycle and proliferation, cancer pathways, DNA damage, cell growth and differentiation (Fig. 1B). Thus, Alb-Cre:Hdac3−/− mice are appropriate for investigating HCC development in a low-HDAC3 expression background.
HDAC3-inactive livers display regional distinct injury
As reported previously, the Alb-Cre:Hdac3−/− liver develops apparent histological and metabolic abnormalities at a very early age and finally HCC (28). We found that the periportal hepatocytes, in which the HDAC3 gene was also deleted, maintained a relatively normal size, shape, hematoxylin and eosin (H&E) staining pattern (Fig. 1C; Supplementary Fig. S2A, B), and metabolic status, as indicated by Oil red O and PAS staining (Fig. 1D). Moreover, along the portal-central axis of the lobule, the hepatocytes gradually increased in size and exhibited a more irregular shape, a more slightly stained cytoplasm (Fig. 1C), more lipid deposition and less glycogen deposition (Fig. 1D). Nuclear disaggregation and apoptosis were frequently observed in the centrilobular hepatocytes (Fig. 1C; Supplementary Fig. S1E), and this effect was further confirmed by transmission electronic microscopy (Supplementary Fig. S2C). GS+ cells play a pivotal role in ammonia detoxification by expressing glutamine synthetase, which catalyzes condensation of ammonia with glutamate to glutamine (4), and are strictly restricted around the central vein; however, rare GS+ cells were observed in the mutant liver, and most intriguingly, GS+ cells were irregularly scattered in the parenchyma (Fig. 1D), demonstrating disarrangement of the lobules. It has been well documented that the deletion of HDAC3 impairs DNA damage repair and genome stability. Consistent with the histological changes, immunofluorescent staining of DSB markers, including γH2A.X and PKAP1 (16, 29), showed few foci in the periportal cells in the Alb-Cre:Hdac3−/− liver; however, the number of foci was notably increased along the portal-central axis and reached its peak in centrilobular cells (Supplementary Fig. S3A, B). These findings suggested that hepatocytes displayed abnormal morphology and function with increasing degrees of DNA damage following HDAC3 ablation.
A compensatory increase in HDAC1 and HDAC2 does not facilitate DNA damage repair
HDAC1 and HDAC2, in contrast to HDAC3/NcoR1 signaling, are widely increased in human HCC tissues, which suggests their compensatory elevation (Fig. 1A). In addition, a previous study showed that the knockdown of HDAC1 or HDAC2 using siRNA increases the number of DNA foci in MEFs (16). However, in accordance with a previous study (16), assessment of whole liver homogenates revealed that neither HDAC1 nor HDAC2 was compensatively increased in the Alb-Cre:Hdac3−/− liver (Fig. S1A). Intriguingly, we noted that their expression also exhibited spatial disparity, which was robustly expressed in periportal cells, including cholangiocytes and small-sized hepatocytes, and gradually decreased along the hepatic cords and missing in the pericentral cells in the Alb-Cre:Hdac3−/− liver (Fig. 1E). Furthermore, we sorted HDAC3-ablated hepatocytes that were similar in size to those of WT mice, which possessed less DNA damage and relatively normal function (Fig. 1F). The expression of HDAC1 and HDAC2 in these sorted HDAC3-inactive cells was substantially increased compared with that in the WT hepatocytes (Fig. 1G).
We subsequently investigated whether HDAC1 and HDAC2 expression increases and compensates for the loss of HDAC3 in response to genotoxic challenge. For the pre-existing lesions in the Alb-cre:Hdac3−/− mouse liver, we constructed liver-specific tamoxifen-inducing HDAC3-ablated mice (Alb-CreERT:Hdac3−/−) as previously described (23) (Fig. S4A, B). In the WT livers, the administration of tamoxifen did not induce liver injury or impair the repair of DNA damage caused by ionizing radiation (IR) (Fig. S4C). In these livers, both HDAC1 and HDAC2 were maintained at stable levels after a single dose of 3-Gy IR, but their levels were slightly increased after IR if HDAC3 was inducibly ablated (Fig. 2A, B). Noticeably, if the mice were further exposed to IR at 1 Gy/day for six consecutive days, both HDAC1 and HDAC2 were robustly increased, regardless of the expression of HDAC3 (Fig. 2A, B).
Fig. 2. The compensatory increase in HDAC1 and HDAC2 does not facilitate DNA damage repair.
(A) Schematic of the experimental protocol for establishing tamoxifen-dependent hepatocyte HDAC3 deletion and subsequent experimental analyses. (B) Immunoblotting was performed using total protein lysates from WT and Alb-CreERT:Hdac3−/−(+ TAM) livers after irradiation. (C) Immunofluorescence analysis of γH2A.X was performed following irradiation. The arrowheads indicate γH2A.X foci. Scale bar, 25 μm. (D) To obtain quantitative data for the γH2A.X foci distribution in (C), the number of foci in 100 cells was manually counted, and the percentages of cells containing 0, 1–10, 10–20, and more than 20 foci per cell were calculated (n = 3 mice per group). The data are presented as the mean ± s.e.m.; the P value was determined by two-tailed Student’s t test.
The role of HDAC1 and HDAC2 in DNA damage repair remains unclear. Compared with the WT liver, the constitutive ablation of HDAC1 or HDAC2 did not significantly increase the number of DNA foci after a single or repeat IR challenge (Fig. 2C). In contrast, in the HDAC3-silenced liver, although HDAC1 and HDAC2 were substantially increased, a high number of foci were observed in the hepatocytes after IR (Fig. 2C, D).
HDAC3 controls DNA damage repair by promoting the H3K9ac/H3K9me3 transition
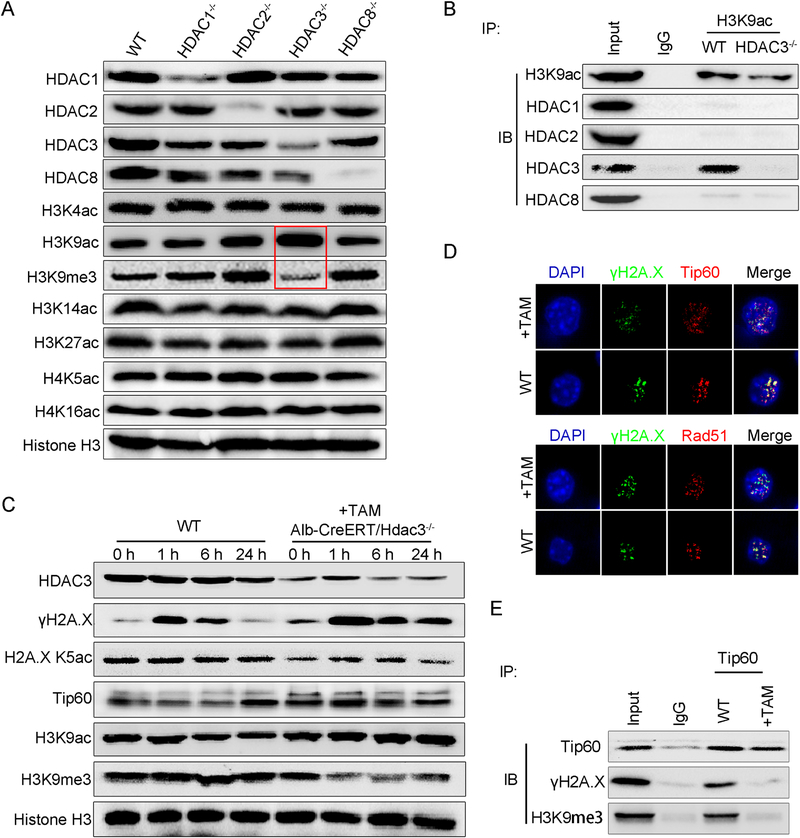
We further evaluated why HDAC3, but not other HDAC members, plays an essential role in DNA damage control. Several lysine residues that can potentially be acetylated are present on the N terminus of core histones; however, the correspondence between individual HDACs and their targets has not yet been fully defined. We examined changes in the expression levels of acetylated residues, including H3K4, H3K9, H3K14, H3K27, H4K5, and H4K16, corresponding to the deficiency of each individual HDAC member. Compared with those in the WT liver, most of these acetylation markers were not significantly increased in the quiescent liver upon the single disruption of hepatic HDAC1, HDAC2, or HDAC8 (Fig. 3A), which indicated the extensive overlap between the catalytic sites of these enzymes. Noticeably, in the HDAC3-inactive liver, H3K9ac was robustly increased, whereas H3k9me3 was correspondingly decreased (Fig. 3A, red box), which indicated that H3K9 might be exclusively targeted by HDAC3. Furthermore, HDAC3 was the unique class I HDAC member that coimmunoprecipitated with H3K9ac (Fig. 3B). The H3K9ac/H3K9me3 transition acts as the first step in DSB repair and allows the binding of Tip60 to DSB foci to recruit other DDR proteins. Because HDAC3 ablation results in H3K9ac/H3K9me3 transition failure, we further examined whether Tip60 is still recruited to sites of DNA damage and acetylates H2A.X. After IR, Tip60 accumulated at DSB foci in the WT liver (Fig. 3C, D). In contrast, in the Alb-CreERT:Hdac3−/− liver, although the protein level was increased (Fig. 3C), Tip60 was dispersed in the nucleus but not concentrated in foci (Fig. 3D, E). Rad51, another essential factor of the DDR complex (30), showed a similar dispersive pattern in the nucleus (Fig. 3D). Correspondently, these livers showed significantly delayed γH2A.X dephosphorylation and a lower level of H2A.XK5 acetylation compared with the WT livers (Fig. 3C).
Fig. 3. HDAC3 deletion impairs the H3K9ac/H3K9me3 transition and assembly of the DNA damage repair complex.
(A) Immunoblotting was performed using total protein lysates from WT, HDAC1−/−, HDAC2−/−, HDAC3−/− and HDAC8−/− livers. Histone H3 served as the loading control. (B) Coimmunoprecipitation showing that H3K9ac combined with HDAC3 but not other HDAC members in the WT liver. (C) Immunoblotting was performed using total protein lysates from WT and Alb-CreERT:Hdac3−/− livers treated with tamoxifen after irradiation. (D) Immunofluorescence staining showing that Tip60 and Rad51 were located in γH2A.X foci in WT hepatocytes but diffused in HDAC3-null cells. (E) Coimmunoprecipitation showing that Tip60 cannot combine with γH2A.X and H3K9me3 in tamoxifen-treated livers after irradiation.
The H3K9ac/H3K9me3 transition is dependent on the HDAC3 enzyme and Suv39h1
In addition to HDAC3 ablation, the accumulation of H3K9ac could also result from the overactivation of PCAF and GCN5, both of which are H3K9-specific acetylases (31, 32). To explore this possibility, we examined the expression of these two acetylases and found that neither was significantly increased in cancer and noncancer tissues in the HDAC3−/− liver (Fig. 4A). To further confirm the role of HDAC3 in the H3K9ac/H3K9me transition, we generated a HepG2 cell line with an enzyme inhibiting mutation (S424A) in the HDAC3 protein (33). As expected, the S424A mutation in HDAC3 led to an increase in H3K9ac and a decrease in H3K9me3, which was similar to the results obtained with HDAC3 knockdown by specific siRNA (Fig. 4B, C) (23). An impaired DNA damage repair ability was also observed in S424A and siHDAC3 cells after 1-Gy IR (Fig. 4D, E).
Fig. 4. The H3K9ac/H3K9me3 transition is dependent on the HDAC3 enzyme and Suv39h1.
(A) The expression of the H3K9 acetylases PCAF and GCN5 was detected in the WT liver, HDAC3−/− HCC (T) and adjacent tissues (NT); (B-C) The enzyme inhibiting mutation of HDAC3 (S424A) and the knockdown of HDAC3 by siRNA led to the elevation of H3K9ac and the reduction of H3K9me3. (D-E) The detection of γH2A.X by immunofluorescence staining (D) and immunoblotting (E) reveals the impairment of the DNA damage repair capacity caused by the deletion or catalytic mutation of HDAC3. (F) Expression of H3K9ac, H3K9me3 and γH2A.X in siHDAC3 and siSuv39h1-treated HepG2 cells with or without X-ray irradiation. (G-H) The detection of γH2A.X by immunofluorescence staining (G) and immunoblotting (H) reveals the impairment of the DNA damage repair capacity caused by HDAC3 and Suv39h1 deletion. The data are presented as the mean ± s.e.m.; the P value was determined by two-tailed Student’s t test.
The deacetylation of H3K9ac mediated by HDAC3 is critical for H3K9 methylation, but the molecular mechanism for H3K9me3 remains unknown. Suv39h1 methylates DSB-specific H3K9me3 and facilitates DNA damage repair (34, 35). To investigate whether Suv39h1 is responsible for H3K9me3, we knocked down Suv39h1 in HepG2 cells. After 1-Gy IR, H3K9ac was substantially reduced whereas H3K9me3 was not elevated in siSuv39h1 cells (Fig. 4F), resulting in deficient DNA damage repair complex resembling that found in siHDAC3 cells (Fig. 4G,H). Thus, we concluded that HDAC3 selectively targets H3K9 in an enzyme-dependent manner and, together with Suv39h1, plays an essential role in controlling the H3K9ac/H3K9me3 transition, which is quite pivotal for the assembly of the DDR complex at the sites of DNA damage.
H3K9ac activates carcinoma-related genes
The role of HDAC3 ablation in transcriptional reprogramming remains unclear. We have revealed that the loss of HDAC3 specifically led to the overacetylation of H3K9, which has long been considered a marker of active gene transcription (36). To identify the genes directly regulated by H3K9ac, we performed a whole-genome chromatin immunoprecipitation sequencing (ChIP-seq) assay. The H3K9ac-targeted genes were highly enriched in the enhancer or promoter regions (defined as −2 kb to +2 kb from the transcriptional start site [TSS]) overlapping the RNA Pol-II binding distribution peak (Fig. 5A). The comparison of the densities of H3K9ac in tumors with those of adjacent tissues revealed that a unique subset of genes exhibited strong H3K9ac enrichment in the tumor tissues (Fig. 5B). A cohort of H3K9ac-targeted sites was identified specifically in tumors (Fig. 5C). These sites are mainly associated with genes known to encode regulators of the cell cycle and differentiation, such as Cdk6, Usp39, Actn4, Ptpn7, Mapre3, and Tfdp2, along with DNA damage repair-related genes, such as Ercc1 and Trip13. Strikingly, we observed that the expression of oncogenic genes, such as Kras, Pbx3, Rpl34 and Mical2, was tightly correlated with direct binding to H3K9ac (Fig. 5D, E).
Fig. 5. H3K9ac enhances carcinoma-related genes.
(A) Anchor plot of the RNA Pol-II binding distribution peak and H3K9ac ChIP-seq peaks in tumor tissues and adjacent normal tissues. (B) Heatmap of the signal intensity of H3K9ac-targeted gene loci in tumor tissues and adjacent nontumor tissues. (C) Venn diagram of H3K9ac-targeted genes detected by ChIP-seq in tumor tissues (T) and adjacent nontumor tissues (N). (D) GO analysis of biological processes corresponding to H3K9ac-targeted genes in tumor tissue. The bars indicate the P value; the threshold of P = 0.05 is shown. (E) H3K9ac-binding profiles on the indicated gene loci in tumor tissues and adjacent nontumor tissues. (F) Heatmap of the distinct tumor-related genes between spontaneous tumor tissues and adjacent nontumor tissues in an Alb-Cre:Hdac3−/− mouse (n = 3 mice per group). The P value indicates the significance of the enrichment score.
Consistently, the expression levels of H3K9ac-target genes, such as Kras, Ercc1, Cdk6, Usp39, and Mapre3, were dramatically increased in the HDAC3-depleted tumors, as revealed by the expression profile analyses (Fig. 5F). These data suggest that H3K9ac transcriptionally targets carcinoma-related genes and activates their expression to promote tumor formation.
HDAC3-ablated progenitors give rise to liver cancer in Fah−/− mice
Similar to the histological changes in the chronically injured human liver, an apparent ductular reaction developed in the Alb-Cre:Hdac3−/− liver, characterized by many highly proliferative cells coexpressing several progenitor markers, in which HDAC3 was also deleted (Fig. 6A, B; Supplementary Fig. S5A–D). Actively proliferating HPCs are considered seed cells of liver cancer. To further validate the role of HDAC3 in cell malignant transformation, we traced the fate of HPCs in a FAH-deficient mouse. HPCs were isolated by CD133 MACS from Alb-Cre:Hdac3−/− mice, and a total of 5 × 105 CD133+ HPCs were injected into the spleen of Fah−/− mice (Fig. 6C). The liver was gradually repopulated by FAH+HDAC3− hepatocytes from the periportal area, and 8 weeks after NTBC withdrawal, approximately 92% of the hepatocytes displayed positive FAH but negative HDAC3 staining, which indicated that these cells were donor-derived (Fig. 6D). Similar to the changes in the Alb-Cre:Hdac3−/− liver, significant histological abnormalities, including severe DNA damage, hepatocyte degeneration, lobular disarrangement and HPC expansion, had developed in the liver (Fig. 6E). More convincing, 10 months after the infusion of HDAC3−/− HPCs, liver cancers developed in the recipient Fah−/− mice (Fig. 6F), and all tumor parenchymal cells were HDAC3-negative and Fah-positive (Fig. 6G), which again demonstrated that they originated from the infused cells. In addition, the HDAC3-absent tumors in Fah−/− mice also showed similar H3K9ac expression and gene profiles to those in the Alb-Cre:Hdac3−/− mice (Fig. 6H, I).
Fig. 6. HDAC3-inactive HPC-derived hepatocytes have a similar fate in Fah−/− mice.
(A) Double immunohistochemistry staining of Ki67 and CK19. The arrowheads indicate proliferating HPCs. Scale bar, 50 μm. (B) Double immunofluorescence staining of the HPC markers CK19 and CD133. Scale bar, 50 μm. (C) Schematic design for CD133+ cell separation, transplantation and analysis. (D) Immunohistochemistry staining showing that HDAC3-inactive HPCs repopulate the Fah−/− liver. (E) Immunofluorescence staining of γH2A.X, GS, CK19 and HDAC3 in the repopulated liver. Scale bar, 50 μm. (F) Gross morphology of the livers. Scale bar, 5 mm. (G) Immunoblotting was performed using total protein lysates from different genotypes or treated livers. Histone H3 served as the loading control. (H) Histology of spontaneous liver cancers in Alb-Cre:Hdac3−/− and Fah−/− mice. Note that in the recipient Fah−/− mice, the tumor cells are HDAC3-negative and FAH-positive. Scale bar, 50 μm. (I) Pearson correlation coefficient showing the similarity of the gene profiles in spontaneous liver cancers in Alb-Cre:Hdac3−/− mice (T1–T3) with those in the recipient Fah−/− mice (Ta-Tc) (n = 3 samples per group).
Human HCCs display distinct gene profiles according to the HDAC3 levels
Because HDAC3 expression can be either increased or decreased in human HCC tissues, we examined the levels of H3K9ac in 20 HCC cases. As expected, immunoblotting and immunohistochemistry analyses showed that H3K9ac was negatively correlated with the HDAC3 levels (Fig. 7A, B). To further confirm the involvement of HDAC3 in the development of HCC through a different transcription pattern, we compared the gene expression profiles between the HDAC3low/H3K9achigh and HDAC3high/H3K9aclow cases. Many genes, including genes related to DNA damage repair, tumor development and cell proliferation, were differentially expressed (Fig. 7C). Furthermore, a GSEA revealed that the transcriptome profiles of the HDAC3low tumors significantly matched the features of H3K9ac targets in the tumors of HDAC3-inactive mice (Fig. 7D).
Fig. 7. Human HCCs display distinct gene profiles according to the HDAC3 levels.
(A) Microphotograph of H3K9ac and HDAC3 expression in tumor tissues as determined by immunohistochemistry. Scale bar, 100 μm. (B) Immunoblotting of HDAC3 and H3K9ac was performed in tumor tissues and adjacent nontumor tissues. (C) Heatmap showing the patterns of pathway activity between HDAC3low/H3K9achigh and HDAC3high/H3K9aclow human HCCs (n = 3 samples per group). (D) GSEA enrichment plots showing the comparison of gene profiles in human HDAC3low-HCC and HDAC3high-HCC with the H3K9ac targets in HDAC3-null mouse tumors as indicated. NES, normalized enrichment score; the P value indicates the significance of the enrichment score. (E) Model showing that the hepatic loss of HDAC3 specifically disrupts the H3K9ac/H3K9me transition and promotes HCC development.
Discussion
Herein, we reveal that HDAC3 selectively targets histone H3K9 and plays an indispensable role in promoting the H3K9ac/H3K9me3 transition. Failure to accomplish this transition impairs DNA damage repair, leading to the accumulation of DNA damage. Moreover, the overacetylation of H3K9 enhances the expression of many tumor-related genes that drive malignant transformation. Our work demonstrated an intrinsic link between DNA damage and the initiation of transcriptional reprogramming and thus provides novel insights into the primary tumorigenesis trigger.
Cells are constantly challenged by various genotoxic stresses, which highlights the critical role of DNA damage repair mechanisms in the maintenance of chromatin and genomic stability (2, 5). Histone modulation constitutes the major epigenetic mechanism involved in the remodeling of the chromatin structure and the control of gene activities. Any disarrangement in histone modulation can either disturb gene expression or impair DSB repair. HDACs, among which class I HDAC members are the most studied, remove acetyl groups from lysine residues, leading to the formation of condensed and transcriptionally silenced chromatin. Increased HDAC levels are widely observed in diverse cancers and might contribute to the inactivation of tumor-suppressor genes, and thus, these increased HDAC levels are potential targets for tumor therapy (24, 37, 38). However, the significance of aberrant HDAC levels remains unclear.
Despite having similar enzymatic activities, class I HDAC members play distinct roles in liver pathophysiology. It has been previously reported that single or combined HDAC1 and HDAC2 disruption in mice does not noticeably disturb the liver structure and function and does not promote tumor development (25). Similarly, the silencing of HDAC8 has no substantial impact on liver pathophysiology (Supplementary Fig. S6A, B). HDAC3 has been demonstrated to play a critical role in DNA damage control (16, 17), but its signaling cascade is far from clear. Using mice in which individual HDACs were ablated, we identified that among the numerous acetylation sites in histones, H3K9 was exclusively targeted by HDAC3. The defect in the H3K9ac/H3K9me3 transition following HDAC3 inactivation impairs Tip60 binding to DSB foci, the assembly of the DDR complex upon DNA damage, and thereby results in DNA damage accumulation. HDAC1 and HDAC2 reportedly play roles in DNA damage repair (39), but their inactivation did not impair DNA damage repair in the mouse livers. In addition, elevated HDAC1 and HDAC2 levels in the liver did not facilitate DNA repair in response to ion exposure, most likely because they do not target H3K9 in the liver.
By regulating histone modulation, HDAC3 plays various roles in metabolic regulation (28, 40). Our findings demonstrated that deteriorated metabolic disorders exhibit apparent lobular distribution and appear to be closely associated with the degree of DNA damage. Although HDAC3 plays an important role in mitotic control, we did not observe an obvious regeneration defect in the inducible HDAC3-deficient liver, despite some delay in liver recovery after partial hepatectomy (PH) (23). In addition, hepatic progenitors, which do not exhibit severe DNA damage, can repopulate the FAH-deficient liver. Thus, we demonstrate that HDAC3-inactive hepatocytes can maintain relatively normal morphology and function before the accumulation of severe DNA damage, and this conclusion is logically consistent with the finding that only those cells that survive an increasing number of mutations have the potential to undergo transformation (41, 42).
Transcriptional reprogramming, which is characterized by the aberrant expression of numerous tumor-suppressor genes and oncogenes, is one of the most essential features of malignances (9). However, the factors that trigger and accelerate transcriptional reprogramming remain obscure. Histone acetylation leads to a looser chromatin structure and thereby allows transcription factor access. Indeed, the overexpression of H3K9ac, which serves as a marker of active transcription (36), enhances the transcription of numerous tumor-related genes. Thus, the impact of inactive HDAC3 on gene expression is substantially dependent on H3K9 overacetylation. Our study at least partially reveals the intrinsic link between DNA damage and the initiation of transcription programming. The accumulation of DNA damage itself can result from primary defects in DNA damage repair mechanisms, such as insufficient HDAC3/NCORI signaling in our transgenic mice and in some clinical HCC cases. Conversely, decreased HDAC3 levels might stimulate the expression of other HDAC members, such as HDAC1, and this finding was further confirmed by our ChIP-seq assay, which showed that HDAC1 is potentially targeted by H3K9ac. Therefore, HDACs reciprocally regulate each other, which complicates the signaling pathways in transcriptional reprogramming and suggests that any therapeutic strategy targeting HDACs in the clinic should fully consider the balance between each HDAC member.
Conversely, we showed that chronic and severe DNA damage can stimulate the aberrant activation of HDACs, which can lead to the abnormal expression of numerous genes. Cells either die because of severe DNA damage due to insufficient repair or survive when they aberrantly express a certain group of genes, such as genes that overactivate c-Myc and Ras signaling, and the surviving cells are vulnerable to malignant transformation. Although HCCs display similar changes in gene profiles, many critical genes are differentially expressed due to the different levels of HDAC3, and this finding offers a molecular basis for tumor heterogeneity.
Inconsistent with the findings obtained using Cre-dependent cell lineage tracing strategies, which revealed that HPCs do not differentiate into mature hepatocytes or give rise to HCC (43), we observed that HPCs are the seeds of the liver when mature hepatocytes suffer severe injury and lose their capacity for self-renewal. The assessment of why HPCs are exempt from DNA damage would be interesting. One possible explanation is that HPCs are transcriptionally inactive, with chromatin that is tightly condensed and shielded from genotoxic stress. Our data support the hypothesis that HCC more likely develops from mature hepatocytes that are derived from either parental cells or HPCs because they are transcriptionally active and more susceptible to genotoxic stress. This hypothesis might also be supported by the evidence that some HCCs develop in livers lacking a cirrhotic background and ductular reaction (44).
In summary, we report that the H3K9ac/H3K9me3 transition, which is exclusively targeted by HDAC3, serves as a critical intersection that controls both DNA damage repair and the transcription of many tumor-related genes (Fig. 7E). Our observations provide novel insights into the linkage between DNA damage and transcriptional reprogramming in tumorigenesis promotion, and these insights will be helpful for the development of novel strategies for HCC prevention and treatment.
Supplementary Material
Significance:
Findings show that HDAC3 exclusively regulates H3K9ac in response to DNA damage, and loss of HDAC3 activity shifts the balance from DNA damage control to protumorigenic transcriptional activity.
Acknowledgment
We thank Li Li and Fei Chen for the assistance provided with the pathology techniques. We would also like to acknowledge Guang Yang and Guangneng Liao for their help with the mice management and breeding. We acknowledge the support provided by Biotechservice Incorporation with the chip-sequence assay and the support provided by Genechem Co., Ltd. (Shanghai) and Beijing Genomic Institution (Shenzhen) with the microarray experiments.
Financial Support: This work was supported by grants from the Natural Science Foundation of China (No. 81872019 and 81472303), the Major Science and Technology Project of Science & Technology Department of Sichuan Province (2017SZ0003), “1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC18008 and ZY2017308)”, and “West China Hospital -Jefferson Clinical Cooperation Research Fund”.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- [1].Lord C, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287–94. [DOI] [PubMed] [Google Scholar]
- [2].Roos W, Thomas A, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 2016;16:20–33. [DOI] [PubMed] [Google Scholar]
- [3].Yu MC, Yuan JM. Environmental factors and risk for hepatocellular carcinoma. Gastroenterology 2004;127:S72–8. [DOI] [PubMed] [Google Scholar]
- [4].Olinski R, Gackowski D, Cooke MS. Endogenously generated DNA nucleobase modifications source, and significance as possible biomarkers of malignant transformation risk, and role in anticancer therapy. Biochim Biophys Acta 2018;1869:29–41. [DOI] [PubMed] [Google Scholar]
- [5].Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, et al. High-resolution characterization of a hepatocellular carcinoma genome. Nature Genetics 2011;43:464–69. [DOI] [PubMed] [Google Scholar]
- [6].Fernandez-Capetillo O, Nussenzweig A. Linking histone deacetylation with the repair of DNA breaks. Proc Natl Acad Sci U S A 2004;101:1427–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 2008;453:682–6. [DOI] [PubMed] [Google Scholar]
- [8].Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, et al. Histone H3 methylation 14 links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 2009;11:1376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Raggi C, Factor V, Seo D, Holczbauer A, Gillen M, Marquardt J, et al. Epigenetic reprogramming modulates malignant properties of human liver cancer. Hepatology 2014;59:2251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Khan A, Kuttikrishnan S, Siveen K, Prabhu K, Shanmugakonar M, Al Naemi H, et al. RAS-mediated oncogenic signaling pathways in human malignancies. Semin Cancer Biol 2018;S1044-579X;30002–6. [DOI] [PubMed] [Google Scholar]
- [11].Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun 2018;9:1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci U S A 2000;97:13708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jenuwein T, Allis CD. Translating the histone code. Science 2001;293:1074–80. [DOI] [PubMed] [Google Scholar]
- [14].Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res 2007;17:195–211. [DOI] [PubMed] [Google Scholar]
- [15].Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 2008;118:3588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008;30:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010;18:436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bhaskara S, Hiebert SW. Role for histone deacetylase 3 in maintenance of genome stability. Cell Cycle 2011;10:727–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 2000;97:10014–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 2006;20:2566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jiang Y, Hsieh J. HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc Natl Acad Sci U S A 2014;111:13541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vidallaliena M, Gallastegui E, Mateo F, Martínezbalbás M, Pujol MJ, Bachs O. Histone deacetylase 3 regulates cyclin A stability. J Biol Chem 2013;288:21096–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu X, Cao X, Zhu Y, Wu Z, Zhuang X, Shao M, et al. Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell Death Dis 2018;9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R, et al. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch 2011;459:129–39. [DOI] [PubMed] [Google Scholar]
- [25].Xia J, Zhou Y, Ji H, Wang Y, Wu Q, Bao J, et al. Loss of histone deacetylases 1 and 2 in hepatocytes impairs murine liver regeneration through Ki67 depletion. Hepatology 2013;58:2089–98. [DOI] [PubMed] [Google Scholar]
- [26].Qi J, Singh S, Hua WK, Cai Q, Chao SW, Li L, et al. HDAC8 Inhibition Specifically Targets Inv(16) Acute Myeloid Leukemic Stem Cells by Restoring p53 Acetylation. Cell Stem Cell, 2015:S1934590915003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fontburgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, et al. Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell 2015;162:766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 2008;27:1017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 2006;8:870–6. [DOI] [PubMed] [Google Scholar]
- [30].West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 2003;4:435–45. [DOI] [PubMed] [Google Scholar]
- [31].Zhang P, Liu Y, Jin C, Zhang M, Lv L, Zhang X, et al. Histone H3K9 acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may be involved in osteoporosis. Stem Cells, 2016, 34(9): 2332–2341. [DOI] [PubMed] [Google Scholar]
- [32].Guo R, Chen J, Mitchell DL, Johnson DG.. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. NUCLEIC ACIDS RES, 2010, 39(4): 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, et al. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. GENE DEV, 2005, 19(7): 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, et al. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. PNAS, 2014, 111(25): 9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shirai A, Kawaguchi T, Shimojo H, Muramatsu D, Ishida-Yonetani M, Nishimura Y, et al. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. Elife, 2017, 6: e25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pokholok D, Harbison C, Levine S, Cole M, Hannett N, Lee T, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005;122:517–27. [DOI] [PubMed] [Google Scholar]
- [37].Spurling C, Godman C, Noonan E, Rasmussen T,et al. HDAC3 overexpression and colon cancer cell proliferation and differentiation. Mol Carcinog 2008;47:137–47. [DOI] [PubMed] [Google Scholar]
- [38].Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, et al. Histone Deacetylase 3 (HDAC3) and Other Class I HDACs Regulate Colon Cell Maturation and p21 Expression and Are Deregulated in Human Colon Cancer. J Biol Chem 2006;281:13548–58. [DOI] [PubMed] [Google Scholar]
- [39].Krumm A, Barckhausen C, Kücük P, Tomaszowski K, Loquai C, Fahrer J, et al. Enhanced Histone Deacetylase Activity in Malignant Melanoma Provokes RAD51 and FANCD2-Triggered Drug Resistance. Cancer Res. 2016;76:3067–77. [DOI] [PubMed] [Google Scholar]
- [40].Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med 2012;18:934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science, 2015, 349(6255):1483–1489. [DOI] [PubMed] [Google Scholar]
- [42].Gerlinger M, Mcgranahan N, Dewhurst SM, et al. Cancer: Evolution Within a Lifetime. Annual Review of Genetics, 2014, 48(1):215–236. [DOI] [PubMed] [Google Scholar]
- [43].Mu X, Español-Suñer R, Mederacke I, Affò S, Manco R, Sempoux C, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. The J Clinic Inv. 2015;125:3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wörns Marcus A., Bosslet T, Victor A, et al. Prognostic factors and outcomes of patients with hepatocellular carcinoma in non-cirrhotic liver. Scand J Gastroenterol, 2012, 47(6):718–728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.