Abstract
Spliceosome mutations are common in MDS and AML, yet the oncogenic changes due to these mutations have not been identified. A global analysis of exon usage in AML samples revealed distinct molecular subsets containing alternative spliced isoforms of inflammatory and immune genes. IRAK4 was the dominant alternatively spliced isoform in MDS/AML and is characterized by a longer isoform that retains exon 4, encoding a protein, IRAK4-Long (L) that assembles with the Myddosome, results in maximal activation of NF-κB, and is essential for leukemic cell function. Expression of IRAK4-L is mediated by mutant U2AF1 and is associated with oncogenic signaling in MDS/AML. Inhibition of IRAK4-L abrogates leukemic growth, particularly in AML cells with higher expression of the IRAK4-L isoform. Collectively, mutations in U2AF1 induce expression of therapeutically targetable “active” IRAK4 isoforms and provide a genetic link to activation of chronic innate immune signaling in MDS and AML.
Introduction
The majority of human genes express more than one RNA isoform resulting from alternative sites of splicing, and transcriptional initiation and termination. Alternative splicing can generate diversity at the RNA and protein levels, which is important for many biological processes.1 Aberrations in alternative splicing are also implicated in human cancers, resulting from somatic mutations in genes encoding RNA splicing proteins or in splicing regulatory motifs within exons and introns, and/or altered expression of RNA-binding proteins. The importance of optimal RNA splicing and isoform expression in chronic lymphocytic leukemia, myelodysplastic syndromes (MDS), and acute myeloid leukemia (AML) is evident by the frequent occurrence of mutations in genes encoding RNA splicing factors.2–9 Although relatively uncommon in solid tumors, ~50% of MDS patients harbor mutations in RNA splicing factors.10,11 Despite significant advances in understanding the role of RNA splicing factors in hematopoiesis, how RNA splicing gene mutations contribute to cancer is unclear.
A subset of leukemia-associated genes is regulated at the expression level, which results from the coordinated change in all of a gene’s individual isoforms, typically due to usage of alternative transcriptional start sites. However, we posited that a subset of genes is regulated exclusively at the isoform level in leukemia, which results in anti-correlated expression of the individual RNA isoforms. Herein, a global analysis of exon usage in AML revealed enrichment of genes associated with inflammatory and immune pathways that are regulated by RNA isoform changes. Of the inflammatory and immune pathway genes undergoing RNA isoform switching in MDS/AML, isoform expression of interleukin-1 receptor-associated kinase 4 (IRAK4) was the most significantly altered. This isoform encodes a longer protein (IRAK4-L) that physically associates with MyD88 and results in activation of NF-κB and MAPK. We also demonstrated that expression of IRAK4-L is directly mediated by the mutant U2AF1 splicing factor in MDS/AML. Inhibition of IRAK4 abrogates leukemic growth and is more efficacious in AML cells with U2AF1 mutations and/or higher expression of IRAK4-L. Taken together, we determined that mutations in U2AF1 induce expression of therapeutically targetable “active” IRAK4 isoforms and provide a genetic link to activation of chronic innate immune signaling in MDS and AML.
Results
RNA isoforms define a subset of AML associated with innate immune pathway activation.
Molecularly distinct subsets of AML have been largely defined on the basis of somatic mutations and changes in global gene expression.10,12,13 As a complementary approach, we investigated whether differential RNA isoform expression and alternative splicing is sufficient to distinguish clinically-relevant molecular subtypes of AML and uncover oncogenic signaling dependencies (Supplemental Figure 1A). To determine the prevalence of differential RNA isoform usage in AML, we analyzed the RNA sequencing data from The Cancer Genome Atlas (TCGA). First, we identified genes that were explicitly regulated at the level of mRNA isoform switching. Regulation at the level of mRNA isoform switching for a gene was defined when at least 2 of its RNA isoforms exhibited a mutually exclusive expression pattern (expression correlation < −0.4) and higher isoform-level expression variance as compared to gene-level variance (Supplemental Figure 1B). Based on these criteria, we identified 887 genes that showed patterns of RNA isoform switching (Figure 1A, Supplemental Table 1). A semi-unsupervised hierarchical cluster analysis revealed 3 groupings of AML characterized by distinct patterns of differential isoform usage for these genes. The three groups have distinct clinical outcomes, with Group 2 having the worst prognosis (Figure 1B, Supplemental Figure 1C). To specify alternative exon-level expression events that confer worse prognosis, individual exon inclusion/exclusion events for these genes were correlated with the overall survival using a multivariate Cox regression analysis. We identified a subset of genes undergoing mRNA isoform switching with a least one differential exon inclusion/exclusion event that significantly correlated with AML survival in patients from Group 2 (Supplemental Table 2). Functional enrichment analysis revealed that the genes whose differential exon usage correlates with worse AML prognosis is enriched for RNA splicing pathways and innate immune and NF-κB signaling (Figure 1C, Supplemental Table 3).
Figure 1. Differential RNA isoform usage correlates with AML prognosis and oncogenic IRAK4 isoforms.
(A) Scatterplot of cumulative and isoform variances of genes in AML samples from TCGA (n = 160). Every point represents a gene, colored by the expression correlation of the most negatively correlated pair of isoforms for that gene. Blue-colored genes have at least one pair of isoforms with mutually exclusive expression pattern. (B) Hierarchical clustering analysis and relative expression of mRNA isoforms in AML samples. Relative expression is such that a value of 1 indicates that the isoform is the only isoform expressed for the given gene, and 0 indicates that the isoform is not expressed. (C) Pathway analysis of genes in Group 2 (from Figure 1B and Supplemental Figure 1C) associated with worse clinical outcome (n = 347 genes) determined by hypergeometric distribution test. (D) Kaplan-meier analysis of AML patients stratified on IRAK4-L (exon 4 included) or IRAK4-S (exon 4 excluded) expression. (E) Exon architecture of IRAK4 and protein domains (below). Sashimi plots represent junction reads in representative AML samples. Exon 4 reads were normalized to the total number of reads in each sample and plotted as a ratio (inclusion/total and exclusion/total). (F) Cumulative expression of IRAK4-L and IRAK4-S in individual AML patients (TCGA). Patients are ordered according the relative expression of IRAK4-L versus IRAK4-S. (G) Relative expression of IRAK4-L to IRAK4-S in normal BM (n = 4) and AML samples (TCGA; n = 160) is shown as box plots including the center (mean) and top and bottom quartiles. Wilcoxon Test, P = 0.07. (H) RT-PCR of IRAK4-L/S using primers flanking exon 4. Densitometric quantification of IRAK4 exon 4 inclusion calculated as the ratio of IRAK4-L versus both isoforms is shown below (three independent experiments). (I) Immunoblot of IRAK4 using an N-terminal antibody that recognizes IRAK4-L and a C-terminal antibody that recognizes IRAK4-L and IRAK4-S (three independent experiments). (J) Immunoblot of IRAK4 in AML patient samples (AML1 and AML2) and healthy samples (cord blood (CB), bone marrow CD34+ (BM-CD34+), BM mononuclear cells (BM-MNC)). The healthy samples are collected from independent donors. The percent of IRAK4-L of total IRAK4 isoforms is shown below.
Dysregulation of immune-related genes is widely reported in MDS and AML, however the precise alterations that drive innate immune signaling in MDS or AML HSC remain largely unknown14. Therefore, we decided to focus on RNA isoform switching of genes that belong to innate immune and NF-κB signaling. Among these genes, IRAK4 emerged as the leading candidate since the magnitude of IRAK4 isoform switching was highly significant among AML samples and the inclusion of exon 4 alone correlated with worse outcome (Figure 1D, Supplemental Figure 1D). IRAK4 is a serine/threonine kinase that mediates signaling downstream of the Toll-like receptor (TLR) superfamily, resulting in NF-κB and MAPK activation15. IRAK4 protein consists of the death domain (aa 1–125), a hinge domain (aa 140–150), and kinase domain (aa 150–460). IRAK4 in AML is encoded by two major protein isoforms, defined by the inclusion or the exclusion of exon 4 in the mRNA (based on NCBI Refseq annotation, or exon 6 according to AltAnalyze Ensembl 72 annotation)(Figure 1E). Inclusion of exon 4 is predicted to encode a longer 460 aa IRAK4 protein, which contains the 3 main functional domains (IRAK4-L). In contrast, exclusion of exon 4 is predicted to result in utilization of an alternative translational start site and encode a shorter 336 aa IRAK4 protein lacking the N-terminal death domain (IRAK4-S) (Figure 1E).
Expression of IRAK4-L is associated with AML.
We next compared IRAK4 exon 4 usage in AML and normal hematopoietic cells utilizing RNA-sequencing data obtained from AML samples and normal BM samples. Although the cumulative expression of both IRAK4 isoforms did not significantly differ among the patients, >50% of AML patients preferentially expressed IRAK4-L (>1.25-fold), while 18% preferentially expressed IRAK4-S (Figure 1F). As compared to normal BM cells, AML samples exhibit a significant proportion of IRAK4-L expression relative to IRAK4-S (Figure 1G). Breast, colon, and lung cancer cells also express a higher ratio of IRAK4-L relative to IRAK4-S as compared to normal tissues, indicating that expression of IRAK4-L is associated with oncogenic states (Supplemental Figure 1E). To independently verify IRAK4 RNA isoform expression in AML, we examined human AML and MDS cell lines. Sanger sequencing confirmed that both IRAK4 RNA isoforms are expressed (Figure 1H, Supplemental Figure 1F). Consistent with the finding that AML patients express 2 major isoforms of IRAK4; MDSL, TF1, and THP1 exhibit preferential expression of IRAK4-L, while HL60 and F36P preferentially express IRAK4-S (Figure 1H). In contrast, healthy cord-blood and BM CD34+ cells preferentially express IRAK4-S (Figure 1H). As such, a subset of AML is associated with expression of IRAK4-L resulting from inclusion of exon 4, while normal BM cells primarily express IRAK4-S as a result of exon 4 exclusion.
To determine whether both IRAK4 RNA isoforms encode proteins in AML, we examined IRAK4 protein expression in MDS/AML cell lines using an antibody that recognizes a C-terminal epitope present on both IRAK4 protein isoforms. In agreement with the RNA isoform expression analysis, MDSL, TF1, and THP1 exhibit preferential expression of the longer IRAK4 protein isoform (IRAK4-L at ~50 kDa) while HL60 and F36P primarily express a shorter IRAK4 protein isoform (IRAK4-S at ~37 kDa), that isn’t recognized by the N-terminal antibody (Figure 1I). F36P cells express a moderately smaller IRAK4 protein (~32 kDa) that corresponds to an IRAK4 isoform utilizing a minor second downstream open reading frame. Although expression of IRAK4-L differed between the AML cell lines, expression of MyD88 and IRAK1 was comparable across the AML cell lines (Figure 1I). Primary AML cells also express IRAK4-L, while normal BM-derived CD34+ hematopoietic cells and mononuclear cells predominantly express IRAK4-S (Figure 1J). These findings indicate that a significant proportion of AML patients primarily express the longer IRAK4 RNA and protein isoforms.
IRAK4-L results in maximal activation of innate immune signaling.
To investigate whether IRAK4-L expression is correlated with distinct gene regulatory networks in AML, RNA-sequencing data was examined in AML patients stratified based on the expression of IRAK4-L and IRAK4-S. Gene ontology and pathway analysis revealed that AML samples expressing IRAK4-L (top 25%) are significantly enriched in inflammatory and TLR signaling signatures as compared to AML samples expressing IRAK4-S (bottom 25%), suggesting that IRAK4-L could be a feed-forward activation node of oncogenic innate immune signaling (Figure 2A, Supplemental Figure 2A). To test whether the IRAK4 isoforms exert differential effects on innate immune signaling, IRAK4-L or IRAK4-S were expressed in HEK293 cells and evaluated for NF-κB and MAPK activation. Expression of IRAK4-L resulted in increased phosphorylation of IRAK1, p65, p38, and JNK (Figure 2B,C and Supplemental Figure 2B,C). In contrast, expression of IRAK4-S resulted in phosphorylation of p38, and JNK, but was less efficient at inducing phosphorylation of IRAK1 and p65. IRAK4-L expression also induced NF-κB- and AP1- dependent transcriptional activation as compared to control cells, while IRAK4-S induced AP1- dependent activation, but was less efficient at inducing NF-κB-dependent activation (Figure 2D,E). Expression of IRAK4-L in HL60, which primarily express IRAK4-S, resulted in activation of IRAK1 and p65, suggesting that IRAK4-L is sufficient to activate NF-κB in AML cells (Supplemental Figure 2D). To determine whether IRAK4-L and/or IRAK4-S are required to activate NF-κB activation, THP1 cells expressing an NF-κB reporter were transduced with shRNAs targeting IRAK4-L (shIRAK4-L) or IRAK4-L and IRAK4-S (shIRAK4-L/S)(Figure 2F). Knockdown of IRAK4-L resulted in suppression of NF-κB activation at levels similar to knockdown of IRAK4-L/S, indicating that IRAK4-S does not contribute significantly to NF-κB activation in AML (Figure 2G). These findings show that MAPK signaling is mediated by the C-terminal domain of IRAK4-S and IRAK4-L, while NF-κB signaling is mediated by the N- and C-terminal domains of IRAK4-L (Figure 2H).
Figure 2. IRAK4-L results in Myddosome assembly and maximal activation of innate immune signaling.
(A) Pathway analysis of enriched genes in AML patients preferentially expressing IRAK4-L (top) or IRAK4-S (bottom). The pathway scores were calculated by NetWalker51 based on a random walk method52. The scores directly reflect the enrichment of the indicated pathways for high (positive scores) or low (negative scores) expression of the IRAK4 Long isoform. The pathways shown here were the highest-scoring pathways. (B) HEK293 cells transfected with vector (pcDNA3.1), or FLAG-tagged IRAK4-L or IRAK4-S for 24 hours and immunoblotted for the indicated NF-κB and MAPK proteins (three independent experiments). (C) Densiometric analysis of panel (B). One-sided t-tests were used for statistical analyses. (D-E) NF-κB (D) and AP1 (E) activation was measured by κB-site and AP1-site containing reporter assays in HEK293 cells transfected with empty vector, IRAK4-L, or IRAK4-S. Values are normalized to Renilla-luciferase and empty vector (1.0) (3 or 6 independent experiments, respectively). Two- sided t-tests were used for statistical analyses. (F) Representative PCR and immunoblot of THP1 cells following knockdown of IRAK4-L and IRAK4-S (shIRAK4-L/S) or just IRAK4-L (shIRAK4-L) (three independent experiments). (G) NF-κB activation was measured in THP1 cells stably expressing a κB-site containing reporter and shRNAs targeting IRAK4-L (left) or IRAK4-L/S (right) after stimulation with PAM3CSK4 (+, 500 ng/ml or ++, 1000 ng/ml). (3 or 5 independent experiments, respectively). Two-sided t-tests were used for statistical analyses. (H) Overview of IRAK4-L and IRAK4-S downstream signaling based on Figure 2A–E. (I) Sequence-based prediction of protein-protein and domain-domain interactions using all isoforms of IRAK1, IRAK4, and MyD88. Superscripts indicate distinct RNA isoforms encoding the indicated long or short proteins. (J) HEK293 cells transfected with HA-MyD88 and either FLAG-IRAK4-L or FLAG-IRAK4- S were immunoblotted for HA-MyD88 after immunoprecipitation of FLAG. (representative of two independent experiments). (K) TF1 and HL60 cells were immunoblotted for IRAK4 with C-terminal antibody after immunoprecipitation of MyD88 (representative of two independent experiments). *, indicate IgG bands. All data represent the mean ± s.e.m.
To further explore NF-κB signaling differences between IRAK4-L and IRAK4-S, we performed a sequence-based prediction of protein-protein and domain-domain interactions.16 IRAK4-L protein isoforms, which retain the N-terminal death-domain, are predicted to directly bind MyD88 and IRAK1 (Figure 2I). Death domain interactions between MyD88 and IRAK4 initiate Myddosome formation, which promotes IRAK4 trans-autophosphorylation and activation.17,18 IRAK4 then activates IRAK1, enabling recruitment of TRAF6 and NF-κB activation. Based on these predictions, IRAK4-S protein isoforms, which lack the N-terminal death-domain, are predicted not to bind MyD88 (Figure 2I). To confirm this, FLAG-IRAK4-L or FLAG-IRAK4-S were expressed in HA-MyD88-expressing HEK293 cells followed by immunoprecipitation of IRAK4-L or IRAK4-S (anti-FLAG). MyD88 binds IRAK4-L, however is less efficient at binding to IRAK4-S (Figure 2J). The interaction between IRAK4 isoforms and MyD88 was also confirmed in AML cells that primarily express IRAK4-L (TF1) or IRAK4-S (HL60). Immunoprecipitation of endogenous MyD88 from TF1 and HL60 cells resulted in binding of MyD88 to IRAK4-L (Figure 2K). In contrast, IRAK4-S was not immunoprecipitated with MyD88 in HL60 cells (Figure 2K, Supplemental Figure 2E). Therefore, IRAK4-L mediates maximal activation of NF-κB signaling through stochastic assembly of the Myddosome complex in AML, while IRAK4-S exhibits reduced NF-κB activation resulting from diminished MyD88 binding.
IRAK4-L expression and kinase activity is required for leukemic cell function.
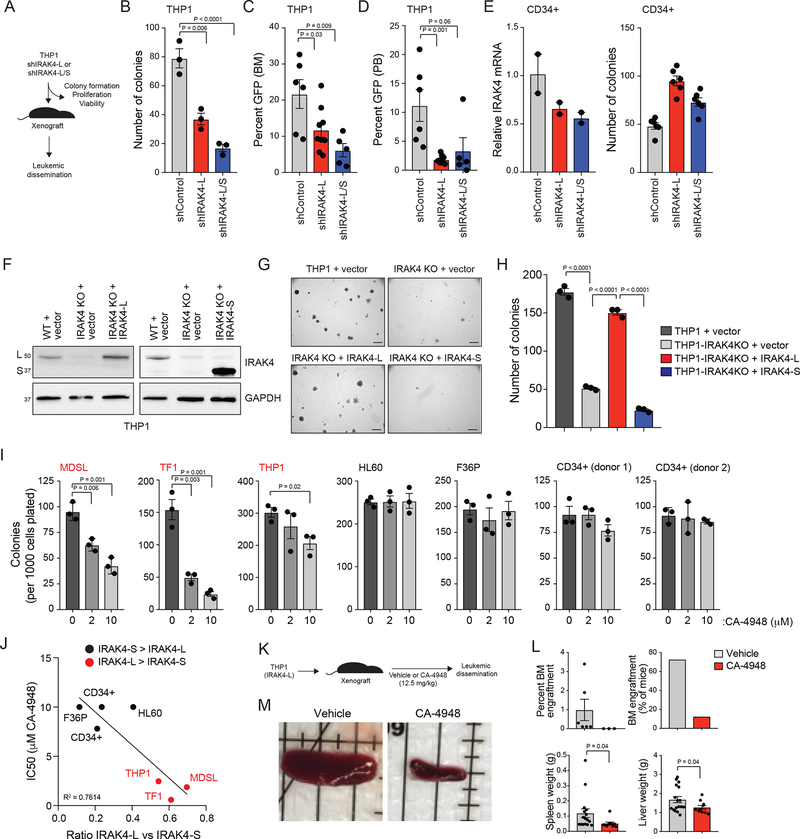
The requirement of IRAK4-L in AML was determined by utilizing shRNAs that target IRAK4- L (shIRAK4-L) or both isoforms (shIRAK4-L/S) (Figure 3A). THP1 cells, which express both IRAK4 isoforms, were transduced with shIRAK4-L or shIRAK4-L/S (Figure 2F). Knockdown of IRAK4-L is sufficient to impair the leukemic progenitor function of THP1 cells as evident by formation of fewer colonies in methylcellulose (Figure 3B). Moreover, THP1 cells expressing shIRAK4-L or shIRAK4- L/S were xenografted into NSG mice and examined for leukemic burden (Figure 3A). Knockdown of IRAK4-L in THP1 cells is sufficient to reduce leukemic burden in xenografted mice (Figure 3C,D). Importantly, the anti-leukemic effects of knocking down IRAK4-L/S was similar to knockdown of IRAK4-L alone, suggesting that IRAK4-L is the dominant isoform in leukemic cells (Figure 3B–D). As an orthogonal approach, all IRAK4 isoforms were deleted in THP1 cells using CRISPR/Cas9 gene editing (Supplemental Figure 3A,B). Deletion of IRAK4-L/S in THP1 cells (THP1-IRAK4KO) resulted in reduction of leukemic colony formation in methylcellulose-based media (Supplemental Figure 3C). Although knockdown or knockout of IRAK4 isoforms did not affect the growth and/or viability of THP1 cells when cultured in liquid media (Supplemental Figure 3D–F), deletion of IRAK4 resulted in increased expression of the monocyte differentiation marker CD14 and a corresponding decrease in the hematopoietic stem/progenitor marker CD34 (Supplemental Figure 3G), suggesting that IRAK4-L is required for maintaining a primitive hematopoietic state in THP1 cells. Reduced expression of IRAK4-L (shIRAK4-L) or in combination with IRAK4-S (shIRAK-L/S) did not impair the progenitor function of normal CD34+ cells, but rather resulted in a moderate increase in myeloid and erythroid progenitor colonies (Figure 3E). These findings demonstrate the requirement of IRAK4-L specifically in leukemia-propagating/initiating cells.
Figure 3. IRAK4-L is required for leukemic cell function.
(A) Overview of experimental design. (B) Colony formation of THP1 cells expressing shIRAK4-L, shIRAK4-L/S, or a control shRNA (shControl) (three independent experiments). Two-sided t-tests were used for statistical analyses. (C-D) THP1 cells expressing shIRAK4-L (n = 9 animals), shIRAK4-L/S (n = 5 animals), or a control shRNA (shControl; n = 6 animals) were engrafted into sublethally-irradiated NSG mice. After 6 weeks, leukemic burden (GFP+ cells) in the BM (C) and PB (D) was determined. Two-sided t-tests were used for statistical analyses. (E) Cord-blood CD34+ cells were transduced with shIRAK4-L, shIRAK4-L/S, or a control shRNA (shControl) and examined for IRAK4 knockdown (left; two independent experiments) and progenitor colony formation in methylcellulose (right; six independent experiments). Two-sided t-tests were used for statistical analyses. (F) THP1 cells transduced with empty retroviral vector (MSCV-IRES-GFP as a control) or THP1 IRAK4-KO cells transduced with retroviral vectors encoding IRAK4-L (left panel) or IRAK4-S (right panel) and then immunoblotted with the N-terminal IRAK4 antibody (left panel) and C-terminal IRAK4 antibody (right panel). (G-H) Representative images of parental and IRAK4-KO THP1 cells examined for leukemic progenitor function in methylcellulose (three biological replicates). Scale bar, 700 microns. Two-sided t-tests were used for statistical analyses. (I) Colony formation of MDS/AML cell lines and control CD34+ cells (2 independent donors) treated with DMSO or CA-4948 for 7–10 days (three independent experiments). Two-sided t-tests were used for statistical analyses. (J) Graph representing the IC50 relative to the ratio of IRAK4-L to IRAK4-S expression in the indicated cell lines and CD34+ cells summarized from Figure 1I and 3I. (K) THP1 cells were xenografted in NSG mice (n = 10) and treated with IRAK4 inhibitor (CA-4948) or vehicle at 12.5 mg/kg/5d/week for 5 weeks. (L) Mice xenografted with THP1 cells were assessed for leukemic engraftment in BM, and spleen and liver weight. Two-sided t-tests were used for statistical analyses. (M) Representative spleens are shown isolated from mice xenografted with THP1 cells at time of death after treatment with vehicle or CA-4948. All data represent the mean ± s.e.m.
To corroborate the requirement of IRAK4 isoforms in AML, IRAK4-L or IRAK4-S were re- expressed into THP1-IRAK4KO cells (Figure 3F). Expression of IRAK4-L in THP1-IRAK4KO cells restored the leukemic progenitor function as comparable to control THP1 cells (THP1 + vector)(Figure 3G,H). In contrast, IRAK4-S was unable to rescue the leukemic progenitor defect of THP1-IRAK4KO cells (Figure 3G,H). As a complimentary approach, restoration of IRAK4-L in THP1 cells expressing shRNAs targeting both IRAK4 isoforms (shIRAK4-L/S) resulted in leukemic colony formation comparable to control-transduced THP1 cells, while IRAK4-S was unable to rescue the leukemic progenitor defect of THP1 cells expressing shIRAK4-L/S (Supplemental Figure 4A–C). We next asked whether IRAK4 kinase activity was required for leukemic cells expressing IRAK4-L by using an ATP-competitive IRAK4 kinase inhibitor (CA-4948) approved for clinical use ()19. MDSL, TF1, and THP1 cells, which preferentially express IRAK4-L, formed fewer leukemic progenitor colonies with CA-4948 (Figure 3I). In contrast, HL60 and F36P, or normal CD34+ cells, which express IRAK4-S, were not affected by treatment with CA-4948 (Figure 3I). Thus, inhibition of IRAK4 kinase activity in MDS/AML cell lines that express IRAK4-L resulted in diminished leukemic function in vitro as compared to cells that preferentially express IRAK4-S (Figure 3J). To demonstrate that IRAK4 inhibition can suppress leukemic cell function in vivo, THP1 were xenografted into NSG mice. Following treatment with IRAK4 inhibitor (Figure 3K), THP1 infiltration in the BM, spleen, and liver was reduced as compared to mice receiving vehicle control (Figure 3L,M). Collectively, these results establish that IRAK4-L, but not IRAK4-S, signaling programs are required for leukemia-propagating cells, and that inhibition of IRAK4 abrogates leukemic growth in AML cells expressing IRAK4-L.
IRAK4-L expression is associated with U2AF1 mutations in MDS/AML.
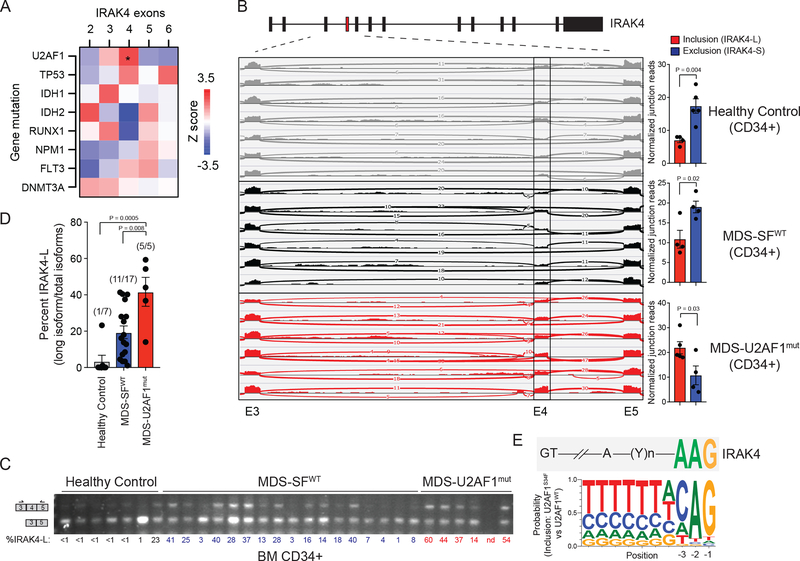
We next sought to identify the genetic alterations associated with IRAK4 exon 4 inclusion in AML. Among all AML patients examined in TCGA, inclusion of IRAK4 exon 4 was significantly correlated with mutations in U2 small nuclear RNA auxiliary factor 1 (U2AF1), the gene encoding an RNA-binding protein critical for 3’ splice-acceptor site recognition in pre-mRNAs (Figure 4A). Given the prevalence of U2AF1 mutations in MDS, we examined IRAK4 exon 4 usage in MDS CD34+ cells. All of the MDS patients with U2AF1 mutations (S34F, R156H, and Q157P/R) expressed IRAK4-L (6/6 by RNA-sequencing; and 5/5 by RT-PCR), while 9 of 17 (52%) MDS patients without splicing factor mutations expressed IRAK4-L (MDS-SF wild-type) (Figure 4B–D). In contrast, less than 20% of healthy age-matched BM CD34+ cells expressed IRAK4-L (Figure 4B–D). As observed in AML, MDS patients expressing increased levels of IRAK4-L had a worse prognosis as compared to MDS patients with lower levels of IRAK4-L (Supplemental Figure 4D).20
Figure 4. IRAK4-L expression is associated with U2AF1 mutations in MDS and AML.
(A) Heatmap was generated based on the Z-score correlation of individual exon’s expression to all somatic mutations. Each indicated mutation was regressed against the relative expression (after adjusting for total IRAK4 expression) of each IRAK4 exon (RPKM) in a multiple linear regression model. Shown are z-scores of partial correlations. A z-score of greater than |1.96| corresponds to P < 0.05. Six out of 160 AML patients exhibit U2AF1 mutations (Z = 2.43; *. P = 0.01). (B) Sashimi plots representing IRAK4 exon 4 inclusion or exclusion in healthy CD34+ cells (n = 7 samples), CD34+ cells isolated from MDS patients with no splicing factor (SFWT) mutations (MDS-SFWT; n = 6 samples), and from patients with mutation (mut) in U2AF1 (MDS-U2AF1mut; n = 6 samples) based on RNA-sequencing junction reads. Quantification of the junction reads for exon 4 are shown for healthy controls (n = 7), MDS-SFWT (n = 6), and MDS-U2AF1mut (n = 6). Exon 4 inclusion and exclusion reads were normalized to the total number of reads in each sample and plotted as a ratio (inclusion/total and exclusion/total). Two-sided t-tests were used for statistical analyses. (C) RT- PCR analysis of healthy CD34+ cells (n = 7 samples), CD34+ cells isolated from MDS patients with no splicing factor mutations (MDS-SFWT; n = 17 samples), and from patients with mutation in U2AF1 (MDS-U2AF1mut; n = 5 samples) using primers flanking IRAK4 exon 4. Densitometric quantification of IRAK4 exon 4 inclusion calculated as the ratio of the long isoform versus both isoforms is shown below each sample. (D) Densitometric quantification of IRAK4 exon 4 inclusion is summarized from panel (C). The number in parentheses indicates the number of samples exhibiting >10% expression of IRAK4-L relative to all IRAK4 isoforms. Two-sided t-tests were used for statistical analyses. (E) Consensus sequence motifs identified at the distal 3’ splice sites of exon exclusion events in U2AF1-S34F cells as compared to U2AF1 using publicly available data and visualized with weblogo software. Above the U2AF1-S34F consensus motifs is the sequence of the distal 3’ splice site of IRAK4 exon 4. All data represent the mean ± s.e.m.
The consensus sequence flanking the adenine-guanine dinucleotide at the 3’ splice acceptor site is recognized by U2AF1 and impacts exon usage by the U2AF1 mutants.21,22 A cytosine (C) or adenine (A) at the −3 position correlates with exon inclusion by U2AF1-S34F as compared to wild-type U2AF1.23,24 The −3 position of IRAK4 exon 4 contains an adenine, supporting the model in which U2AF1-S34F mediates exon inclusion (Figure 4E). Moreover, the +1 position of IRAK4 exon 4 contains a guanine, which is associated with exon inclusion of U2AF1- Q157X as compared to wild-type U2AF1.21 These findings suggest that mutant U2AF1 correlates with inclusion of exon 4 and preferential expression of IRAK4-L in AML and MDS CD34+ cells.
U2AF1-S34F induces IRAK4-L and innate immune pathway activation.
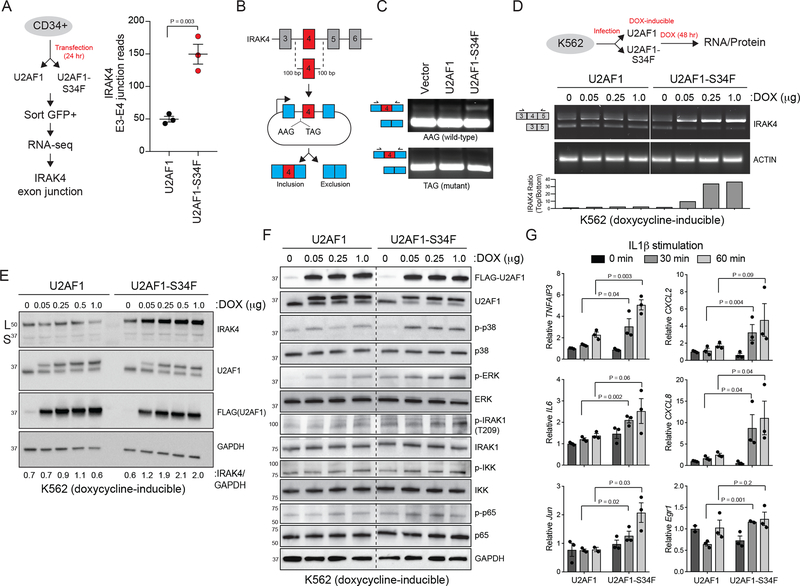
To investigate whether mutant U2AF1 directly regulates IRAK4 exon 4 usage and expression of IRAK4-L, RNA-sequencing was performed on healthy CD34+ cells expressing wild-type or U2AF1-S34F (Figure 5A). Importantly, expression of U2AF1-S34F resulted in increased reads at the IRAK4 exon 3–4 junction as compared to U2AF1-WT, indicating that mutant U2AF1 mediates inclusion of exon 4 (Figure 5A).25 To gain further insight into the regulation of IRAK4 exon 4, we generated an IRAK4 exon 4 cassette splicing reporter (HEK293-IRAK4exon4)(Figure 5B). A mutant version of the IRAK4 exon 4 cassette (“TAG”) was also generated containing a thymine (T) in place of adenine (A) at the −3 position (“AAG”) since U2AF1-S34F mediates exon inclusion in the presence of AAG but not TAG (Figure 5B). Transfection of U2AF1-S34F into HEK293-IRAK4exon4 cells resulted in inclusion of IRAK4 exon 4 from the WT “AAG” splicing reporter (11.7% exon 4 inclusion) compared to U2AF1-WT (1.3 % exon 4 inclusion) or vector- transfected HEK293-IRAK4exon4 cells (1.4% exon 4 inclusion)(Figure 5C). Conversely, U2AF1- S34F did not induce inclusion of IRAK4 exon 4 from the mutant “TAG” splicing reporter (0.4% exon 4 inclusion; n = 2). Therefore, U2AF1-S34F likely mediates inclusion of IRAK4 exon 4 by direct binding. To extend these observations to human AML cells, we utilized K562 erythroleukemia cells that express doxycycline-inducible FLAG-tagged U2AF1-S34F or U2AF1-WT.26 Expression of U2AF1-WT with doxycycline did not affect IRAK4 exon 4 usage or protein expression (Figure 5D,E). In contrast, expression of U2AF1-S34F after doxycycline resulted in robust and dose-dependent IRAK4 exon 4 inclusion (Figure 5D), which coincided with increased expression of IRAK4-L protein (Figure 5E).
Figure 5. U2AF1-S34F induces expression of IRAK4-L and increased innate immune pathway activation.
(A) Experimental design to analyze RNA splicing changes in normal CD34+ cells expressing U2AF1 or U2AF1-S34F (left). Exon 3–4 junction reads of IRAK4 in U2AF1 and U2AF1-S34F CD34+ cells (three independent samples) (right). Two-sided t-tests were used for statistical analyses. (B) Schematic of IRAK4 exon 4 splicing reporter. Wild-type IRAK4 exon 4 (AAG at −3 position) or mutant IRAK4 exon 4 (TAG at −3 position) and 100 bp of flanking introns were cloned into a splicing reporter (pFlare5A)53 and expressed into HEK293 cells (HEK293-IRAK4exon4). (C) Representative PCR of IRAK4 exon 4 splicing in HEK293-IRAK4exon4 expressing U2AF1 and U2AF1-S34F (five independent experiments for AAG; two independent experiments for TAG). (D) (Top) Experimental design to measure IRAK4 RNA and protein isoform expression in K562 cell lines that have stably integrated doxycycline-inducible FLAG-U2AF1 or FLAG-U2AF1-S34F. (Bottom) IRAK4 RNA isoform expression was measured using primers flanking exon 4 in doxycycline (DOX)-induced K562 cells expressing FLAG-U2AF1 or FLAG- U2AF1-S34F. Top band indicates IRAK4-L and bottom band indicates IRAK4-S (five independent experiments). (E) IRAK4 protein isoform expression analyzed with a C-terminal IRAK4 antibody in DOX-induced K562 cell expressing FLAG-U2AF1 or FLAG-U2AF1-S34F (three independent experiments). (F) The indicated NF-κB and MAPK protein expression was measured in DOX- induced K562 cells expressing FLAG-U2AF1 or FLAG-U2AF1-S34F by immunoblotting (two independent experiments). (G) Expression of the indicated NF-κB (TNFAIP3, IL6, CXCL2, and CXCL8) and MAPK (Jun and Egr1) target genes was measured in DOX-induced K562 cells expressing FLAG-U2AF1 or FLAG-U2AF1-S34F after IL-1β stimulation (10 ng/ml) for 30 or 60 min (three independent experiments). Two-sided t-tests were used for statistical analyses. All data represent the mean ± s.e.m.
Since U2AF1-S34F expression results in expression of IRAK4-L, and expression of IRAK4-L alone is sufficient to mediate activation of innate immune and NF-κB signaling, we next investigated the consequences of U2AF1-S34F on NF-κB and MAPK activation in AML cells. Expression of U2AF1-S34F in K562 cells resulted in activation of MAPK signaling (p-p38 and p- ERK) and NF-κB signaling (p-IRAK1, p-IKK, and p-p65) as compared to wild-type U2AF1- expressing K562 cells (Figure 5F). U2AF1-S34F-expressing K562 cells are also hypersensitive to IL1b stimulation as indicated by increased expression of NF-κB (TNFAIP3, IL6, CXCL2, and CXCL8) and MAPK (Jun and Egr1) target genes (Figure 5G). As such, U2AF1-S34F is sufficient to induce expression of IRAK4-L and consequently innate immune pathway activation.
U2AF1-S34F AML cells are sensitive to IRAK4 inhibitors.
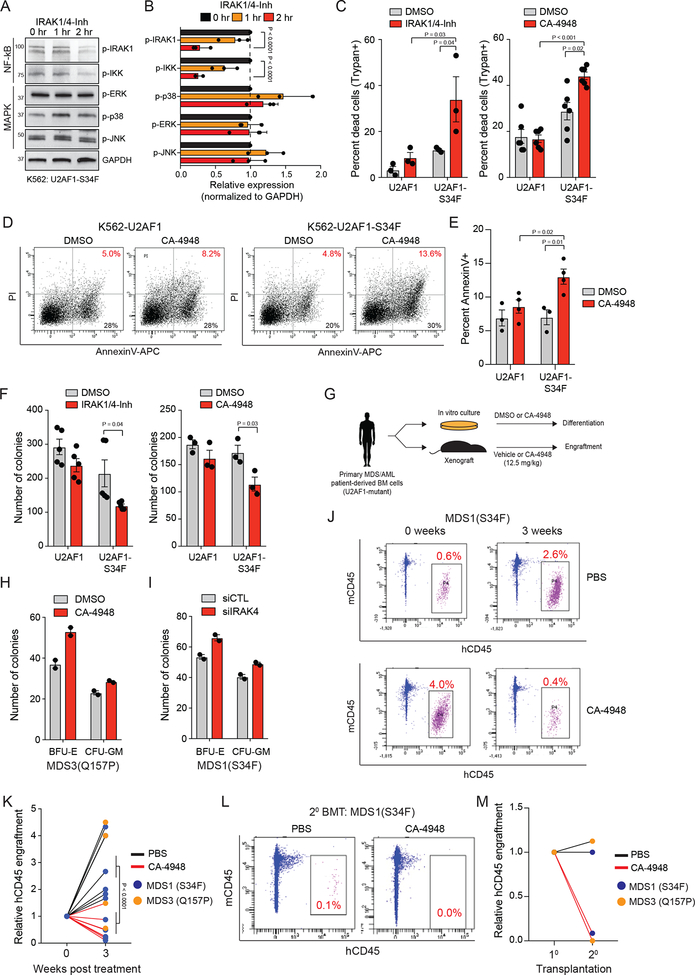
We determined whether inhibiting IRAK4 kinase activity was sufficient for suppressing NF-κB and/or MAPK signaling in U2AF1-mutant AML cells expressing IRAK4-L. To inhibit IRAK4 kinase activity, we evaluated two structurally-unrelated IRAK4 inhibitors, IRAK1/4-Inh27 and CA-4948.19 When K562 cells expressing U2AF1-S34F (K562-U2AF1-S34F) were treated with IRAK1/4-Inh, the activation of NF-κB signaling was inhibited as indicated by reduced p-IRAK1 and p-IKK (Figure 6A,B, and Supplemental Figure 5). In contrast, MAPK signaling was not diminished following treatment with IRAK1/4-Inh. These findings suggest that MAPK activation not only occurs downstream of IRAK4-L, but also secondary to RNA splicing changes.28 Based on these observations, we reasoned that inhibition of IRAK4-L catalytic function will suppress pro-survival signaling via NF-κB, thus exposing a therapeutic vulnerability in U2AF1-mutant AML/MDS. K562-U2AF1-S34F cells treated with IRAK4 inhibitors (IRAK1/4-Inh or CA-4948) exhibited increased cytotoxicity (Trypan blue-positive cells) as compared to vehicle-treated cells (Figure 6C), which correlated with elevated AnnexinV/PI-positive cells (Figure 6D,E). In contrast, K562- U2AF1 were not sensitive to the IRAK4 inhibitors (Figure 6C–E). In addition, K562-U2AF1-S34F cells formed fewer leukemic progenitor colonies when treated with IRAK4 inhibitors as compared to vehicle-treated cells (Figure 6F). Next, primary patient-derived cells from MDS and AML patients with U2AF1 mutations were grown in clonogenic assays after siRNA mediated knockdown or pharmacologic inhibition of IRAK4. Both IRAK4 knockdown and inhibition with CA-4948 led to enhanced erythroid and myeloid differentiation (Figure 6G–I; Supplemental Figure 6A–C). In contrast, normal CD34+ cells treated with CA-4948 did not exhibit altered differentiation (Supplemental Figure 6D).
Figure 6. U2AF1-S34F AML cells are sensitive to IRAK4 inhibitors.
(A) K562-U2AF1-S34F cells were treated with 10 mM IRAK1/4-inhibitor for 1 or 2 hours and immunoblotted for NF-κB and MAPK activation. (B) Densitometric analysis of panel (A) summarized from three independent biological replicates. Two-sided t-tests were used for statistical analyses. (C) K562-U2AF1 cells were treated with DMSO, 10 μM IRAK1/4-Inh, or 10 μM CA-4948 for 7 days and assessed for viability by Trypan Blue exclusivity (three independent experiments). One-sided t-tests were used for statistical analyses. (D) Representative images of K562 cells expressing wild-type U2AF1 or U2AF1-S34F were treated with DMSO or 10 μM CA-4948 for 48 hours days and then analyzed by flow cytometry for AnnexinV and Propidium Iodide (PI) staining (three independent experiments). (E) Summary of (D). Two-sided t-tests were used for statistical analyses.(F) K562 cells treated with 10 mM IRAK1/4-Inh (five independent experiments) or CA-4948 (three independent experiments) were evaluated after 7 days for colony formation in methylcellulose. Two-sided t-tests were used for statistical analyses. (G) Schematic of experimental design. (H) MDS patient-derived BM cells were evaluated after 7 days for colony formation in methylcellulose treated with 0.5 μM CA-4948 or vehicle (two independent experiments). (I) MDS patient-derived BM cells were transfected with siIRAK4 or control siRNA and then evaluated after 7 days for colony formation in methylcellulose (two independent experiments). (J) NSG mice were xenografted with U2AF1-mutant MDS BM cells. After engraftment, mice were treated with CA-4948 or vehicle and human cell engraftment was assessed by flow cytometry on BM aspirates. (K) Summary of BM cell engraftment (hCD45) for independent mice and patient-derived samples. Two-sided t-tests were used for statistical analyses. (L) MDS BM cells were isolated from mice treated with vehicle (PBS) or CA-4948 treatment after 3 weeks (panel J,K) and then xenografted into secondary NSG mice. Human cell engraftment was assessed by flow cytometry on BM aspirates. (M) Summary of BM cell engraftment (hCD45) for mice and 2 independent patient-derived samples. Data represent the mean ± s.e.m. for all panels except H and I where data represent the mean ± s.d.
To determine the efficacy of IRAK4 inhibitors on primary U2AF1-mutant cells in vivo, xenografts from MDS patients with U2AF1 mutations were established in NSG mice and treated with CA-4948 (12.5 mg/kg/d) or vehicle (Figure 6G). While vehicle-treated mice exhibited a 3.2- fold increased expansion of xenografted MDS cells in the BM, IRAK4 inhibition led to a 50% decrease in MDS cell engraftment after 3 weeks of CA-4948 treatment (Figure 6J,K). To establish that inhibition of IRAK4 suppresses the function of disease-propagating MDS cells, BM cells were isolated from primary xenografted mice treated with vehicle or CA-4948 for 3 weeks (from Figure 6J,K), and then transplanted into secondary recipient NSG mice. Mice transplanted with vehicle-treated MDS cells engrafted into secondary recipient mice (Figure 6L,M). In contrast, the engraftment of CA-4948-treated MDS cells was negligible in secondary recipients (Figure 6L,M), suggesting that inhibition of IRAK4-L impairs U2AF1-mutant MDS propagating cells in vivo. In parallel, we evaluated potential hematologic toxicity after repeated administration of CA-4948 in mice. Administration of CA-4948 for 12 weeks in NSG mice did not affect body weight (not shown) and hematologic blood parameters (Supplemental Figure 6E). These findings indicate that U2AF1-S34F-expressing MDS/AML cells have acquired a dependency on IRAK4-L and are sensitive to IRAK4 inhibition.
Discussion
Clinically-relevant molecular subtypes of AML were identified based on differential RNA isoform usage. One major subtype of AML was enriched for RNA isoform changes affecting innate immune pathway genes. Among these genes, an isoform of IRAK4 that retains exon 4 and encodes IRAK4-L is expressed in AML and U2AF1-mutant MDS/AML. While consistent with the current paradigm of Myddosome signaling, our findings uncovered a mechanism by which IRAK4-L is generated by alternative splicing in MDS/AML to mediate maximal activation of NF-κB through MyD88 binding. In contrast, healthy hematopoietic cells primarily express IRAK4-S and have diminished binding to MyD88 and lower NF-κB activation potential. Thus, IRAK4-S is normally expressed to mitigate spurious and/or excessive NF-κB activation as a result of its diminished ability to bind MyD88. This observation may reveal an evolutionarily-conserved mechanism to regulate innate immune responses. Overexpression of immune-related genes is widely reported in MDS and AML,29,30 and chronic innate immune pathway activation, predominantly via TLRs, increases the risk of developing myeloid malignancies.31 Evidence that innate immune pathway activation is causal in MDS or AML is supported by work related to miR-146a and TIFAB, which reside within the deleted region in del(5q) MDS/AML. Deletion of miR-146a and/or TIFAB results in derepression of their targets, TRAF6 and IRAK1, and impaired hematopoiesis and BM failure in mice32–34. Phosphorylation of IRAK1 is observed in non-del(5q) MDS/AML and other cancers, however the mechanism of IRAK1 activation remains unidentified.35–41 In some of these cases, we propose that isoform switching from IRAK4-S to IRAK4-L is the molecular basis of IRAK1 activation. While there are examples of indirect mechanisms that result in activation of innate immune signaling in MDS/AML, the present study provides direct genetic evidence, via mutations in the splicing factor U2AF1, that links oncogenic innate immune signaling and IRAK4 activation in MDS/AML. Although all U2AF1-mutant MDS and AML exhibit expression of IRAK4-L, approximately 50% of remaining patients express IRAK4-L indicating that other genetic and/or molecular mechanisms regulate alternative splicing of IRAK4 and expression of IRAK4-L. IRAK4-L correlated with mutations in U2AF1, but it did not correlate with other splicing factor mutations in MDS or AML. For example, mutations in SRSF2 also result in activation of NF-κB signaling, but these effects are independent of IRAK4 isoform regulation42.
Mutations in RNA splicing factors are the most frequent class of mutation in MDS and common in AML.2,5,9,43–45 Not surprisingly, mouse models with splicing factor gene mutations develop features of MDS.3,26,46–49 Mutations affecting the genes encoding core spliceosome machinery, such as in SF3B1, U2AF1, and SRSF2 do not result in loss-of-function phenotypes, but rather in selective splicing changes due to aberrant splice site recognition.3,5,21,25,48–50 Although many RNA splicing changes in U2AF1-mutant cells result in aberrant RNA splicing, non-functional transcripts, and/or nonsense-mediated decay, IRAK4 is an example in which mutant U2AF1 generates an isoform with oncogenic activity. Given the prevalence of RNA splicing gene mutations in hematologic malignancies, effective strategies to therapeutically target spliceosome mutant malignancies are needed. Small molecule modulators of the spliceosome target normal splicing by interfering with the splicing machinery and altering 3’ splice site recognition. Based on our reported findings, an alternative strategy to target U2AF1-mutant MDS/AML is by exposing the therapeutic vulnerability of IRAK4-L expression with IRAK1/4 inhibitors. However, elucidating the mechanism of IRAK4 isoform control during normal development, inflammation, and in non-splicing factor mutant cancers will better inform of disease conditions and patients most suitable for treatment with IRAK1.4 inhibitors. Taken together, we report that mutations in U2AF1 induce expression of targetable “active” IRAK4 isoforms and provide a genetic link to activation of oncogenic innate immune signaling in MDS and AML.
Supplementary Material
Acknowledgments
This work was supported by Cincinnati Children’s Hospital Research Foundation, Leukemia Lymphoma Society, and National Institute of Health (R35HL135787, RO1DK102759, RO1DK113639), and Edward P. Evans Foundation grants to DTS. AV is supported by National Institute of Health (R01HL139487 and R01DK103961), Leukemia and Lymphoma Society, and EvansMDS grants. AP and JB are supported by Bloodwise (UK, grant 13042). MAS is supported by a National Institute of Health Research Training and Career Development Grant (F31HL132420). IDCF is supported by the Intramural Research Program of NIAID. We also thank Robert Booher (Curis Inc.) for suggestions. We thank Melissa Scott and Yueh-Chiang Hu from the Gene Editing Core at CCHMC for their assistance.
Conflicts of Interest
A.V. has received research funding from GlaxoSmithKline, Incyte, MedPacto, Novartis, Curis and Eli Lilly and Company, has received compensation as a scientific advisor to Novartis, Stelexis Therapeutics, Acceleron Pharma, and Celgene, and has equity ownership in Stelexis Therapeutics.
References
- 1.Vitting-Seerup K & Sandelin A The Landscape of Isoform Switches in Human Cancers. Mol Cancer Res 15, 1206–1220 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Inoue D, Bradley RK & Abdel-Wahab O Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev 30, 989–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim E, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A 112, E4726–4734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez B, Walter MJ & Graubert TA Splicing factor gene mutations in hematologic malignancies. Blood 129, 1260–1269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 44, 53–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch CM, et al. Molecular features of early onset adult myelodysplastic syndrome. Haematologica 102, 1028–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellagatti A & Boultwood J Splicing factor gene mutations in the myelodysplastic syndromes: impact on disease phenotype and therapeutic applications. Adv Biol Regul 63, 59–70 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627; quiz 3699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindsley RC, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125, 1367–1376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363, 2424–2433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreyro L, Chlon TM & Starczynowski DT Chronic immune response dysregulation in MDS pathogenesis. Blood 132, 1553–1560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patra MC & Choi S Recent Progress in the Molecular Recognition and Therapeutic Importance of Interleukin-1 Receptor-Associated Kinase 4. Molecules 21(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 164, 805–817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dossang AC, et al. The N-terminal loop of IRAK-4 death domain regulates ordered assembly of the Myddosome signalling scaffold. Sci Rep 6, 37267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrao R, et al. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Mol Cell 55, 891–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Booher RN SM, Xu G, Cheng H, Tuck DP. Efficacy of the IRAK4 inhibitor CA-4948 in patient-derived xenograft models of diffuse large B cell lymphoma in American Association for Cancer Research (Washington DC, 2017). [Google Scholar]
- 20.Gerstung M, et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun 6, 5901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25, 14–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks AN, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 9, e87361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fei DL, et al. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and Is Required for Cell Survival. PLoS Genet 12, e1006384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yip BH, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest 127, 2206–2221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okeyo-Owuor T, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 29, 909–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirai CL, et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat Commun 8, 14060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powers JP, et al. Discovery and initial SAR of inhibitors of interleukin-1 receptor- associated kinase-4. Bioorg Med Chem Lett 16, 2842–2845 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Maimon A, et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep 7, 501–513 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Hofmann WK, et al. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood 100, 3553–3560 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Pellagatti A, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Kristinsson SY, et al. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. J Clin Oncol 29, 2897–2903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med 16, 49–58 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Varney ME, et al. Epistasis between TIFAB and miR-146a: neighboring genes in del(5q) myelodysplastic syndrome. Leukemia 31, 1659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varney ME, et al. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J Exp Med 212, 1967–1985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhyasen GW, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell 24, 90–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beverly LJ & Starczynowski DT IRAK1: oncotarget in MDS and AML. Oncotarget 5, 1699–1700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dussiau C, et al. Targeting IRAK1 in T-cell acute lymphoblastic leukemia. Oncotarget 6, 18956–18965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, et al. Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J Clin Invest 125, 1081–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goh JY, et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat Med 23, 1319–1330 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Adams AK, et al. IRAK1 is a novel DEK transcriptional target and is essential for head and neck cancer cell survival. Oncotarget 6, 43395–43407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wee ZN, et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance to paclitaxel. Nat Commun 6, 8746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SC, et al. Synthetic Lethal and Convergent Biological Effects of Cancer-Associated Spliceosomal Gene Mutations. Cancer Cell 34, 225–241 e228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Makishima H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa S Splicing factor mutations in AML. Blood 123, 3216–3217 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Kon A, et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood 131, 621–635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komeno Y, et al. SRSF2 Is Essential for Hematopoiesis, and Its Myelodysplastic Syndrome-Related Mutations Dysregulate Alternative Pre-mRNA Splicing. Mol Cell Biol 35, 3071–3082 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obeng EA, et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 30, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mupo A, et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 31, 720–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirai CL, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell 27, 631–643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komurov K, Dursun S, Erdin S & Ram PT NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics 13, 282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komurov K, White MA & Ram PT Use of data-biased random walks on graphs for the retrieval of context-specific networks from genomic data. PLoS Comput Biol 6(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoilov P, Lin CH, Damoiseaux R, Nikolic J & Black DL A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci U S A 105, 11218–11223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.