Abstract
The circadian clock is a biological mechanism that dictates an array of rhythmic physiological processes. Virtually all cells contain a functional clock whose disruption results in altered timekeeping and detrimental systemic effects, including cancer. Recent advances have connected genetic disruption of the clock with multiple transcriptional and signaling networks controlling tumor initiation and progression. An additional feature of this circadian control relies on cellular metabolism both within the tumor microenvironment and the organism systemically. A discussion of major advances related to cancer metabolism and the circadian clock will be outlined, including new efforts related to metabolic flux of transformed cells, metabolic heterogeneity of tumors, and the implications of circadian control of these pathways.
The circadian clock maintains cell autonomous oscillations with a 24-hour periodicity (Figure 1, Box 1). Yet, the clock also utilizes external synchronization by cues such as light, temperature and food intake, the so-called zeitgebers or time-givers that can adapt circadian timekeeping. The mammalian pacemaker governs rhythms in sleep/wake cycles, feeding/fasting control, metabolism, hormone secretion, and immune function. Disruptions in biological rhythms result in numerous physiological disorders in organismal homeostasis, the consequences of which have been linked to several pathologies, including endocrine disruption, metabolic syndrome and cancer [1]. Specifically, clinical and laboratory evidence has long suggested that a relationship exists between the circadian clock and tumorigenesis [2], yet the precise molecular mechanisms of this connection are not fully elucidated. Interestingly, epidemiological evidence shows a link between hormone-dependent cancers and circadian environmental disruption by shift-work, light at night exposure, and late-night eating behavior [3–6]. To further support this epidemiological data, genetic disruption of the circadian clock in mouse models has revealed a strong link with specific cancers [7–9], though the precise mechanisms related to cancer initiation versus progression remain unknown. Specifically, the role of cellular metabolism in driving these transformation events, and how this is controlled by the clock and therefore disrupted during tumorigenesis, still requires further investigation.
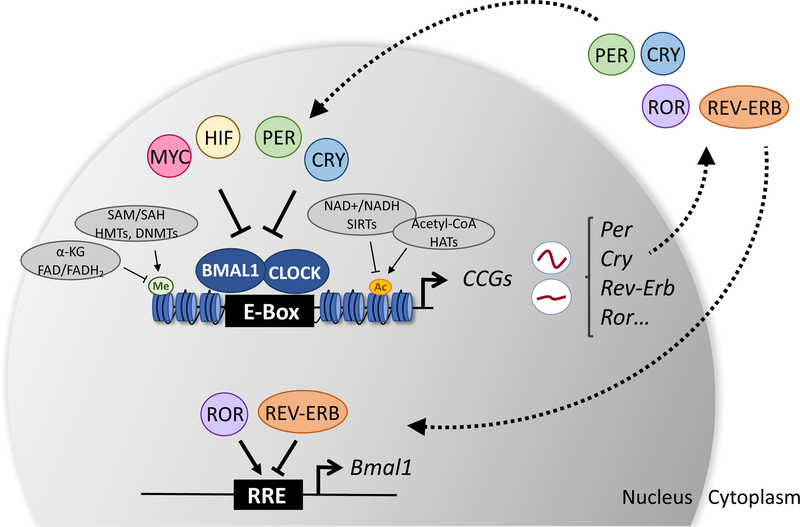
Figure 1: The transcriptional/translational feedback loop driving the mammalian circadian clock.
The circadian transcriptional/translational feedback loop (TTFL) operates within a precisely timed period of 24-hours in mammalian cells. The core clock machinery consists of the bHLH DNA-binding transcription factors, CLOCK and BMAL1 [51, 151]. These transcription factors heterodimerize and bind E-Box sequences to control the rhythmic expression of ~10–15% of core clock and clock-controlled genes (CCGs) [13, 104–106]. CLOCK:BMAL1-dependent transcription of CCGs peaks during the day and is inhibited by the circadian repressors, Period (PER) and Cryptochrome (CRY), at night [107–109]. An additional level of circadian regulation exists with the nuclear receptors RORα and REV-ERBα that activate and repress transcription of Bmal1, respectively [113, 114]. This secondary feedback loop offers an additional level of control outside the core TTFL to modulate circadian gene expression. Additional transcriptional crosstalk between the circadian machinery with MYC and HIF is also shown. Inputs of regulatory metabolites and their influence on circadian gene expression through epigenetic control are highlighted. Abbreviations: nicotinamide adenine dinucleotide (NAD+), flavin adenine dinucleotide (FAD), s-adenosylmethionine (SAM), s-adenosylhomocysteine (SAH), alpha-ketoglutarate (α-KG).
Box 1: The Cogs and Gears that Drive the Mammalian Circadian Clock.
The circadian molecular machinery consists of the core DNA-binding transcription factors, circadian locomotor cycles output kaput (CLOCK) and aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL or BMAL1), that heterodimerize and drive the oscillation of ~10–15% of genes in a defined tissue-specific program [13, 51, 104–106] by binding E-Box sequences within the genome [51, 107] (Figure 1). CLOCK:BMAL1-dependent transcription of core clock and clock-controlled genes (CCGs) peaks during the day, while transcription is inhibited by the circadian repressors, Period (PER) and Cryptochrome (CRY), at night [107, 108]. The PER and CRY repressive complexes of the circadian clock have been an active area of research for several years and key structural features of these complexes have been elucidated [109–112]. An additional level of circadian regulation exists with the orphan nuclear receptors RORα and REV-ERBα which activate and repress transcription of the Bmal1 gene, respectively [113–116].
Moreover, regulation of circadian transcription also includes a large array of epigenetic modifications, including those modulated by histone acetyltransferases (HATs), histone deacetylases (HDACs), histone and DNA methyl transferases (HMTs and DNMTs) and demethylases, which establishes a mechanism by which the circadian transcriptional circuit can be fine-tuned over the day/night cycle (Figure 1) [80, 117–122]. For instance, the acetylation state of histone H3 (H3K9/K14) is critical in dictating circadian gene expression through CLOCK, CBP, p300 and PCAF [123–125], and these marks are conversely regulating by the HDAC activity of the mammalian sirtuins, SIRT1 and SIRT6 [80, 117, 118, 126], and HDAC1 and HDAC3 [109, 122, 127]. In addition to acetylation, histone methylation has been also implicated in circadian chromatin remodeling through the histone methyltransferases (myeloid/lymphoid or mixed-lineage leukemia 1) MLL1 [120] and MLL3 [119] which are permissive for circadian gene expression. Previous studies have also described the role of EZH2 in the regulation of H3K27 methylation [128] and the role of demethylases in proper regulation of circadian gene expression [121, 129]. Lastly, mono-ubiquitination of histone H2B has also been described as a critical aspect of crosstalk between the positive and negative limbs of the circadian transcriptional/translational feedback loop [130]. Interestingly, the extent of epigenetic regulation within the context of the circadian system remains largely unexplored. This is especially important given the large array of acylation modifications that have been recently described such as propionylation, butyrylation, and crotonylation and their implication in regulation of gene expression specifically linked to cellular metabolism [131, 132].
This review serves to summarize the current state of knowledge regarding the links between the biological pacemaker and cancer in an effort to highlight new avenues for therapeutic intervention. Specifically, we focus on several facets of tumor metabolism that can be rewired in response to circadian disruption. We extend our discussion of cancer metabolism to address differences in metabolic demand of tumors, and potential heterogeneity in fuel preference. Both clinical and laboratory data has recently relied on use of metabolic tracing in vivo to address these complex questions, and we highlight these efforts in terms of the power of metabolic flux and metabolic fate tracing using stable isotopes. Additionally, we discuss recent advances in teasing out how the circadian clock is implicated in regulating several pathways linked to oncogenes and tumor suppressors. Ultimately, our goal is to point to new directions where further research emphasis is required to fully understand how clock disruption and tumor initiation and progression converge at the molecular level.
Disruption of the Circadian Axis is Linked to Cancer
The circadian clock is the biological pacemaker that controls rhythmic processes related to endocrinology, sleep/wake cycles, mood regulation, feeding/fasting rhythms and overall metabolism. These circadian rhythms are cell autonomous and perpetuated in nearly all cells within a 24-hour period (Figure 1) [10]. Yet, the circadian clock is heavily dependent on external cues, or zeitgebers (light, nutritional cues, temperature), that can synchronize the otherwise self-sustained rhythms driven by the clock [11, 12]. Light provides synchrony to the organism through the central circadian clock, housed within the suprachiasmatic nucleus (SCN), which is able to transmit signals to the peripheral clocks, such as liver, muscle, skin, to maintain their rhythms in a tissue-specific manner [12, 13].
Early findings in the circadian field identified that mice with an ablation of the central clock located within the SCN exhibit increased growth of tumor xenografts as compared to mice with an intact circadian pacemaker [14]. Bilateral electrolytic lesions of the SCN enhanced tumor growth of implanted Glasgow osteosarcoma and pancreatic ductal adenocarcinoma (PDAC) versus sham operated mice [14]. Yet, the molecular mechanisms by which disruption of the central pacemaker results in enhanced tumor growth is unknown. These findings suggest that disruption of the synchrony between the SCN and peripheral clocks may have important physiological consequences for tumorigenesis.
To further address the role of physiological disruption of the clock and the impact of ‘desynchronization’ on tumor initiation and progression, environmental disruption paradigms have been utilized in rodent models. Chronic Jet Lag (CJL) refers to repeated 8-hour phase advances in the light schedule every 2 days for several weeks. This paradigm is known to simulate harsh jet lag conditions similar to humans and results in detrimental physiological effects in rodent models through disruption of circadian rhythms and acceleration of tumor growth in vivo. For instance, wild-type (WT) mice undergoing repeated jet lag manipulation display disrupted circadian gene expression, resulting in increased growth of Glasgow osteosarcoma [15] as well as enhanced incidence of lymphoma and hepatocellular carcinoma (HCC) [9, 16]. Similar experiments have been performed with mice harboring mutations in clock genes, specifically Cry1/2−/−, Per2−/−, or Per1−/−;Per2m/m. These mutant mice display accelerated incidence of lymphoma, osteosarcoma and HCC when subjected to severe CJL versus WT mice [9]. Furthermore, environmental disruption of the circadian clock can also be performed by light-at-night (LaN) exposure or dim LaN (dLaN), which is an increasing concern in modern society. Current protocols for LaN exposure involve changing the circadian subjective night, so that the dark period becomes shortened from a 12-hour light/12-hour dark (12:12) cycle to a 16:8 light/dark cycle for several weeks [17]. LaN and dLaN exposure (light intensity of 5 lux) are known to alter feeding behavior, circadian gene expression and body weight in rodents [17–19], therefore defining how altered light exposure drives tumor formation is essential. These concerns regarding circadian environmental disruption are particularly relevant to human behavior and activity patterns in developed society.
Clinical Evidence Linking the Clock with Cancer
Epidemiological studies have linked circadian clock disruption through night shift work or light-at-night exposure with hormone-dependent cancers, and this evidence has been recently reviewed [2]. Additionally, clinical evidence connects methylation of clock gene promoters or single nucleotide polymorphisms (SNPs) with multiple cancer types, including breast [20, 21], prostate [22, 23], lung [24], colorectal [25, 26], HCC [27] and other tumor types. To understand the global changes in core clock genes or CCGs, recent bioinformatics approaches have compared patient data from The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Genomics of Drug Sensitivity in Cancer (GDSC), and the Cancer Therapeutics Response Portal (CTRP). Comparison of 14 different tumor types identified that 90.2% of ‘clock genes’ (both core clock genes and CCGs) were differentially expressed in at least one tumor type [28]. Interestingly, the Arntl2 gene was up-regulated in multiple cancer types, while the circadian repressors (Per, Cry, and ROR) were strongly down-regulated as determined by RNA-sequencing analysis [28]. Moreover, these circadian repressors were significantly associated with inhibition of gene expression regulating apoptosis, cell cycle control, and DNA damage response pathways [28]. Further analysis similarly found that a coordinated signature of 12 clock genes was significantly deregulated in multiple tumors types when compared to expression in normal human tissue [29]. This omics approach reinforces the importance of circadian synchrony in maintaining cellular homeostasis and further implies that disruption of this network of precisely timed clock genes is likely involved in several human cancers.
Oncogenes and Tumor Suppressors Crosstalk with the Clock
In support of this clinical evidence, laboratory studies have helped to understand the molecular crosstalk between the circadian clock and signaling pathways regulating survival and proliferation. The role of the circadian clock and its molecular connection with several oncogenes, tumor suppressors and other transcriptional regulatory factors will be highlighted (Figure 2).
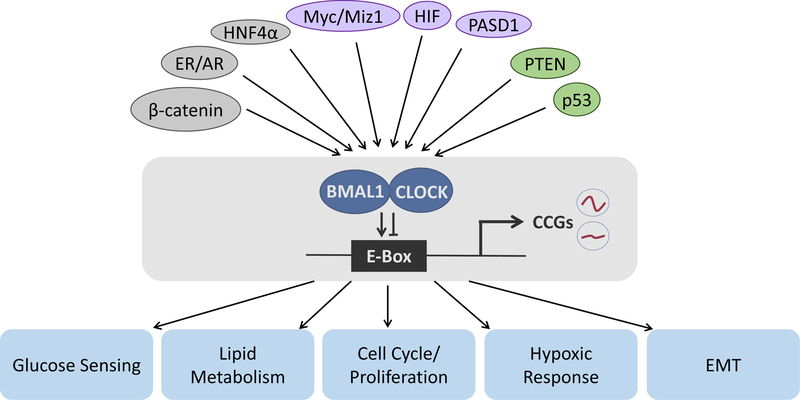
Figure 2: Oncogenes and tumor suppressors crosstalk with the circadian clock.
An overview of the input pathways of oncogenes and tumor suppressors which are known to deregulate the circadian clock. Several nuclear hormone receptors are implicated in regulating circadian gene expression in breast [42], prostate [23] and hepatocellular carcinoma (HCC) [67]. Also, the circadian clock machinery is known to crosstalk with the MYC oncogene to regulate cell proliferation and metabolism [52, 68, 72]. PTEN and p53 tumor suppressors also regulate the clock, including downstream signaling cascades involving PI3K/AKT [7, 56, 75]. The output of this circadian crosstalk with these oncogenes and tumor suppressors results in altered circadian gene expression regulating key pathways involved in glucose sensing, lipid metabolism, cell cycle and proliferation control, modulation of the hypoxic response, and control of the epithelial to mesenchymal transition (EMT). Nuclear hormone receptors and transcriptional co-regulatory proteins are shown in grey, transcription factors that directly interfere with CLOCK:BMAL1 transcription through E-Box binding are shown in purple, and tumor suppressors are indicated in green.
Ras-mediated Transformation
Ras-dependent cellular transformation models have been used to dissect the different roles of the positive and negative limbs of the circadian transcriptional/translational feedback loop in cancer. H-Ras-transformed human keratinocytes display a loss of Per2 and Bmal1 gene expression, while Cry1 and Clock expression are up-regulated in synchronized cells [30]. Using the Bmal1-luciferase reporter to determine rhythmic gene expression, induction of H-Ras or KRAS resulted in a significant lengthening of the circadian period, indicating that Ras-dependent transformation perturbs circadian gene expression [30]. Interestingly, Per2 mutant or Cry1/2-deficent cells are transformed by H-Ras, while Bmal1-null or Clock mutant mouse embryonic fibroblasts (MEFs) are resistant to oncogene-induced transformation [31]. This Ras-mediated transformation was found to be dependent on ATF4 activation that represses cell cycle regulators and tumor suppressors p16INK4a and p19ARF. These findings were further confirmed in vivo using a chemical carcinogen DMBA-dependent model of skin carcinogenesis in clock mutant mice [32]. Yet, use of genetically engineered mouse models (GEMMs) whereby tumor development occurs over several months reveals that both the positive and negative components of the circadian machinery accelerate tumor formation. Crossing Per2m/m or Bmal1fl/fl mice with a KrasLSL-G12D/+;p53fl/fl GEMM of lung adenocarcinoma resulted in increased tumor burden, more aggressive Grade 3 and 4 lung tumors, and subsequent decreased overall survival [7]. These findings suggest that disruption or abnormal function of the circadian molecular machinery could differentially impact Ras-mediated transformation. These results could be tumor type specific and therefore, cell type could differentially impact oncogene-induced transformation both in vivo and in vitro.
Wnt/β-catenin signaling
A bi-directional crosstalk has been reported between the circadian clock machinery and Wnt/β-catenin signaling, specifically involved in colorectal cancer (CRC). Per2m/m mice, when crossed with the ApcMin/+ model of multiple intestinal neoplasia [33, 34], developed increased numbers of intestinal and colonic polyps, as well as increased frequency of adenomas as compared to ApcMin/+ mice alone [35]. Furthermore, it has been reported that heightened β-catenin signaling in the ApcMin/+ mouse model destabilized PER2 protein levels in the intestinal mucosa through interaction with an F-Box protein of the SCF ubiquitin E3 ligase family called β-TrCP [36]. This bi-directional crosstalk links β-catenin signaling with the circadian molecular machinery in the intestine. The role of the clock in other cancer types that are dependent on Wnt/β-catenin signaling, such as gastric, breast, prostate and melanoma, is unknown. Moreover, Wnt signaling is critically involved in development and cell fate determination. In normal intestinal stem cells, the circadian clock was found to gate cell cycle progression in the mouse epithelium through clock-dependent secretion of Wnt using 3D in vitro organoid models [37]. Furthermore, disruption of the circadian clock results in arrhythmic cell division of normal intestinal stem cells in drosophila [38]. Therefore, disruption of the circadian clock could deregulate Wnt signaling and alter proliferative control of intestinal stem cells. What remains unknown is if circadian disruption could also impinge on Wnt signaling and cancer-initiating cells (CICs) resulting in tumorigenesis in the intestine and other tissues. Though the circadian clock is implicated in cancer stem cells [39, 40], the precise role and detailed molecular mechanism in different cancer types is undefined.
Nuclear Hormone Receptors and Cancer
Of the 49 nuclear hormone receptors expressed in fat, liver and skeletal muscle, 25 exhibit a rhythmic profile in gene expression [41]. One of these receptors, estrogen receptor alpha (ERα), is critically involved in hormone-dependent breast cancer and has been linked to the circadian clock. CLOCK interacts with ERα and this interaction is enhanced by estrogen. Estrogen stimulates the sumoylation of CLOCK, which results in increased CLOCK-dependent transcriptional activity, enhanced ERα-mediated transcription, and proliferation of MCF7 and T47D breast cancer cell lines [42]. BMAL1 is also sumoylated [43], but the role of ER/estrogenic signaling in this event is undefined. Interestingly, PER2 is also reported to regulate the stability of ERα protein levels, and Per2 expression is estrogen-inducible, suggesting a controlled feedback mechanism [44]. Moreover, PER1 is reported to inhibit the transactivation of androgen receptor (AR) by direct interaction and ectopic expression of Per1 in LNCaP prostate cancer cells decreased the expression of known AR-target genes [23]. Though these findings are compelling, the role of circadian clock crosstalk with nuclear hormone receptors in vivo is unknown. Mounting epidemiological evidence implicates shift work and light-at-night exposure with hormone-dependent cancers, specifically breast and prostate [3, 5, 45], suggesting a need to further elucidate the molecular mechanisms involved in tumorigenesis. Several additional connections likely exist between the clock, rhythmic hormone secretion, and oscillations in nuclear hormone receptors, but these links remain unexplored in several hormone-dependent cancers. For instance, the contribution of circadian regulation of progesterone receptor in breast cancer is unexplored. Also, secretion of thyroid hormone is constant over the day/night cycle, yet the expression of thyroid hormone receptors alpha and beta are dynamically rhythmic [41], and this establishes an interesting question that remains undefined.
c-Myc Oncogene
The c-Myc oncogene is a ‘master regulator’ of both cellular growth and metabolism in transformed cells [46, 47]. The metabolic connections between c-Myc and the clock will be discussed in a later section, but we focus on the transcriptional regulation that is shared between these pathways. The X-ray crystal structures of the CLOCK:BMAL1 and MYC-MAX heterodimers reveal a basic helix-loop-helix (bHLH) domain [48, 49] that is required to recognize E-Box sequences [50, 51]. This common E-Box sequence could suggest that a rewiring may take place during tumorigenesis whereby the balance of clock-controlled transcription can be lost and consequently compensated by oncogenic MYC-dependent transcription (Figure 3). In support of this notion, MYC and its binding partner MIZ1 are responsible for forming a repressive complex which down-regulates core clock gene expression [52]. Conversely, this transcriptional crosstalk can be extended in that the circadian repressor CRY2 has been reported to promote MYC degradation through the FBXL3-containing E3 ligase, and Cry2 deletion resulted in enhanced Myc-driven lymphomas in mice [53]. Additionally, the expression of c-Myc is negatively controlled by direct promoter binding of CLOCK:BMAL1, and MYC can repress CLOCK:BMAL1-dependent transactivation of Per1 expression [54]. In further support of these findings, the expression of BMAL1 was found to be inversely correlated with MYC in 102 human lymphoma samples [52]. Also, Per2 mutant mice (Per2m/m) are susceptible to lymphoma and these mice exhibit an up-regulation of c-Myc expression and its target gene Ccnd1 [8]. Collectively, this data suggests that an inverse relationship exists between MYC and the circadian transcriptional axes that controls cellular survival and proliferation pathways. Interestingly, other examples exist whereby a transcriptional crosstalk has been demonstrated between the clock and alternative pathways in cancer. For example, the cancer/testis antigen PASD1, which is normally restricted in expression to the germline, is induced upon oncogenic transformation. PASD1 has been shown to directly interact with the CLOCK:BMAL1 complex to repress clock-dependent transcription [55]. Additional evidence of transcriptional crosstalk exists between the clock complex and HIF, and the implications of this work will be discussed in a later section.
Figure 3: Involvement of the MYC superfamily and the circadian clock in cancer.
Several reports implicate a transcriptional competition between several members of the MYC superfamily with the circadian clock. MYC/MIZ are involved in regulating cell proliferation and nutrient utilization of glucose and glutamine by repressing the circadian clock [52, 68, 72]. Also, the circadian repressor CRY2 has been reported to promote MYC degradation through the FBXL3-containing E3 ligase [53]. The MondoA/MLX transcriptional pathway is involved in nutrient sensing and downstream regulation of lipogenesis and glutaminolysis in different cancer types [72]. To date, the direct connection between the circadian clock and the nutrient sensing arm of MondoA remains to be defined. The consequences of this transcriptional competition between CLOCK:BMAL1 and the MYC superfamily result in regulation of gene expression programs involved in controlling the cell cycle, proliferation, and cellular metabolism.
The Tumor Suppressor p53
The tumor suppressor p53 is commonly mutated and subsequently inactivated in over 50% of human cancers [46, 47]. The circadian clock is regulated by p53 as Per2 expression is transcriptionally controlled by p53 [56]. The p53 response element overlaps with the E-Box sequence in the Per2 promoter, thereby p53 occupancy blocks CLOCK:BMAL1 promoter recruitment [56]. Is this the case for other core clock genes or CCGs, and to what extent is this co-occupancy of p53 and the clock machinery occurring genome-wide? Conversely, PER2 forms a stable complex with p53, which subsequently prevents MDM2-mediated ubiquitination and proteasomal degradation of p53 [57, 58]. Therefore, modulating cellular levels of the clock proteins directly controls p53 stability, and this is supported by the finding that ectopic expression of PER2 results in nuclear shuttling of p53 [59]. Recent evidence also supports the notion that PER2 stability is regulated through an atypical circadian mechanism by which PER2 is a direct target of the ubiquitin ligase MDM2 [60]. As previously discussed, crossing Per2m/m mice with a KrasLSL-G12D/+;p53fl/fl GEMM of lung adenocarcinoma resulted in increased tumor burden, more aggressive lung tumors, and subsequent decreased overall survival [7]. Also, expression of Per2 in A549 lung cancer cells suppresses growth and metastasis in vitro and in vivo [24]. Remarkably, the connection between the clock and p53 has mostly focused on PER2, yet the role of other clock proteins remains unclear. However, recent work has identified that knockdown of Bmal1 in pancreatic cancer cells promotes cell growth while ectopic expression of Bmal1 inhibits cell cycle and proliferative control in a p53-dependent manner [61]. Conversely, crossing p53−/− mice with Cry1−/−;Cry2−/− mutant mice resulted in a 50% extension of median lifespan as compared to p53−/− mice alone [62]. This data suggests differential roles of the clock proteins in regulating p53. These differences in p53 activity linked to the clock could be attributed to the multiple functions of p53 linked to UV stress, DNA damage response, DNA repair, or apoptosis pathways in a cell type dependent manner [63–66].
Linking the Clock, Metabolism and Cancer
The circadian clock regulates an array of metabolic processes that are critical for maintaining homeostasis (Box 2). Interestingly, several avenues have been explored to unravel how circadian alterations may impinge on metabolic pathways needed to sustain cell survival. Using environmental disruption paradigms of CJL, non-alcoholic fatty liver disease (NAFLD) has been reported in WT mice, which can progress to steatohepatitis and eventually HCC [16]. Mechanistically, CJL disrupts hepatic circadian gene expression as well as rhythmic metabolism, resulting in induction of hepatic cholesterol and bile acid levels that activate the oncogenic program of the nuclear receptor constitutive androstane receptor (CAR), and downstream activation of β-catenin [16]. Additionally, hepatocyte nuclear factor 4 alpha (HNF4α) is a master regulator of liver-specific gene expression. A specific isoform of HNF4α (P2-HNF4α) is expressed in human HCC and P2-HNF4α represses BMAL1 transcriptional activity, including genes involved in EMT [67]. Also, overexpression of BMAL1 in HNF4α-positive HCC inhibits tumor growth in vivo [67]. Properly controlled rhythmic metabolism is important for cancer prevention, which is specifically highlighted in this example of HCC and CJL-mediated disruption of hepatic metabolism. The extent of these findings is likely not limited to HCC and therefore requires further investigation in other tumor types.
Box 2: Circadian Control of Metabolism.
The circadian clock controls a wide array of metabolic processes such as glycolysis, the Krebs cycle, gluconeogenesis, mitochondrial function, lipogenesis, lipolysis, amino acid metabolism and nucleotide synthesis. Untargeted mass spectrometry relying on metabolomics or lipidomics has identified 30–60% of lipids, amino acids, nucleotides, carbohydrates and cofactors/coenzymes are rhythmic [78, 90, 133, 134]. This includes rhythmic metabolites such as NAD+, FAD, acetyl-CoA, SAM/SAH, which have important implications in redox biology, mitochondrial processes, and histone/non-histone protein modifications influencing gene expression [86, 135–138] (Figure 1). Moreover, recent studies demonstrate that high fat diet (HFD) feeding in mice phase shifts 40–60% of circadian metabolites in liver, muscle, adipose tissues and serum [78, 90, 139, 140]. Also, food availability as evidenced by time-restricted feeding or fasting can rapidly and dynamically shift circadian gene expression [90, 141–144]. Conceptually, this experimental evidence has established that circadian metabolism can exhibit plasticity, and reprogramming of transcriptional and metabolic circuits occurs in response to nutritional perturbation or challenge [78].
Furthermore, genetic disruption of the circadian clock is also linked with metabolic dysfunction in rodent models. Clock mutant (ClockΔ19) mice are obese compared to their WT littermates and display hyperlipidemia, hepatic steatosis, hyperglycemia, and hypoinsulinemia [145]. Also, hepatic glucose production through gluconeogenesis is controlled by the circadian repressor CRY1, that subsequently regulates the activity of cAMP response element-binding protein (CREB) [146]. In addition, the hepatic clock also controls levels of SREBP-dependent fatty acid, cholesterol and bile acid production through the functions of the circadian transcriptional repressors REV-ERBα [147], and the mammalian histone deacetylase sirtuin 6 (SIRT6) [80]. Also, the conditional knockout of the pancreatic clock also results in glucose intolerance and diabetes mellitus [148]. Recent work has elucidated the role of the circadian clock in pancreatic function. In both mouse [149] and human [150] pancreatic islets, the clock was found to directly regulate circadian insulin secretion, by regulating the expression of genes involved in the secretory machinery and signaling factors that control insulin release. Collectively, these findings establish a strong link between the circadian clock and control of cellular and tissue-specific metabolic pathways. The implications of these circadian metabolic pathways in sustaining tumor metabolism will be discussed.
Myc and Cancer Metabolism
Up-regulation of MYC has been shown to disrupt circadian gene expression and perturb circadian glucose and glutamine metabolism in cancer cells both in vitro [68] and in vivo [7]. This activity of MYC was initially reported to be dependent on the negative transcriptional arm of the clock through REV-ERBα that inhibits expression of Bmal1 and also deregulates oscillations in metabolic sensing through AMPK and hexokinases HK1 and HK2 [68]. Recent evidence also suggests that MYC-dependent activity may not be solely dependent on REV-ERBα [69, 70]. The nutrient-sensing function of the MYC superfamily plays an important role in tumor metabolism (Figure 3). In addition to MYC, MondoA/Mlx are E-Box binding transcription factors that sense glycolytic intermediates such as glucose 6-phosphate. Depending on tumor type and metabolic fuel preference, MondoA either antagonizes MYC-dependent regulation of glycolytic genes in triple negative breast cancer [71] or coordinately regulates glutaminolysis and lipogenesis in neuroblastoma [72]. The role of MondoA is tumor metabolism involving the circadian clock machinery is currently unknown, especially related to how MondoA may transcriptionally converge with CLOCK:BMAL1 activity. Yet, evidence from drosophila suggest that these transcriptional networks likely intersect [73], and therefore could be important to explore within the context of tumor metabolism. Additionally, given that tumor heterogeneity in metabolic fuel preference is an important consideration [74], MYC-dependent nutrient sensing is likely far more complex than our current understanding.
PTEN, PI3K/AKT and mTOR signaling
Emerging evidence has started to connect the circadian clock with the tumor suppressor PTEN and additional related pathways. PTEN controls the PI3K/AKT signaling pathway, and disruption of this pathway by oxidative stress results in activation of BMAL1 in a mTOR-dependent manner [75]. Though this study identified a potential role of both TORC1 and TORC2 in controlling the clock, further molecular studies are still required to better define this link. Yet, BMAL1 is a known substrate of the mTOR effector kinase S6K1, and this phosphorylation of BMAL1 is important for interaction with the translation machinery and subsequent rhythmic protein synthesis [76]. Also, mTOR activation results in increased expression of several members of the core clock machinery in fibroblasts which regulates circadian gene expression [77]. These results are particularly exciting and warrant further investigation given that amino acids, cellular energy levels, and growth factors, which are sensed by the mTOR pathway, are rhythmic over the day/night cycle [78–80]. Indeed, mTOR activity has been reported to be rhythmic and follow food intake, and this can be uncoupled from the light-dependent circadian mechanism [81], suggesting a central role for the circadian metabolic clock linked to mTOR activity. Recent studies reveal that PER2 functions as a scaffold protein to suppress the activity of the mTORC1 complex, and loss of PER2 enhances protein synthesis and cell proliferation [82]. The implications of rhythmic mTOR-dependent nutrient sensing and proliferation in cancer merit further investigation, as this suggests permissive time windows for optimal druggability and therapeutic targeting.
Hypoxia and HIF signaling
Oxygen is critical for cellular respiration and the hypoxic tumor microenvironment regulates tumor metabolism and angiogenesis [46, 47]. A transcriptional crosstalk has been reported between the clock and the transcription factor hypoxia-inducible factor (HIF) in non-transformed cells, though the implications of these studies in tumorigenesis are possible but currently unknown. The hypoxia-response element (HRE) is an E-Box like sequence that is recognized and bound by HIF under low oxygen conditions to regulate gene expression. The HRE is recognized by HIF heterodimers consisting of two similar proteins, HIF1α and HIF1β. Interestingly, the hypoxic response is gated by the circadian clock as the Hif1α promoter is directly controlled by the CLOCK:BMAL1 transcriptional complex, which drives rhythmic gene expression of clock genes and hypoxia-dependent genes [83]. Also, blood oxygen levels exhibit daily rhythms which influences expression of core clock genes in a HIF1α-dependent manner in several tissue types [84]. The transcriptional crosstalk between the clock and HIF is further supported by genetic data illustrating that Bmal1−/− in C2C12 myotubes reduced anaerobic glycolysis in a HIF1α-dependent manner [85]. An intriguing concept emerges from these findings linking HIF with a circadian transcriptional crosstalk. If the clock gates the hypoxic response by regulating gene expression, how would the oxygen-deplete tumor microenvironment be regulated by CLOCK:BMAL1-dependent transcription? Yet, if circadian gene expression is deregulated during tumorigenesis, the impact on HIF-dependent signaling could be altered in the absence of a functional clock mechanism. Additionally, oxygen levels control cellular respiration in a circadian manner [86], and how disruption of these metabolic rhythms could further drive tumor progression are unknown.
The Tumor Microenvironment and Macroenvironment
To sustain their heightened proliferation, cancer cells need to generate energy, provide reducing equivalents to drive enzymatic reactions for energy production, and produce macromolecular precursors for rapidly proliferating cells [87]. As a consequence of this heightened metabolism, tumors excrete metabolic intermediates into the microenvironment but also into circulation. Emerging evidence suggests that tumor metabolism extends beyond the niche of the tumor microenvironment, and this so-called tumor macroenvironment may play an important role in driving cancer proliferation [88, 89]. Given that the circadian metabolic clock has the unique ability to exhibit plasticity and adapt to new metabolic perturbations via reprogramming [78, 90], the question arises if tumor-dependent metabolism can alter organismal homeostasis. Indeed, recent findings demonstrate that circadian metabolism in the liver is distally reprogrammed by lung adenocarcinoma, suggesting that tumor-instructed cues can mediate circadian disruption of host metabolism [91]. This tumor-dependent distal rewiring of hepatic metabolism includes deregulation of lipid metabolism, and insulin/glucose-dependent signaling [91]. Similar results have been reported in a mouse model of triple negative breast cancer, where rewiring of circadian gene expression was distally observed in the liver, resulting in increased oxidative stress [92]. The question that arises from these studies is what is the tumor-specific advantage to distally rewire circadian metabolism? One speculation is that this rewiring of circadian metabolism can feedback and supply additional or alternative fuel sources to support heightened metabolic demand of cancer cells. Additionally, another possibility is that deregulation of the circadian clock and metabolism could support cites of distal metastasis, and these concepts will need to be explored experimentally.
Metabolic heterogeneity in fuel preference of different tumor types has become quite apparent, which underscores the complexity of cancer metabolism. First, the environment plays an important role in dictating fuel preference, and tumors in vivo display differences in fuel utilization versus cultured cancer cells [74]. These differences in tumor cell type and metabolic preference are being elucidated, and dependence on glucose, glutamine, aspartate, arginine and other nutrients is becoming apparent in a cell-type dependent manner [93–96]. Yet, nutrient addiction of a tumor subtype can differ. For instance, when glutamine levels are limiting, cell proliferation can be alternatively driven by aspartate or asparagine [97, 98]. This establishes an interesting paradox whereby limiting the availably of a specific nutrient may not inhibit cancer cell proliferation, as tumors can switch their fuel preference. An additional factor to consider is that abundance of these nutrients is rhythmic over the circadian cycle [78–80], which raises the possibility that fuel utilization of tumors may differ based on rhythmic availability of nutrients.
Additionally, tumors can also utilize non-traditional metabolites as fuel. For instance, patients with non-small cell lung cancer (NSCLC) exhibit metabolic heterogeneity in fuel utilization where, even though glucose oxidation is metabolically imperative, lactate is used as a carbon source to fuel the TCA cycle [99, 100]. Metabolic flux studies in GEMMs using uniformly labeled [U-13C] lactate (where all three carbons are heavy labeled) have demonstrated that lactate is the major carbon source for tumor TCA cycle intermediates in lung adenocarcinoma [101]. Strikingly, blood perfusion of [U-13C] lactate into two different lung adenocarcinoma GEMMs, KrasLSL-G12D/+;Trp53−/− and KrasG12D/+;Stk11−/−, illustrated preferential utilization of [U-13C]lactate over both [U-13C] glucose and [U-13C] glutamine, suggesting that circulating lactate can be a major energy source for lung tumors. These findings are quite intriguing especially given that lactate is a metabolic byproduct of glycolytic metabolism, therefore tumors can rely on their metabolic intermediates to fuel cell proliferation. Importantly, these findings appear to be tumor-type specific as a GEMM of pancreatic ductal adenocarcinoma (PDAC) instead displayed a strong preference for [U-13C] glutamine as a carbon feedstock of the tumor TCA cycle over glucose and lactate [101]. Moreover, in terms of nitrogen sources, a similar example has been reported in breast cancer cells where the enzymatic activity of glutamate dehydrogenase (GDH) can recycle ammonia waste into central amino acid metabolism [102]. Overall, these findings demonstrate the complexity of tumor metabolism and illustrate that certain types of metabolic byproducts can be repurposed for intra-tumoral energy and building block production.
Concluding Remarks and Future Directions for Therapy
The precise biological timing of the circadian clock is imperative for maintaining organismal homeostasis. This review outlines several connections between the circadian biological clock and pathways regulating cell proliferation, endocrinology and metabolism. Disruption of these pathways is linked to tumor initiation and progression, and we highlight the molecular links between the clock and oncogenes and tumor suppressors such as Ras, c-Myc, Wnt/β-Catenin, p53, PTEN and mTOR. Also, the circadian clock is intimately linked with intracellular and organismal metabolism. Therefore, we discuss exciting avenues that require further experimental investigation, specifically linked with tumor heterogeneity, metabolic fuel preference of cancer cells, and the consequences of tumor-derived metabolic byproducts and fuel repurposing. Recent advances highlight the potential for therapeutic targeting of the circadian clock for cancer treatment [2, 103]. Yet, the molecular mechanisms of these approaches still require further examination in terms of targeting the molecular clock for cancer therapy. Several cancers highlighted throughout this piece exhibit a down-regulation of the circadian machinery, therefore inhibition of the circadian clock in cancer therapy may be tumor type specific. Another point of consideration is chronotherapy, which is the optimal druggability window over the circadian cycle for effective treatment, and this approach warrants better investigation. This is especially relevant in targeting circadian metabolic pathways that can be optimally drugged based on peak metabolite levels, circadian protein expression or enzymatic activity. Overall, the circadian clock represents an underexplored area of cancer research and treatment that is a vital connection linking tumor metabolism and oncogenic signaling networks.
Acknowledgements
The Masri laboratory is supported by a K22 Transition Career Development Award through the National Cancer Institute (NCI), the Concern Foundation, and the V Foundation for Cancer Research. A.V. is supported by the Hitachi-Nomura Postdoctoral Fellowship through the Department of Biological Chemistry at UC Irvine.
Footnotes
The authors declare no conflicts of interest.
References:
- 1.Asher G and Sassone-Corsi P (2015) Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161 (1), 84–92. [DOI] [PubMed] [Google Scholar]
- 2.Masri S and Sassone-Corsi P (2018) The emerging link between cancer, metabolism, and circadian rhythms. Nat Med 24 (12), 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schernhammer ES et al. (2001) Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst 93 (20), 1563–8. [DOI] [PubMed] [Google Scholar]
- 4.Srour B et al. (2018) Circadian nutritional behaviours and cancer risk: New insights from the NutriNet-sante prospective cohort study: Disclaimers. Int J Cancer [DOI] [PubMed]
- 5.Papantoniou K et al. (2015) Night shift work, chronotype and prostate cancer risk in the MCC-Spain case-control study. Int J Cancer 137 (5), 1147–57. [DOI] [PubMed] [Google Scholar]
- 6.Knutsson A et al. (2013) Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand J Work Environ Health 39 (2), 170–7. [DOI] [PubMed] [Google Scholar]
- 7.Papagiannakopoulos T et al. (2016) Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab 24 (2), 324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu L et al. (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111 (1), 41–50. [DOI] [PubMed] [Google Scholar]
- 9.Lee S et al. (2010) Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5 (6), e10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18 (3), 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsh DK et al. (2010) Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki S et al. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288 (5466), 682–5. [DOI] [PubMed] [Google Scholar]
- 13.Masri S and Sassone-Corsi P (2010) Plasticity and specificity of the circadian epigenome. Nat Neurosci 13 (11), 1324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filipski E et al. (2002) Host circadian clock as a control point in tumor progression. J Natl Cancer Inst 94 (9), 690–7. [DOI] [PubMed] [Google Scholar]
- 15.Filipski E et al. (2004) Effects of chronic jet lag on tumor progression in mice. Cancer Res 64 (21), 7879–85. [DOI] [PubMed] [Google Scholar]
- 16.Kettner NM et al. (2016) Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 30 (6), 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonken LK et al. (2010) Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A 107 (43), 18664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonken LK et al. (2013) Dim light at night disrupts molecular circadian rhythms and increases body weight. J Biol Rhythms 28 (4), 262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonken LK et al. (2013) Dim light at night exaggerates weight gain and inflammation associated with a high-fat diet in male mice. Endocrinology 154 (10), 3817–25. [DOI] [PubMed] [Google Scholar]
- 20.Chen ST et al. (2005) Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26 (7), 1241–6. [DOI] [PubMed] [Google Scholar]
- 21.Lesicka M et al. (2018) Altered circadian genes expression in breast cancer tissue according to the clinical characteristics. PLoS One 13 (6), e0199622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y et al. (2009) Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res 69 (24), 9315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Q et al. (2009) A role for the clock gene per1 in prostate cancer. Cancer Res 69 (19), 7619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang R et al. (2018) Circadian clock gene Per2 downregulation in nonsmall cell lung cancer is associated with tumour progression and metastasis. Oncol Rep 40 (5), 3040–3048. [DOI] [PubMed] [Google Scholar]
- 25.Mazzoccoli G et al. (2016) Deregulated expression of cryptochrome genes in human colorectal cancer. Mol Cancer 15, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzoccoli G et al. (2011) Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int 28 (10), 841–51. [DOI] [PubMed] [Google Scholar]
- 27.Lin YM et al. (2008) Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog 47 (12), 925–33. [DOI] [PubMed] [Google Scholar]
- 28.Ye Y et al. (2018) The Genomic Landscape and Pharmacogenomic Interactions of Clock Genes in Cancer Chronotherapy. Cell Syst 6 (3), 314–328 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shilts J et al. (2018) Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ 6, e4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relogio A et al. (2014) Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet 10 (5), e1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katamune C et al. (2016) Different Roles of Negative and Positive Components of the Circadian Clock in Oncogene-induced Neoplastic Transformation. J Biol Chem 291 (20), 10541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashikawa KI et al. (2017) Dysfunction of the circadian transcriptional factor CLOCK in mice resists chemical carcinogen-induced tumorigenesis. Sci Rep 7 (1), 9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moser AR et al. (1990) A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247 (4940), 322–4. [DOI] [PubMed] [Google Scholar]
- 34.Morin PJ et al. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275 (5307), 1787–90. [DOI] [PubMed] [Google Scholar]
- 35.Wood PA et al. (2008) Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res 6 (11), 1786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X et al. (2009) Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem 145 (3), 289–97. [DOI] [PubMed] [Google Scholar]
- 37.Matsu-Ura T et al. (2016) Intercellular Coupling of the Cell Cycle and Circadian Clock in Adult Stem Cell Culture. Mol Cell 64 (5), 900–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karpowicz P et al. (2013) The circadian clock gates the intestinal stem cell regenerative state. Cell Rep 3 (4), 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puram RV et al. (2016) Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell 165 (2), 303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janich P et al. (2011) The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480 (7376), 209–14. [DOI] [PubMed] [Google Scholar]
- 41.Yang X et al. (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126 (4), 801–10. [DOI] [PubMed] [Google Scholar]
- 42.Li S et al. (2013) CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha. Oncogene 32 (41), 4883–91. [DOI] [PubMed] [Google Scholar]
- 43.Cardone L et al. (2005) Circadian clock control by SUMOylation of BMAL1. Science 309 (5739), 1390–4. [DOI] [PubMed] [Google Scholar]
- 44.Gery S et al. (2007) The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 26 (57), 7916–20. [DOI] [PubMed] [Google Scholar]
- 45.Lie JA et al. (2006) Breast cancer and night work among Norwegian nurses. Cancer Causes Control 17 (1), 39–44. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D and Weinberg RA (2000) The hallmarks of cancer. Cell 100 (1), 57–70. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5), 646–74. [DOI] [PubMed] [Google Scholar]
- 48.Nair SK and Burley SK (2003) X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112 (2), 193–205. [DOI] [PubMed] [Google Scholar]
- 49.Huang N et al. (2012) Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337 (6091), 189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walhout AJ et al. (1997) c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res 25 (8), 1493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ripperger JA and Schibler U (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38 (3), 369–74. [DOI] [PubMed] [Google Scholar]
- 52.Shostak A et al. (2016) MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat Commun 7, 11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber AL et al. (2016) CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol Cell 64 (4), 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Repouskou A and Prombona A (2016) c-MYC targets the central oscillator gene Per1 and is regulated by the circadian clock at the post-transcriptional level. Biochim Biophys Acta 1859 (4), 541–52. [DOI] [PubMed] [Google Scholar]
- 55.Michael AK et al. (2015) Cancer/Testis Antigen PASD1 Silences the Circadian Clock. Mol Cell 58 (5), 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miki T et al. (2013) p53 regulates Period2 expression and the circadian clock. Nat Commun 4, 2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gotoh T et al. (2015) Association of the circadian factor Period 2 to p53 influences p53’s function in DNA-damage signaling. Mol Biol Cell 26 (2), 359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gotoh T et al. (2014) The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol Biol Cell 25 (19), 3081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gotoh T et al. (2016) Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor Period 2. Proc Natl Acad Sci U S A 113 (47), 13516–13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J et al. (2018) Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci Signal 11 (556). [DOI] [PubMed] [Google Scholar]
- 61.Jiang W et al. (2016) The circadian clock gene Bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett 371 (2), 314–25. [DOI] [PubMed] [Google Scholar]
- 62.Ozturk N et al. (2009) Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci U S A 106 (8), 2841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Y et al. (2018) BMAL1 and CLOCK proteins in regulating UVB-induced apoptosis and DNA damage responses in human keratinocytes. J Cell Physiol 233 (12), 9563–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawamura G et al. (2018) Cooperative interaction among BMAL1, HSF1, and p53 protects mammalian cells from UV stress. Commun Biol 1, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J et al. (2016) Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol 18 (5), 480–90. [DOI] [PubMed] [Google Scholar]
- 66.Horiguchi M et al. (2013) Rhythmic control of the ARF-MDM2 pathway by ATF4 underlies circadian accumulation of p53 in malignant cells. Cancer Res 73 (8), 2639–49. [DOI] [PubMed] [Google Scholar]
- 67.Fekry B et al. (2018) Incompatibility of the circadian protein BMAL1 and HNF4alpha in hepatocellular carcinoma. Nat Commun 9 (1), 4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altman BJ et al. (2015) MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab 22 (6), 1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shostak A et al. (2017) Correspondence: Reply to ‘Oncogenic MYC persistently upregulates the molecular clock component REV-ERBalpha’. Nat Commun 8, 14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altman BJ et al. (2017) Correspondence: Oncogenic MYC persistently upregulates the molecular clock component REV-ERBalpha. Nat Commun 8, 14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen L et al. (2015) Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci U S A 112 (17), 5425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carroll PA et al. (2015) Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell 27 (2), 271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bartok O et al. (2015) The transcription factor Cabut coordinates energy metabolism and the circadian clock in response to sugar sensing. EMBO J 34 (11), 1538–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davidson SM et al. (2016) Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 23 (3), 517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsumoto CS et al. (2016) PI3K-PTEN dysregulation leads to mTOR-driven upregulation of the core clock gene BMAL1 in normal and malignant epithelial cells. Oncotarget 7 (27), 42393–42407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lipton JO et al. (2015) The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell 161 (5), 1138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramanathan C et al. (2018) mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet 14 (5), e1007369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eckel-Mahan KL et al. (2013) Reprogramming of the Circadian Clock by Nutritional Challenge. Cell 155 (7), 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eckel-Mahan KL et al. (2012) Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A 109 (14), 5541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masri S et al. (2014) Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158 (3), 659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khapre RV et al. (2014) Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany NY) 6 (8), 675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu R et al. (2019) The Circadian Protein Period2 Suppresses mTORC1 Activity via Recruiting Tsc1 to mTORC1 Complex. Cell Metab 29 (3), 653–667 e6. [DOI] [PubMed] [Google Scholar]
- 83.Wu Y et al. (2017) Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab 25 (1), 73–85. [DOI] [PubMed] [Google Scholar]
- 84.Adamovich Y et al. (2017) Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab 25 (1), 93–101. [DOI] [PubMed] [Google Scholar]
- 85.Peek CB et al. (2017) Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab 25 (1), 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peek C et al. (2013) Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science 341 (6152). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vander Heiden MG and DeBerardinis RJ (2017) Understanding the Intersections between Metabolism and Cancer Biology. Cell 168 (4), 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutkowski MR et al. (2015) The Tumor Macroenvironment: Cancer-Promoting Networks Beyond Tumor Beds. Adv Cancer Res 128, 235–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Al-Zoughbi W et al. (2014) Tumor macroenvironment and metabolism. Semin Oncol 41 (2), 281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hatori M et al. (2012) Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab 15 (6), 848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Masri S et al. (2016) Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell 165 (4), 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hojo H et al. (2017) Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget 8 (21), 34128–34140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sullivan LB et al. (2018) Aspartate is an endogenous metabolic limitation for tumour growth. Nat Cell Biol 20 (7), 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garcia-Bermudez J et al. (2018) Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol 20 (7), 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Poillet-Perez L et al. (2018) Autophagy maintains tumour growth through circulating arginine. Nature 563 (7732), 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wise DR and Thompson CB (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 35 (8), 427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alkan HF et al. (2018) Cytosolic Aspartate Availability Determines Cell Survival When Glutamine Is Limiting. Cell Metab 28 (5), 706–720 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pavlova NN et al. (2018) As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid. Cell Metab 27 (2), 428–438 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hensley CT et al. (2016) Metabolic Heterogeneity in Human Lung Tumors. Cell 164 (4), 681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Faubert B et al. (2017) Lactate Metabolism in Human Lung Tumors. Cell 171 (2), 358–371 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hui S et al. (2017) Glucose feeds the TCA cycle via circulating lactate. Nature 551 (7678), 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spinelli JB et al. (2017) Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science [DOI] [PMC free article] [PubMed]
- 103.Sulli G et al. (2018) Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553 (7688), 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Partch CL et al. (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24 (2), 90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Akhtar RA et al. (2002) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12 (7), 540–50. [DOI] [PubMed] [Google Scholar]
- 106.Panda S et al. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109 (3), 307–20. [DOI] [PubMed] [Google Scholar]
- 107.Rey G et al. (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9 (2), e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koike N et al. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338 (6105), 349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Duong HA et al. (2011) A molecular mechanism for circadian clock negative feedback. Science 332 (6036), 1436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duong HA and Weitz CJ (2014) Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 21 (2), 126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Michael AK et al. (2017) Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci U S A 114 (7), 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Narasimamurthy R et al. (2018) CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci U S A 115 (23), 5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sato TK et al. (2004) A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43 (4), 527–37. [DOI] [PubMed] [Google Scholar]
- 114.Preitner N et al. (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110 (2), 251–60. [DOI] [PubMed] [Google Scholar]
- 115.Cho H et al. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485 (7396), 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yin L et al. (2006) Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311 (5763), 1002–5. [DOI] [PubMed] [Google Scholar]
- 117.Nakahata Y et al. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134 (2), 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Asher G et al. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134 (2), 317–28. [DOI] [PubMed] [Google Scholar]
- 119.Valekunja UK et al. (2013) Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A 110 (4), 1554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Katada S and Sassone-Corsi P (2010) The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 17 (12), 1414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DiTacchio L et al. (2011) Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333 (6051), 1881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Alenghat T et al. (2008) Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456 (7224), 997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hung HC et al. (2007) Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J Biol Chem 282 (43), 31349–57. [DOI] [PubMed] [Google Scholar]
- 124.Hosoda H et al. (2009) CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Doi M et al. (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125 (3), 497–508. [DOI] [PubMed] [Google Scholar]
- 126.Chang HC and Guarente L (2013) SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153 (7), 1448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feng D et al. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331 (6022), 1315–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Etchegaray JP et al. (2006) The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem 281 (30), 21209–15. [DOI] [PubMed] [Google Scholar]
- 129.Jones MA et al. (2010) Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A 107 (50), 21623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tamayo AG et al. (2015) Histone monoubiquitination by Clock-Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat Struct Mol Biol 22 (10), 759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sabari BR et al. (2017) Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol 18 (2), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dutta A et al. (2016) Diverse Activities of Histone Acylations Connect Metabolism to Chromatin Function. Mol Cell 63 (4), 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aviram R et al. (2016) Lipidomics Analyses Reveal Temporal and Spatial Lipid Organization and Uncover Daily Oscillations in Intracellular Organelles. Mol Cell 62 (4), 636–48. [DOI] [PubMed] [Google Scholar]
- 134.Minami Y et al. (2009) Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A 106 (24), 9890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakahata Y et al. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324 (5927), 654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramsey KM et al. (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324 (5927), 651–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sahar S et al. (2014) Circadian Control of Fatty Acid Elongation by SIRT1-mediated Deacetylation of Acetyl-CoA Synthetase 1. J Biol Chem [DOI] [PMC free article] [PubMed]
- 138.Krishnaiah SY et al. (2017) Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab 25 (4), 961–974 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dyar K et al. (2018) Atlas of Circadian Metabolism Reveals System-Wide Coordination and Communication between ClocksAtlas of Circadian Metabolism Reveals System-Wide Coordination and Communication between Clocks. Cell 174 (6), 1571–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kohsaka A et al. (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6 (5), 414–21. [DOI] [PubMed] [Google Scholar]
- 141.Chaix A et al. (2014) Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 20 (6), 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stokkan KA et al. (2001) Entrainment of the circadian clock in the liver by feeding. Science 291 (5503), 490–3. [DOI] [PubMed] [Google Scholar]
- 143.Damiola F et al. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14 (23), 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kinouchi K et al. (2018) Fasting Imparts a Switch to Alternative Daily Pathways in Liver and Muscle. Cell Rep 25 (12), 3299–3314 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Turek FW et al. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308 (5724), 1043–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang EE et al. (2010) Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16 (10), 1152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Le Martelot G et al. (2009) REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7 (9), e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Marcheva B et al. (2010) Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature [DOI] [PMC free article] [PubMed]
- 149.Perelis M et al. (2015) Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350 (6261), aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saini C et al. (2015) A functional circadian clock is required for proper insulin secretion by human pancreatic islet cells. Diabetes Obes Metab [DOI] [PubMed]
- 151.Gekakis N et al. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280 (5369), 1564–9. [DOI] [PubMed] [Google Scholar]