Abstract
Background
In a four-generation Caucasian family variably diagnosed with autosomal dominant (AD) Stickler or Wagner disease, commercial gene screening failed to identify a mutation in COL2A1 or VCAN. We utilized linkage mapping and exome sequencing to identify the causal variant.
Materials and methods
Genomic DNA samples collected from 40 family members were analyzed. A whole-genome linkage scan was performed using Illumina HumanLinkage-24 BeadChip followed by two-point and multipoint linkage analyses using FASTLINK and MERLIN. Exome sequencing was performed on two affected individuals, followed by co-segregation analysis.
Results
Parametric multipoint linkage analysis using an AD inheritance model demonstrated HLOD scores > 2.00 at chromosomes 1p36.13–1p36.11 and 12q12–12q14.1. SIMWALK multipoint analysis replicated the peak in chromosome 12q (peak LOD = 1.975). FASTLINK two-point analysis highlighted several clustered chromosome 12q SNPs with HLOD > 1.0. Exome sequencing revealed a novel nonsense mutation (c.115C>T, p.Gln39*) in exon 2 of COL2A1 that is expected to result in nonsense-mediated decay of the RNA transcript. This mutation co-segregated with all clinically affected individuals and seven individuals who were clinically unaffected.
Conclusions
The utility of combining traditional linkage mapping and exome sequencing is highlighted to identify gene mutations in large families displaying a Mendelian inheritance of disease. Historically, nonsense mutations in exon 2 of COL2A1 have been reported to cause a fully penetrant ocular-only Stickler phenotype with few or no systemic manifestations. We report a novel nonsense mutation in exon 2 of COL2A1 that displays incomplete penetrance and/or variable age of onset with extraocular manifestations.
Keywords: Linkage, penetrance, Stickler syndrome, Wagner syndrome
Introduction
Stickler syndrome (MIM 108300) is a hereditary arthro-ophthalmopathy that was first described by Gunnar Stickler in 1965.1 The genetically heterogeneous disease has been characterized predominantly by abnormalities in ocular, skeletal, orofacial, and auditory systems, and clinical manifestations can vary within families.2,3 Prevalence rates among neonates range from 1:7500 to 1:9000.4
To date, five subtypes of Stickler syndrome have been delineated. Type I Stickler syndrome (STL1, MIM 108300) accounts for the majority of cases and is caused by mutations in the gene encoding the alpha 1 procollagen chain of type II collagen, COL2A1.5 Alternative splicing effectively removes exon 2 from all COL2A1 mRNA transcripts in non-ocular tissues, so mutations in this exon result in an ocular-only phenotype, with individuals at a high risk of retinal detachment with little or no systemic manifestations.6,7 Disease is attributed to haploinsufficiency for normal COL2A1 protein products, often the result of loss-of-function missense mutations or nonsense mutations leading to decay of the mRNA transcript.8 Mutations in the COL11A1 or COL11A2 genes, which encode the alpha 1 and alpha 2 chains of type XI collagen, are associated with type 2 (STL2, MIM 604841) and type 3 (STL3, MIM 184840) Stickler syndrome, respectively. Ocular abnormalities with a “beaded” vitreous may be found in individuals with COL11A1 mutations, whereas individuals with COL11A2 mutations typically present with craniofacial, joint, and hearing loss manifestations, but no ocular features.2,3 Recently, autosomal recessive type 4 (STL4, MIM 614134) and type 5 (STL5, MIM 614284) Stickler syndromes have been discovered and are caused by mutations in the COL9A1 and COL9A2 genes, respectively.9,10
Wagner disease, regarded as a vitreoretinal degeneration and erosive vitreoretinopathy, was first described by Wagner in 1938.11 It was once diagnosed synonymously with ocularonly Stickler syndrome, but the identification of mutations in the versican-encoding gene, VCAN, enabled Wagner syndrome to be more clearly differentiated first by molecular genetics and then by clinical phenotype.12–14 The most notable differentiating clinical features of Wagner syndrome are an empty vitreous with avascular veils and chorioretinal atrophy.15,16
Early diagnosis of individuals through a combination of molecular genetic testing and clinical examination can lead to early treatment and care. Recent advances in sequencing technologies serve as efficient tools for both researchers and clinicians to identify implicated genes of various ocular diseases.17–20 The efficacy of exome sequencing to identify pathogenic coding mutations has shed new light into a cost-effective method to identify mutations in phenotypes demonstrating locus heterogeneity.21
Herein, we present a large multigenerational Caucasian pedigree for which the phenotype of Wagner or Stickler syndrome appears to segregate in an autosomal dominant manner. After commercial gene screening failed to identify a causal mutation in COL2A1 or VCAN, we utilized linkage mapping and exome sequencing to identify the disease-causing mutation.
Methods
Patients and controls
The study family was identified after referral from a retinal specialist (Dr. Max Johnson, Retina Consultants, Fargo, ND). Forty individuals were recruited at Duke University Eye Center (Durham, NC) and/or University of Iowa Carver College of Medicine (Iowa City, IA). Prior to recruitment, all institutional review board human subjects’ research study approvals were obtained. Participating members underwent full ophthalmic examinations, and medical records were obtained. In addition to the standard ophthalmic history, health histories included questions regarding hearing loss, previous repair of hard or soft cleft palate, other midline defects, skeletal or joint abnormalities, and early onset arthritis. Venous blood or saliva was collected from participating family members, and genomic DNA extracted using AutoPure LS DNA Extractor and PUREGENE reagents (Gentra Systems Inc, MN), respectively.
Linkage analysis
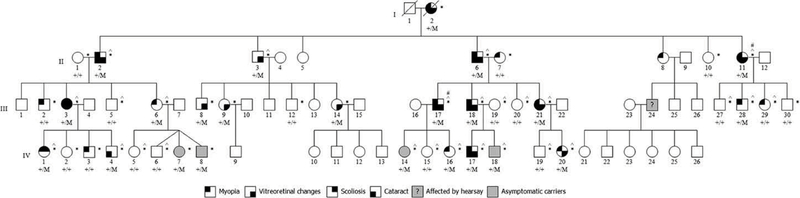
A HumanLinkage-24 BeadChip panel (Illumina, San Diego, CA) consisting of 5913 SNPs was used to conduct a whole-genome linkage scan with 26 individuals (11 clinically affected, 15 unaffected; Figure 1). PEDCHECK was used to determine Mendelian inconsistencies across the family structure, RELPAIR and PREST were used to detect possible errors in sample relationships, and any markers out of Hardy–Weinberg Equilibrium (HWE) were excluded. Data were analyzed using an autosomal dominant model in two-point (FASTLINK) and multipoint (MERLIN and SIMWALK) analyses.
Figure 1.
Study family pedigree. The study family consisted of 70 individuals in four generations. Circles indicate females and squares indicate males. An individual’s affection status is depicted as black-shaded quadrants for the four main phenotypes (myopia, vitreoretinal changes, cataract, and scoliosis), according to the figure key shown. Gray-shaded shapes indicate clinically unaffected individuals who carried the mutation. The gray-shaded individual with a question mark (III:24) is known to be affected with Stickler syndrome by hearsay. * indicates DNA available for this study. ^ indicates samples used for whole-genome linkage analysis. # indicates samples were used for exome sequencing.
Exome sequencing
DNA samples from two affected individuals (II:11, III:17) were chosen for exome sequencing (Figure 1). DNA (5 ug) was submitted to the Hudson Alpha Institute for processing (Huntsville, AL). Samples were prepared using the NimblGen V2 SeqCap EZ exome library capture kit (Madison, WI) and sequenced on an Illumina HiSeq platform to a median coverage of 50×. The resulting 100 nt paired-end reads were aligned to the human reference genome (GRCh37/hg19) using BWA. Reads were sorted and PCR duplicates removed using SAMtools. Variant SNPs and insertion/deletions (indels) were called with GATK. Variants covered by less than eight reads and with a quality score less than 30 were excluded from further analysis.
PCR and Sanger sequencing
Primer3 software was used to design primers (Supplemental Table 1). Standard PCR conditions were run to amplify all coding exons (±10 bp) and untranslated regions. Patient sequences were compared against the reference genome sequence using Sequencher 5.0 software (Gene Codes, Ann Arbor, MI). Variant frequencies in ethnically matched and global populations were determined from the Exome Aggregation Consortium (ExAC) online database.
Results
Clinical summary
Patients were examined at the Department of Ophthalmology (University of Iowa, IA) and/or Retina Specialists Inc. (Fargo, ND). Supplemental Table 2 summarizes the various clinical manifestations observed in all patients where clinical data were available.
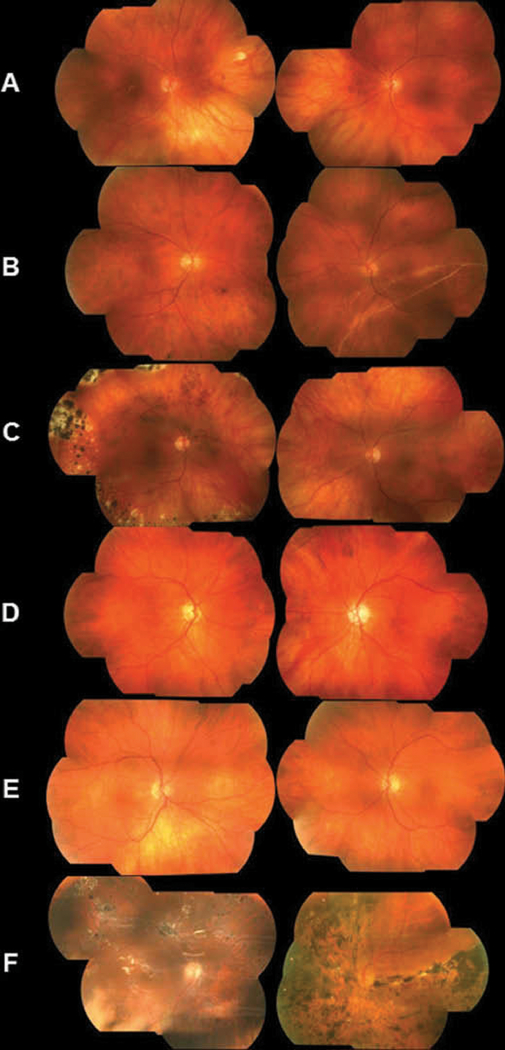
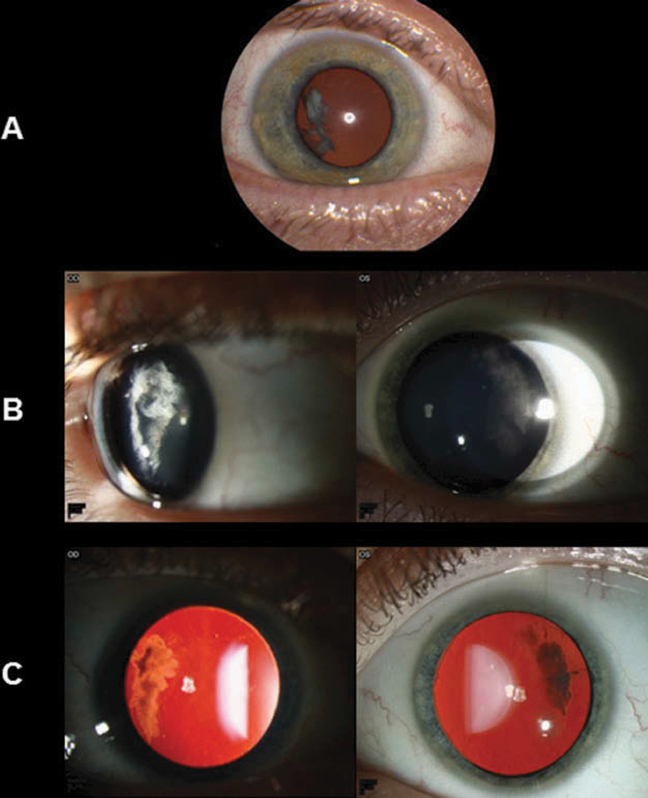
Ocular presentations varied for each patient, and severity of phenotype seemed to increase with age (Table 1). Fundus photos were available on six affected individuals from generations II and III (Figure 2), and all six presented ocular manifestations which included an optically empty vitreous, chorioretinal scarring, and myopia. Cataract (5/6), avascular sheets (5/6), and retinal detachment (5/6) were also widespread (Table 2). The common optically empty vitreous and avascular sheets seen in some patients resulted in an initial diagnosis of Wagner syndrome. Slit-lamp and retro-illumination photos were available for a father (III:18) and his 12-year-old son (IV:17) who presented with a cortical temporal cataract not seen in other family members (Figure 3).
Table 1.
Clinical phenotype and symptoms observed in at least one individual clinically diagnosed with Stickler syndrome.
| Clinical manifestations | Observed in one or more clinically affected individuals |
|---|---|
| Cataract | Yes |
| Nuclear sclerosis | Yes |
| Avascular sheets | Yes |
| Optically empty vitreous | Yes |
| Horse shoe retinal tear | Yes |
| Retinal detachment | Yes |
| Chorioretinal scars | Yes |
| Myopia | Yes |
| Scoliosis | Yes |
| Flexible joint hyper-mobility | No |
| Sensorineural or conductive hearing loss | No |
| Cleft palate | No |
| Mild epiphyseal dysplasia | No |
| Congenital megalophthalmos | No |
Figure 2.
Fundus photos of affected individuals harboring the p.Gln39* mutation. (A) Photos of a 42-year-old female (III:3). The right eye underwent retinal detachment repair with scleral buckle at age 24. The vitreous was optically empty in both eyes. In the right eye, there were focal chorioretinal scars in the macula and mid-peripherally in the 2:00 meridian. Not visualized were inferior and temporal vitreous condensation and vitreous sheets bilaterally, as well as indentation from a scleral buckle in the right eye and chorioretinal scarring bilaterally. (B) Photos of the right eye of a male individual III:17 at age 43 (left image) and at age 46 (right image). At age 43, the right eye had an optically empty vitreous centrally with avascular sheets in the far temporal periphery. At age 46, the patient presented after one week of sudden visual field loss. A large retinal detachment could be observed, extending from the 12:00 to the 8:00 meridian, caused by a peripheral retinal hole at 02:00. (C) Photos of a 45-year-old female (III:21) showed peripheral scarring in the right eye from repair of a superior giant retinal tear occurring at the age of 15. The tear was treated with laser and a scleral buckle. Later, an inferior retinal hole was diagnosed and treated with laser and cryotherapy. Both eyes had an optically empty vitreous. (D) Photos of a 69-year-old male (II:2). The fundus exam was normal for the right eye. The left eye had an optically empty vitreous with avascular sheets in the periphery. Outside the extent of the left image was superior retinal scarring from the treatment of two retinal tears with pneumatic retinopexy. (E) Photos of a 66-year-old male (II:6) showed an optically empty vitreous in both eyes with avascular sheets in the far periphery. There was no central chorioretinal scarring, but the left eye had some scarring in the far superior periphery (not shown). (F) Photos of a 58-year-old male (II:11). The right eye sustained several retinal tears and detachments and was treated on multiple occasions with laser, cryotherapy, scleral buckle, vitrectomy, and finally placement of silicone oil. Multiple retinal scars were observed as well as light reflections from the oil. The left eye exhibited extensive chorioretinal scarring and pigmentation after treatment of the retinal detachment and scleral buckle placement.
Table 2.
Clinical manifestations observed in affected individuals with fundus photos.
| Patient | Cataract | Avascular sheets | Optically empty vitreous | Retinal detachment | Chorioretinal scars | Myopia |
|---|---|---|---|---|---|---|
| III:21 | Yes | No | Yes | Yes | Yes | Yes |
| III:17 | Yes | Yes | Yes | Yes | Yes | Yes |
| III:3 | Yes | Yes | Yes | Yes | Yes | Yes |
| II:2 | Yes | Yes | Yes | Yes | Yes | Yes |
| II:6 | No | Yes | Yes | No | Yes | Yes |
| II:11 | Yes | Yes | Yes | Yes | Yes | Yes |
Figure 3.
Slit-lamp and retro-illumination photos of a father (III:18) and son (IV:17) with lamellar cataract in the temporal cortex of each eye. (A) Slit-lamp photo of the 42-year-old father’s right eye, which shows a temporal cortical cataract. The left eye underwent cataract extraction and intraocular lens placement for a visually significant cataract a year previously. (B) Slit-lamp photos of the 12-year-old son’s eyes, which show temporal cortical cataracts that are not visually significant. (C) Retro-illumination of the son’s eyes shows the distinctive temporal location of the cataracts.
Extraocular manifestations were noted in several individuals (Supplemental Table 2). Of particular note, individuals III:3 and IV:1 both presented with severe scoliosis of 39 or more degrees of curvature, for which they required a brace.
Linkage analysis
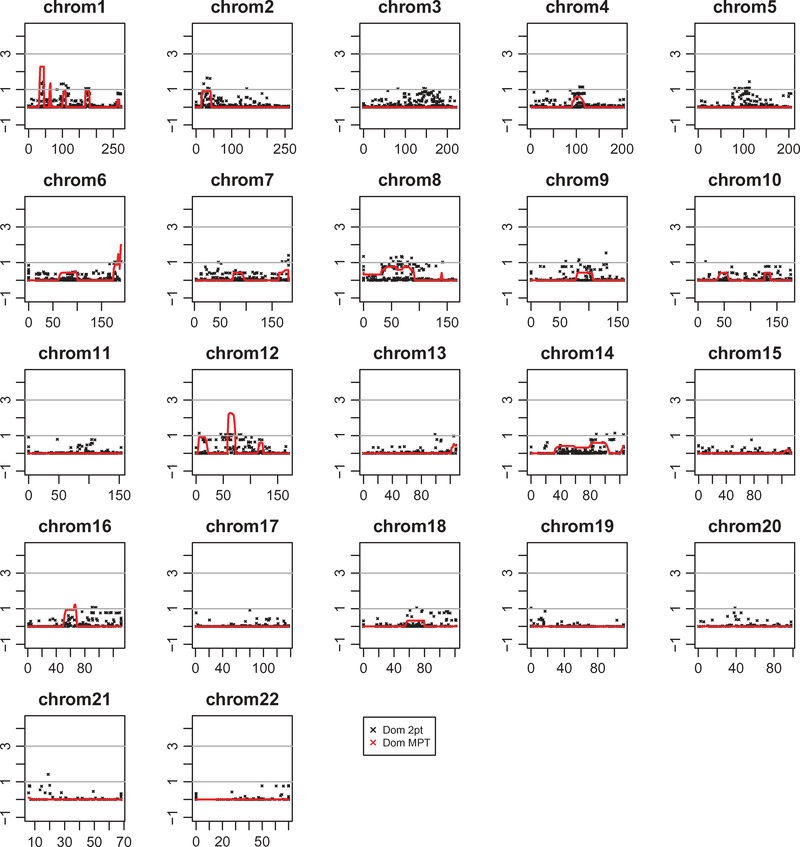
Five thousand nine hundred and thirteen markers were analyzed from the Illumina HumanLinkage-24 panel in 26 DNA samples. The multipoint analysis run under an autosomal dominant model demonstrated HLOD scores greater than 2.00 on chromosomes 1p36.13–1p36.11 (HLOD rs4237, 13 cM) and 12q12–12q14.1 (HLOD rs1107654, 13 cM) (Figure 4). Multipoint analysis with SIMWALK replicated the 12q peak, and a FASTLINK two-point analysis also resulted in clusters of SNPs with LOD scores greater than 1.00 for both regions (Supplemental Table 3).
Figure 4.
Summary of two-point and multipoint linkage analyses using MERLIN. Plots of −log10 chromosome-wide significant p values (−log10P) for autosomal dominant parametric two-point and multipoint linkage analyses in 26 individuals. The maximum peak LOD scores were achieved on chromosomes 1p36.13–1p36.11 and 12q12–12q14.1. Y-axis represents −log10 ratio of the LOD score. X-axis represents chromosome position in centimorgans (cM).
Exome sequencing and Sanger validation
DNA samples of two affected individuals were exome sequenced at a mean coverage depth of 59×, where over 95% of the targeted exons had at least 1× coverage and more than 70% of the exons were captured at 20×.
Using linkage data as a prioritization tool for filtering, all variants annotated within chromosomes 1p and 12q from the linkage interval were analyzed first. SNP and Variation Suite 7.5 software (Golden Helix, Bozeman, MT) were used to filter single nucleotide variants (SNVs) and micro-insertions/deletions (indels) to exclude those present in dbSNP135, NHLBI, and 1000 genomes databases. Novel, non-synonymous heterozygous variants present in both affected individuals were kept in the final list (Supplemental Table 4).
Filtering revealed a novel single base-pair transition (c.115C>T, mRNA reference NM_001844.4) in exon 2 of the gene encoding the alpha 1 chain of type II collagen, COL2A1. The variant resulted in an amino acid change from glutamine to a premature stop codon (p.Gln39*, protein reference NP_001835.3). To confirm co-segregation of the mutation with disease phenotype, the remaining family members were Sanger sequenced for the presence of the exon 2 mutation. Of the 40 family members analyzed, all 15 clinically affected individuals and 7 individuals who were clinically unaffected carried the mutation (Figure 1). Coverage across the COL2A1 genomic region was visually inspected in the exome dataset using GenomeBrowse software (Golden Helix, Bozeman, MT) and revealed several exonic regions with low or absent coverage. We screened the remaining exons of COL2A1 by Sanger sequencing and identified no additional variants that co-segregated with the disease phenotype.
Discussion
Although Stickler syndrome encompasses at least five subgroups, type I is encountered most frequently by ophthalmologists due to the ocular manifestations with minimal to no systemic features.2 In particular, COL2A1 exon 2 mutations have been regarded as an ocular-only fully penetrant phenotype associating with membranous vitreous anomalies and magalophthalmos.22–24 Using a combination of linkage analysis and exome sequencing, we report a novel nonsense mutation (c.115C>T, p.Gln39*) in exon 2 of COL2A1 segregating with disease in a large four-generation Caucasian pedigree. Surprisingly, both ocular and some extraocular phenotypic manifestations were present in our family.
Surveys reveal that 95% of Stickler patients report variable ocular pathological changes such as retinal detachment (60%) and myopia (90%).25 In our family, cataract, avascular sheets, optically empty vitreous, retinal detachment, chorioretinal scarring, and myopia were ocular features found commonly among clinically affected individuals (Table 2). Furthermore, a 42-yearold male (III-18) and his 12-year-old son (IV-17) demonstrated distinct ocular-only Stickler presentation with bilateral temporal cataracts (Figure 3).26 It is important to note that not all family members carrying the mutation had ocular characteristics (Figure 1).
We also report a 35-year-old male (III:28) who carried the mutation, but ophthalmic examination showed a clear vitreous and no retinal degeneration. Though he was anisometropic (−3.50 D; −0.75 D), he had no systemic manifestations. Furthermore, two of the triplets (IV:7 and IV:8) carried the mutation, but ocular findings were also normal (Supplemental Figure 2). Such phenotypic variability among family members may be due to differential expression levels of COL2A1,27,28 possibly as a result of variation within an enhancer domain.29 Furthermore, studies have shown a possible ratio imbalance between the type IIA and type IIB splice isoforms of COL2A1 which may result in dominant isoform expression.8 Though variable expressivity has recently been reported for exon 2 mutations resulting in nonsense-mediated decay or creation of an alternatively spliced protein, incomplete penetrance has not been reported in the literature to date.8,22
A points system has been proposed to diagnose patients based on clinical manifestations, but it may not effectively capture the clinical picture for families in which incomplete penetrance or age of onset is a contributing factor to the pathogenesis of disease.30,31 For example, the ages of the six individuals who were clinically unaffected but carried the mutation ranged from 2 to 15 years at the time of their exam (Supplemental Table 2). Clinically unaffected young children carrying such a mutation without Sticklerlike symptoms suggest a possible presymptomatic stage.31 Furthermore, variable age of onset has been reported in Stickler patients with mutations in COL2A1.32,33 Phenotypic and age-ofonset variability in Stickler patients harboring collagen mutations may challenge proper clinical diagnosis, and a full understanding of allied disorders associated with Stickler syndrome is important for differential diagnosis.31,34,35
Nevertheless, future findings from next-generation sequencing will certainly alleviate difficulties in disease gene identification in heterogeneous diseases with many candidate loci. The recent rapid decrease in cost of whole-genome and exome sequencing has resulted in widespread capability to uncover genetic causality in Mendelian diseases.36 However, there is still debate regarding efficacy of coverage, bioinformatics tools, and data management to effectively use in the clinical setting.37,38 In the field of retinal dystrophies, attempts have been made to create a targeted sequencing approach to mitigate these challenges.39 Finally, until the molecular etiologies of these disorders have been completely resolved, combining linkage analysis and exome sequencing can help aid in the discovery of causative mutations, as has also been shown for familial exudative vitreoretinopathy.40
Lastly, it is unclear how a commercial, CLIA-certified, diagnostic facility failed to identify the causal mutation in COL2A1, but presumably this was the result of human error while analyzing electrophoretogram output from Sanger sequencing. Ultimately, we identified the causal mutation through exome sequencing, a technology that utilizes algorithms to specifically extract and present only the variant information from an individual’s exome sequence, an additional strength of this powerful methodology.
Supplementary Material
Acknowledgments
The authors would like to thank all family members for participation in the study. We wish to also acknowledge the technical contributions of Caldwell Powell, Whitney Call, Valencia Quiett, and Erica Nading.
Funding
This research was funded by the National Institutes of Health Grant R01 EY014685, the Lew Wasserman Award from Research to Prevent Blindness Inc., New York, NY, an unrestricted departmental grant from Research to Prevent Blindness, Inc, New York, NY, and a Duke-National University of Singapore core grant to Terri Young. Vincent Soler was supported by the Toulouse Hospital Young Researcher Fellowship, the Fondation pour la Recherche Médicale, and Fondation de France. Benjamin Bakall is supported by a Foundation Fighting Blindness career development award.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- 1.Stickler GB, Belau PG, Farrell FJ, et al. Hereditary progressive arthro-ophthalmopathy. Mayo Clin Proc 1965;40:433–455. [PubMed] [Google Scholar]
- 2.Snead MP, McNinch AM, Poulson AV, et al. Stickler syndrome, ocular-only variants and a key diagnostic role for the ophthalmologist. Eye (Lond) 2011;25:1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snead MP, Yates JR. Clinical and molecular genetics of stickler syndrome. J Med Genet 1999;36:353–359. [PMC free article] [PubMed] [Google Scholar]
- 4.Printzlau A, Andersen M. Pierre Robin sequence in Denmark: a retrospective population-based epidemiological study. Cleft Palate Craniofac J 2004;41:47–52. [DOI] [PubMed] [Google Scholar]
- 5.Fertala A, Ala-Kokko L, Wiaderkiewicz R, Prockop DJ. Collagen II containing a Cys substitution for arg-alpha1–519. Homotrimeric monomers containing the mutation do not assemble into fibrils but alter the self-assembly of the normal protein. J Biol Chem 1997;272:6457–6464. [DOI] [PubMed] [Google Scholar]
- 6.Donoso LA, Edwards AO, Frost AT, et al. Clinical variability of Stickler syndrome: role of exon 2 of the collagen COL2A1 gene. Surv Ophthalmol 2003;48:191–203. [DOI] [PubMed] [Google Scholar]
- 7.Donoso LA, Edwards AO, Frost AT, et al. Identification of a stop codon mutation in exon 2 of the collagen 2A1 gene in a large stickler syndrome family. Am J Ophthalmol 2002;134:720–727. [DOI] [PubMed] [Google Scholar]
- 8.McAlinden A, Majava M, Bishop PN, et al. Missense and nonsense mutations in the alternatively-spliced exon 2 of COL2A1 cause the ocular variant of Stickler syndrome. Hum Mutat 2008;29:83–90. [DOI] [PubMed] [Google Scholar]
- 9.Van Camp G, Snoeckx RL, Hilgert N, et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet 2006;79:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker S, Booth C, Fillman C, et al. A loss of function mutation in the COL9A2 gene causes autosomal recessive Stickler syndrome. Am J Med Genet Part A 2011;155A:1668–1672. [DOI] [PubMed] [Google Scholar]
- 11.Wagner H Ein bisher unbekanntes des auges (degeneration hyaloideo-retinalis hereditaria), beobachtet im Kanton Zurich. Klin Monbl Augenheilkd (German) 1938;100:840–857. [Google Scholar]
- 12.Liberfarb RM, Hirose T, Holmes LB. The Wagner-Stickler syndrome: a study of 22 families. J Pediatr 1981;99:394–399. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto T, Inoue H, Sakamoto Y, et al. Identification of a novel splice site mutation of the CSPG2 gene in a Japanese family with Wagner syndrome. Invest Ophthalmol Vis Sci 2005;46:2726–2735. [DOI] [PubMed] [Google Scholar]
- 14.Meredith SP, Richards AJ, Flanagan DW, et al. Clinical characterisation and molecular analysis of Wagner syndrome. Br J Ophthalmol 2007;91:655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graemiger RA, Niemeyer G, Schneeberger SA, Messmer EP. Wagner vitreoretinal degeneration – follow-up of the original pedigree. Ophthalmology 1995;102:1830–1839. [DOI] [PubMed] [Google Scholar]
- 16.Brown DM, Graemiger RA, Hergersberg M, et al. Genetic linkage of Wagner disease and erosive vitreoretinopathy to chromosome 5q13–14. Arch Ophthalmol 1995;113:671–675. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Li Y, Zhang D, et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet 2011;7:e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majewski J, Wang Z, Lopez I, et al. A new ocular phenotype associated with an unexpected but known systemic disorder and mutation: novel use of genomic diagnostics and exome sequencing. J Med Genet 2011;48:593–596. [DOI] [PubMed] [Google Scholar]
- 19.Koenekoop RK, Wang H, Majewski J, et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet 2012;44:1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strom SP, Gao YQ, Martinez A, et al. Molecular diagnosis of putative Stargardt disease probands by exome sequencing. BMC Med Genet 2012;13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon-Salazar TJ, Silhavy JL, Udpa N, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med 2012;4:138ra178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robin NH, Moran RT, Ala-Kokko L Stickler syndrome. Synonym: Arthro-ophthalmopathy 2000. June 9 [Updated 2014 Nov 26]. In: Pagon RA, Adam MP, Ardinger HH, et al. , editors. GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2016. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1302/. [Google Scholar]
- 23.Richards AJ, Martin S, Yates JR, et al. COL2A1 exon 2 mutations: relevance to the Stickler and Wagner syndromes. Br J Ophthalmol 2000;84:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards AJ, Snead MP. The influence of pre-mRNA splicing on phenotypic modification in Stickler’s syndrome and other type II collagenopathies. Eye (Lond) 2008;22:1243–1250. [DOI] [PubMed] [Google Scholar]
- 25.Stickler GB, Hughes W, Houchin P. Clinical features of hereditary progressive arthro-ophthalmopathy (Stickler syndrome): a survey. Genet Med 2001;3:192–196. [DOI] [PubMed] [Google Scholar]
- 26.Seery CM, Pruett RC, Liberfarb RM, Cohen BZ. Distinctive cataract in the Stickler syndrome. Am J Ophthalmol 1990;110: 143–148. [DOI] [PubMed] [Google Scholar]
- 27.Dale RM, Topczewski J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol 2011;357:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niebler S, Bosserhoff AK. The transcription factor activating enhancer-binding protein epsilon (AP-2epsilon) regulates the core promoter of type II collagen (COL2A1). FEBS J 2013;280:1397–1408. [DOI] [PubMed] [Google Scholar]
- 29.Shinomura T, Ito K, Hook M, Kimura JH. A newly identified enhancer element responsible for type II collagen gene expression. J Biochem 2012;152:565–575. [DOI] [PubMed] [Google Scholar]
- 30.Antunes RB, Alonso N, Paula RG. Importance of early diagnosis of Stickler syndrome in newborns. J Plast Reconstr Aesthet Surg 2012;65:1029–1034. [DOI] [PubMed] [Google Scholar]
- 31.Hoornaert KP, Vereecke I, Dewinter C, et al. Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur J Hum Genet 2010;18:872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su P, Li R, Liu S, et al. Age at onset-dependent presentations of premature hip osteoarthritis, avascular necrosis of the femoral head, or Legg-Calve-Perthes disease in a single family, consequent upon a p.Gly1170Ser mutation of COL2A1. Arthritis Rheum 2008;58:1701–1706. [DOI] [PubMed] [Google Scholar]
- 33.Kannu P, Bateman JF, Randle S, et al. Premature arthritis is a distinct type II collagen phenotype. Arthritis Rheum 2010;62:1421–1430. [DOI] [PubMed] [Google Scholar]
- 34.Acke FR, Dhooge IJ, Malfait F, De Leenheer EM. Hearing impairment in Stickler syndrome: a systematic review. Orphanet J Rare Dis 2012;7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ang A, Ung T, Puvanachandra N, et al. Vitreous phenotype: a key diagnostic sign in Stickler syndrome types 1 and 2 complicated by double heterozygosity. Am J Med Genet A 2007;143:604–607. [DOI] [PubMed] [Google Scholar]
- 36.Rabbani B, Mahdieh N, Hosomichi K, et al. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet 2012;57:621–632. [DOI] [PubMed] [Google Scholar]
- 37.Xuan J, Yu Y, Qing T, et al. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett 2013;340:284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norton N, Li D, Hershberger RE. Next-generation sequencing to identify genetic causes of cardiomyopathies. Curr Opin Cardiol 2012;27:214–220. [DOI] [PubMed] [Google Scholar]
- 39.Shanks ME, Downes SM, Copley RR, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet 2013;21:274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikopoulos K, Gilissen C, Hoischen A, et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet 2010;86:240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.