Key Points
LILRB1 inhibits BiTE molecule–induced cytolytic activity of human CD8+ T cells.
LILRB1 and PD1 express on distinct human CD8+ T cell subsets in tumors.
LILRB1 and PD1 blockades have synergistic effects to enhance CD8+ T cell activity.
Abstract
Elicitation of tumor cell killing by CD8+ T cells is an effective therapeutic approach for cancer. In addition to using immune checkpoint blockade to reinvigorate existing but unresponsive tumor-specific T cells, alternative therapeutic approaches have been developed, including stimulation of polyclonal T cell cytolytic activity against tumors using bispecific T cell engager (BiTE) molecules that simultaneously engage the TCR complex and a tumor-associated Ag. BiTE molecules are efficacious against hematologic tumors and are currently being explored as an immunotherapy for solid tumors. To understand mechanisms regulating BiTE molecule–mediated CD8+ T cell activity against solid tumors, we sought to define human CD8+ T cell populations that efficiently respond to BiTE molecule stimulation and identify factors regulating their cytolytic activity. We find that human CD45RA+CCR7− CD8+ T cells are highly responsive to BiTE molecule stimulation, are enriched in genes associated with cytolytic effector function, and express multiple unique inhibitory receptors, including leukocyte Ig-like receptor B1 (LILRB1). LILRB1 and programmed cell death protein 1 (PD1) were found to be expressed by distinct CD8+ T cell populations, suggesting different roles in regulating the antitumor response. Engaging LILRB1 with its ligand HLA-G on tumor cells significantly inhibited BiTE molecule–induced CD8+ T cell activation. Blockades of LILRB1 and PD1 induced greater CD8+ T cell activation than either treatment alone. Together, our data suggest that LILRB1 functions as a negative regulator of human CD8+ effector T cells and that blocking LILRB1 represents a unique strategy to enhance BiTE molecule therapeutic activity against solid tumors.
Introduction
T cells, especially Ag-specific cytotoxic T cells, can detect and eliminate cancer cells through the recognition of tumor-associated Ags, such as neoantigens. Neoplastic cells, however, evade immune surveillance through various mechanisms. For example, tumor-infiltrating T cells often fail to eliminate cancer because of an immunosuppressive tumor microenvironment that induces a dysfunctional state, characterized by the expression of multiple inhibitory receptors such as programmed cell death protein 1 (PD1), TIM3, and CTLA4, and designated as exhausted T cells (TEXH). Importantly, Abs targeting CTLA-4 and PD1 have demonstrated dramatic therapeutic benefit in various cancer types, correlating with their ability to enhance effector T cell (TEFF) function (1). However, these immunotherapy strategies are successful in only a subset of patients. Although multiple mechanisms likely account for a failure to respond to immune checkpoint inhibitory therapy, the inherent immunogenicity of a patient’s tumor and corresponding levels of pre-existing tumor-reactive T cells present at the start of therapy have emerged as important factors governing the response (2). For instance, patients with lower mutation burden and/or with scarce T cell infiltration in their tumors generally have poor responses to immune checkpoint inhibitors (3, 4).
Given the potential limitations of immune checkpoint inhibitory therapy in patients with low pre-existing antitumor immunity, an alternative promising therapeutic strategy involves mobilizing polyclonal T cells against tumor cells in an MHC-peptide presentation-independent manner. Two such clinically successful approaches include adoptive cell therapy with chimeric Ag receptor (CAR) T cells (5, 6) and the use of bispecific T cell engager (BiTE) Ab constructs (7). CAR T therapy involves engineering autologous T cells to express a chimeric receptor that is capable of recognizing tumor-associated surface Ag to trigger T cell activation, whereas BiTE Ab constructs are a novel class of immunotherapy molecules engineered to redirect T cells to tumor sites and induce T cell activation, immune synapse formation, and ultimately tumor cell killing, regardless of Ag specificity (5–7). BiTE molecules contain two fused single-chain variable fragments, with one that binds to CD3 on T cells and the other that binds to a tumor-associated Ag (7).
Blinatumomab, a CD19/CD3 BiTE Ab construct, is the first BiTE molecule approved by the U.S. Food and Drug Administration to treat various hematologic malignancies (8). Although this therapy provides evidence that BiTE molecules can induce robust tumor cell killing in humans, the degree to which this activity will translate to the solid tumor setting is largely unknown. PD1-expressing TEXH represent a dominant phenotype among solid tumor–infiltrating CD8+ T cells (1), and PD1 is known to be induced by BiTE molecule treatment in vitro (9, 10). This suggests that the PD1/programmed death ligand 1 (PDL1) pathway may potentially interfere with BiTE molecule activity in solid tumors and provides a rationale for combining BiTE molecules with PD1 inhibitors. However, other CD8+ T cell subsets beyond TEXH are found within solid tumors (11, 12). Further dissecting the function of these populations and their ability to respond to BiTE molecule engagement may lead to additional combination therapy approaches aimed at eradicating solid tumors.
The human peripheral blood CD8+ T cell compartment is comprised of multiple subsets, often distinguished by their expression of the naive/memory marker CD45RA and the chemokine receptor CCR7. CD8+ naive T cells (TN) are CD45RA+CCR7+, whereas CD8+ effector memory T cells (TEM) are CD45RA−CCR7+/−. Previous studies have demonstrated that CD8+ TEM contribute to BiTE molecule activity among peripheral blood CD8+ T cell subsets, whereas CD8+ TN show minimal response to BiTE molecule treatment (13). In addition, blocking PD1 has been shown to mainly expand the intratumoral effector memory subset of CD8+ T cells in patients (14). An additional Ag-experienced CD8+ T cell population is defined as CD45RA+CCR7− (effector memory T cells reexpressing CD45RA [TEMRA]). These effector cells have high cytotoxicity, high sensitivity to apoptosis, low IL-2 production (15), and represent a significant CD8+ T cell population in elderly individuals and cancer patients (16, 17). However, the degree to which TEMRA can efficiently mediate BiTE molecule–induced tumor killing, the function of these cells within solid tumors, and the mechanisms regulating their activity, remain largely unknown.
In this study, we studied the phenotype of Ag-experienced CD8+ T cell subsets isolated from human peripheral blood and solid tumor biopsy specimens, their function following BiTE molecule treatment, and the mechanisms regulating their activity. In blood, we identify TEMRA as the subset with the most potent cytolytic activity following BiTE molecule engagement and find that a subset of these cells expressing high levels of cytolytic effector molecules also expresses the ITIM-containing inhibitory leukocyte Ig-like receptor B1 (LILRB1), which can function as a negative regulator of BiTE molecule–induced tumor cell killing. Importantly, LILRB1 and PD1 showed nonoverlapping expression patterns across CD8+ TEM and TEMRA subsets, and blocking both pathways synergistically enhanced CD8+ T cell function. We extend these data from peripheral blood to tumor-associated T cell populations, demonstrating similar phenotypic and functional properties of TEMRA. These data highlight the potential importance of TEMRA in antitumor immune surveillance and BiTE molecule–mediated activity against solid tumors and suggest that blockade of LILRB1 may be a promising therapeutic approach to enhance their cytolytic activity.
Materials and Methods
Abs and flow cytometry
The following Abs were used: CD3 (Sk7; eBioscience), CD4 (OKT4; BioLegend), CD8 (RPA-T8; BD Biosciences), CCR7 (G043H7; BioLegend), CD45RA (HI100; BioLegend), LILRB1(GHI/75; BioLegend), PD1 (EH12.1; BD Biosciences), CD69 (FN50; BD Biosciences), granzyme B (GB11; BioLegend) and perforin (B-D48; BioLegend), HLA-G (MEM-G/9; Thermo Fisher Scientific). LIVE/DEAD Fixable Viability Dye eFluor 506 (eBioscience), or SYTOX Blue (Thermo Fisher Scientific) was included to mark dead cells. For staining of cell surface proteins, cells were incubated with appropriate Ab mixtures for 30 min at 4°C. To determine expression of intracellular proteins, cells were fixed for 30 min at 4°C using the Foxp3 Transcription Factor Staining Buffer Set (catalog no. 00-5523-00; Thermo Fisher Scientific) following the manufacturer’s protocol. Subset cell population was isolated by cell sorting with a BD FACSAria cytometer (BD Biosciences). Data acquisition of Ab-stained samples was carried out on an LSR II (BD Biosciences) and analyzed with FlowJo software v10.3 (Tree Star).
Primary human cell assays
Human PBMCs from healthy volunteers were obtained after informed consent and authorized by the Amgen Research Blood Donor Program. PBMCs were isolated by density gradient centrifugation (Ficoll-Paque PREMIUM, catalog no. 17-5442-02; GE Healthcare). Cell subsets were isolated from PBMCs with appropriate magnetic beads following manufacturer’s protocol (STEMCELL Technologies) or by cell sorting with a BD FACSAria cytometer. Isolated human primary lymphocytes were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin and streptomycin, and 2-ME (all from Life Technologies) at 37°C. PBMCs were stimulated with 10 ng/ml recombinant human IL-2 (catalog no. 202-IL-010/CF), 10 ng/ml recombinant human IL-15 (catalog no. 247-ILB-025/CF), or 100 ng/ml recombinant human TNF (catalog no. 210-TA-020/CF) (all from R&D Systems) and then subjected to flow cytometry analysis. Isolated T cells were activated with plate-bound anti-CD3 (OKT3, 5 μg/ml; BD Biosciences) plus soluble anti-CD28 (CD28.2, 2 μg/ml; BD Biosciences). When indicated, 10 μg/ml recombinant human PDL1 (catalog no. 762506; BioLegend) or the same amount of human IgG1 isotype control Ab (catalog no. 403501; BioLegend) was added in culture medium. Samples were analyzed by flow cytometry. Cytokine concentrations were measured in cell culture supernatants 48 h after stimulation with Luminex assay (EMD Millipore, Billerica, MA).
Human tumor dissociation
All human tumor specimens were collected under Institutional Review Board approval with appropriate informed consent. In all cases, materials obtained were surplus to standard clinical practice. Patient identity and protected health information/identifying information were redacted from tissues and clinical data. Biopsy specimens were mechanically disrupted and incubated with 200 μg/ml Liberase TL (catalog no. 5401020001; Roche) and 20 U/ml DNase I (catalog no. 4716728001; Roche) at 37°C for 15 min followed by tissue disaggregation using gentleMACS Octo Dissociator (Miltenyi Biotec). Cell suspension was then passed through a 70-μm filter twice and washed with DMEM/F12 containing 10% FBS (Life Technology, Carlsbad, CA).
Cell lines
SK-MEL-2 (SK2; catalog no. HTB-68, SK2; American Type Culture Collection) and SK-MEL-5 (catalog no. HTB-70, SK5; American Type Culture Collection) were maintained EMEM (catalog no. 30-2003; American Type Culture Collection) with 10% FBS and 1% penicillin/streptomycin. HLA-G–expressing SK2 (SK2.HLA-G) cells with stable expression of human HLA-G were established by retroviral transduction. The HLA-G-β2 microglobulin cDNA was inserted into retrovirus vector pLHCX2 (catalog no. 631503; Clontech) and was cotransfected with Lipofectamine 3000 (catalog no. L3000001; Thermo Fisher Scientific) into GP2-293 packaging cells (catalog no. 631458; Clontech) with pVSV-G (catalog no. 631530; Clontech). Supernatants were collected 48 h after transfection and filtered. Then, SK2 cells were transduced with a mixture of viral supernatant with polybrene at 5 μg/ml (catalog no. TR-1003-G; EMD Millipore). After spin transduction for 1.5 h at 1200 g, 32°C, fresh media was added. The human PDL1 cDNA was inserted into expression vector pcDNA3.1/Zeo(+) and was transfected into parental SK2 cells or SK2.HLA-G cells with Lipofectamine 3000 (Thermo Fisher Scientific). PDL1 and HLA-G expression was confirmed by flow cytometry.
MLR
Isolated human total T cells were seeded into round-bottom 96-well plates (Corning) at 1 × 105 per well together with 2 × 104 irradiated allogeneic CD11c+ cells. When indicated, 10 μg/ml of anti-PD1 (clone EH12.2H7, catalog no. 329902; BioLegend) or anti-LILRB1 (clone GHI/75, catalog no. 333704; BioLegend) or both Abs were added to the culture. As control, 10 μg/ml of each matched isotype Abs were used as control. Day 5 supernatants were collected, and IFN-γ level was determined by ELISA (catalog no. 550612; BD Biosciences).
Cytotoxicity assay
Sorted CD8+ T cells from healthy donor PBMCs expressing indicated markers or total CD8+ T cells from non–small cell lung cancer (NSCLC) biopsy specimens were used as TEFF cells and coincubated with tumor target cells at 4:1 ratio for the indicated time period. The BiTE molecule in this assay is CD3/melanoma Ag recognized by T cells 1 (MART-1) BiTE Ab construct (Amgen). When indicated, 25 μg/ml anti-LILRB1 (clone GHI/75; BioLegend) or 10 μg/ml of anti-PD1 (clone EH12.2H7; BioLegend) were added in the assay. After the culture, T cells were removed by washing with warm media, and target cell killing was determined from measuring the number of viable target cells by adding CellTiter-Glo (catalog no. G7570; Promega). Luminescence units (measured in relative light units [RLUs]) were measured by EnVision 2104 multilable reader (PerkinElmer). Percentage of specific lysis was calculated as follows: % Specific Cytotoxicity = [1 − (RLU live target cells + BiTE molecule)/(mean RLU live target cells − BiTE molecule)] × 100.
Cell culture and cytotoxic function assay of MelanA-specific Mel-13 CD8+ T cell line
Mel-13 T cells were expanded as described (18). Expanded Mel-13 T cells (>50% LILRB1+ by flow cytometry) were used as effector cells and cocultured with SK5 tumor target cells (104 per well) at indicated ratio. When indicated, anti-LILRB1 (GHI/75, 20 μg/ml) or same amount of isotype control Ab were added in the culture. After overnight incubation at 37°C, T cells were removed by washing the wells with warm media. Target cell killing was determined by measuring the number of viable target cells using CellTiter-Glo (Catalog no. G7570; Promega). Luminescence, in RLUs, was measured by an EnVision 2104 Multilabel Reader (PerkinElmer). Percentage of specific lysis was calculated as follows: Percentage Specific Cytotoxicity = [1 − (RLU live target cells + Mel-13)/(mean RLU live target cells − Mel-13)] × 100.
RNA sequencing
Global transcript expression in CCR7+ and CCR7− CD8+ T cells isolated from healthy donor PBMCs was assessed by RNA sequencing (RNAseq). RNAseq was performed on a cDNA library prepared from total RNA (2 μg; RNA integrity number >9.5) of each sample isolated using mirVana miRNA Isolation Kits (Ambion, Grand Island, NY) with on-column DNase treatment. Total RNA quality and concentration was determined using the Bioanalzyer (Agilent Technologies, Santa Clara, CA) and NanoDrop (Thermo Fisher Scientific, Wilmington, DE). cDNA was prepared using a modified protocol based on the Illumina Truseq RNA Sample Preparation Kit (Illumina, San Diego, CA) and the published methods for strand-specific RNAseq (19, 20). After size selection of libraries (Pippen Prep; Sage Science, Beverly, MA), dUTP-containing cDNA strands were destroyed by digestion of USER enzymes (New England Biolabs, Ipswich, MA) followed by PCR enrichment for introduction of strand specificity. The enriched cDNA libraries were analyzed in an Agilent Bioanalyser and quantified by Quant-iT PicoGreen assays (Life Technologies). RNA sequencing reads (Illumina HiSeq platform, 75-bp paired end sequencing) were aligned to human genome build 38 and fragments per kb per million were sequenced. Fragments per kilobase of transcript per million mapped reads were determined using Array Suite software (Omicsoft, Cary, NC) and in-house software. Differential gene expression analysis was performed using DESeq V2 (21). The RNAseq data have been submitted to National Center for Biotechnology Information Sequence Read Archive database (https://urldefense.proofpoint.com/v2/url?u=https-3A__www.ncbi.nlm.nih.gov_sra_PRJNA535519&d=DwICaQ&c=Sexio4usKrYWFsrnxgjbcQ&r=isDLOdYR9iBOWpfW7BvglA&m=2FGyR3rJtIJMf51upHaeB-XYgRM7_qjfjweuZN6liT0&s=suTXFXS5v_GXcmv4Ba1ju_LXZLZvdLsJKTVkmSHCdwc&e=) under the accession number PRJNA535519.
Single-cell RNA sequencing
Fresh human CD8+ T cells were sorted into LILRB1+ CCR7− CD45RA+, LILRB1− CCR7−CD45RA+/−, and LILRB1− CCR7+ CD45RA+ subsets on a BD FACSAria (BD Biosciences) to purity >95%. Each sorted cell subset was loaded on the 10× Chromium System (22), and single-cell RNAseq libraries were generated according to the manufacturer’s instructions. Sequencing was conducted on a HiSeq 4000 (sequencing performed by GENEWIZ). Each pooled sample was run in its own lane according to the 10× 3′ v2 Chemistry protocol. The Cell Ranger command line tools (10× Genomics v2.0.0) mkfastq script was used to convert each Illumina BCL file to FASTQ. Paired FASTQ files for each pooled sample (1 Lane) were aligned to GRCh38 human genome (v. 1.2.0 from 10× Genomics) using cellranger count script with default settings. The three CD8+ samples were then aggregated using cellranger aggr script. Using the R software package, Monocle (v2.6.1) data were transformed from raw unique molecular identifiers to normalized counts using Monocle’s negbinomial.size (23) model. Cells were classified by expression of each respective gene (LILRB1, CCR7), positive if expression >1 and negative if expression <1. The classifyCells function in Monocle was used to create the cell classification labels. The q value and p value statistics, along with fold change, were computed across the three cell classifications. Genes with a q value <0.04 were kept. The mean expression of each gene was computed and the top 50 genes by fold change across each pairwise group were kept. The heatmap was created from mean expression values in each group using the python package seaborn “clustermap” function. Gene averages are z-scores across each gene (mean of 0 and variance of 1).
Single-cell RNAseq data visualization
All visualizations are created using R programming language (24) using ggplot2 and ggbeeswarm visualization packages (25, 26). Statistics were computed using function from ggpubr package (27). Each swarm plot (gene expression per cluster across cells) visualizes a single gene by custom cluster assignment with gene expression values represented as binary log plus + 0.1. Previously determined and published clusters for corresponding datasets (11, 12) were used to define clusters: TEM (hepatocellular carcinoma: CD8_Cluster5-GZMK; NSCLC: CD8_C4-GZMK); TEFF (hepatocellular carcinoma: CD8_Cluster2-CX3CR1; NSCLC: CD8_C3-CX3CR1); and TEXH (hepatocellular carcinoma: CD8_Cluster4-LAYN; NSCLC: CD8_C6-LAYN). Boxplots visualize five summary statistics (the median, two hinges, and two whiskers) and are overlaid with swarm plots of cells roughly representing the density distribution of the data points along the gene expression values. A nonparametric Kruskal–Wallis test was used to test for difference in gene expression of each gene between clusters, followed by pairwise Wilxocon rank sum test pairwise comparisons. The p values were adjusted using Benjamini–Hochberg procedure. For per patient boxplot, gene expression (log2(transcripts per million + 0.1)), as measured from previously published NSCLC datasets (12), was averaged per patient for each depicted gene (NSCLC npatients = 14). For gene expression of each gene between clusters, an ANOVA was used to analyze the differences among group means, followed by pairwise t test comparisons. The p values were adjusted using Benjamini–Hochberg procedure. Statistical plots were generated with GraphPad Prism software, and a two-sided paired or nonpaired Student t test was used to obtain p values. Sample sizes were determined based on experimental data distribution pattern.
Results
LILRB1-expressing effector CD8+ T cells can efficiently mediate BiTE molecule–induced cytolytic activity
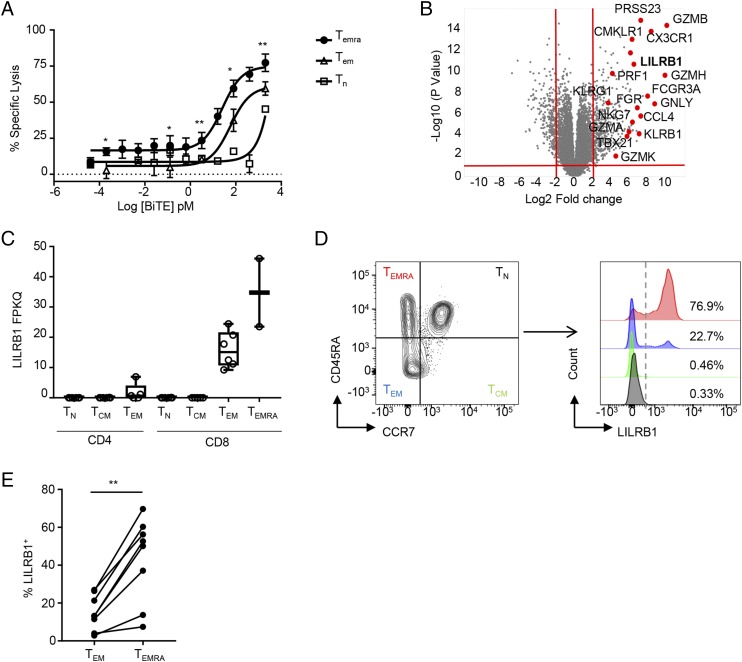
To understand which human CD8+ T cell subset most efficiently mediates BiTE molecule–induced T cell–dependent cellular cytotoxicity, we isolated human CD8+ TN, TEM, and TEMRA subsets from peripheral blood based on expression of surface markers CD45RA and CCR7 (15). The cytolytic activity of isolated CD8+ T cell subsets was determined using SK2 cells endogenously expressing the MART-1 tumor Ag and a MART-1–specific BiTE molecule. As shown in Fig. 1A, TEMRA were the most potent at BiTE molecule–induced killing of target cancer cells (EC50 = 25.51 pM), followed by CD45RA−CCR7− TEM (EC50 = 58.11 pM) and CD45RA+CCR7+ TN (EC50 = 7948 pM). TEMRA also had the highest maximum percentage of tumor cell lysis (Fig. 1A). These data suggest that enhancing the effector function of TEMRA may further enhance BiTE molecule–mediated tumor killing.
FIGURE 1.
CD8+ TEMRA show potent BiTE molecule–mediated tumor cell killing and preferentially express the LILRB1 inhibitory receptor. (A) Human CD8+ T cells from healthy donors were sorted into TN, TEM, and TEMRA subsets and incubated with SK2 tumor cells in the presence of indicated amount of MART-1–specific BiTE molecule. Specific cytotoxicity was determined after 45 h. Results (mean ± SEM) are shown from four independent experiments using three healthy donors as the source of CD8+ T cells. (B) Volcano plot showing differentially expressed genes between CCR7− and CCR7+ human CD8+ T cells with p value <0.05 and fold change >−4 (red lines). (C) Box plot showing LILRB1 mRNA expression of RNAseq analysis of TN, TCM, TEM, and TEMRA subsets of CD8+ T cells (n = 2–6 individual donor samples per cell type). (D) Representative FACS plots showing preferential LILRB1 expression on the surface of CD8+ TEMRA. (E) Quantitation of LILRB1 expression in indicated CD8+ subsets determined by FACS analysis across multiple donors (n = 8). *p < 0.05, **p < 0.01, paired Student t test.
To identify regulatory pathways that modulate CTL activity, we evaluated the gene transcriptional profiles of sorted TN, TEM, central memory T cell (TCM) and TEMRA subsets. Differential expression analysis comparing CCR7+ TN and TCM with CCR7− TEM and TEMRA populations revealed expression of various genes associated with CD8+ T cell function, including PRF1, GZMB, and CX3CR1 (Fig. 1B, Supplemental Table I). In addition, LILRB1, an ITIM-containing inhibitory receptor that signals downstream of interactions with HLA-G and several classical MHC class I alleles (28), was also found to be highly expressed by the TEMRA and TEM populations (Fig. 1B, Supplemental Table I), with preferential expression in TEMRA (Fig. 1C). Analysis of CD8+ T cell phenotypes by flow cytometry confirmed that the majority of TEMRA but only a small population of TEM express LILRB1 (Fig. 1D, 1E).
LILRB1 is a marker of CD8+ T cell with potent effector function
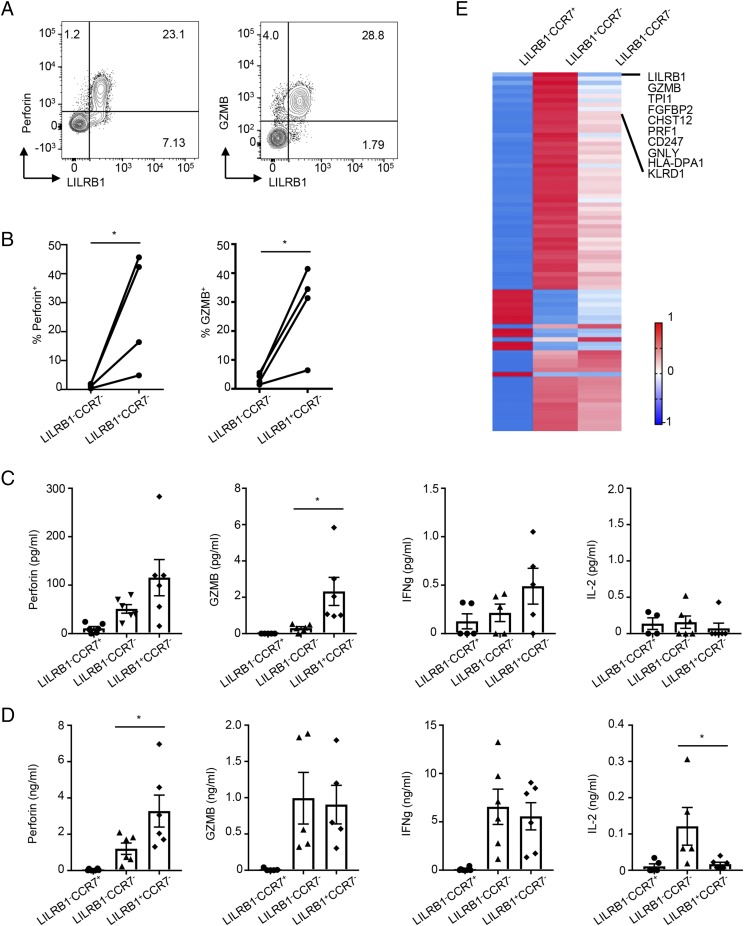
The above data suggested that LILRB1 expression may be associated with acquisition of CD8+ T cell effector function. Indeed, ex vivo FACS analysis of peripheral blood T cells from multiple donors confirmed that the majority of CD8+LILRB1+ T cells constitutively express perforin and granzyme B (Fig. 2A), whereas very few perforin- or granzyme B–expressing cells are found in LILRB1− TEMRA and CD8+ TEM (Fig. 2B). Expression of effector molecules by LILRB1+CCR7−, LILRB1−CCR7−, and LILRB1−CCR7+ CD8+ T cell subsets was also evaluated in supernatants from cultured-sorted T cells either without (Fig. 2C) or with (Fig. 2D) activation. In the absence of stimulation, LILRB1+CCR7− T cells were found to secrete perforin and granzyme B to a greater extent than the other two subsets, consistent with the above FACS data. In contrast, upon TCR activation, both LILRB1+CCR7− and LILRB1−CCR7− T cells were induced to produce an equal amount of granzyme B and IFN-γ; however, the LILRB1+ subset still produced higher quantities of perforin compared with the LILRB1− population (Fig. 2D). Interestingly, LILRB1+CCR7− cells produced minimal amounts of IL-2 relative to the LILRB1−CCR7− subset. Finally, single-cell RNAseq was used to further characterize total transcripts in sorted CD8+ LILRB1+ and LILRB1− T cell subsets. We found that LILRB1+ cells show preferential expression of multiple genes known to be associated with TEFF, including GNLY, PRF1, GZMB, and KLRD1 (Fig. 2E), further supporting the finding that LILRB1 is a marker of an effector CD8+ T cell population.
FIGURE 2.
LILRB1 marks CD8+ cells with higher effector molecule expression. (A) Representative dot plots show LILRB1 coexpression with perforin (left panel) and granzyme B (GZMB, right panel) in gated CD8+ T cell by ex vivo FACS analysis. (B) Quantitation of perforin (left panel) and GZMB (right panel) expression in LILRB1−CCR7− and LILRB1+CCR7− CD8+ subsets from multiple donors (n = 4). (C and D) Human CD8+ T cells from each donor were sorted into three subsets based on LILRB1 and CCR7 expression. Quantitation of effector molecules in supernatants were performed using a Luminex assay from resting (C) and TCR-activated (D) CD8+ T cell subsets after 48 h (n = 6). (E) Single-cell RNAseq results show differential gene expression between LILRB1+ and LILRB1− memory CD8+ T cells. *p < 0.05, paired Student t test.
LILRB1 inhibits BiTE molecule–mediated CD8+ T cell effector function
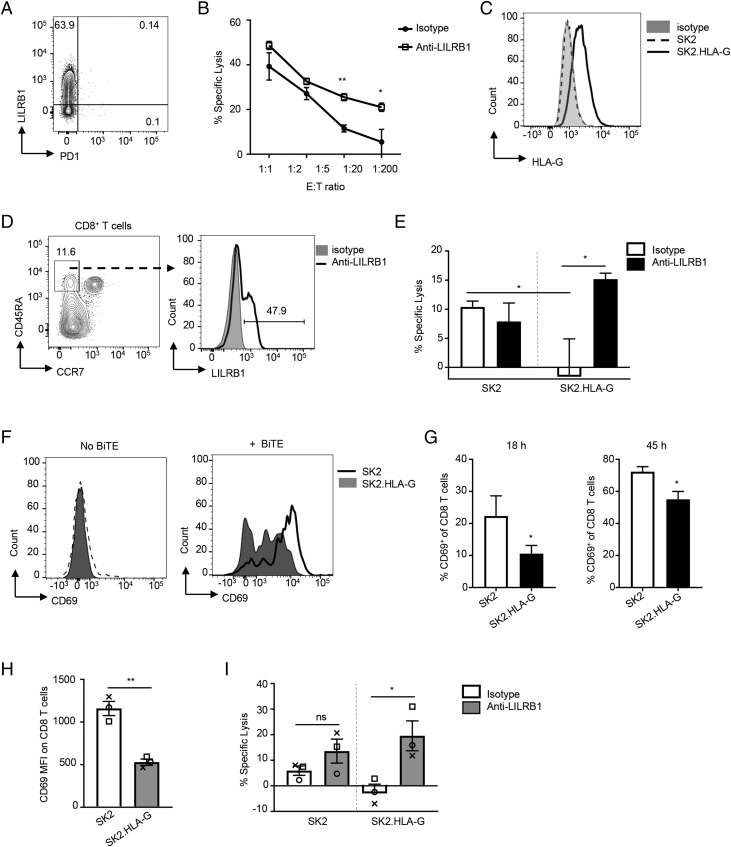
LILRB1 has been previously shown to negatively regulate the function of NK and CD8+ T cells (29, 30). Mel-13 is a cytolytic CD8+ T cell line that is specific for HLA A*0201 (HLA-A2)–restricted Melan-A (also known as MART-1)–derived epitope and may lyse melanoma tumor cells (18). These cells upregulated LILRB1 expression upon expansion with appropriate feeder cells (Fig. 3A). Blocking LILRB1 significantly enhanced Mel-13 CD8+ T cell cytolytic activity of SK-MEL-5 (HLA-A2+) melanoma cells (Fig. 3B). These data suggest that LILRB1 may function to inhibit CTL effector function and their ability to kill tumor cells. To determine if LILRB1 can regulate the function of CD8+ TEFF following stimulation with BiTE molecules, we performed a CD8+ T cell–mediated tumor cell–killing assay using SK2 target cells engineered to express HLA-G, the ligand for LILRB1, and a MART-1–specific BiTE molecule (Fig. 3C). TEMRA were enriched from healthy donor PBMCs by flow sorting (Fig. 3D) and were cocultured with parental SK2 cells or SK2.HLA-G cells in the presence of anti-LILRB1– blocking or control Abs. TEFF function was induced by a suboptimal concentration of the MART-1 BiTE molecule and measured as specific target cell lysis. These data demonstrate that HLA-G expression on target cells can dramatically reduce BiTE molecule–mediated T cell cytolytic activity and that this effect is reversed by LILRB1 blockade (Fig. 3E). The presence of HLA-G on tumor cells also significantly inhibited BiTE molecule–mediated upregulation of CD69 on effector CD8+ T cells (Fig. 3F, 3G). These data indicate that LILRB1 functions as a negative regulator of CD8+ T cell effector function and that blocking LILRB1 can be used to enhance BiTE molecule–mediated tumor cell killing.
FIGURE 3.
LILRB1 inhibits cytotoxic CD8+ T cell effector function in vitro. (A) Representative dot plot shows LILRB1 expression on expanded Mel-13 cytolytic CD8+ T cells. (B) In vitro Ag-specific CTL cytotoxicity assay with expanded Mel-13 cytolytic CD8+ T cells as effector cells and SK5 cells as target cells. Indicated ratio of effector cells and target cells were coincubated for 20 h in the presence of anti-LILRB1 blocking Ab (open squares) or isotype control Ab (filled circles). Results shown as mean ± SD of triplicate wells for each indicated ratio, and data are a representative of three independent experiments. (C) Representative histogram for HLA-G expression on SK2 cells (dotted line) and HLA-G–transfected SK2 cells (SK2.HLA-G, solid line) analyzed by FACS. Gray-filled histogram represents isotype control. (D) Representative FACS plots to show gating strategy for CD8+ TEMRA sorting from healthy donor PBMCs (left panel) and LILRB1 expression on sorted TEMRA (right panel). (E) BiTE molecule–mediated cytotoxicity of isolated CD8+ TEMRA to indicated target cells in the presence of anti-LILRB1 blocking Ab (filled bars) or isotype control Ab (open bars). T cell and tumor cell coculture with no BiTE Ab construct was used as baseline for specific lysis calculation. Results shown as mean ± SEM of T cells isolated from three donors (n = 3). (F) Representative histograms for CD69 expression on TEMRA cocultured with SK2 cells (line histograms) or SK2.HLA-G cells (gray-filled histograms) for 45 h in the presence of 0.4 nM BiTE molecule (right panel) or without BiTE molecule (left panel). (G) Isolated CD8+ TEMRA were cocultured with indicated tumor cells for 18 (left panel) or 45 h (right panel) in the presence of 0.4 nM BiTE molecule. Percentage of CD69+ cells (mean ± SD) were determined by FACS analysis. Data are representative of three independent experiments with two individual donors as T cell source. (H) Isolated tumor-associated CD8+ T cells were added to SK2 or SK2.HLA-G cells and treated with a MART-1–specific BiTE molecule for 45 h. Expression of CD69 measured as mean florescence intensity (MFI) on CD8+ T cells was determined by FACS analysis. Each type of symbol represents data obtained from an individual donor with mean ± SEM shown. (I) BiTE molecule–mediated cytotoxicity of isolated tumor CD8+ T cells to indicated target cells in the presence of anti-LILRB1 blocking Ab (gray-filled bars) or isotype control Ab (open bars). Each type of symbol represents data obtained from an individual donor with mean ± SEM shown. *p < 0.05, **p < 0.01, paired Student t test. ns, not significant.
We next wanted to confirm that LILRB1 was capable of inhibiting tumor-associated CD8+ T cells similar to its effect on T cells derived from peripheral blood of health donors. We isolated tumor-infiltrating CD8+ T cells from NSCLC biopsy specimens and evaluated their activity against SK2 and SK2.HLA-G target cells upon treatment with a MART-1–specific BiTE Ab construct. We found that the presence of HLA-G on the target cell significantly inhibits BiTE molecule–induced T cell activation (Fig. 3H) and cytolytic activity (Fig. 3I). Inhibition of cytolytic activity can be restored with LILRB1 blockade (Fig. 3I). These results demonstrate that LILRB1 can function as an immune checkpoint inhibitory receptor for tumor-infiltrating CD8+ TEFF and suggests that blocking LILRB1 may enhance BiTE molecule–mediated tumor CD8+ T cell cytolytic activity in patients.
Differential regulation of LILRB1 and PD1 expression in CD8+ T cells
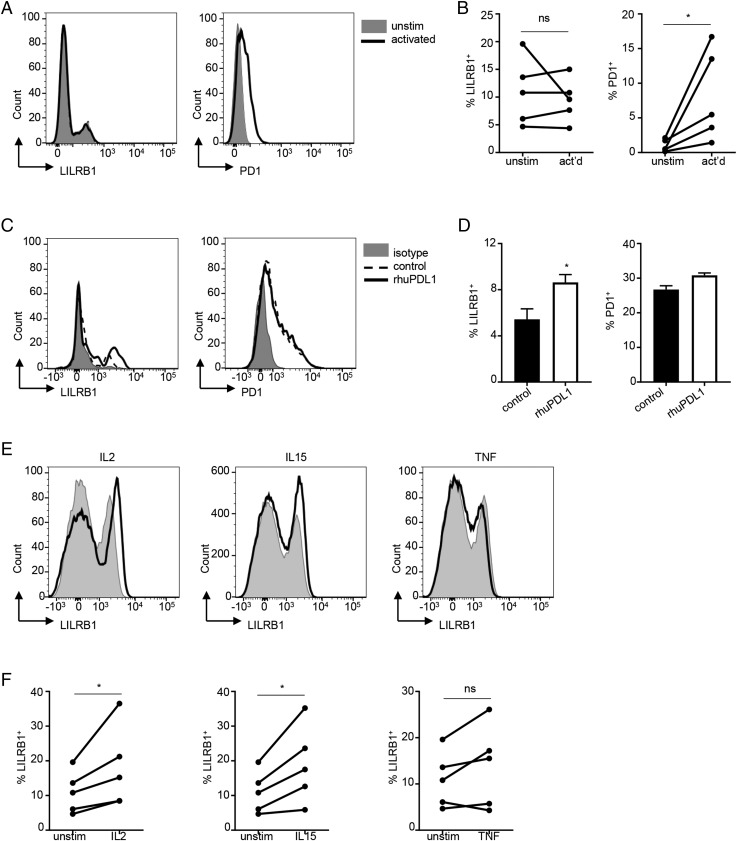
PD1 is known to exert potent repressive activity on tumor-infiltrating CD8+ T cells, and therapeutic Abs blocking the PD1 pathway can significantly enhance T cell effector function, providing therapeutic benefit in multiple cancers (31). Given the finding that LILRB1 can also function as negative regulator of CD8+ T cell function, we examined the regulation of LILRB1 and PD1 expression on total CD8+ T cells. We first examined LILRB1 and PD1 expression downstream of TCR signaling, confirming that PD1 is rapidly upregulated upon T cell activation. In contrast, LILRB1 expression was not sensitive to TCR signaling (Fig. 4A, 4B). As blocking the PD1 pathway has been shown to further promote effector function of CD8+ T cells (31, 32), we next examined whether LILRB1 expression can be regulated by PD1 signaling. Activation of CD8+ TEM in the presence of PD1 blockade resulted in an increase in the frequency of LILRB1+ effector cells, whereas the expression of PD1 was not significantly altered (Fig. 4C, 4D). Furthermore, cytokines known to promote CD8+ T cell effector differentiation and function, such as IL-2 and IL-15 but not TNF-α, also significantly increased the frequency of LILRB1+ T cells (Fig. 4E, 4F). Thus, signals that enhance CD8+ T cell effector function but not TCR activation alone can promote differentiation of LILRB1+ TEFF.
FIGURE 4.
LILRB1 expression on CD8+ T cell surface is upregulated by effector cytokines and anti-PD1 blockade. (A) FACS analysis of LILRB1 (left panel) and PD1 (right panel) expression on the surface of CD8+ T cells upon TCR activation (solid lines). Gray-filled histograms are control CD8+ T cells without stimulation. (B) Quantitation of the results from (A) across multiple donors (n = 5). (C) Purified human CD8+ TEM cells were activated with anti-CD3 plus anti-CD28 in the presence of 10 μg/ml recombinant human PDL1 (rhuPDL1, solid lines) or human IgG1 (control, dashed lines). After 48 h, LILRB1 (left panel) and PD1 (right panel) expression was determined by FACS analysis. Gray-filled histograms are isotype staining controls. (D) Quantitation results of (C) across multiple donors (n = 3). Representative data shown as mean ± SD. (E) Human PBMCs were stimulated with rIL-2, IL-15, or TNF for 48 h (solid lines) and subjected to FACS analysis. Gray-filled histograms represent CD8+ T cells gated from unstimulated PBMCs. (F) Quantitation of (E) across multiple donors (n = 5). *p < 0.05, Student t test. ns, not significant.
PD1 is preferentially upregulated on LILRB1− CD8+ T cells
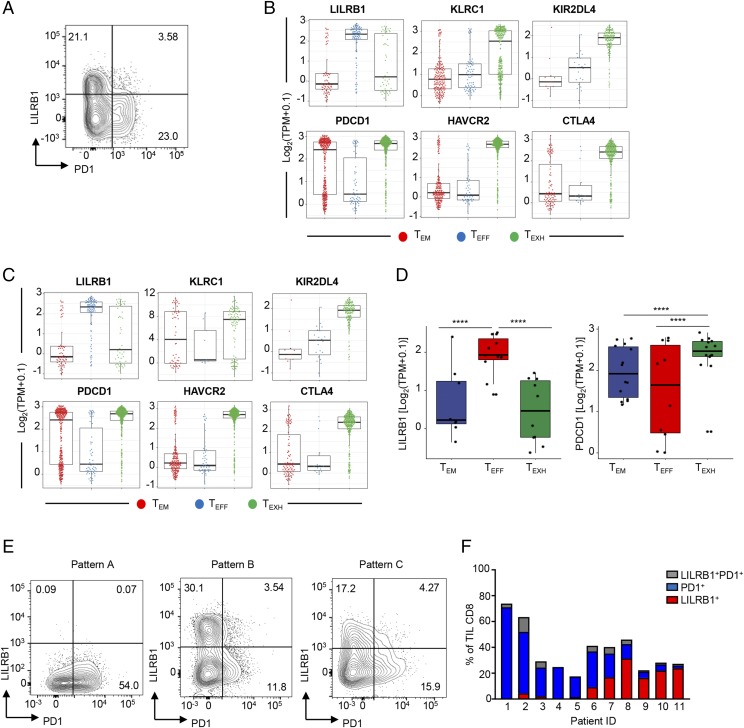
Given our findings that PD1 but not LILRB1 is upregulated on CD8+ T cells upon TCR activation, we next asked whether there is a difference in the expression of PD1 on either LILRB1+ or LILRB1− CD8+ T cell subsets. Upon TCR stimulation, PD1 expression was found to be preferentially upregulated on LILRB1− CD8+ T cells isolated from peripheral blood (Fig. 5A). These data suggest that LILRB1+ T cells represent a unique population that may not be subject to regulation by the PD1 pathway. To extend these data to tumor-associated T cells, we analyzed publicly available single–T cell RNAseq data from hepatocellular carcinoma (11) and NSCLC (12). In both datasets, LILRB1 and PDCD1 showed mutually exclusive expression patterns in tumor CD8+ T cells (Fig. 5B–D). In contrast, expression patterns of other inhibitory receptor genes, such as HAVCR2 and CTLA-4, were largely overlapping with PDCD1 (Fig. 5B, 5C). Notably, expression levels of another HLA-G receptor KIR2DL4 (33) (Fig.5B, 5C), as well as many other NK cell receptors (NKR) (e.g., KLRC1 and KIRs, data not shown), are low and/or less frequent in effector CD8+ T cell clusters in tumor.
FIGURE 5.
LILRB1 and PD1 are expressed by distinct CD8+ T cell subsets in tumor. (A) Representative FACS plot to show LILRB1 and PD1 expression on TCR-activated human CD8+ T cells. Percentage of cells in each quadrant is indicated. (B and C) Single-cell RNAseq swarm plots showing the expression of indicated genes in tumor-infiltrated CD8+ T cell clusters isolated from NSCLC (B) or hepatocellular carcinoma (C). (D) Boxplots of LILRB1 and PD1 gene expression in tumor-infiltrated CD8+ T cell clusters from NSCLC patients. Each dot represents average gene expression from a given patient. ****p < 0.0001, paired Student t test. (E) Representative FACS plots to show LILRB1 and PD1 expression on tumor-infiltrating CD8+ T cells. Numbers represent percentage of cells in a given quadrant. (F) Quantitation of (E) from 11 NSCLC tumor patients. Percentage of total tumor CD8+ T cells expressing LILRB1 (in red), PD1 (in blue), or both (in gray) in each tumor.
We also evaluated PD1 and LILRB1 expression by flow cytometry on CD8+ T cells isolated from additional NSCLC biopsy specimens. PD1 and LILRB1 showed a heterogenous pattern of expression across tumor samples, with some samples having CD8+ T cells predominantly expressing either PD1 or LILRB1 and others having similar frequencies of LILRB1+ or PD1+ CD8+ T cells (Fig. 5E, 5F). Nevertheless, in all the cases we examined, PD1 and LILRB1 showed a mutually exclusive expression pattern consistent with data from healthy donor T cells. Importantly, LILRB1+ CD8+ T cells represented a dominant phenotype in 4 out of 11 NSCLC tumors (Fig. 5F), suggesting that across patients, different inhibitory mechanisms may function to preferentially restrict antitumor T cell responses.
LILRB1 and PD1 have independent roles in restricting CD8+ TEFF function
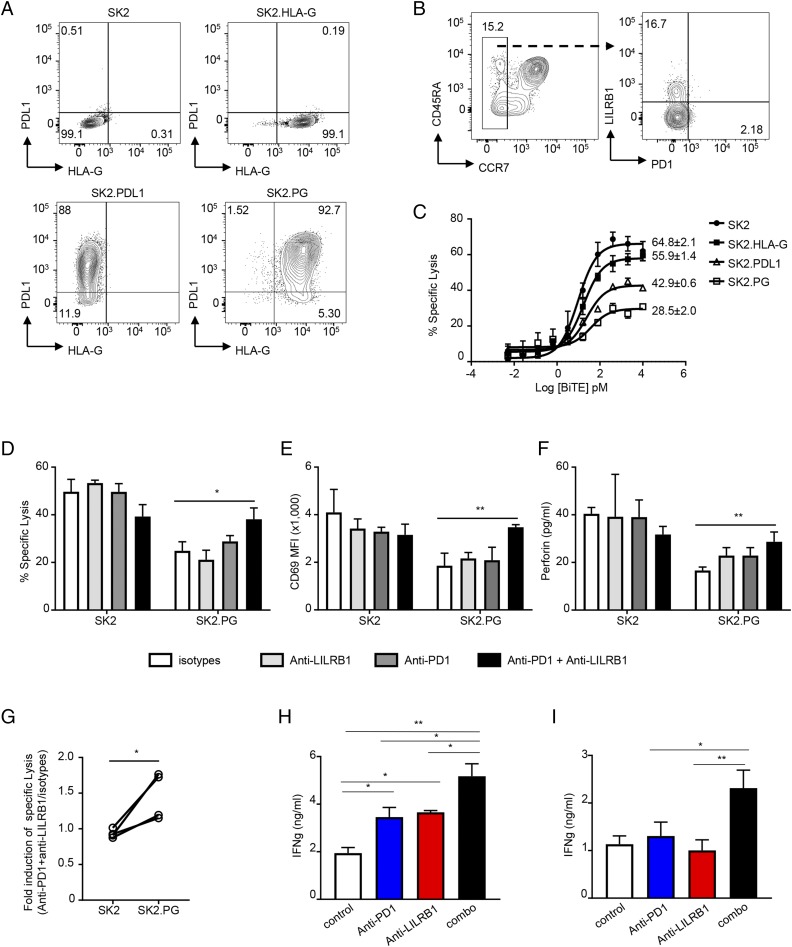
Given the finding that LILRB1 and PD1 are expressed on different populations of CD8+ T cells and that PD1 blockade may promote differentiation of LILRB1+ T cells, we hypothesized that simultaneous blockade of LILRB1 and PD1 could lead to greater T cell activation than blockade of either pathway alone. To test this hypothesis, we engineered SK2 tumor cell lines stably expressing PDL1 (SK2.PDL1), HLA-G (SK2.HLA-G), or both molecules (SK2.PG) (Fig. 6A). CCR7− CD8+ TEFF containing both LILRB1+ TEMRA and PD1+ TEM were enriched from healthy donor PBMCs by flow sorting (Fig. 6B). As expected, target cells expressing both HLA-G and PDL1 are more resistant to BiTE molecule–induced CCR7− CD8+ T cell cytolytic activity than parental SK2 cells or SK2 cells expressing either PDL1 or HLA-G alone (Fig. 6C). Consistent with these data, only the combination of anti-PD1 and anti-LILRB1 was able to fully restore CTL effector cell–killing activity against SK2.PG tumor cells expressing both HLA-G and PDL1 (Fig. 6D–G).
FIGURE 6.
Anti-LILRB1 synergize with anti-PD1 to promote CD8+ T cell effector function. (A) Dot plots to show HLA-G and PDL1 expression on established tumor cells lines. (B) Representative FACS plots to show gating strategy for CCR7− CD8+ T cell sorting from healthy donor PBMCs (left panel) and LILRB1 and PD1 expression on sorted CCR7− CD8+ T cells (right panel). (C) Sorted CCR7− CD8+ T cells were incubated with indicated tumor cells in the presence of indicated amount of MART-1–specific BiTE molecule. Specific cytotoxicity was determined after 45 h. Results shown as mean ± SD from triplicated wells and are a representative from three independent experiments using two healthy donors as the source of CD8+ T cells. Numbers are maximum percentage of specific lysis. (D–F) BiTE molecule–mediated cytotoxicity of isolated CCR7− CD8+ T cells to indicated target cells in the presence of anti-LILRB1 blocking Ab (light gray bars), anti-PD1 blocking Ab (dark gray bars), combination of anti-LILRB1 and anti-PD1 (black bars), or isotype control Ab (open bars). T cell and tumor cells coculture with no BiTE Ab construct was used as baseline for specific lysis calculation. Percentage of specific lysis (D), T cell activation represented by CD69 upregulation measured by FACS (E), and perforin production by Luminex (F) was determined and plotted. Results shown as mean ± SD of triplicated wells. Data are a representative of three independent experiments using T cells isolated from two donors. (G) Quantitation of anti-LILRB1 plus anti-PD1 in promoting CCR7− CD8+ T cell cytolytic activity. Each dot represents data obtained from an individual donor. *p < 0.05, paired Student t test. (H and I) Isolated total T cells were cocultured with allogeneic CD11c+ cells in the presence of indicated Abs. After 5 d, culture supernatants were collected, and IFN-γ levels were determined by ELISA. Data are plotted as mean ± SD. Results is representative of three independent experiments with T cells from two individual donors and CD11c+ cells from three individual donors. *p < 0.05, **p < 0.01, Student t test.
In addition, evaluating the role of LILRB1 and PD1 in BiTE molecule–mediated T cell activation assays, we also performed MLR assays, comparing the effects of anti-LILRB1 or anti-PD1 alone and in combination on T cell activation. We found that the combination of anti-PD1 and anti-LILRB1 blockade leads to significantly increased IFN-γ production compared with either treatment alone (Fig. 6H, 6I). Taken together, these data indicate that LILRB1 functions as an inhibitory receptor for a unique population of PD1− TEFF and suggest that the combination of LILRB1 and PD1 blockade may be required to mobilize the full complement of tumor-associated CD8+ T cells to participate in the antitumor response.
Discussion
BiTE molecules have been shown to activate CD8+ T cells and enhance CTL function in vitro and in vivo (34). The BiTE Ab construct showed translating to inhibition of solid tumor growth in preclinical mouse models (35). However, the ability of BiTE molecules to effectively treat solid tumors in patients is still under evaluation. Major challenges associated with applying BiTE molecules to the treatment of solid tumors are the suppressive tumor microenvironment and the expression of multiple checkpoint inhibitory receptors by tumor-associated T cells that function to reduce BiTE molecule activity and thus limit their therapeutic efficacy. For example, the PD1 pathway and the presence of TGF-β or IL-10 can reduce BiTE molecule–mediated CD8+ cytolytic T cell–killing activity in vitro (35, 36). Identifying additional regulatory pathways capable of restricting CD8+ T cell function is critical for our ability to unlock the full potential of BiTE molecule therapies and other T cell–targeted approaches in solid tumors.
In this study we identify LILRB1 as a novel checkpoint inhibitory molecule capable of restricting BiTE molecule–mediated CD8+ T cell effector function. Given the following factors: 1) LILRB1 is highly expressed by the CD8+ TEMRA subset, which is the most potent population for BiTE molecule–induced toxicity, 2) LILRB1–expressing CD8+ T cells infiltrate solid tumors, 3) HLA-G overexpression has been reported in many solid tumors, and its expression is positively associated with poor prognosis (37), and 4) LILRB1 blockade increases CD8+ T cell cytolytic activity in vitro, these findings suggest that blocking LILRB1 may enhance BiTE molecule–mediated efficacy against solid tumors. Blocking LILRB1 may be especially important in the context of HLA-G+ solid tumors, in which LILRB1-mediated inhibitory signals may be a dominant mechanism restricting cytolytic CD8+ T cell effector function. Similar to previous observations (38), we find that LILRB1 is coexpressed with perforin and granzyme B in human CD8+ T cells. We extend these findings by performing RNAseq analysis on CD8+ T cells that were sorted based on LILRB1 expression, demonstrating that LILRB1+ CD8+ T cells preferentially express multiple effector molecules beyond perforin and granzymes. These data suggest functional specialization of the LILRB1+ T cell subset for cytolytic activity, a hypothesis consistent with findings from our BiTE molecule–mediated cytotoxicity studies. In addition to marking effector CD8+ T cells with potent cytolytic activity, we also demonstrate that LILRB1 functions as an immune checkpoint receptor, effectively inhibiting both Ag-specific CTL cytolytic activity and BiTE molecule–induced polyclonal activation of effector CD8+ T cells.
Importantly, we find that LILRB1 and PD1 are preferentially expressed by distinct CD8+ T cell subsets in both tumor tissue and in healthy donor blood. The nonoverlapping expression of LILRB1 and PD1 in tumor-infiltrating CD8+ T cells is also observed by single-cell RNAseq analysis in multiple tumor types, including hepatocellular carcinoma (11), NSCLC (12), and colorectal cancer (39). Although many well-studied inhibitory receptors, such as TIM3 and CTLA-4, are coexpressed with PD1 on CD8+ TEXH in tumor, LILRB1+ CD8+ T cells are largely devoid of these inhibitory receptors. These data suggest that LILRB1+ cells are not subject to the same regulatory pathways as TEXH. Previous studies have demonstrated that NKR, such as KLRG-1, KLRC-1, and KIRs, are upregulated during CD8+ T cell differentiation toward an effector phenotype. Further analysis of published single-cell RNAseq data demonstrated that these NKRs either have low expression levels on very few CD8+ T cells, such as many KIRs, or their expression is largely not overlapping with LILRB1, such as KLRC-1. These findings suggest that LILRB1 is unique in terms of its expression and function on CD8+ T cells.
The regulation of LILRB1 gene expression is largely unknown (40). We find that cytokines that promote T cell effector function (such as IL-2 and IL-15) but not proinflammatory cytokines, such as TNF-α, increase LILRB1 expression. Interestingly, blocking PD1 also enhances LILRB1 expression, possibly because of increased IL-2 production by T cells upon PD1 blockade. Interestingly, expression of KLRG-1, another inhibitory receptor that is highly expressed by subsets of effector CD8+ T cells, is also upregulated by IL-2 (41). Thus, LILRB1 upregulation may serve as a negative feedback regulator for effector CD8+ T cells in response to signals that boost CD8+ T cell effector function. An implication of these data is that blocking LILRB1 will be additive/synergistic with PD1 blockade both by virtue of the ability of this therapeutic combination to activate two different CD8+ T cell subsets and the potential for anti-PD1/PDL1 therapy to increase differentiation and accumulation of LILRB1+ effector T cells, whose activity would be potentiated by anti-LILRB1.
Our study identifies LILRB1 as an inhibitory pathway that is highly differentiated from current immune checkpoint inhibitor combinations and has clear relevance as a therapeutic target capable of enhancing the efficacy BiTE molecules or other T cell–targeted approaches that are currently undergoing clinical evaluation. Moreover, our findings also generate, to our knowledge, new hypotheses beyond BiTE Ab construct therapy, suggesting that further characterization of T cell heterogeneity in tumors with respect to function and expression of inhibitory pathways may identify orthogonal mechanisms regulating distinct aspects of the antitumor T cell response, thus representing attractive targets for combination therapy approaches.
Supplementary Material
Acknowledgments
We sincerely thank the Amgen flow core laboratory for supporting immune cell sorting. We greatly thank Julie Bailis, Shyun Li, and Natalie Mariano for providing reagents and technical help about BiTE molecule–mediated T cell–dependent cellular cytotoxicity assay and Hajime Hiraragi, Yi Liu, Daniel Wu, Sarah Sun, and Saravanan Kaliyaperumal for pathology support. We also greatly thank Jennifer Hawkins, Karen Snyder, Leslie Marshall, and Eduardo Laifung for human tumor biopsy specimen collection. We thank Dev Bhatt and Chi-Ming Li for preparing for single-cell RNAseq libraries.
The RNA sequencing data presented in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive database under accession number PRJNA535519.
The online version of this article contains supplemental material.
- BiTE molecule
- bispecific T cell engager
- LILRB1
- leukocyte Ig-like receptor B1
- Mart-1
- melanoma Ag recognized by T cells 1
- NKR
- NK cell receptor
- NSCLC
- non–small cell lung cancer
- PD1
- programmed cell death protein 1
- PDL1
- programmed death ligand 1
- RLU
- relative light unit
- RNAseq
- RNA sequencing
- SK2
- SK-MEL-2
- SK2.HLA-G
- HLA-G–expressing SK2
- TCM
- central memory T cell
- TEFF
- effector T cell
- TEM
- effector memory T cell
- TEMRA
- effector memory T cells reexpressing CD45RA, TEXH, exhausted T cell
- TN
- naive T cell.
Disclosures
A.K., C.-J.H., I.D., A.O., W.O., J.G.E., and X.Y. are full-time employees of Amgen, Inc., a for-profit company.
References
- 1.Pardoll D. M. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumeh P. C., Harview C. L., Yearley J. H., Shintaku I. P., Taylor E. J., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., et al. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515: 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizvi N. A., Hellmann M. D., Snyder A., Kvistborg P., Makarov V., Havel J. J., Lee W., Yuan J., Wong P., Ho T. S., et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian S. L., Taube J. M., Anders R. A., Pardoll D. M. 2016. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maus M. V., Grupp S. A., Porter D. L., June C. H. 2014. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123: 2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chmielewski M., Hombach A. A., Abken H. 2013. Antigen-specific T-cell activation independently of the MHC: chimeric antigen receptor-redirected T cells. Front. Immunol. 4: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huehls A. M., Coupet T. A., Sentman C. L. 2015. Bispecific T-cell engagers for cancer immunotherapy. Immunol. Cell Biol. 93: 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullard A. 2015. FDA approves first bispecific. Nat. Rev. Drug Discov. 14: 7. [DOI] [PubMed] [Google Scholar]
- 9.Krupka C., Kufer P., Kischel R., Zugmaier G., Lichtenegger F. S., Köhnke T., Vick B., Jeremias I., Metzeler K. H., Altmann T., et al. 2016. Blockade of the PD-1/PD-L1 axis augments lysis of AML cells by the CD33/CD3 BiTE antibody construct AMG 330: reversing a T-cell-induced immune escape mechanism. Leukemia 30: 484–491. [DOI] [PubMed] [Google Scholar]
- 10.Osada T., Patel S. P., Hammond S. A., Osada K., Morse M. A., Lyerly H. K. 2015. CEA/CD3-bispecific T cell-engaging (BiTE) antibody-mediated T lymphocyte cytotoxicity maximized by inhibition of both PD1 and PD-L1. Cancer Immunol. Immunother. 64: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng C., Zheng L., Yoo J. K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J. Y., Zhang Q., et al. 2017. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169: 1342–1356.e16. [DOI] [PubMed] [Google Scholar]
- 12.Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. [Published erratum appears in 2018 Nat. Med. 24: 1628.] Nat. Med. 24: 978–985. [DOI] [PubMed] [Google Scholar]
- 13.Dreier T., Lorenczewski G., Brandl C., Hoffmann P., Syring U., Hanakam F., Kufer P., Riethmuller G., Bargou R., Baeuerle P. A. 2002. Extremely potent, rapid and costimulation-independent cytotoxic T-cell response against lymphoma cells catalyzed by a single-chain bispecific antibody. Int. J. Cancer 100: 690–697. [DOI] [PubMed] [Google Scholar]
- 14.Ribas A., Shin D. S., Zaretsky J., Frederiksen J., Cornish A., Avramis E., Seja E., Kivork C., Siebert J., Kaplan-Lefko P., et al. 2016. PD-1 blockade expands intratumoral memory T cells. Cancer Immunol. Res. 4: 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casado J. G., Soto R., DelaRosa O., Peralbo E., del Carmen Muñoz-Villanueva M., Rioja L., Peña J., Solana R., Tarazona R. 2005. CD8 T cells expressing NK associated receptors are increased in melanoma patients and display an effector phenotype. Cancer Immunol. Immunother. 54: 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang X., Kim J., Deng M., John S., Chen H., Wu G., Phan H., Zhang C. C. 2016. Inhibitory leukocyte immunoglobulin-like receptors: immune checkpoint proteins and tumor sustaining factors. Cell Cycle 15: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pita-Lopez M. L., Gayoso I., DelaRosa O., Casado J. G., Alonso C., Muñoz-Gomariz E., Tarazona R., Solana R. 2009. Effect of ageing on CMV-specific CD8 T cells from CMV seropositive healthy donors. Immun. Ageing 6: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legut M., Dolton G., Mian A. A., Ottmann O. G., Sewell A. K. 2018. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 131: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sultan M., Schulz M. H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960. [DOI] [PubMed] [Google Scholar]
- 20.Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M. I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng G. X., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A., Trapnell C. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14: 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: Available at: https://www.R-project.org/. Accessed: December 20, 2018. [Google Scholar]
- 25.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- 26.Clarke E., Sherrill-Mix S. 2017. ggbeeswarm: categorical scatter (violin point) plots. R package version 0.6.0. Available at: https://CRAN.R-project.org/package=ggbeeswarm. Accessed: August 7, 2017.
- 27.Kassambara A. 2018. ggpubr: 'ggplot2' based publication ready plots. R package version 0.1.8. Available at: https://CRAN.R-project.org/package=ggpubr. Accessed: September 5, 2018.
- 28.Burshtyn D. N., Morcos C. 2016. The expanding spectrum of ligands for leukocyte ig-like receptors. J. Immunol. 196: 947–955. [DOI] [PubMed] [Google Scholar]
- 29.Favier B., Lemaoult J., Lesport E., Carosella E. D. 2010. ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. FASEB J. 24: 689–699. [DOI] [PubMed] [Google Scholar]
- 30.Ince M. N., Harnisch B., Xu Z., Lee S. K., Lange C., Moretta L., Lederman M., Lieberman J. 2004. Increased expression of the natural killer cell inhibitory receptor CD85j/ILT2 on antigen-specific effector CD8 T cells and its impact on CD8 T-cell function. Immunology 112: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas A., Wolchok J. D. 2018. Cancer immunotherapy using checkpoint blockade. Science 359: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barber D. L., Wherry E. J., Masopust D., Zhu B., Allison J. P., Sharpe A. H., Freeman G. J., Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan S., Long E. O. 1999. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. [Published erratum appears in 2000 J. Exp. Med. 191: following 2027.] J. Exp. Med. 189: 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dao T., Pankov D., Scott A., Korontsvit T., Zakhaleva V., Xu Y., Xiang J., Yan S., de Morais Guerreiro M. D., Veomett N., et al. 2015. Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat. Biotechnol. 33: 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amann M., Brischwein K., Lutterbuese P., Parr L., Petersen L., Lorenczewski G., Krinner E., Bruckmeier S., Lippold S., Kischel R., et al. 2008. Therapeutic window of MuS110, a single-chain antibody construct bispecific for murine EpCAM and murine CD3. Cancer Res. 68: 143–151. [DOI] [PubMed] [Google Scholar]
- 36.Deisting W., Raum T., Kufer P., Baeuerle P. A., Münz M. 2015. Impact of diverse immune evasion mechanisms of cancer cells on T cells engaged by EpCAM/CD3-bispecific antibody construct AMG 110. PLoS One 10: e0141669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin A., Yan W. H. 2015. Human Leukocyte Antigen-G (HLA-G) expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol. Med. 21: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anfossi N., Doisne J. M., Peyrat M. A., Ugolini S., Bonnaud O., Bossy D., Pitard V., Merville P., Moreau J. F., Delfraissy J. F., et al. 2004. Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J. Immunol. 173: 7223–7229. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J. Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564: 268–272. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima H., Asai A., Okada A., Ping L., Hamajima F., Sata T., Isobe K. 2003. Transcriptional regulation of ILT family receptors. J. Immunol. 171: 6611–6620. [DOI] [PubMed] [Google Scholar]
- 41.Mathieu C., Beltra J. C., Charpentier T., Bourbonnais S., Di Santo J. P., Lamarre A., Decaluwe H. 2015. IL-2 and IL-15 regulate CD8+ memory T-cell differentiation but are dispensable for protective recall responses. Eur. J. Immunol. 45: 3324–3338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.