Abstract
Depression is a common and disabling disorder, representing a major social and economic health issue. Moreover, depression is associated with the progression of diseases with an inflammatory etiology including many inflammatory-related disorders. At the molecular level, the mechanisms by which depression might promote the onset of these diseases and associated immune-dysfunction are not well understood. In this study we assessed genome-wide patterns of DNA methylation in whole blood-derived DNA obtained from individuals with a self-reported history of depression (n = 100) and individuals without a history of depression (n = 100) using the Illumina 450K microarray. Our analysis identified six significant (Šidák corrected P < 0.05) depression-associated differentially methylated regions (DMRs); the top-ranked DMR was located in exon 1 of the LTB4R2 gene (Šidák corrected P = 1.27 × 10−14). Polygenic risk scores (PRS) for depression were generated and known biological markers of inflammation, telomere length (TL) and IL-6, were measured in DNA and serum samples, respectively. Next, we employed a systems-level approach to identify networks of co-methylated loci associated with a history of depression, in addition to depression PRS, TL and IL-6 levels. Our analysis identified one depression-associated co-methylation module (P = 0.04). Interestingly, the depression-associated module was highly enriched for pathways related to immune function and was also associated with TL and IL-6 cytokine levels. In summary, our genome-wide DNA methylation analysis of individuals with and without a self-reported history of depression identified several candidate DMRs of potential relevance to the pathogenesis of depression and its associated immune-dysfunction phenotype.
Introduction
An estimated 300 million people are affected by depression worldwide (1), representing a major social and economic health burden. Depression is associated with a reduced life expectancy and predicts the incidence and progression of diseases associated with an inflammatory etiology such as cardiovascular disease and many autoimmune disorders (2). Emerging data suggests that depression is associated with a low grade, chronic inflammatory response including an increase in inflammatory mediators such as cytokines and chemokines (3). Moreover, depression is associated with telomere shortening (4,5), a marker of biological aging, which is thought to be influenced by inflammation and cellular stress (6). The most recent genome-wide association study (GWAS) meta-analysis of depression by the Psychiatric Genomics Consortium (PGC), which identified 44 robust depression-associated loci, strongly implicated immune-related genes (e.g. LRFN5) and biological pathways associated with cytokine and immune responses in the etiology of depression (7). Although these epidemiological data suggest a link between depression and inflammation, the mechanisms by which depression might promote these diseases and associated immune-dysfunction at a molecular level are not well understood.
Epigenetic processes—which act to developmentally regulate gene expression via modifications to DNA, histone proteins and chromatin, independently of DNA sequence variation—have been implicated in the etiology of both depression (8–10) and inflammatory-related diseases (11,12). DNA methylation, the most widely studied epigenetic mark, can be influenced by environmental factors [e.g. smoking (13)] and stressful exposures [e.g. childhood adversity (14)] that are associated with both depression (2,15) and inflammatory diseases (15,16). It is therefore plausible that epigenetic dysregulation of immune-related genes may contribute to the immune-dysfunction phenotype associated with depression.
In this study, we quantified genome-wide patterns of DNA methylation in whole blood-derived DNA obtained from individuals with a self-reported history of depression (n = 100) and individuals without a history of depression (n = 100). The study was designed to ensure that 50% of cases and controls also had a self-reported history of an inflammatory disorder. We further integrated our epigenetic analysis with genetic data and known biological markers of inflammation, including telomere length (TL) and serum interleukin 6 levels (IL-6) (17,18). We identify evidence for altered DNA methylation at multiple immune-related loci in individuals with a history of depression, with depression-associated co-methylation modules also correlated with TL and IL-6.
Results
Examining the association between a history of depression, immune cells and IL-6
Given the postulated role of immune-dysfunction in depression, we first examined differences in predicted immune cell composition between our self-reported history of depression cases and controls. Individuals with a history of depression show significantly increased CD4+ T cells (P = 0.036) and reduced levels of plasma blast cells (P = 0.024), granulocytes (P = 0.036) and memory and effector T cells (CD8pCD28nCD45RAn) (P = 0.02). Interestingly, when we stratified the depression individuals by a history of inflammation, individuals with a history of depression and a history of an inflammatory disorder had significantly higher CD4+ T cells and reduced levels of plasma blast cells, memory and effector T cells and granulocytes compared to the other three groups (see Supplementary Material, Fig. S1). In contrast, the depression only group did not differ significantly from the other groups, suggesting that individuals with both a history of depression and a history of an inflammatory disorder have distinct immune profiles (e.g. Th1/Th2 cell activation) compared to individuals with a history of either depression or an inflammatory condition alone. Previously, it has been shown that individuals with depression have increased levels of the pro-inflammatory cytokine, IL-6 (19). We did not observe increased serum IL-6 levels in our study between groups (P = 0.848), or between individuals with and without a history of depression (P = 0.583).
Individuals with a history of depression show evidence of shortened telomeres but not DNA methylation age acceleration
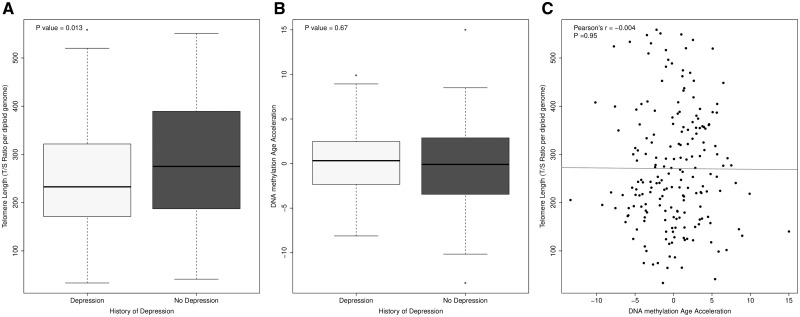
TL has been widely implicated as a marker of biological age (6), and is influenced by genetic and epigenetic regulation, as well as by inflammation and cellular stress (20). Furthermore, shorter telomeres are robustly associated with depression (21). We examined differences in TL between individuals with and without a self-reported history of depression using logistic regression analysis, while controlling for a history of an inflammatory condition and potential confounders (age, sex and estimated blood cell composition). A self-reported history of depression was significantly associated with shortened TL after controlling for confounders (P = 0.013, see Fig. 1A). Another hypothesized proxy of biological age is the epigenetic clock (22), with elevated DNA methylation age (DNAm Age) also linked to depression (23). Although we found a large and highly significant positive correlation between chronological age and DNAm Age calculated using an epigenetic clock based on DNA methylation values (22) (Pearson’s r = 0.89, P < 2.2e-16; Supplementary Material, Fig. S2), we found no evidence for accelerated ‘epigenetic aging’ among individuals with depression (P = 0.67; Fig. 1B). As reported previously (24–26), we found no correlation between DNAm Age acceleration and TL (Pearson’s r = −0.004, P = 0.95; Fig. 1C). Taken together, these results indicate that a history of depression is associated with shortened TL but is not associated with DNAm Age acceleration in this study.
Figure 1.
A self-reported history of depression is associated with shortened TL but not DNAm Age acceleration. (A) A self-reported history of depression is associated with shortened TL (P = 0.013), TL was measured by determining the ratio of telomere repeat copy number to single-copy gene copy number (T/S ratio) per diploid genome in study samples. (B) A self-reported history of depression was not associated with DNAm Age acceleration. (C) We observed no correlation between TL and DNAm Age acceleration in our study.
Methylomic differences between individuals with a history of depression and controls—differentially methylated positions and regions
An overview of the methodological approach used in this study is given in Supplementary Material, Figure S3. First, we assessed genome-wide patterns of DNA methylation in individuals with a self-reported history of depression (cases) compared with controls using linear regression, controlling for potential confounders (see Materials and Methods). Although no differentially methylated positions (DMP) were identified at a stringent experiment-wide significance threshold (P < 1.66 × 10−7) estimated from permutation analysis in a large dataset generated previously by our group (27), several DMPs showed evidence for association at a more relaxed ‘discovery’ threshold (P < 2 × 10−5). The 10 top-ranked DMPs between cases and controls are listed in Supplementary Material, Table S1; of note these include probes annotated to a number of loci previously implicated in neuropsychiatric phenotypes. Most notably, the top-ranked DMP (cg141959258) is located in intron 4 of the RNA-binding protein fox-1 homolog 3 gene (RBFOX3) gene (also known as HRNBP3), which is implicated in the largest depression GWAS meta-analysis to date (7).
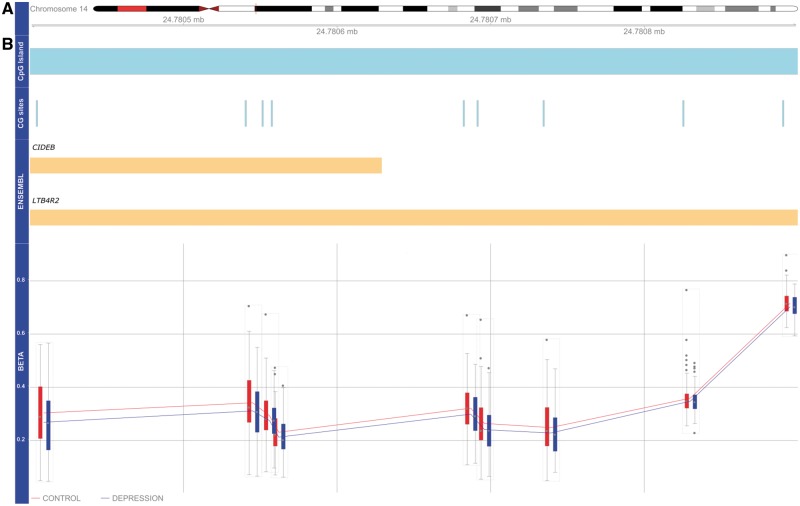
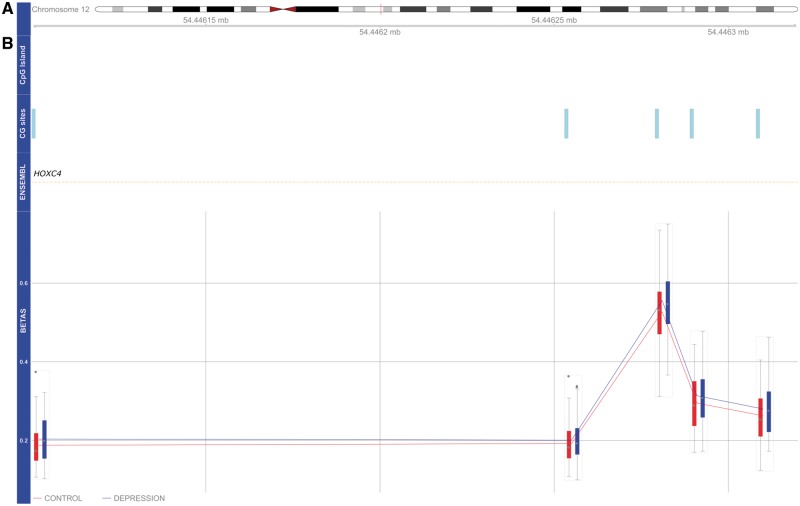
We next used Comb-p (28) to identify DMRs associated with a history of depression. Our analysis identified six significant (Šidák corrected P-value < 0.05) DMRs (Table 1); the top-ranked DMR (spanning nine CpG sites) is located in the promoter region of the LTB4R gene (Šidák corrected P-value = 1.27 × 10−14). The LTB4R2-associated DMR is hypomethylated across all CpG sites in depression cases compared to controls (Fig. 2). The second ranked DMR is located in intron 1 of the HOXC4 gene (Šidák corrected P-value = 1.21 × 10−9). The HOXC4-associated DMR is hypermethylated across all CpG sites in cases compared to controls (Fig. 3). In addition, we identified a DMR located in the third exon of the TRIM39 gene (Šidák corrected P-value = 0.00016); a DMR located upstream in intron 10 of the DNAJC17 gene (Šidák corrected P-value = 0.00023); a DMR located upstream in the promoter region of the PNPLA2 gene (Šidák corrected P-value = 0.00024) and DMR located in intron 1 of the MEF2A gene (Šidák corrected P-value = 0.00027).
Table 1.
Comb-p differentially methylated regional analysis
| Probe IDs | Hg19 | Annotated gene (UCSC) | No. of probes | Slk P-value | Slk Šidák P-value |
|---|---|---|---|---|---|
| cg12853742, cg20007021, cg21886367, cg26310551, cg25576711, cg10193721, cg06796435, cg06823034 | chr14: 24780404-24780891 | LTB4R2; CIDEB; LTB4R | 9 | 1.27E-17 | 1.12E-14 |
| cg15244786, cg22370252, cg26201952, cg27138204, cg05992786 | chr12: 54446100-54446309 | HOXC4 | 5 | 5.88E-13 | 1.21E-09 |
| cg04425551, cg07905808, cg13079571, cg11327408, cg17080697, cg09020199 | chr6: 30297257-30297390 | TRIM39; TRIM39-RPP21 | 6 | 4.87E-08 | 0.0001576 |
| cg17113968, cg22079077, cg16764778 | chr15: 41061384-41061528 | DNAJC17 | 3 | 7.84E-08 | 0.0002344 |
| cg24427660, cg18313182, cg22016649, cg14522803, cg06196145 | chr11: 818752-818918 | PNPLA2 | 5 | 9.32E-08 | 0.0002416 |
| cg01229787, cg16214653, cg25885684 | chr15: 100048371-100048501 | MEF2A | 3 | 8.10E-08 | 0.0002683 |
Stouffer–Liptak–Kechris correction (slk); one-step Šidák (1967) multiple-testing correction. University of California, Santa Cruz Human Genome Browser (UCSC).
Figure 2.
DNA hypomethylation of DMR in exon 1 of the LTB4R2 gene. (A) Idiogram of chromosome 14 with genomic coordinates of DMR illustrated. The DMR—spanning 9 CpG sites—overlaps a CpG island (shown in blue). (B) LTB4R2-associated DMR is hypomethylated across all nine CpG sites in depression cases compared with controls.
Figure 3.
DNA hypermethylation of DMR located in intron 1 of the HOXC4 gene. (A) Idiogram of chromosome 12 with genomic coordinates of DMR illustrated. (B) HOXC4-associated DMR is hypermethylated across all five CpG sites in depression cases compared with controls.
Interaction between a history of depression and an inflammatory disorder on depression-associated DMRs
To explore the role of epigenetic variation in modifying the association between depression and chronic inflammatory disorders we tested for statistical interactions between a self-reported history of depression and history of an inflammatory disorder on DNA methylation at depression-associated DMPs and DMRs. We observed a significant interaction (P < 0.05) between history of depression and a history of inflammation on DNA methylation at 6 of the top 10 depression-associated DMPs. We also observed a significant interaction between a history of depression and a history of an inflammatory disorder on DNA methylation at more than one probe in depression-associated DMRs, LTB4R2 and MEF2A (Table 2).
Table 2.
DMRs associated with a history of depression
| DMR | Probe ID | Mean Δβ | P-value | Interaction P-value |
|---|---|---|---|---|
| LTB4R2 | ||||
| cg12853742 | −0.04194 | 0.0001 | 0.0195 | |
| cg25576711 | −0.06655 | 0.0004 | 0.0235 | |
| cg26310551 | −0.10142 | 0.0004 | 0.0349 | |
| cg06823034 | −0.08362 | 0.0006 | 0.0433 | |
| cg20007021 | −0.11113 | 0.0006 | 0.0400 | |
| cg21886367 | −0.04719 | 0.0009 | 0.0127 | |
| cg06796435 | −0.07357 | 0.0009 | 0.0311 | |
| cg15364618 | −0.07088 | 0.0010 | 0.0572 | |
| cg10193721 | −0.07892 | 0.0011 | 0.0402 | |
| HOXC4 | ||||
| cg15244786 | 0.08099 | 0.0000 | 0.1128 | |
| cg22370252 | 0.06029 | 0.0001 | 0.1007 | |
| cg26201952 | 0.04552 | 0.0002 | 0.0235 | |
| cg27138204 | 0.04620 | 0.0005 | 0.0855 | |
| cg05992786 | 0.04530 | 0.0032 | 0.6545 | |
| TRIM39 | ||||
| cg04425551 | 0.01938 | 0.0015 | 0.3644 | |
| cg07905808 | 0.01936 | 0.0029 | 0.3694 | |
| cg13079571 | 0.02125 | 0.0054 | 0.4970 | |
| cg11327408 | 0.01720 | 0.0063 | 0.6202 | |
| cg17080697 | 0.01588 | 0.0137 | 0.9874 | |
| cg09020199 | 0.01379 | 0.0435 | 0.9476 | |
| DNAJC17 | ||||
| cg17113968 | 0.02027 | 0.0004 | 0.1187 | |
| cg22079077 | 0.01806 | 0.0004 | 0.2361 | |
| cg16764778 | 0.01619 | 0.0024 | 0.3670 | |
| PNPLA2 | ||||
| cg24427660 | 0.01786 | 0.0014 | 0.2711 | |
| cg18313182 | 0.01371 | 0.0015 | 0.0779 | |
| cg22016649 | 0.01186 | 0.0022 | 0.4433 | |
| cg14522803 | 0.01210 | 0.0188 | 0.2174 | |
| cg06196145 | 0.01298 | 0.0253 | 0.0557 | |
| MEF2A | ||||
| cg01229787 | 0.04985 | 0.0001 | 0.0375 | |
| cg16214653 | 0.02818 | 0.0012 | 0.0296 | |
| cg25885684 | 0.04698 | 0.0023 | 0.0602 |
DMR, differentially methylated region, Mean Δβ; difference between individuals with a history of depression compared to those without, Interaction P-value; P-value of interaction between a history of depression and a history of inflammation, Hg19; Human Genome version 19, GREAT, Genomic Regions Enrichment of Annotations Tool, TSS, transcription start site.
Individuals with a self-reported history of depression have higher polygenic burden for the depression
To test whether individuals with a self-reported history of depression had higher polygenic burden for the depression, we generated a polygenic risk scores (PRS) for depression in each sample using findings from the most recent PGC GWAS meta-analysis of depression (7). Depression PRS scores were significantly higher (P = 0.0018) in our history of depression cases compared with controls (Supplementary Material, Fig. S4), highlighting the utility of this score.
Modules of co-methylated loci are associated with a history of depression and markers of inflammation
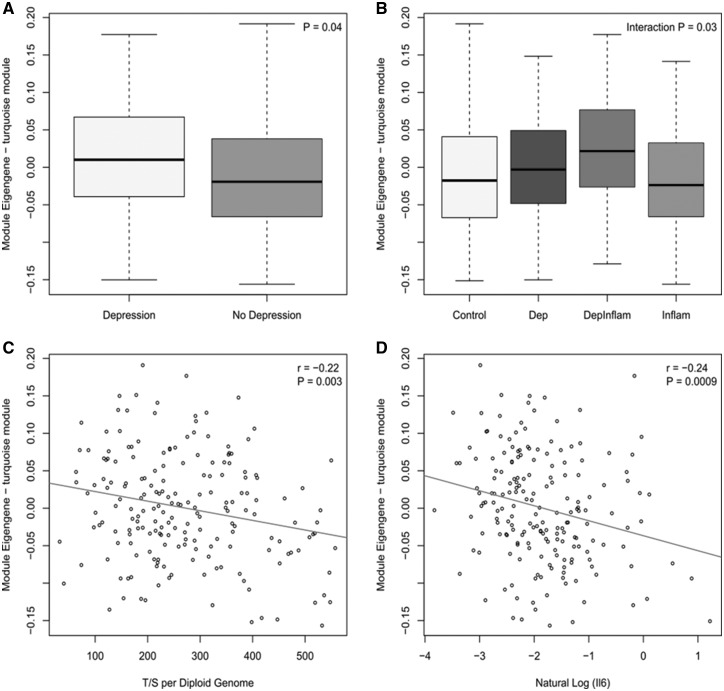
We employed weighted gene co-methylation network analysis (WGCNA) (29) to undertake a systems-level approach to identify networks of co-methylated positions associated with a history of depression and to integrate our DNA methylation data with available genetic data and known biomarkers of inflammation, TL and IL-6. WGCNA identified 20 modules comprising co-methylated loci. The module eigengene (ME) was used to assess the association between DNA methylation modules and history of depression, identifying two co-methylation modules that were nominally significantly associated with a history of depression, of which only one module (Turquoise, P = 0.04) remained significant after controlling for confounders. Interestingly we observed an interaction between the history of depression and history of an inflammatory disorder (interaction P = 0.03), with the ‘turquoise’ ME differing between individuals with both a history of depression and a history of inflammatory disorder compared with a history of depression only (Fig. 4). This co-methylation module was also associated with both TL and IL-6 serum levels (Fig. 4C and D) but not PRS score for depression. To further facilitate biological interpretation of this depression-associated co-methylation module we performed gene ontology enrichment analysis on genes annotated to probes in the module using a logistic regression method finding that the ‘turquoise’ module is highly enriched for pathways related to immune function (see Supplementary Material, Table S2 for details). These analyses implicate a role for coordinated changes in DNA methylation at immune-related loci in individuals with a history of depression.
Figure 4.
Boxplots and scatterplots of Module Eigengene (ME) against (A) diagnosis (depression versus no depression), (B) group [Depression (Dep), history of depression and inflammatory disorder only (DepInflam), history of inflammatory disorder only (Inflam), healthy control (Control)], (C) Telomere length (T/S per diploid genome) and (D) natural log of IL-6 serum levels.
Discussion
Although a number of studies have previously examined DNA methylation differences in peripheral tissue associated with both depression (8,9,30,31) and inflammatory conditions (11,12), this represents the first study exploring overlaps in patterns of epigenetic variation associated with a history of depression, a history of an inflammatory disorder and measures of inflammation markers, TL and IL-6.
Given that much attention has been given to the potential role of the immune system in pathophysiology of major depression (32), we first examined the association between a history of depression, predicted immune cell composition and IL-6 levels. Interestingly, individuals with a history of depression and a history of an inflammatory disorder had significantly higher CD4+ T cells and reduced levels of plasma blast cells, memory and effector T cells and granulocytes compared to the other three groups. In contrast, the depression only group did not differ significantly from the other groups. Suggesting that individuals with both a history of depression and a history of an inflammatory disorder have distinct immune profiles (e.g. Th1/Th2 cell activation) compared to individuals with a history of either depression or an inflammatory condition alone. Of note, T cells may play an important neuroprotective role in the context of stress and inflammation (32–34). Previously, it has been shown that depressed individuals have increased levels of the pro-inflammatory cytokine, IL-6, although both positive and negative results have been reported in individual studies (19). In the current study, we did not observe increased serum IL-6 levels between groups (P = 0.848), or between individuals with and without a history of depression (P = 0.583). Many clinical variables (e.g. medication use, infection and current depressive symptoms) may have affected the relationship between IL-6 levels and depression in our study.
Next, we examined site-specific genome-wide patterns of DNA methylation in depression cases compared with controls. While no DMP reached experiment-wide significance, several DMPs identified have been previously implicated in depression. Most notably, the top-ranked DMP (cg141959258) is in intron 4 of the RBFOX3 gene, which was recently implicated in a large-scale depression GWAS meta-analysis (7). Pathway analysis implicated RBFOX1, RBFOX2 and RBFOX3 regulatory networks in depression (7) and mutations in RBFOX3 have been previously linked to sleep latency (35)—a known risk factor for depression (36). RBFOX3 belongs to the Fox-1 family of genes and shows high homology to RBFOX1 and RBFOX2, and codes for the neuronal nuclei protein (37). The Fox proteins are a highly conserved family of tissue-specific splicing regulators and RBFOX3 is thought to have a role in neuron-specific alternative splicing (35). Moreover, this gene has been previously shown to be differentially methylated in twins discordant for adolescent depression (9). Taken together our data supports the role for the RBFOX3 gene in depression.
To increase the power of our study to identify regionally coordinated changes in DNA methylation between cases and controls, we employed comb-p to identify differentially methylated regions (DMRs). Our analysis identified six DMRs; two DMRs are associated with genes previously implicated in mental illness. The DNAJC17 gene is a negative regulator of transcription from RNA polymerase II promoter and has been previously associated with autism in a small study (38). The MEF2A gene encodes a transcription factor and plays a key role in skeletal, cardiac, smooth muscle and neuronal cell differentiation (39). Genetic variation in MEF2A has been previously associated with formal thought disorder (40), a major feature of schizophrenia and other psychotic disorders, and Alzheimer’s disease (39). The remaining four depression-associated DMRs are associated with immune-related genes. The top-ranked DMR, spanning nine CpG sites, is located within a CpG island in exon 1 of the LTB4R2 gene and overlaps the 5′ UTR of both the CIDEB and LTB4R genes. Both LTB4R and LTB4R2 are leukotriene B-4 receptors and are thought to play a role in inflammatory responses (41). Of note, DNA methylation at this region showed a significant depression * inflammatory disorder interaction. The HOXC4 gene, which is located on chromosome 12, encodes the Homeobox protein HOX-C4. The homeobox genes encode a highly conserved family of transcription factors that play a key role in morphogenesis in all multicellular organisms. HOXC4 has been shown to be an important regulator of transcription in B cell differentiation (42). TRIM39-RPP21 is a protein-coding gene and overlaps the third exon of TRIM39. TRIM39-RPP21 encodes a putative PKGI interactor that is a novel variant of TRIM39 (43). TRIM39-RPP21 regulates the type I interferon pathway and is thus important in regulation of viral immunity (44). The TRIM39-RPP21–associated DMR identified in this study is hypermethylated in depression cases compared with controls. Of interest, this DMR overlaps a previously reported larger inflammatory bowel disease (IBD)-associated DMR (11), which is differentially methylated in IBD cases compared with controls. We observed no evidence of a depression * inflammatory disorder interaction at this DMR, suggesting that DNA methylation at this locus is independently associated with a history of depression. PNPLA2 encodes the adipose triglyceride lipase and mutations in this gene have been implicated in neutral lipid storage disease, which is marked by triglyceride-containing droplets in the cytoplasm of neutrophils (45). Taken together our results provide evidence for a potential role for epigenetic-dysregulation of immune-related genes in depression.
Next, we used WGCNA analysis to identify networks of co-methylated modules associated with a history of depression, depression PRS, TL and serum IL-6 levels. Our analysis identified one depression-associated module (‘Turquoise module’), which remained nominally significant after controlling for confounders (P = 0.04). Interestingly, we also observed a history of depression * history of an inflammatory disorder interaction (interaction P = 0.03), where the Turquoise ME differs between individuals with both a history of depression and a history of inflammatory disorder compared with a history of depression only. This supports the hypothesis that individuals with a history of both depression and inflammatory disorders have distinct epigenetic signatures compared to those with a history of depression alone. Moreover, the ‘Turquoise’ co-methylation module was associated with markers of inflammation, TL and IL-6 serum levels. This co-methylation module was not associated with PRS for depression, suggesting that genetic risk for depression is not underlying the observed changes in DNA methylation. Furthermore, this depression-associated co-methylation module was highly enriched for pathways related to immune function. These analyses implicate a role for coordinated changes in DNA methylation at immune-related loci, which are associated with a history of depression, TL and IL-6 serum levels.
Despite the power of the methodological approaches used in this study, there are several limitations. First, our analysis was based on a relatively small number of samples (n = 200); although we had 80% power (see 46 for details) to detect DNA methylation difference >∼10% between cases and controls, we are relatively underpowered to detect smaller changes (<10%) in DNA methylation, which are frequently observed in DNA methylation studies of complex diseases, including depression. Second, in this study we used DNA derived from whole blood and cellular heterogeneity is a potentially important confounder in DNA methylation studies (47–49). However, we used a previously reported in silico method to estimate the blood cell composition in each sample and included these estimates in our statistical models. Third, we used a measure of self-reported history of depression in our study. Previous studies have shown the utility of using a self-reported measure of depression in genetic studies. For example, the recent depression GWAS showed that common-variant genetic architecture of self-reported depression strongly overlapped with that of current depressive symptoms (7). Moreover, our depression cases showed higher PRS for depression than controls and have shortened TL, which provides further evidence of the self-reported measure of depression in our study. Anti-depressant medication is often a confounder in many DNA methylation studies of depression, in this study ∼50% of our depression cases were currently taking anti-depressants, thus we could control for current anti-depressant use in our study. Moreover, there were more females than males in our depression cases. This is likely due to the high prevalence of depression observed in females compared with males (50). In this study, controls and cases were matched for gender. Finally, although our study presents evidence for novel DNA methylation changes at immune-related loci associated with depression, further replication using a larger sample size is required to support these results. In addition, future studies could also examine the transcriptional consequences of the observed DNA methylation changes.
Conclusion
In summary, genome-wide DNA methylation analysis in individuals with and without a self-reported history of depression has identified several candidate DMRs of potential relevance to the pathogenesis of depression and its associated immune-dysfunction phenotype.
Materials and Methods
Samples
DNA samples, extracted using standard protocols from whole blood (n = 200), and serum samples were provided by the Royal Devon and Exeter (RDE) Tissue Bank, part of the NIHR Exeter Clinical Research Facility. Samples selected for this study included: (a) 50 individuals with a self-reported history of depression (individuals answering yes to the question ‘Has a doctor ever diagnosed you with depression requiring regular medical treatment?’) and a self-reported history of a chronic inflammatory condition (individuals answering yes to the questions ‘Has a doctor ever diagnosed you with (1) irritable bowel/ulcerative colitis/crohns/diverticulitis or (2) rheumatoid arthritis/lupus?’), (b) 50 individuals with a self-reported history of depression but no history of an inflammatory condition, (c) 50 individuals with a self-reported history of a chronic inflammatory condition but no self-reported history of depression or diagnosed mental health problem and (d) 50 healthy controls. Groups were matched for gender, age and body mass index (BMI) (see Supplementary Material, Table S3 for further details). All samples included in this study were of white ethnicity. This study was approved by the University of Exeter Medical School Research Ethics Board (REB).
Telomere length measurements
Quantitative real-time PCR (QRT-PCR) was used to measure TL as described previously (51) by determining the ratio of telomere repeat copy number to single-copy gene copy number (T/S ratio) per diploid genome in each of the DNA samples included in this study. A difference in TL between individuals with a self-reported history of depression and individuals without a history of depression was assessed using logistic regression, controlling for a history of an inflammatory condition and potential confounders (i.e. age, gender and estimated blood cell composition).
Serum interleukin-6 measurements
Serum was separated from whole blood by centrifugation at 2500g for 10 min at 4°C. Serum was collected using a transfer pipette and stored long term at −80°C. The Meso Scale V-plex plus Human IL-6 kit (Meso Scale Diagnostics, Rockville, Maryland, USA), a highly sensitive multiplex enzyme-linked immunosorbent assay (ELISA), was used to quantify serum levels of IL-6 at the Immunoassay Biomarker Core Laboratory, University of Dundee. The lower limit of detection for these assays was 0.06 pg/ml. All samples were assayed in duplicate and the coefficients of variation for these assays were <30%. After quality control (QC), we obtained serum IL-6 measurements for 171 samples.
Methylomic profiling
Genomic DNA (500 ng) was treated with sodium bisulfite using the Zymo Gold-kit and DNA methylation levels quantified using the Illumina Infinium HumanMethylation450 BeadChip (“Illumina 450K array”) array. The methylumi package (52) was used to extract signal intensities for each CpG probe and perform initial QC with data normalization and pre-processing using the WateRmelon package (53). Additional QC (age checks, data quality checks) were performed using the online epigenetic clock calculator (http://labs.genetics.ucla.edu/horvath/dnamage/; date last accessed March 7, 2017) (22). Non-specific probes on the Illumina 450K array were removed (54). Six out of 200 samples did not pass initial stringent QC checks (samples having 1% of sites with a detection P-value >0.01 and/or bisulfite metric <80%) and were excluded. In total, 750 CpG probes with a bead count < 3 in 5% of samples and a further 1969 CpG probes with a detection P-value >0.05 in 1% of samples were removed. Quantile normalization of raw β was performed using DASEN (for further details, see 53). The final analyses included 430 574 probes, and 194 samples. Raw Illumina 450K array data is deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE113725).
Estimating DNAm Age and differential blood cell counts
Using the online epigenetic clock calculator (http://labs.genetics.ucla.edu/horvath/dnamage/) (55), we obtained DNAm Age estimates, derived cell-type proportion estimates and age acceleration estimates (the residuals from a linear regression of DNAm Age on chronological age) for each sample. A student’s t-test was used to examine the difference in DNAm Age acceleration and a history of depression. Pearson’s correlation was used to examine the correlation between DNAm Age acceleration and TL.
Data analyses of genome-wide DNA methylation
Statistical analyses were performed using R (version 3.2.1). The beta value (β) is a ratio between methylated probe intensity and total probe intensities (sum of methylated and unmethylated probe intensities) and ranges from 0 to 1. Linear regression was used to examine differences in DNA methylation scores [reported as change in beta value (Δβ)] between individuals with a self-reported history of depression and individuals without a history of depression at each CpG site, controlling for potential confounders (history of an inflammatory disorder, age, gender, anti-depressant use, chip and estimated blood cell composition).
To identify DMRs in our data we used the Python module Comb-p (28) that groups spatially correlated DMPs (seed P-value < 1 ×10−3, minimum of three probes) at a maximum distance of 500 bp. DMR P-values were corrected for multiple testing using Šidák correction (56).
Genotyping and MDD polygenic risk scoring
Genomic DNA was genotyped using the Infinium® Global Screening Array-24 v1.0 (Illumina), according to manufacturer’s instructions. PLINK (57) was used to remove samples with a call rate < 0.975 and filter variants with a call rate < 0.975, Hardy-Weinberg equilibrium P < 0.0001 or minor allele frequency of < 1%. Samples were filtered to European Ancestry only. PRS were calculated using the P-values and log odds ratios from the most recent GWAS from the PGC MDD working group (7), without external meta-analysis study 23andMe. PRS were constructed from summary statistics of imputed single-nucleotide polymorphisms (SNPs) from the 29 PGC depression studies analysed with external meta-analysis studies (excluding 23andMe) [cases (n = 84 891), controls (n = 224 116)]. Linkage disequilibrium (LD) clumping was performed using default settings in PRSice, to retain the strongest signals for association. Standard thresholds for significance (P-value threshold [PT] 5 × 10−8, 1 × 10−6, 1 × 10−4, 0.001, 0.01, 0.05, 0.1, 0.2, 0.5, 1.0) were used, and the most predictive PT threshold (PT < 0.5) for our cohort, as applied in PRSice (58), was assessed (see Supplementary Material, Fig. S5). A depression PRS (PT < 0.5) was constructed in 173 individuals with genotyping data available from the EXETER 10000 epidemiological cohort using PRSice. The number of genetic variants included for the depression PRS was 56 411. Logistic regression, with two ancestry-information dimensions as covariates, was used to test for a difference in PRSmean between MDD cases and controls.
Weighted gene co-methylation network analysis (WGCNA)
Network analysis was performed on normalized beta values using WGCNA (29). Pair-wise correlations were used to identify modules of highly co-methylated probes, independent of disease status as described previously (59). Briefly, an unsigned network was created using the blockwiseModules function based on a block size of 5000 and using a soft threshold parameter of 6. Each module was then labeled with a unique color identifier and the ME for each module (a weighted average methylation profile) was calculated based on the first principal component (PC) of the methylation matrix. To identify modules associated with a history of depression, a t-test was used to compare mean ME values between cases (individuals with a history of depression) and controls (individuals with no history of depression). Pearson’s correlation coefficients were used to examine the association between ME values and continuous variables (TL, natural log of IL-6, cell types, age and depression PRS). Linear regression was used to examine the association of two depression-associated co-methylation modules while controlling for known confounders. Next, gene ontology enrichment analysis implementing a previously described logistic regression approach (60) was used to test if genes (Illumina UCSC gene annotation) annotated to probes (n = 18 180) in the depression-associated module predicted pathway membership, while controlling for the number of probes annotated to each gene. Briefly, pathways were downloaded from the Gene Ontology (GO) website (http://geneontology.org/) and genes with at least one Illumina probe annotated to it and which mapped to at least one GO pathway were included. Pathways were filtered to those containing between 10 and 2000 genes and a list of significant pathways (P < 0.05) was identified as described previously (60).
Supplementary Material
Acknowledgements
We are grateful to all participants of EXETER 10000.
Conflict of Interest statement. None declared.
Funding
The authors would like to acknowledge funding support for the project from the Brain and Behaviour Research Foundation (BBF) through a NARSAD Young Investigator Grant to TMM. This project was also supported by the National Institute for Health Research (NIHR) Exeter Clinical Research Facility. The views expressed are those of the authors and not necessarily those of the BBF, NHS, the NIHR or the Department of Health.
References
- 1. WHO. (2017) http://www.who.int/mediacentre/factsheets/fs369/en/; date last accessed July 25, 2017.
- 2. Berk M., Williams L.J., Jacka F.N., O'Neil A., Pasco J.A., Moylan S., Allen N.B., Stuart A.L., Hayley A.C., Byrne M.L.. et al. (2013) So depression is an inflammatory disease, but where does the inflammation come from? BMC Med., 11, 200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller A.H., Raison C.L. (2016) The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat. Rev. Immunol., 16, 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henje Blom E., Han L.K., Connolly C.G., Ho T.C., Lin J., LeWinn K.Z., Simmons A.N., Sacchet M.D., Mobayed N., Luna M.E.. et al. (2015) Peripheral telomere length and hippocampal volume in adolescents with major depressive disorder. Transl. Psychiatry, 5, e676.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mamdani F., Rollins B., Morgan L., Myers R.M., Barchas J.D., Schatzberg A.F., Watson S.J., Akil H., Potkin S.G., Bunney W.E.. et al. (2015) Variable telomere length across post-mortem human brain regions and specific reduction in the hippocampus of major depressive disorder. Transl. Psychiatry, 5, e636.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aubert G., Lansdorp P.M. (2008) Telomeres and aging. Physiol. Rev., 88, 557–579. [DOI] [PubMed] [Google Scholar]
- 7. Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., Adams M.J., Agerbo E., Air T.M., Andlauer T.M.F.. et al. (2018) Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet., 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies M.N., Krause L., Bell J.T., Gao F., Ward K.J., Wu H., Lu H., Liu Y., Tsai P.C., Collier D.A.. et al. (2014) Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol., 15, R56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dempster E.L., Wong C.C., Lester K.J., Burrage J., Gregory A.M., Mill J., Eley T.C. (2014) Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biol. Psychiatry, 76, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murphy T.M., Crawford B., Dempster E.L., Hannon E., Burrage J., Turecki G., Kaminsky Z., Mill J. (2017) Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl. Psychiatry, 7, e989.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDermott E., Ryan E.J., Tosetto M., Gibson D., Burrage J., Keegan D., Byrne K., Crowe E., Sexton G., Malone K.. et al. (2016) DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J. Crohns Colitis, 10, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Aryee M.J., Padyukov L., Fallin M.D., Hesselberg E., Runarsson A., Reinius L., Acevedo N., Taub M., Ronninger M.. et al. (2013) Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol., 31, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeilinger S., Kuhnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A.. et al. (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One, 8, e63812.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Labonte B., Suderman M., Maussion G., Navaro L., Yerko V., Mahar I., Bureau A., Mechawar N., Szyf M., Meaney M.J.. et al. (2012) Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry, 69, 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller G.E., Cole S.W. (2012) Clustering of depression and inflammation in adolescents previously exposed to childhood adversity. Biol. Psychiatry, 72, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costenbader K.H., Karlson E.W. (2006) Cigarette smoking and autoimmune disease: what can we learn from epidemiology? Lupus, 15, 737–745. [DOI] [PubMed] [Google Scholar]
- 17. Hohensinner P.J., Goronzy J.J., Weyand C.M. (2011) Telomere dysfunction, autoimmunity and aging. Aging Dis., 2, 524–537. [PMC free article] [PubMed] [Google Scholar]
- 18. Gabay C. (2006) Interleukin-6 and chronic inflammation. Arthritis Res. Ther., 8(Suppl 2), S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dowlati Y., Herrmann N., Swardfager W., Liu H., Sham L., Reim E.K., Lanctot K.L. (2010) A meta-analysis of cytokines in major depression. Biol. Psychiatry, 67, 446–457. [DOI] [PubMed] [Google Scholar]
- 20. Ridout S.J., Ridout K.K., Kao H.T., Carpenter L.L., Philip N.S., Tyrka A.R., Price L.H. (2015) Telomeres, early-life stress and mental illness. Adv. Psychosom. Med., 34, 92–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin P.Y., Huang Y.C., Hung C.F. (2016) Shortened telomere length in patients with depression: a meta-analytic study. J. Psychiatr. Res., 76, 84–93. [DOI] [PubMed] [Google Scholar]
- 22. Horvath S. (2013) DNA methylation age of human tissues and cell types. Genome Biol., 14, R115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whalley H.C., Gibson J., Marioni R., Walker R.M., Clarke T.-K., Howard D.M., Adams M.J., Hall L., Morris S., Deary I.J.. et al. (2017) Accelerated epigenetic ageing in major depressive disorder. bioRxiv, doi: https://doi.org/10.1101/210666.
- 24. Breitling L.P., Saum K.U., Perna L., Schottker B., Holleczek B., Brenner H. (2016) Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenetics, 8, 21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marioni R.E., Harris S.E., Shah S., McRae A.F., von Zglinicki T., Martin-Ruiz C., Wray N.R., Visscher P.M., Deary I.J. (2016) The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int. J. Epidemiol., 47, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Belsky D.W., Moffitt T.E., Cohen A.A., Corcoran D.L., Levine M.E., Prinz J.A., Schaefer J., Sugden K., Williams B., Poulton R.. et al. (2017) Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: do they measure the same thing? Am. J. Epidemiol., 187, 1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viana J., Hannon E., Dempster E., Pidsley R., Macdonald R., Knox O., Spiers H., Troakes C., Al-Saraj S., Turecki G.. et al. (2017) Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet., 26, 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pedersen B.S., Schwartz D.A., Yang I.V., Kechris K.J. (2012) Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics, 28, 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuan P.F., Waszczuk M.A., Kotov R., Marsit C.J., Guffanti G., Gonzalez A., Yang X., Koenen K., Bromet E., Luft B.J. (2017) An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl. Psychiatry, 7, e1158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uddin M., Koenen K.C., Aiello A.E., Wildman D.E., de los Santos R., Galea S. (2011) Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol. Med., 41, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller A.H. (2010) Depression and immunity: a role for T cells? Brain Behav. Immun., 24, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewitus G.M., Schwartz M. (2009) Behavioral immunization: immunity to self-antigens contributes to psychological stress resilience. Mol. Psychiatry, 14, 532–536. [DOI] [PubMed] [Google Scholar]
- 34. Lewitus G.M., Wilf-Yarkoni A., Ziv Y., Shabat-Simon M., Gersner R., Zangen A., Schwartz M. (2009) Vaccination as a novel approach for treating depressive behavior. Biol. Psychiatry, 65, 283–288. [DOI] [PubMed] [Google Scholar]
- 35. Amin N., Allebrandt K.V., van der Spek A., Muller-Myhsok B., Hek K., Teder-Laving M., Hayward C., Esko T., van Mill J.G., Mbarek H.. et al. (2016) Genetic variants in RBFOX3 are associated with sleep latency. Eur. J. Hum. Genet., 24, 1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nutt D., Wilson S., Paterson L. (2008) Sleep disorders as core symptoms of depression. Dialog. Clin. Neurosci., 10, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim K.K., Adelstein R.S., Kawamoto S. (2009) Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem., 284, 31052–31061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stamova B.S., Tian Y., Nordahl C.W., Shen M.D., Rogers S., Amaral D.G., Sharp F.R. (2013) Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders. Mol. Autism, 4, 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gonzalez P., Alvarez V., Menendez M., Lahoz C.H., Martinez C., Corao A.I., Calatayud M.T., Pena J., Garcia-Castro M., Coto E. (2007) Myocyte enhancing factor-2A in Alzheimer's disease: genetic analysis and association with MEF2A-polymorphisms. Neurosci. Lett., 411, 47–51. [DOI] [PubMed] [Google Scholar]
- 40. Thygesen J.H., Zambach S.K., Ingason A., Lundin P., Hansen T., Bertalan M., Rosengren A., Bjerre D., Ferrero-Miliani L., Rasmussen H.B.. et al. (2015) Linkage and whole genome sequencing identify a locus on 6q25-26 for formal thought disorder and implicate MEF2A regulation. Schizophr. Res., 169, 441–446. [DOI] [PubMed] [Google Scholar]
- 41. Hashimoto A., Endo H., Hayashi I., Murakami Y., Kitasato H., Kono S., Matsui T., Tanaka S., Nishimura A., Urabe K.. et al. (2003) Differential expression of leukotriene B4 receptor subtypes (BLT1 and BLT2) in human synovial tissues and synovial fluid leukocytes of patients with rheumatoid arthritis. J. Rheumatol., 30, 1712–1718. [PubMed] [Google Scholar]
- 42. Kim E.C., Edmonston C.R., Wu X., Schaffer A., Casali P. (2004) The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3' hs1, 2 enhancer in human B cells. Relevance to class switch DNA recombination. J. Biol. Chem., 279, 42258–42269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vlantis K., Polykratis A., Welz P.S., van Loo G., Pasparakis M., Wullaert A. (2016) TLR-independent anti-inflammatory function of intestinal epithelial TRAF6 signalling prevents DSS-induced colitis in mice. Gut, 65, 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts J.D. Jr, Chiche J.D., Kolpa E.M., Bloch D.B., Bloch K.D. (2007) cGMP-dependent protein kinase I interacts with TRIM39R, a novel Rpp21 domain-containing TRIM protein. Am. J. Physiol. Lung Cell Mol. Physiol., 293, L903–L912. [DOI] [PubMed] [Google Scholar]
- 45. Missaglia S., Maggi L., Mora M., Gibertini S., Blasevich F., Agostoni P., Moro L., Cassandrini D., Santorelli F.M., Gerevini S.. et al. (2017) Late onset of neutral lipid storage disease due to novel PNPLA2 mutations causing total loss of lipase activity in a patient with myopathy and slight cardiac involvement. Neuromuscul. Disord., 27, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai P.C., Bell J.T. (2015) Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol., 44, 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guintivano J., Aryee M.J., Kaminsky Z.A. (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics, 8, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heijmans B.T., Mill J. (2012) Commentary: the seven plagues of epigenetic epidemiology. Int. J. Epidemiol., 41, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Birney E., Smith G.D., Greally J.M. (2016) Epigenome-wide association studies and the interpretation of disease -omics. PLoS Genet., 12, e1006105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Albert P.R. (2015) Why is depression more prevalent in women? J. Psychiatry Neurosci., 40, 219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Callaghan N.J., Fenech M. (2011) A quantitative PCR method for measuring absolute telomere length. Biol. Proced. Online, 13, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davis S.D.P., Bilke S., Triche T. Jr, Bootwalla M. (2015) methylumi: Handle Illumina methylation data. R package version 2.14.0.
- 53. Pidsley R., Y Wong C.C., Volta M., Lunnon K., Mill J., Schalkwyk L.C. (2013) A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics, 14, 293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Price M.E., Cotton A.M., Lam L.L., Farre P., Emberly E., Brown C.J., Robinson W.P., Kobor M.S. (2013) Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin, 6, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Horvath S., Erhart W., Brosch M., Ammerpohl O., von Schonfels W., Ahrens M., Heits N., Bell J.T., Tsai P.-C., Spector T.D.. et al. (2014) Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. U.S.A., 111, 15538–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Šidák Z. (1967) Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Statist. Assoc., 62, 626–633. [Google Scholar]
- 57. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J.. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Euesden J., Lewis C.M., O'Reilly P.F. (2015) PRSice: Polygenic Risk Score software. Bioinformatics, 31, 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pidsley R., Viana J., Hannon E., Spiers H., Troakes C., Al-Saraj S., Mechawar N., Turecki G., Schalkwyk L.C., Bray N.J.. et al. (2014) Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol., 15, 483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lunnon K., Hannon E., Smith R.G., Dempster E., Wong C., Burrage J., Troakes C., Al-Sarraj S., Kepa A., Schalkwyk L.. et al. (2016) Variation in 5-hydroxymethylcytosine across human cortex and cerebellum. Genome Biol., 17, 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.