Summary
Heteromorphic flower development in Primula is controlled by the S locus. The S locus genes, which control anther position, pistil length and pollen size in pin and thrum flowers, have not yet been characterized. We have integrated S‐linked genes, marker sequences and mutant phenotypes to create a map of the P. vulgaris S locus region that will facilitate the identification of key S locus genes. We have generated, sequenced and annotated BAC sequences spanning the S locus, and identified its chromosomal location.
We have employed a combination of classical genetics and three‐point crosses with molecular genetic analysis of recombinants to generate the map. We have characterized this region by Illumina sequencing and bioinformatic analysis, together with chromosome in situ hybridization.
We present an integrated genetic and physical map across the P. vulgaris S locus flanked by phenotypic and DNA sequence markers. BAC contigs encompass a 1.5‐Mb genomic region with 1 Mb of sequence containing 82 S‐linked genes anchored to overlapping BACs. The S locus is located close to the centromere of the largest metacentric chromosome pair.
These data will facilitate the identification of the genes that orchestrate heterostyly in Primula and enable evolutionary analyses of the S locus.
Keywords: chromosome in situ, genetic map, heterostyly, Primula vulgaris, S locus
Introduction
Heterostyly is found in over 160 genera across 24 families (Ganders, 1979), suggesting a polyphyletic origin (Barrett, 1990). Despite different origins and therefore potentially different mechanisms, the locus controlling heterostyly is uniformly known as the S locus (Lewis & Jones, 1993), and is diallelic in Primula vulgaris (Bateson & Gregory, 1905), Turnera subulata (Shore & Barrett, 1985), Fagopyrum esculentum (Garber & Quisenberry, 1927) and Linum grandiflorum (Ushijima et al., 2012). Molecular genetic studies on floral heteromorphy have focused on species from four families: Turneraceae (T. subulata) (Athanasiou et al., 2003; Labonne et al., 2008, 2009; Labonne & Shore, 2011); Polygonaceae (F. esculentum) (Wang et al., 2005; Yasui et al., 2008, 2012); Primulaceae (P. vulgaris) (Manfield et al., 2005; McCubbin et al., 2006; Li et al., 2007, 2008, 2010; Cocker et al., 2015); and Linaceae (L. grandiflorum) (Ushijima et al., 2012).
In Primula, flowers have either a long style, low anthers and small pollen, or a short style, high anthers and large pollen (Darwin, 1862). Fagopyrum esculentum (Yasui et al., 2012) and T. subulata (Labonne & Shore, 2011) also have long‐styled and short‐styled flowers. In these species, the anther filament length determines the height of the anthers in different floral morphs; in Primula, it is the point of attachment to the corolla tube that differs between pin and thrum flowers (Darwin, 1862; Webster & Gilmartin, 2003). Linum grandifolium shows stigma–height dimorphism with flowers that differ in stigma height, but not the position of the anthers (Darwin, 1863; Barrett, 2010; Ushijima et al., 2012).
In Turnera, progress towards the identification of the S locus includes a high‐resolution genetic map (Labonne et al., 2009) and the identification of deletion mutants (Labonne et al., 2010). This approach enabled the assembly of three BAC contigs spanning the S locus which, in combination with deletion mutants, enabled the positional cloning of the recessive s allele in T. subulata (Labonne & Shore, 2011). Although the key genes have not yet been described, these studies represent a significant step towards the identification of the molecular mechanisms of floral heteromorphy in this species. Similar map‐based approaches have been used in F. esculentum (Yasui et al., 2004, 2008; Konishi et al., 2006), where next‐generation sequencing and in silico analysis have identified a candidate S locus gene, S‐ELF3 (Yasui et al., 2012). The analysis of a short‐styled chromosome deletion mutant, which produces long‐styled flowers, revealed that S‐ELF3 had been lost in the deletion. Although the large deletion may contain other genes, mutations in S‐ELF3 in the other homomorphic cultivars suggest that S‐ELF3 is a candidate regulator of heteromorphic flower development in Fagopyrum. Approaches to study floral heteromorphy in L. grandiflorum (Ushijima et al., 2012) used a combination of suppressive subtractive hybridization and two‐dimensional‐polyacrylamide gel electrophoresis (2D‐PAGE) analysis to reveal 12 floral morph‐related genes. Four genes implicated in the control of style length (Ushijima et al., 2012) include a Myb transcription factor, LgMYB21, which, when constitutively overexpressed in Arabidopsis, reduces style length and anther height.
Building on the work of Darwin, early studies on the genetics and control of heterostyly in Primula revealed the dominance of the thrum phenotype and defined S and s alleles (Bateson & Gregory, 1905; Gregory, 1911). Subsequent analysis (Ernst, 1925, 1936b; Pellow, 1928; Haldane, 1933; Dowrick, 1956; Lewis & Jones, 1993) defined three diallelic genes, G/g, P/p and A/a, at the S locus with thrums heterozygous GPA/gpa and pins homozygous recessive gpa/gpa. The rare occurrence of homostyles was predicted to arise via mutation (Ernst, 1928b, 1936a), and subsequently interpreted as a result of crossovers within the S locus gene cluster (Dowrick, 1956; Lewis & Jones, 1993). Subsequent studies expanded the linkage group to include genes involved in pollen size dominance (Kurian & Richards, 1997) and pollen and style self‐incompatibility behaviour (Dowrick, 1956; Lewis & Jones, 1993; Richards, 1997).
Early reports of S‐linked genes not involved in heterostyly include Hose in Hose (Ernst, 1928a, 1942) and four loci in P. sinensis: magenta (b), red stigma (g), red leaf back (l) and double (x) (De Winton, 1928; De Winton & Haldane, 1935); these plants are no longer available. More recently, quantitative trait locus (QTL) analysis of floral morphology in P. sieboldii provided a genome‐wide linkage map with four markers within 1 cM of the S locus (Yoshida et al., 2011). We have characterized previously Hose in Hose (Webster & Grant, 1990), sepaloid (Li et al., 2008) and Oakleaf (Webster, 2005; Cocker et al., 2015) as S‐linked phenotypes, and identified S locus markers by random amplified polymorphic DNA (RAPD) analysis (Manfield et al., 2005), fluorescent differential display (Li et al., 2007) and analysis of Hose in Hose (Li et al., 2010). Here, we combine these studies into an integrated genetic map of the S locus; we describe the assembly and sequence of a BAC contig spanning the region, and use in situ hybridization to define the chromosomal location of the Primula S locus.
Materials and Methods
Growth of plants and genetic crosses
The plants used in this study are cultivated varieties of Primula vulgaris Huds. Primula vulgaris cv Blue Jeans was obtained from Thompson and Morgan (http://www.thompson-morgan.com/). Hose in Hose (Gerard, 1597; Webster & Grant, 1990), sepaloid (Webster, 2005; Li et al., 2008) and Oakleaf (Webster, 2005; Cocker et al., 2015) from the National Collection of Primula, British Floral Variants, maintained by Margaret Webster, were used to generate parental genotypes for three‐point crosses by pollination under insect‐free conditions, with the plants grown as described previously (Webster & Gilmartin, 2006).
Southern analysis
Genomic DNA was isolated from P. vulgaris leaves by a Nucleon Phyto‐Pure Genomic DNA Extraction kit (GE‐LifeSciences, Little Chalfont, Buckinghamshire, UK; www.gelifesciences.com) according to the manufacturer's instructions, and Southern analysis was performed as described previously (Manfield et al., 2005; Li et al., 2010).
Chromosome in situ hybridization
BACs were labelled with biotin‐dUTP or digoxigenin‐dUTP and detected with fluorescein or Alexa‐594 conjugates. In situ hybridization to root‐tip metaphase chromosomes, detection, counterstaining with 4′,6‐diamidino‐2‐phenylindole (DAPI), microscopy and imaging were performed as described previously (Heslop Harrison & Schwarzacher, 2002). Unlabelled P. vulgaris DNA (1 μg, 25 × probe amount) was added to each slide (40 μl) before hybridization to limit the hybridization of repetitive probe sequences. After hybridization, the most stringent wash in 0.05 × saline sodium citrate (SSC) at 43°C corresponded to a hybridization stringency of 78% (high‐stringency hybridization), and in 1 × SSC at 42°C to low‐stringency hybridization.
BAC sequencing, assembly and annotation
BAC library construction and screening have been described previously (Li et al., 2011). Seven of 42 BACs were sequenced individually by 454GSFLX at the Centre for Genomic Research (Liverpool, UK); four were sequenced as HiSeq2000 paired‐end reads at The Genome Analysis Centre (Norwich, UK). The remaining BACs were sequenced in two pools of 19 and 18 by 454GSFLX (Liverpool, UK). Five of the BACs sequenced individually were also included in the pools. To facilitate BAC contig assembly, we used a draft genome assembly of Illumina paired‐end reads from thrum genomic DNA (83× coverage), scaffolded with a 9‐kb thrum genomic DNA mate‐pair library (32× coverage). The assembly was curated using The Genome Analysis Centre (TGAC) browser http://www.tgac.ac.uk/tgac-browser/. Contig assembly and annotation details are given in Supporting Information Methods S1. All sequences have been deposited at the National Center for Biotechnology Information (NCBI) under Bioproject number PRJEB7311. BAC contigs and annotations are available at http://browser.tgac.ac.uk/primula_vulgaris_slocus/.
Results
Analysis of allelic variants of S locus‐linked genes
We have identified previously four S‐linked markers in P. vulgaris cv Blue Jeans. PvSLP1 was identified by RAPD analysis (Manfield et al., 2005), PvSLL1 and PvSLL2 by fluorescent differential display (Li et al., 2007) and PvGlo as Hose in Hose (Li et al., 2010). In each case, markers were analysed independently in different F1 and F2 individuals. Here, we show an integrated analysis of these markers in the same five plants (Fig. 1). Figure 1(a) shows restriction fragment length polymorphisms (RFLPs) for PvSLP1. The 4.2‐ and 3.5‐kb bands are common to pins and thrums; the 2.6‐ and 1.4‐kb bands are thrum specific (Manfield et al., 2005). RFLP analysis of the same five plants for PvSLL1 (Li et al., 2007) (Fig. 1b) reveals two different pin alleles defined by 5.0‐kb (P 1) and 2.9‐kb (P 2) bands; a 3.0‐kb band (T) represents the thrum allele. A P 1 P 1 homozygote, a P 1 P 2 heterozygote and a P 2 P 2 homozygote with respect to PvSLL1 are shown. The two thrum plants have genotypes P 1 T and P 2 T (Fig. 1b). RFLP analysis of PvSLL2 (Li et al., 2007) using the same plants reveals two pin alleles distinguished by 7.0‐kb (P 1) and > 12‐kb (P 2) bands; a 4.0‐kb band represents the thrum allele (T) (Fig. 1c). This example shows individuals with P 1 P 1, P 1 P 2, P 2 P 2, P 1 T and P 2 T genotypes with respect to PvSLL2.
Figure 1.
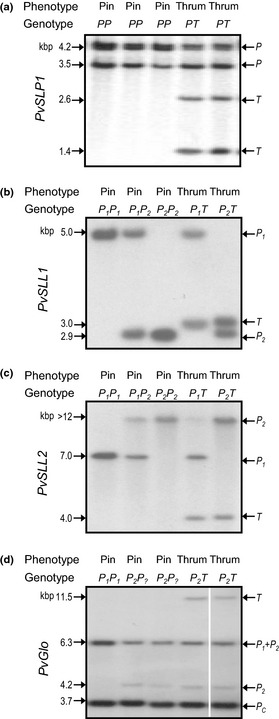
Allelic variation of S locus‐linked sequences in Primula vulgaris cv Blue Jeans. Autoradiographs following gel blot analysis of pin and thrum genomic DNA. The phenotypes of pin and thrum individuals are indicated. The genotypes of plants in relation to pin alleles (P) and thrum alleles (T) are shown and refer to the specific locus. Allele‐specific restriction fragment length polymorphism (RFLP) bands are indicted on the right and sizes in kbp on the left. Where pin alleles can be distinguished, these are indicated as P1 and P2; where unknown, as P?. (a) KpnI‐digested genomic DNA with PvSLP1 as probe. (b) Hind III‐digested genomic DNA with PvSLL1 as probe. (c) XbaI‐digested genomic DNA with PvSLL2 as probe. (d) XbaI‐digested genomic DNA with PvGlo as probe (lane 5 has been positioned next to lanes 1–4 to maintain sample order with parts (a–c), all five lanes are from the same gel). [Correction added after online publication 9 April 2015: panel (d) amended to include missing white line between lanes 4 and 5.]
Figure 1(d) illustrates the RFLP profile obtained with PvGlo (Li et al., 2010) as a probe using the same five plants. A 3.7‐kb hybridization band (P c) is common to pin and thrum alleles of PvGlo. A 6.3‐kb band, also found in pins and thrums, derives from both P 1 and P 2 alleles, and is designated P 1 + P 2. The plant in lane 1 lacks the 4.2‐kb P 2 allele, but shows a stronger signal for the 6.3‐kb band compared with other plants, and is a P 1 P 1 homozygote. The pin plants in lanes 2 and 3 are labelled as P 2 P ? as it is not possible to determine whether the 6.3‐kb band derives from the P 1 or P 2 allele. Thrum plants have a thrum‐specific 11.5‐kb band. We assigned P 1 and P 2 to the different pin alleles of PvSLL1, PvSLL2 and PvGlo before genetic analysis to determine their recombination relationships to the S locus and each other.
The data presented in Fig. 1 define the different alleles used in our genotyping study. We extended these analyses by PCR and DNA gel blot analysis to monitor the segregation of the four S‐linked markers with a larger number of F2 P. vulgaris cv Blue Jeans’ progeny. Between 144 and 193 plants were used in each assay, as shown in Table 1. We did not detect recombinants between the S locus and PvSLL1, PvSLP1 or PvGlo in any of the F2 progeny tested. However, we did observe two recombination events between the S locus and PvSLL2. The progeny numbers are small, but enabled us to determine minimum map distances based on the absence of recombination (Table 1).
Table 1.
Summary recombination data for S‐linked markers in Primula vulgaris cv Blue Jeans
| S‐linked marker | Pin plants | Thrum plants | Total assayed | Recombinants observed | Map distance (cM) |
|---|---|---|---|---|---|
| PvSLP1 | 92 | 99 | 191 | 0 | < 0.52 |
| PvSLL1 | 74 | 100 | 174 | 0 | < 0.57 |
| PvSLL2 | 56 | 90 | 146 | 2 | 1.37 |
| PvGlo | 64 | 93 | 157 | 0 | < 0.64 |
Where no recombinants were found, the map distance is the theoretical maximum value based on one hypothetical recombinant in the population.
Classical genetic analysis of the S locus using three‐point crosses
We initiated classical genetic analyses using the S‐linked phenotypes Hose in Hose (Webster & Grant, 1990; Li et al., 2010), sepaloid (Webster, 2005; Li et al., 2008) and Oakleaf (Webster, 2005; Cocker et al., 2015) to determine linkage relationships through two three‐point crosses. The first crossed wild‐type pin plants with thrum Hose in Hose Oakleaf plants with Hose in Hose in coupling and Oakleaf in repulsion to the dominant S allele (Fig. 2). Six crosses were established using two Hose in Hose Oakleaf thrum parents and five wild‐type pin parents; these crosses yielded 2075 progeny which revealed single‐ and double‐crossover events (Fig. 2). Progeny numbers were pooled to determine gene order. From these combined data, we defined the smallest progeny group, Hose in Hose pin and Oakleaf thrum plants, as double‐crossover progeny; this defines the gene order as Oakleaf–S locus–Hose in Hose. A further unexpected progeny class was represented by a single self‐fertile short homostyle Hose in Hose plant with large pollen.
Figure 2.
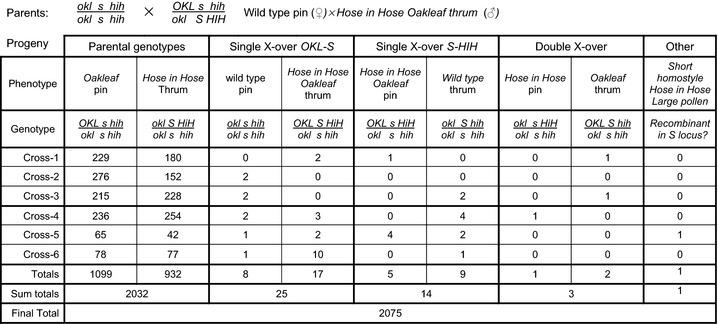
Three‐point cross to define gene order for the S locus, Primula vulgaris Oakleaf and Hose in Hose. Analysis of a three‐point cross between pin female parents (s/s), homozygous for wild‐type recessive alleles of oakleaf (okl) and hose in hose (hih), and two thrum (S/s) male parents, heterozygous for the dominant Oakleaf (OKL) and Hose in Hose (HIH) alleles, with HIH in coupling to S and HIH in repulsion to OAK. Phenotypes and genotypes of the different progeny classes are indicated with progeny numbers from each of six crosses shown. Crosses 1, 2 and 3 used one thrum male parent and crosses 4, 5 and 6 used a second thrum male parent. Each cross had a different pin mother. The number in each class (Totals) and numbers in each recombination category (Sum totals) are indicated. A total of 2075 progeny were characterized.
To determine the gene order between Oakleaf, the S locus and sepaloid, we used a three‐point cross with a pin sepaloid as the female parent and four different Oakleaf thrums carrying a recessive sepaloid allele in repulsion to the dominant S allele, which was in coupling to Oakleaf (Fig. 3); these crosses yielded 601 progeny, and pooled progeny numbers were used to define the gene order. Oakleaf thrums and sepaloid pins represent nonrecombinant parental genotypes. The six Oakleaf sepaloid pin and wild‐type thrum individuals represent single‐crossovers between Oakleaf and S. The progeny groups of wild‐type pins and Oakleaf sepaloid thrums comprised two individuals; the Oakleaf pin and sepaloid thrum progeny group contained a single individual. Based on these numbers, we assigned Oakleaf pin and sepaloid thrum as the double‐crossovers; this gave a gene order of Oakleaf–S locus–sepaloid. These data place Hose in Hose and sepaloid on the same side of the S locus, opposite Oakleaf.
Figure 3.
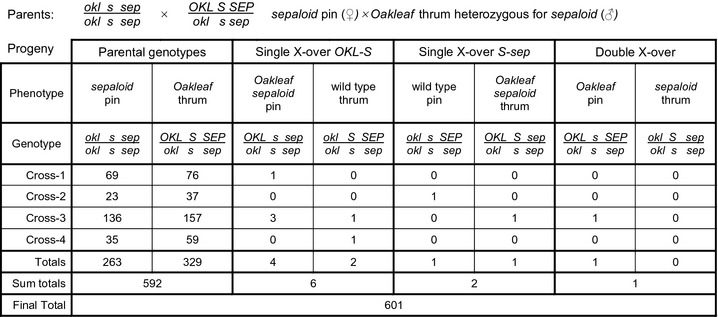
Three‐point cross to define gene order for the S locus, Primula vulgaris Oakleaf and sepaloid. Progeny analysis of a three‐point cross between pin female parents (s/s), homozygous for the wild‐type recessive alleles of oakleaf (okl) and sepaloid (sep), and thrum (S/s) male plants, heterozygous for the dominant Oakleaf (OKL) and recessive sepaloid (sep) alleles, with OAK in coupling with S and sep in coupling with s. Phenotypes and genotypes of different progeny classes are indicated with progeny numbers from each of four crosses shown. The number in each class (Totals) and numbers in each recombination category (Sum totals) are indicated. A total of 601 progeny were characterized.
We predicted the gene order from these crosses from the combined progeny number. However, for the determination of map distances, we analysed recombinants from individual heterozygous parents separately as the recombination frequency is genotype dependent. For the Oakleaf–S locus–Hose in Hose cross, we used five wild‐type pin plants and two Oakleaf, Hose in Hose thrum plants in six crosses, as described previously (Fig. 2). As recombination events are only evident in chromosomes from the heterozygous thrum parents, we combined mapping data into two groups arising from crosses involving thrum‐1 and thrum‐2 (Table 2; Fig. 2); recombination in the wild‐type pin parent has no impact on progeny class and can therefore be pooled. Meiotic recombination events in thrum‐1 yielded eight Oakleaf–S locus recombinants and six S locus–Hose in Hose recombinants. Meiotic recombination events in thrum‐2 yielded 20 Oakleaf–S locus recombinants and 12 S locus–Hose in Hose recombinants (Table 2). These data give map distances of 0.62–2.55 cM between Oakleaf and the S locus and 0.39–1.53 cM between the S locus and Hose in Hose for the two pools. In both cases, we observed negative crossover interference (Auger & Sheridan, 2001) with a higher than anticipated occurrence of double‐crossovers; recombination in thrum‐1 gave a coefficient of coincidence of 103.8 and in thrum‐2 of 8.6 to give negative interference values of −102.8 and −7.6, respectively.
Table 2.
Three‐point cross between the Primula vulgaris Oakleaf (OKL), S locus and Hose in Hose (HIH)
| Mapping OKL–S–HIH | ♀ parent | ♂ parent | Total progeny | OKL to S recombinants | Distance (cM) | S to HIH recombinants | Distance (cM) | Short homostyle | Distance (cM) |
|---|---|---|---|---|---|---|---|---|---|
| Cross 1 | Pin‐1 | Thrum‐1 | 413 | 3 | – | 2 | – | 0 | – |
| Cross 2 | Pin‐2 | Thrum‐1 | 430 | 2 | – | 0 | – | 0 | – |
| Cross 3 | Thrum‐1 | Pin‐3 | 448 | 3 | – | 3 | – | 0 | – |
| Totals from Cross 1–3 | 1291 | 8 | 0.62 | 6 | 0.39 | 0 | 0.00 | ||
| Cross 4 | Pin‐4 | Thrum‐2 | 500 | 6 | – | 5 | – | 0 | – |
| Cross 5 | Pin‐5 | Thrum‐2 | 116 | 3 | – | 6 | – | 1 | – |
| Cross 6 | Thrum‐2 | Pin‐4 | 167 | 11 | – | 1 | – | 0 | – |
| Totals from Cross 4–6 | 784 | 20 | 2.55 | 12 | 1.53 | 1 | 0.13 | ||
| Combined data | 2075 | 28 | 0.62–2.55 | 17 | 0.39–1.53 | 1 | 0.05 |
Similar analyses of the Oakleaf–S locus–sepaloid three‐point cross provided mapping data for the S locus. The four crosses that contributed to the determination of gene order (Fig. 3) used one pin sepaloid pollen recipient and four different heterozygous Oakleaf thrum plants carrying a recessive sepaloid allele as pollen donor. We calculated the map distances for each of the four crosses, which provided a range of 0.68–1.67 cM between Oakleaf and the S locus, and 0.67–1.64 cM between the S locus and sepaloid (Table 3). Again, we observed negative interference in Cross 3, with a coefficient of coincidence of 50.1, producing a negative interference of −49.1.
Table 3.
Three‐point cross between the Primula vulgaris Oakleaf (OKL), the S locus and sepaloid (sep)
| Mapping OKL–S–sep | ♀ parent | ♂ parent | Total progeny | OKL to S recombinants | Distance (cM) | S to sep recombinants | Distance (cM) |
|---|---|---|---|---|---|---|---|
| Cross 1 | Pin‐1 | Thrum‐1 | 146 | 1 | 0.68 | 0 | < 0.68 |
| Cross 2 | Pin‐1 | Thrum‐2 | 61 | 0 | <1.64 | 1 | 1.64 |
| Cross 3 | Pin‐1 | Thrum‐3 | 299 | 5 | 1.67 | 2 | 0.67 |
| Cross 4 | Pin‐1 | Thrum‐4 | 95 | 1 | 1.05 | 0 | < 1.05 |
| Combined data | 601 | 7 | 0.68–1.67 | 3 | 0.67–1.64 |
Integrated mapping using S locus recombinants and molecular markers
Analysis of recombination rates by PCR or RFLP analysis with the S locus‐linked markers (Table 1) was limited by the logistics of screening larger numbers of plants. We therefore capitalized on recombinants from Oakleaf–S locus–Hose in Hose three‐point crosses. DNA was extracted from 29 Oakleaf to S locus and S locus to Hose in Hose recombinant progeny (Fig. 2). Recombinants were examined by gel blot analysis using PvSLL1 and PvSLL2 to determine their location with respect to Oakleaf, Hose in Hose and S. If all progeny resulting from the recombination of Oakleaf or Hose in Hose also showed recombination for PvSLL1 and/or PvSLL2, this would place these markers outside Oakleaf or Hose in Hose with respect to S. If recombination of Oakleaf or Hose in Hose also leads to recombination of PvSLL1 and/or PvSLL2 in all cases, this would place these genes between S and the phenotypic markers.
Eighteen of the 25 Oakleaf–S recombinants and 11 of the 14 Hose in Hose–S recombinants (Fig. 2) were analysed by gel blot analysis. Data from 12 representative plants, four recombinants between the S locus and Hose in Hose (S–H) and eight from recombination between Oakleaf and the S locus (O–S), are shown in Fig. 4. The PvSLL1 allele in coupling with the s allele is represented by a 3.0‐kb RFLP (Fig. 4a); the thrum allele, in coupling to the S allele, is represented by a 2.8‐kb RFLP. None of the eight plants arising from recombination between Oakleaf and S show assortment for PvSLL1; all are thrum and heterozygous for 3.0‐kb pin and 2.8‐kb thrum alleles of PvSLL1. These data show that Oakleaf has recombined independently of PvSLL1; PvSLL1 is therefore not distal to Oakleaf with respect to the S locus. Three of the four progeny showing recombination between the S locus and Hose in Hose (plants 1, 4 and 7) also show that recombination of Hose in Hose does not affect PvSLL1. The two pin plants (plants 1 and 7) are homozygous for the 3.0‐kb pin allele of PvSLL1, and the thrum plant (plant 4) is heterozygous for the pin and thrum alleles of PvSLL1. PvSLL1 cannot therefore be outside Hose in Hose with respect to the S locus. Interestingly, plant 6 reveals a 2.5‐kb PvSLL1 allele in addition to the 3.0‐kb pin allele. This band either represents a deletion or a point mutation that affects the RFLP, or a recombination event between the S locus and PvSLL1 that affects the size of the RFLP. If this band represents a recombination event, this places PvSLL1 between the S locus and Hose in Hose. Subsequent analysis of the BAC contig spanning the S locus confirmed this location (see later).
Figure 4.
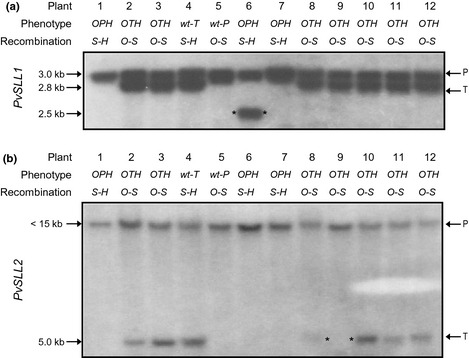
Mapping Primula vulgaris S locus‐linked genes onto three‐point cross recombinants. Twelve plants (1–12) obtained from the Oakleaf–S locus–Hose in Hose three‐point cross, that represent single‐crossover recombination events between either Oakleaf and the S locus or Hose in Hose and the S locus, are shown. The phenotypes of individuals (O, Oakleaf; P, pin; T, thrum; H, Hose in Hose; wt, wild‐type) are indicated, as well as the location of the crossover in each case in relation to the S locus (S) and the phenotypic markers Oakleaf (O) and Hose in Hose (H). (a) Autoradiograph of gel blot analysis of Hind III‐digested genomic DNA using PvSLL1 as probe. Pin (P)‐ and thrum (T)‐specific alleles of PvSLL1, as revealed by restriction fragment length polymorphisms (RFLPs), are indicated. An inconsistent RFLP obtained from plant 6 is identified by asterisks. (b) Autoradiograph of gel blot analysis of XbaI‐digested genomic DNA using PvSLL2 as probe. P‐ and T‐specific alleles of PvSLL1, as revealed by RFLPs, are indicated. The absence of a thrum‐specific RFLP in plant 9 is identified by asterisks.
Similarly, PvSLL2 is represented in this population by two alleles, a < 15‐kb band in coupling to the s allele, and a 5.0‐kb band in coupling with the S allele (Fig. 4b). None of the four plants showing recombination between S and Hose in Hose show assortment of PvSLL2 alleles. PvSLL2 cannot therefore be distal to Hose in Hose with respect to the S locus. Of the eight plants shown resulting from recombination between Oakleaf and S, seven show heterozygosity for the two PvSLL2 alleles. Recombination of Oakleaf from the s chromosome onto the S chromosome does not consistently bring with it the < 15‐kb pin allele of PvSLL2. This marker is therefore not distal to Oakleaf with respect to the S locus. However, one Oakleaf–Thrum–Hose in Hose recombinant, plant 9, is homozygous for the < 15‐kb pin allele of PvSLL2 and lacks the 5.0‐kb thrum allele. This individual represents a recombination event between PvSLL2 and the S locus, and places PvSLL2 between Oakleaf and the S locus. None of the other 17 plants analysed by Southern analysis (data not shown) showed recombination between either PvSLL1 and S or PvSLL2 and S. Only 28 plants showing recombination between Oakleaf and S were identified in 2075 progeny; of these, only one had recombined for PvSLL2. Based on these data, we assign a map distance between PvSLL2 and the S locus of 0.05 cM. Similarly, if the 2.5‐kb PvSLL1 RFLP in plant 6 (Fig. 4) arose by recombination, this would produce a map distance of 0.05 cM between PvSLL1 and the S locus (Table 4). It remains possible that this 2.5‐kb band represents a mutation rather than recombination; if this is the case, the map distance must be < 0.05 cM. As discussed below, the sequence of the BAC contig surrounding the S locus confirms that PvSLL1 is located between the S locus and Hose in Hose.
Table 4.
Recombinants for PvSLL1 and PvSLL2 mapped onto Primula vulgaris three‐point cross recombinants
| OKL–S–HIH three‐point cross | PvSLL1 | PvSLL2 |
|---|---|---|
| Total progeny | 2075 | 2075 |
| OKL to S single‐crossover events | 28 | – |
| Recombinants analysed by blot | 18 | – |
| S–PvSLL1 recombinants | 1a | – |
| Map distance (cM) | 0.05 | – |
| S to HiH single‐crossover events | – | 17 |
| Recombinants analysed by blot | – | 11 |
| S–PvSLL2 recombinants | – | 1 |
| Map distance (cM) | – | 0.05 |
HiH, Hose in Hose; OKL, Oakleaf; S, S locus.
Plant identified in Fig. 5(a) that gave an aberrant restriction fragment length polymorphism (RFLP) profile and may represent a recombinant.
Data from the three‐point crosses, and analysis of assortment of pin and thrum alleles for molecular markers, generated a map of the S locus (Fig. 5a) which includes the order and relative position of the mutants, Oakleaf (Webster, 2005; Cocker et al., 2015), Hose in Hose (PvGlo) (Li et al., 2010) and sepaloid (Li et al., 2008), alongside three molecular markers, PvSLL1 (Webster & Grant, 1990; Li et al., 2007), PvSLL2 (Li et al., 2007) and PvSLP1 (Manfield et al., 2005). PvSLP1 was identified as a RAPD marker specific to P. vulgaris Blue Jeans, and so we could not analyse its segregation in these crosses. However, BAC contig assembly and sequencing (see later) defined the location of PvSLP1 on the map (Fig. 5b).
Figure 5.
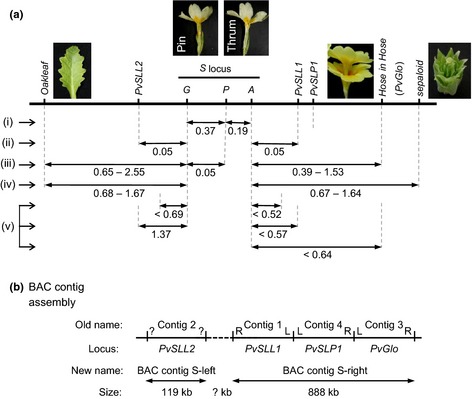
A genetic map of the Primula vulgaris S locus. (a) The relative positions of the mutants Oakleaf, Hose in Hose and sepaloid are indicated in relation to the S locus and its constituent genes G, P and A. The mapped locations of PvSLL1 and PVSLL2 and PvSLP1 and PvSLP2 are indicated. Two potential locations for PvSLP2 (light grey) are shown, as the precise map position is not defined. Map distances are shown in cM and were determined by: (i) Lewis & Jones (1993); (ii) data from Fig. 4 and Table 4; (iii) data from Fig. 2 and Table 2; (iv) data from Fig. 4 and Table 3; (v) data from Table 1. Images of phenotypic markers are included next to the relevant loci. (b) Summary of BAC contig assembly results. The resolved order and current assembly of the four BAC assembly contigs described previously (Li et al., 2011) are indicated (Old name), and their orientation is given by L and R, or ‘?’ if unknown. The S locus‐linked marker associated with each of the four previous BAC contigs is indicated. The revised BAC assembly contig name (New name) is indicated, together with the size estimated from the assembly. The region with no BAC coverage is shown as a dotted line.
Localization of the S locus by chromosome in situ hybridization
Previous cytogenetic studies have suggested that the S locus is located close to the centromere in some species of Primula (Darlington, 1931; Dowrick, 1956; Lewis & Jones, 1993). The availability of BAC clones from sequences flanking the S locus has enabled us to directly visualize the location. Initially, we used two overlapping BACs (BAC56H19 (red) and BAC81I19 (green)) (Fig. 6a–f) from the PvSLL2 side of the S locus (Fig. S1) for chromosome in situ hybridization on metaphase chromosomes. The 22 chromosomes were visualized with DAPI (Fig. 6a,d). A merged DAPI–fluorescence image (Fig. 6c,f) reveals the location of the S locus as centromeric on the largest metacentric chromosome pair. Data from two independent experiments are shown (Fig. 6a–f).
Figure 6.
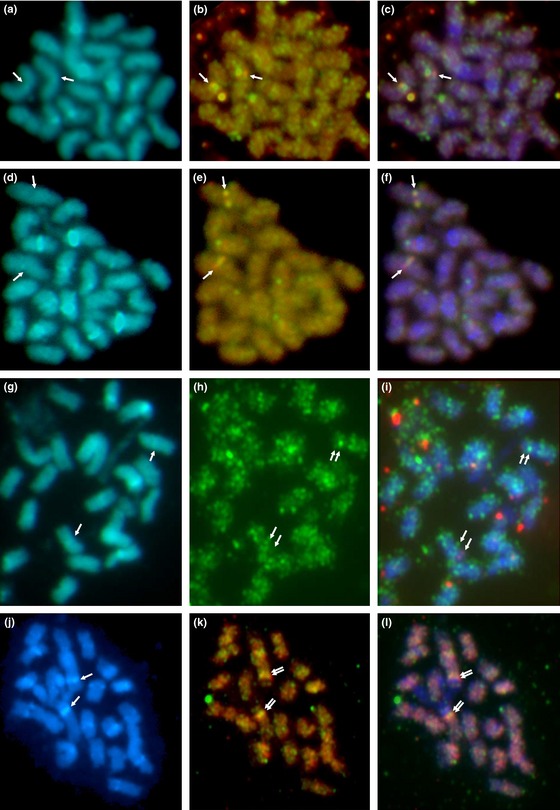
Chromosome in situ hybridization reveals the location of the Primula S locus. Hybridization to metaphase chromosomes of P. vulgaris (2n = 22; fluorescing blue with 4′,6‐diamidino‐2‐phenylindole, DAPI) using S locus‐linked BACs. (a–f) BAC56H19 (detected by red fluorescence) and BAC81I19 (green fluorescence). (g–i) BAC56H19 (red) and BAC13A4 (green) showing colocalization (yellow–orange when overlaid). (j–l) BAC13A4 (red) and BAC81B15 (green). (a, d, g, j) Chromosomes with intercalary DAPI‐positive bands on some chromosomes, and less diffuse labelling in the terminal region, suggesting differentiation of repeat content along the chromosome. (b, e, h, k) Hybridization with S locus‐linked BACs detected by red and green fluorescence; overlapping signals are yellow–orange or green–purple depending on the intensity of the fluorochromes. Arrows indicate: (a, d, g, j) centromeric regions; (b, c, e, f) superimposed fluorescence signals; (h, i, k, l) adjacent fluorescence signals. (c, f, i, l) Overlapping probe and DAPI fluorescence images. In (h, i, k, l), the green signal is proximal to the centromere.
A second analysis using BAC56H19 (red) and BAC13A4 (green), which map to either side of the S locus (Fig. S1), is shown in Fig. 6g–i. The signal obtained from BAC13A4 is quite diffuse (Fig. 6h); however, as indicated by the arrows (Fig. 6i), these probes colocalize. Comparison with DAPI‐stained chromosomes (Fig. 6g) reveals that BAC56H19 is distal and BAC13A4 is proximal to the centromere. We also analysed BAC13A4 (red) and BAC81B15 (green), which are both on the PvGlo side of the S locus (Fig. S1). These data (Fig. 6j–l) show that BAC81B15 is closer than BAC13A4 to the centromere, and support the observations with BAC56H19 and BAC13A4 (Fig 6i) that the PvGlo side of the S locus is proximal to the centromere.
Assembly and sequencing of BAC contigs surrounding the S locus
We have described previously the construction of two P. vulgaris BAC libraries and their screening using four S locus‐linked probes, PvSLL1 and PvSLL2 (Li et al., 2007), PvSLP1 (Manfield et al., 2005) and PvGlo (Li et al., 2010), and the assembly of four BAC contigs around these probes (Li et al., 2011). We have extended and completed this assembly by further BAC‐end screening and walking. These analyses identified several new BACs which clarified existing ambiguities in the contig assemblies, and provided new sequence to bridge gaps in the earlier assembly. The order and integration of the previous four contigs (Li et al., 2011) is shown in Fig. 5(b). Contig 3 sits to the right of the map; our new data show that the left‐hand end of contig 3 overlaps with the right‐hand end of contig 4, and the left‐hand end of contig 4 now links to the left‐hand end of contig 1 (Fig. 5b). The new BAC clones which facilitated the assembly and enabled the integration of three contigs into one are shown in bold and identified by plus signs in Fig. S1. This integrated contig covers 888 kb and was renamed Contig S‐right (Fig. 5b). This contig links, and provides a gene order for, PvSLL1, PvSLP1 and PvGlo (Fig. S1). We have not been able to join contig 3 (Li et al., 2011), renamed Contig S‐left, to the larger assembly (Fig. 5b). BAC‐end sequences from the right‐hand end of contig 1 and both ends of contig 2 are highly repetitive and identified multiple BACs in subsequent screening rounds. We have aligned the assembled BAC contigs to our genetic map based on the location of PvSLL1, PvSLL2, PvSLP1 and PvGlo to integrate genetic and physical maps (Fig. 5a). These data are summarized in Fig. 5(b) and expanded in detail in Fig. S1.
The sequencing of 10 individual BACs from across the contig assemblies enabled us to assemble sequence contigs associated with the BAC contigs. We also sequenced two pools of BACs. Five BACs sequenced individually were also included in the pools (Fig. S1). Pool 1 contained 19 BACs from contigs 1 and 2 (Li et al., 2011), and pool 2 contained 18 BACs from contigs 3 and 4 (Li et al., 2011); the relative positions of these contigs are shown in Fig. 5 and are fully expanded in Fig. S1. The assembly of individually sequenced BACs, together with sequence from pooled BACs, generated four sets of sequence contigs. Group‐A: overlapping DNA sequence anchored to known BACs from Contig S‐left yielded 17 contigs covering 119 kb (Fig. S1). Group‐B: overlapping DNA sequence anchored to BACs from Contig S‐right yielded 58 contigs covering 888 kb. Group‐C: unanchored sequence contigs from pool 1 BACs comprised 56 contigs covering 325 kb. Group‐D: unanchored sequence contigs from pool 2 comprised 99 contigs covering 178 kb. In total, 1.5 Mb of sequence flanking the S locus was assembled; 1 Mb is anchored to specific BACs (Fig. S1). It was not possible to generate a contiguous assembly across the entire region, and this is reflected by the assembly gaps in Group‐A (17 contigs, 16 gaps) and Group‐B (58 contigs, 57 gaps), and the unanchored contigs in Group‐C and Group‐D. The 17 contigs in Group‐A (Contig S‐left) and 58 contigs in Group‐B (Contig S‐right) were ordered relative to the BAC tiling path (Fig. S1); contigs residing between BAC‐ends could not be oriented or ordered relative to each other. Although it is not possible to assign an unambiguous order for all intervening contigs from specific BACs, a high level order of these contigs is shown in Table S1. Contigs containing BAC‐ends are highlighted in bold. The order of intervening contigs remains to be confirmed.
Gene annotation within the BAC contigs flanking the S locus
We undertook de novo annotation of BAC sequence contigs (Fig. S1; Table S1). The number of predicted genes and gene fragments in each of the four BAC sequence groups is summarized in Table S2; 266 potential genes or gene fragments were identified. Of these sequences, 119 identified known proteins in BlastX searches; the remaining 147 predicted gene fragments gave no database similarities. We searched the Arabidopsis TAIR10 database and, after removal of duplicates caused by gene models on the same contig matching the same Arabidopsis gene, or the same Arabidopsis locus matching gene predictions on neighbouring contigs, we found 82 related Arabidopsis genes.
Annotation data for genes associated with Contig S‐left and Contig S‐right are presented in Table S2. Confirmation of gene order on internal BAC contigs remains to be confirmed. Within these contigs, we identified PvSLL1 on S_locus_groupB_ctg13 and PvGlo on S_locus_groupB_ctg58; we also located PvSLP1 to S_locus_groupB_ctg36. These data confirm the order of S‐linked markers on BAC contig assemblies (Fig. 5) and unequivocally demonstrate the order as, S locus–PvSLL1–PvSLP1–PvGlo, within 888 kb of assembled sequence. We also identified PvSLL2 on contig S_locus_groupA_ctg9 within Contig S‐left. The full annotation and orientation of predicted gene models in the assembled 1.5‐Mb sequence will require manual curation and integration with the P. vulgaris genome sequence.
Discussion
Primula provides one of the earliest examples, after Mendel's peas (Bateson, 1902; Bhattacharyya et al., 1990), of a model for genetic analysis. Bateson & Gregory (1905) revealed the dominance relationship of pin and thrum flowers which led to the identification of S‐linked phenotypes in P. sinensis (Gregory et al., 1923; De Winton & Haldane, 1935). This work provided one of the first examples of linkage in plants and one of the first linkage maps (Gregory, 1911; Bridges, 1914; Altenburg, 1916). Yoshida et al. (2011) recently took a different approach and used QTL analysis to develop a genome‐wide linkage map that identified S locus‐linked markers in P. sieboldii.
We pursued a classical approach to generate a genetic map for P. vulgaris and used Hose in Hose (Ernst, 1942; Webster & Grant, 1990; Li et al., 2010) with two other S locus‐linked mutant phenotypes, sepaloid (Webster, 2005; Li et al., 2008) and Oakleaf (Webster, 2005; Cocker et al., 2015), together with the heterostyly phenotypes, to generate a linkage map. We have not found phenotypes corresponding to the P. sinensis S‐linked loci, magenta, red stigma, red leaf back or double (De Winton, 1928; De Winton & Haldane, 1935), in P. vulgaris.
Three‐point cross analysis enabled us to establish a gene order of Oakleaf–S locus–Hose in Hose, with sepaloid predicted to be on the same side as Hose in Hose (Figs 3, 4). Map distances for Oakleaf to S range from 0.62 to 2.55 cM, and for S to Hose in Hose from 0.39 to 1.53 cM. This range reflects different recombination rates in different individual parents. Given the limited number of phenotypic markers, we were fortunate to find that Oakleaf and Hose in Hose mapped to either side of the S locus (Fig. 2). The three‐point cross involving sepaloid involved fewer progeny (Fig. 3); assignment of the double recombinants was therefore less obvious than for the Hose in Hose cross. However, our data indicate that sepaloid is on the same side of the S locus as Hose in Hose, with a gene order of Oakleaf–S locus–sepaloid; the map distance ranges from 0.68 to 1.67 cM for Oakleaf to S, and from 0.67 to 1.64 cM for S to sepaloid. These combined data enabled us to establish a genetic map with phenotypic markers flanking the S locus (Fig. 5). In both three‐point crosses, we observed negative interference, revealed by a higher than expected rate of double recombinants. It is interesting to note that de Winton and coworkers reported differences in male and female recombination rates in P. sinensis (Gregory et al., 1923; De Winton & Haldane, 1933, 1935). Like ours, their data also came from different crosses, and it may be that different recombination rates for male and female parents simply reflect differences between individuals, as seen in our data (Table 2), rather than gender differences.
Our previous studies defined S locus linkage of four sequences (Fig. 1) by RFLP and PCR analysis using modest numbers of plants (Manfield et al., 2005; Li et al., 2008, 2010). We also previously defined Hose in Hose as a mutation in PvGlo (Li et al., 2010). Combined data from these different studies are integrated in Fig. 1 and Table 1. We therefore took advantage of the recombinants from large three‐point crosses segregating for Oakleaf, Hose in Hose and heterostyly to increase the resolution of these map distances and position the markers with respect to phenotypic markers. We were again fortunate that our sequence markers mapped to either side of the S locus (Fig. 5). The RAPD marker PvSLP1 was identified in P. vulgaris Blue Jeans, but is not detectable in all cultivars. Therefore, we could not map PvSLP1 in three‐point cross progeny. However, sequence analysis of the S locus BAC contigs provides an unambiguous location for PvSLP1 between PvSLL1 and PvGlo (Fig. S1; Tables S1, S2), as summarized in Fig. 5.
The appearance of a self‐fertile short homostyle in a mapping cross (Fig. 2) was a surprise. Although Ernst (1925, 1933, 1936b) identified and characterized homostyles in P. viscose and P. hortensis, De Winton & Haldane (1935) did not find any homostyles in 18 000 plants during their genetic studies of P. sinensis. Dowrick's analysis of diploid P. obconica did not identify any homostyles in 5000 plants (Dowrick, 1956), and neither did Ernst (1928b) in 8000 plants studied. Homostyles are therefore very rare. They are known in P. vulgaris and were noted by Darwin (1877), and have since been studied in populations in Somerset (Bodmer, 1960; Piper et al., 1984; Curtis & Curtis, 1985) and the Chilterns (Crosby, 1940, 1948). Both of these populations contain only long homostyles. Ernst (1928b) originally considered mutations to be responsible for the breakdown of heterostyly, but Dowrick (1956) and Lewis & Jones (1993) interpreted homostyles as resulting from recombination within the S locus that disrupts the coupling of dominant alleles within the S locus, and also leads to the breakdown of the self‐incompatibility system to generate self‐fertile homostyles.
It is possible that low levels of recombination within the S locus could reflect a genome rearrangement between the two alleles, for example an inversion (De Winton & Haldane, 1935; Mather, 1950), or may be a consequence of proximity to the centromere (Dowrick, 1956). Evidence from double reduction (Darlington, 1931) in tetraploid plants suggests that the S locus is located close to the centromere in P. sinensis (De Winton & Haldane, 1935) and P. obconica (Dowrick, 1956). We have shown directly by double‐labelling chromosome in situ hybridization using overlapping BACs (Fig. 6) that the S locus in P. vulgaris is located close to the centromere of the largest metacentric chromosome. This direct visualization confirms the prediction made over 80 yr ago by cytogenetic analysis (Darlington, 1931). Darlington (1931) also speculated that the chromosome carrying the S locus in P. sinensis might be the largest, as it carried the greatest number of S‐linked loci; again, our chromosome in situ data confirm his prediction. Our in situ analyses also orientate the S locus map with respect to the centromere, and show that the PvGlo side of the S locus is proximal to the centromere (Fig. 6).
The assembly of over 1 Mb of sequence anchored to BACs surrounding the S locus (Fig. S1; Table S1) has identified at least 82 new S locus‐linked genes (Table S2); a further 500 kb of S‐linked sequences remain to be anchored, and this will be facilitated with a more complete Primula genome assembly. Although we cannot yet predict the size of the gap between the two contigs, our data suggest that PvSLL2 and PvGlo are at least 1.5 Mb apart. PvGlo is at least 888 kb from the S locus (Fig. S1; Table S2), and Hose in Hose (PvGlo) is between 0.39 and 1.53 cM from the S locus; this suggests a relationship of 1 cM to 580–2277 kb.
It remains to be seen whether the assembled BAC contigs include the key S locus genes or whether these reside within the gap between contigs. PvSLL2 is located within a contig flanked by 66.6 and 27.1 kb on either side (Table S1). None of the predicted genes in this contig are obvious candidates for Oakleaf (Webster, 2005; Cocker et al., 2015), and none of the genes upregulated in Oakleaf (Cocker et al., 2015) are found in this contig. This is perhaps not surprising as PvSLL2 is only 0.05 cM from the S locus, and Oakleaf is between 0.56 and 2.55 cM away (Fig. 5). The identification of Oakleaf will require the analysis of the full genome sequences and demonstration of linkage between candidate genes and the S locus. The de novo annotation of the BAC assembly identified 266 gene models, 82 of which correspond to known Arabidopsis genes (Table S2); ongoing annotation of the P. vulgaris genome and transcript‐driven gene model definition will resolve the number of genes. We have not found sequences corresponding to those identified in F. esculentum and L. grandiflorum in our gene annotations (Table S2), and this possibly reflects the polyphyletic origins for floral heteromorphy.
Primula has a long history as a model for studies on floral heteromorphy, built on Darwin's landmark studies (Darwin, 1862) and the various historical observations on heterostyly before his work (van Dijk, 1943; P. M. Gilmartin, unpublished); this plant has also played a fundamental role in the establishment of Mendelian genetic analysis through the attention of Bateson, Haldane, Bridges (Bateson & Gregory, 1905; Bridges, 1914; Haldane, 1933) and others in the early 1900s. One hundred years later, we have generated an integrated genetic and physical map of the P. vulgaris S locus, localized and orientated it by in situ chromosome hybridization, and identified 82 new S locus‐linked genes. Based on this study, and our ongoing annotation of the P. vulgaris genome, we are poised to identify the S locus genes which control floral heteromorphy in Primula.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Map of the S locus showing assembly of the BAC contig flanking the regions.
Methods S1 Bioinformatic supplemental methods.
Table S1 Sequence contig assemblies derived from S locus‐associated BAC sequencing
Table S2 Annotation of sequence contig assemblies derived from S locus‐associated BACs
Acknowledgements
We thank Martin Lappage, Mike Hughes and Pam Wells for horticultural support; colleagues at The Genome Analysis Centre (TGAC) Norwich for Illumina sequencing, and Anil Thanki for support with the TGAC browser; Centre for Genomic Research (CGR) Liverpool for 454 sequencing, and Neil Hall for help and advice; Biotechnology and Biological Sciences Research Council (BBSRC) (BB/H019278/2) for financial support, and University of Leeds, Durham University and Gatsby Foundation for early‐stage funding. We are grateful to the University of East Anglia (UEA) for support and the John Innes Centre (JIC) for hosting P.M.G.'s laboratory under the UEA–JIC Norwich Research Park collaboration.
References
- Altenburg E. 1916. Linkage in Primula sinensis . Genetics 1: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou A, Khosravi D, Tamari F, Shore JS. 2003. Characterization and localization of short‐specific polygalacturonase in distylous Turnera subulata (Turneraceae). American Journal of Botany 90: 675–682. [DOI] [PubMed] [Google Scholar]
- Auger DL, Sheridan WF. 2001. Negative crossover interference in maize translocation heterozygotes. Genetics 159: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH. 1990. The evolution and adaptive significance of heterostyly. Trends in Ecology & Evolution 5: 144–148. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. 2010. Darwin's legacy: the forms, function and sexual diversity of flowers. Philosophical Transactions of the Royal Society of London. Series B, Biological Science 365: 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. 1902. Mendel's principles of heredity. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Bateson W, Gregory RP. 1905. On the inheritance of heterostylism in Primula. Proceedings of the Royal Society of London B Series 76: 581–586. [Google Scholar]
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C. 1990. The wrinkled seed character of pea described by Mendel is caused by a transposon‐like insertion in a gene encoding starch branching enzyme. Cell 60: 115–122. [DOI] [PubMed] [Google Scholar]
- Bodmer WF. 1960. The genetics of homostyly in populations of Primula vulgaris . Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 242: 517–549. [Google Scholar]
- Bridges CB. 1914. The chromosome hypothesis of linkage applied to cases in sweetpeas and Primula . The American Naturalist 48: 524–534. [Google Scholar]
- Cocker JM, Webster MA, Li J, Wright J, Kaithakottil G, Swarbreck D, Gilmartin PM. 2015. Oakleaf: an S locus‐linked mutation of Primula vulgaris that affects leaf and flower development. New Phytologist 208: 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby JL. 1940. High proportions of homostyle plants in populations of Primula vulgaris . Nature 145: 672–673. [Google Scholar]
- Crosby JL. 1948. Population genetics in the genus Primula. PhD thesis, University of Cambridge, Cambridge, UK. [Google Scholar]
- Curtis J, Curtis CF. 1985. Homostyle primroses re‐visited. 1. Variation in time and space. Heredity 54: 227–234. [Google Scholar]
- Darlington CD. 1931. Meiosis in diploid and tetraploid Primula sinensis . Journal of Genetics 24: 65–95. [Google Scholar]
- Darwin CR. 1862. On the two forms or dimorphic condition in the species of Primula, and on their remarkable sexual relations. Journal of the Proceedings of the Linnean Society, Botany 6: 77–96. [Google Scholar]
- Darwin CR. 1863. On the existence of two forms, and on their reciprocal sexual relation, in several species of the genus Linum . Journal of the Proceedings of the Linnean Society, Botany 7: 69–83. [Google Scholar]
- Darwin CR. 1877. The different forms of flowers on plants of the same species. London, UK: John Murray. [Google Scholar]
- De Winton D. 1928. Inheritance in Primula sinensis. The Fourth Primula Conference 1928 London, UK: Ballantyne & Co. Ltd, 84–90. [Google Scholar]
- De Winton D, Haldane JBS. 1933. The genetics of Primula sinensis. II. Segregation and interaction of factors in the diploid. Journal of Genetics 27: 1–44. [Google Scholar]
- De Winton D, Haldane JBS. 1935. The genetics of Primula sinensis. III. Linkage in the diploid. Journal of Genetics 31: 67–100. [Google Scholar]
- van Dijk W. 1943. La decóuverte de l'hétérostylie chez Primula par Ch. de l'Écluse et P. Reneaulme. Nedelandsch Kruidkundig Archief 53: 81–85. [Google Scholar]
- Dowrick VPJ. 1956. Heterostyly and homostyly in Primula obconica . Heredity 10: 219–236. [Google Scholar]
- Ernst A. 1925. Genetische Studien uber Heterostylie bei Primula . Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 1: 13–62. [PubMed] [Google Scholar]
- Ernst A. 1928a. Genetische Studien über Calycanthemie bei Primula . Vierteljahrsschrift der Naturforschenden Gesellschaft in Zurich 15: 665–704. [Google Scholar]
- Ernst A. 1928b. Zur Vererbung der morphologischen Heterostylie merkmale. Berichte der Deutschen Botanischen Gesellschaft 46: 573–588. [Google Scholar]
- Ernst A. 1933. Weitere Untersuchungen zur phananalyse zum fertilitatsproblem und zur genetik heterostyler Primeln. I. Primula viscose . Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 8: 1–215. [Google Scholar]
- Ernst A. 1936a. Heterostylie‐Forschung Versuche zur genetischen analyse eines organisations und ‘Anpassungs’ merkmales. Zeitschrift für Induktive Abstammungs und Vererbungslehre 71: 156–230. [Google Scholar]
- Ernst A. 1936b. Weitere untersuchungen zur Phänanalyse zum Fertilitätsproblem und zur Genetik heterostyler Primeln. II. Primula hortensis . Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 11: 1–280. [Google Scholar]
- Ernst A. 1942. Vererbung durch labile gene. Archive der Julius Klaus Stiftung für Vererbungsforschung Sozialanthropologie und Rassenhygiene 17: 1–567. [PubMed] [Google Scholar]
- Ganders FR. 1979. The biology of heterostyly. New Zealand Journal of Botany 17: 607–635. [Google Scholar]
- Garber R, Quisenberry KS. 1927. Self‐fertilization in buckwheat. Journal of Agricultural Research 34: 185–190. [Google Scholar]
- Gerard J. 1597. The herball or generall historie of plantes. London, UK: John Norton. [Google Scholar]
- Gregory RP. 1911. Experiments with Primula sinensis . Journal of Genetics 1: 73–132. [Google Scholar]
- Gregory RP, De Winton D, Bateson MA. 1923. Genetics of Primula sinensis . Journal of Genetics 13: 219–253. [Google Scholar]
- Haldane JBS. 1933. Two new allelomorphs for heterostylism in primula. The American Naturalist 67: 559–560. [Google Scholar]
- Heslop Harrison JS, Schwarzacher T. 2002. In situ hybridisation to plants chromosomes and DNA fibres In: Gilmartin PM, Bowler CP, eds. Molecular plant biology. Oxford, UK: Oxford University Press, 159–178. [Google Scholar]
- Konishi T, Iwata H, Yashiro K, Tsumura Y, Ohsawa R, Yasui Y, Ohnishi O. 2006. Development and characterization of microsatellite markers for common buckwheat. Breeding Science 56: 277–285. [Google Scholar]
- Kurian V, Richards AJ. 1997. A new recombinant in the heteromorphy ‘S’ supergene in Primula . Heredity 78: 383–390. [Google Scholar]
- Labonne JDJ, Shore JS. 2011. Positional cloning of the s haplotype determining the floral and incompatibility phenotype of the long‐styled morph of distylous Turnera subulata . Molecular Genetics and Genomics 285: 101–111. [DOI] [PubMed] [Google Scholar]
- Labonne JDJ, Tamari F, Shore JS. 2010. Characterization of X‐ray‐generated floral mutants carrying deletions at the S locus of distylous Turnera subulata . Heredity 105: 235–243. [DOI] [PubMed] [Google Scholar]
- Labonne JDJ, Vaisman A, Shore JS. 2008. Construction of a first genetic map of distylous Turnera and a fine‐scale map of the S locus region. Genome 51: 471–478. [DOI] [PubMed] [Google Scholar]
- Labonne JDJ, Goultiaeva A, Shore JS. 2009. High‐resolution mapping of the S locus in Turnera leads to the discovery of three genes tightly associated with the S alleles. Molecular Genetics and Genomics 281: 673–685. [DOI] [PubMed] [Google Scholar]
- Lewis D, Jones DA. 1993. The genetics of heterostyly In: Barrett SCH, ed. Evolution and function of heterostyly. Berlin, Germany: Springer Verlag, 129–150. [Google Scholar]
- Li J, Dudas B, Webster MA, Cook HE, Davies BH, Gilmartin PM. 2010. Hose in Hose, an S locus‐linked mutant of Primula vulgaris is caused by an unstable mutation at the Globosa locus. Proceedings of the National Academy of Sciences, USA 107: 5664–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Webster M, Dudas B, Cook H, Manfield I, Davies B, Gilmartin PM. 2008. The S locus‐linked Primula homeotic mutant sepaloid shows characteristics of a B‐function mutant but does not result from mutation in a B‐function gene. Plant Journal 56: 1–12. [DOI] [PubMed] [Google Scholar]
- Li J, Webster MA, Furuya M, Gilmartin PM. 2007. Identification and characterization of pin and thrum alleles of two genes that co‐segregate with the Primula S locus. Plant Journal 51: 18–31. [DOI] [PubMed] [Google Scholar]
- Li J, Webster MA, Smith MC, Gilmartin PM. 2011. Floral heteromorphy in Primula vulgaris: progress towards isolation and characterization of the S locus. Annals of Botany 108: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Pavlov VK, Li JH, Cook HE, Hummel F, Gilmartin PM. 2005. Molecular characterization of DNA sequences from the Primula vulgaris S locus. Journal of Experimental Botany 56: 1177–1188. [DOI] [PubMed] [Google Scholar]
- Mather K. 1950. The genetical architecture of heterostyly in Primula sinensis . Evolution 4: 340–352. [Google Scholar]
- McCubbin AG, Lee C, Hetrick A. 2006. Identification of genes showing differential expression between morphs in developing flowers of Primula vulgaris . Sexual Plant Reproduction 19: 63–72. [Google Scholar]
- Pellow C. 1928. Report for the year 1928. Merton, UK: The John Innes Horticultural Institution. [Google Scholar]
- Piper JG, Charlesworth B, Charlesworth D. 1984. A high‐rate of self‐fertilization and increased seed fertility of homostyle primroses. Nature 310: 50–51. [Google Scholar]
- Richards AJ. 1997. Plant breeding systems, 2nd edn London, UK: Chapman & Hall. [Google Scholar]
- Shore JS, Barrett SCH. 1985. The genetics of distyly and homostyly In Turnera ulmifolia L (Turneraceae). Heredity 55: 167–174. [Google Scholar]
- Ushijima K, Nakano R, Bando M, Shigezane Y, Ikeda K, Namba Y, Kume S, Kitabata T, Mori H, Kubo Y. 2012. Isolation of the floral morph‐related genes in heterostylous flax (Linum grandiflorum): the genetic polymorphism and the transcriptional and post‐transcriptional regulations of the S locus. Plant Journal 69: 317–331. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Scarth R, Campbell C. 2005. S‐h and S‐c‐two complementary dominant genes that control self‐compatibility in buckwheat. Crop Science 45: 1229–1234. [Google Scholar]
- Webster MA 2005. Floral morphogenesis in Primula: inheritance of mutant phenotypes, heteromorphy, and linkage analysis. PhD thesis, University of Leeds, Leeds, UK. [Google Scholar]
- Webster MA, Gilmartin PM. 2003. A comparison of early floral ontogeny in wild‐type and floral homeotic mutant phenotypes of Primula . Planta 216: 903–917. [DOI] [PubMed] [Google Scholar]
- Webster MA, Gilmartin PM. 2006. Analysis of late stage flower development in Primula vulgaris reveals novel differences in cell morphology and temporal aspects of floral heteromorphy. New Phytologist 171: 591–603. [DOI] [PubMed] [Google Scholar]
- Webster MA, Grant CJ. 1990. The inheritance of calyx morph variants in Primula vulgaris (Huds). Heredity 64: 121–124. [Google Scholar]
- Yasui Y, Mori M, Aii J, Abe T, Matsumoto D, Sato S, Hayashi Y, Ohnishi O, Ota T. 2012. S‐LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short‐styled buckwheat plants that exhibit heteromorphic self‐incompatibility. PLoS ONE 7: e31264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y, Mori M, Matsumoto D, Ohnishi O, Campbell CG, Ota T. 2008. Construction of a BAC library for buckwheat genome research – an application to positional cloning of agriculturally valuable traits. Genes & Genetic Systems 83: 393–401. [DOI] [PubMed] [Google Scholar]
- Yasui Y, Wang YJ, Ohnishi O, Campbell CG. 2004. Amplified fragment length polymorphism linkage analysis of common buckwheat (Fagopyrum esculentum) and its wild self‐pollinated relative Fagopyrum homotropicum . Genome 47: 345–351. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Ueno S, Honjo M, Kitamoto N, Nagai M, Washitani I, Tsumura Y, Yasui Y, Ohsawa R. 2011. QTL analysis of heterostyly in Primula sieboldii and its application for morph identification in wild populations. Annals of Botany 108: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Map of the S locus showing assembly of the BAC contig flanking the regions.
Methods S1 Bioinformatic supplemental methods.
Table S1 Sequence contig assemblies derived from S locus‐associated BAC sequencing
Table S2 Annotation of sequence contig assemblies derived from S locus‐associated BACs


