Abstract
Neurogenesis initiates during early development and it continues through later developmental stages and in adult animals to enable expansion, remodeling, and homeostasis of the nervous system. The generation of nerve cells has been analyzed in detail in few bilaterian model organisms, leaving open many questions about the evolution of this process. As the sister group to bilaterians, cnidarians occupy an informative phylogenetic position to address the early evolution of cellular and molecular aspects of neurogenesis and to understand common principles of neural development. Here we review studies in several cnidarian model systems that have revealed significant similarities and interesting differences compared to neurogenesis in bilaterian species, and between different cnidarian taxa. Cnidarian neurogenesis is currently best understood in the sea anemone Nematostella vectensis, where it includes epithelial neural progenitor cells that express transcription factors of the soxB and atonal families. Notch signaling regulates the number of these neural progenitor cells, achaete‐scute and dmrt genes are required for their further development and Wnt and BMP signaling appear to be involved in the patterning of the nervous system. In contrast to many vertebrates and Drosophila, cnidarians have a high capacity to generate neurons throughout their lifetime and during regeneration. Utilizing this feature of cnidarian biology will likely allow gaining new insights into the similarities and differences of embryonic and regenerative neurogenesis. The use of different cnidarian model systems and their expanding experimental toolkits will thus continue to provide a better understanding of evolutionary and developmental aspects of nervous system formation. WIREs Dev Biol 2017, 6:e257. doi: 10.1002/wdev.257
This article is categorized under:
-
1
Gene Expression and Transcriptional Hierarchies > Cellular Differentiation
-
2
Signaling Pathways > Cell Fate Signaling
-
3
Comparative Development and Evolution > Organ System Comparisons Between Species
INTRODUCTION
Cnidarians are an early offshoot in the evolution of animals, having separated from the lineage that led to the emergence of bilaterians more than 600 million years ago.1 Their importance for understanding the evolution of nervous systems has long been recognized, based both on their phylogenetic position as an outgroup to bilaterians (Figure 1) and on the structure of their nervous system, which is predominantly organized as nerve nets. The cnidarians comprise two major clades, the anthozoans and the medusozoans (Figure 1),2, 3 which separated before 550 million years ago and thus not long after the cnidarian lineage separated from all other animals.4 Two distinct body forms can be found in cnidarians: sessile polyps, which are tube‐shaped animals with a single terminal body opening (called the mouth) surrounded by prey‐catching tentacles; and free‐swimming medusae, which swim by rhythmic contractions of their bell. Polyps are present in both cnidarian clades, while medusae are only present in the medusozoans (Figure 1), suggesting that a polyp stage was present in the last common ancestor of cnidarians, whereas parsimony favors the evolution of the medusa stage after the separation of the two clades. The nervous system of polyps can in first approximation be described as a nerve net with more or less pronounced regionalisation, including in some taxa instances of local condensations of neurites (in the form of nerve cords or nerve rings), for example in the oral region, or along the internal musculature‐bearing mesenteries.5, 6, 7 Medusae often possess sensory organs, in particular eyes and gravity sensors (lacking in polyps), associated with a considerable degree of centralized signal processing, best understood in hydrozoan medusae where it involves two peripheral and parallel nerve rings.8, 9, 10, 11 These sense organs and CNS‐like nervous system elements offer an excellent opportunity to study the independent evolution of advanced signal receiving and integrating structures and how they control the behavior and the unique mode of locomotion of jellyfish (see also Boxes 1 and 2).
Figure 1.
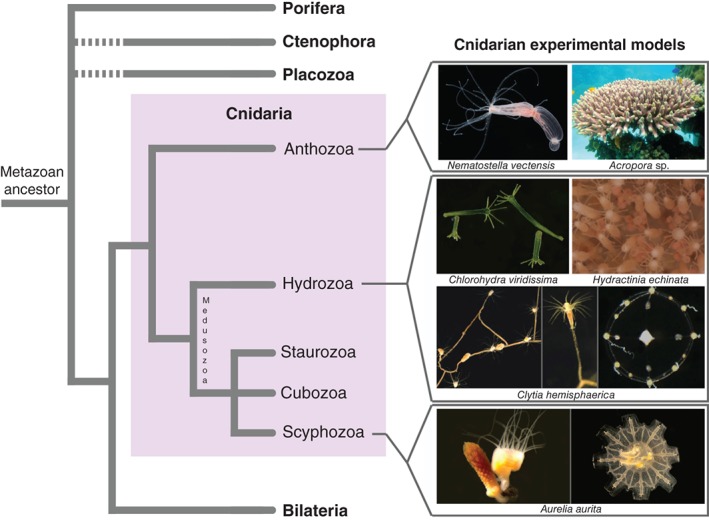
Position of cnidarians in the animal tree of life and phylogenetic diversity within the phylum, with a selection of experimental models used for developmental biology studies. Photo credits: Nematostella vectensis: Chiara Sinigaglia; Clytia hemisphaerica (medusa): Michaël Manuel; Acropora sp.: www.aquaportail.com; Chlorohydra viridissima and Aurelia aurita: Thomas Condamine; Hydractinia echinata: Uri Frank; C. hemisphaerica (polyps): Muriel Jager.
BOX 1. SENSE ORGANS IN CNIDARIANS.
In the medusae of scyphozoans and cubozoans, the multifunctional neuro‐sensory organ known as the rhopalia (which is a highly modified tentacle) contains at least seven groups of sensory cells arranged in complex bilateral pattern.150 The rhopalia nervous system is primarily ectodermal and its development is a highly ordered process.151 Rhopalia are involved in gravity sensing, chemoreception, and in some cases, photoreception. Hydrozoan medusae do not have rhopalia; they either lack sensory organs or possess isolated statocysts (equilibration organs) and in some instances, simple eyes (e.g., in Cladonema).
Apical organs in anthozoan planulae are characterized by a tuft of long cilia, which are thought to have a chemo‐ and/or mechanosensory function that might be required for the transition from planula to polyp stage.36, 140 The tuft region, however, does not contain neurons6, 152 and apical tuft formation in Nematostella is not affected by knockdown of NvSoxB(2) or NvAth‐like. 79, 89 Developmentally, there is thus no evidence that the cells of the apical tuft region are part of the nervous system.
BOX 2. NEUROGENESIS IN MEDUSAE.
In hydrozoan medusae, nematogenesis appears restricted to the manubrium and the tentacle bulbs.153 Tentacle bulbs are basal swellings of the tentacles, attached to the medusa bell periphery. Stem cell marker genes are expressed in two restricted symmetrical areas at the proximal extremity of the tentacle bulb ectoderm (presumably populations of interstitial stem cells) in Clytia hemisphaerica 76, 153 and in Podocoryne carnea. 154 There is some degree of spatial sorting of nematogenesis stages along the tentacle bulb axis, reflected by staggered expression of nematogenesis genes associated with different stages from stem cells to early differentiation and maturation (the latter restricted to the distal third of the bulb).24, 77, 153 Therefore, the tentacle bulb ectoderm behaves as a cellular conveyor belt,155, 156 which can facilitate experimental approaches to understand the temporal progression of nematocyte development. Formation of sensory and ganglion cells has not been described in hydrozoan medusae.
In cnidarians, the nerve cell concept embraces three different but related classes of cells. Sensory (or sensory‐motor) cells generally have an elongated cell body, and always bear an apical cilium that emerges at the body surface. Ganglion cells have their cell body located in a deep, basi‐epithelial position; they are often considered equivalent to interneurons, but they can also synapse on muscle cells and nematocytes.12 Nematocytes are the cnidarian‐specific stinging cells, characterized by a complex intracytoplasmic capsule (nematocyst) housing a coiled tubule, and an apical sensory ciliary cone. That nematocytes are modified nerve cells is supported by a vast array of data relating to their neurophysiological properties, ultrastructural features, and expression of neurogenic genes in nematocyte precursors.13, 14 Sensory cells, ganglion cells, and some nematocytes bear neurites and establish synaptic contacts with other cells. Both morphological and molecular observations show that each of the three general neural cell types consists of several or many subtypes that can be characterized for example by the number of neurites or the expression of various neuropeptides.15, 16 Glial cells have not been identified in cnidarians.
Today, several cnidarian model species can be reared in the laboratory to provide access to embryos and the availability of new molecular tools allows addressing longstanding questions about the development and architecture of their nervous system (Table 1). Transgenic reporter lines can be used to label the nervous system in unprecedented detail and to trace the progeny of cells that express a given gene at a particular time point.6, 25, 30, 35 Inhibition of gene function by RNA interference or morpholino antisense oligonucleotides has been established and can be complemented by overexpression of in vitro synthesized mRNAs.23, 26, 31, 36, 37, 38, 43 Heritable genome manipulations by TALENs and the CRISPR/Cas9 system have been reported39 and the availability of genome and transcriptome resources allows analyzing the effects of gene manipulations and identifying new regulators of neural development beyond candidate genes.
Table 1.
The Experimental Toolkit for Cnidarian Model Systems
| Species | Genome/Transcriptome | Gain‐of‐Function | Loss‐of‐Function | Trans‐genics | Other | Refs |
|---|---|---|---|---|---|---|
| Acropora millepora | No/yes | No | No | No | 17, 18, 19 | |
| Aurelia aurita | In progress/yes | No | dsRNA | No | 20, 21, 22 | |
| Clytia hemispherica | In progress/yes | mRNA, plasmid | MO | No | 23, 24 | |
| Hydra magnipapillata | Yes/yes | Transgene | RNAi | Yes | FACS | 25, 26, 27, 28, 29 |
| Hydractinia echinata | In progress/yes | Transgene | MO, RNAi | Yes | FACS | 30, 31, 32, 33, 34 |
| Nematostella vectensis | Yes/yes | mRNA, plasmid | MO | Yes | BiFC, histone modifications, CRISPR/Cas9, TALEN | 35, 36, 37, 38, 39, 40, 41, 42 |
Note that references for transcriptome resources are not comprehensive.
BiFC, bimolecular complementation fluorescence; CRISPR, clustered regularly interspaced short palindromic repeats; FACS, fluorescence activated cell sorting; MO, morpholino antisense oligonucleotide; TALEN, transcription activator‐like endonucleases.
In this review, we focus on the generation of neurons and the patterning of the nervous system in cnidarians during embryogenesis and in adult polyps and we briefly discuss aspects of the establishment of neural connectivity. Among cnidarian model organisms, neural development is currently best understood in the anthozoan Nematostella vectensis, and accordingly this sea anemone is central to our discussion on the cellular and molecular basis of neurogenesis, particularly during embryonic development.
THE GENETIC TOOLKIT FOR NEUROGENESIS IS CONSERVED IN CNIDARIANS
The genetic toolkit that controls the generation of nerve cells and the patterning of the nervous system in bilaterian animals is represented in cnidarian genomes by an almost full repertoire of orthologues. A few families of neurogenic genes are of more ancient origin than the metazoan lineage itself as deduced from their presence in unicellular holozoan genomes, e.g., several families of bZIP transcription factors (TFs) including CREB; putative members of the paired‐like, POU and LIM classes of homeoboxes, and of the Sox/TCF family of TFs.44, 45 However, the vast majority of genes with regulatory functions in neural development of bilaterians belong to gene families that were established either in a common metazoan ancestor after divergence of choanoflagellates but before divergence of sponges, or in an exclusive ancestor of cnidarians and bilaterians after divergence of sponges46, 47 (leaving here apart ctenophores and placozoans, whose position in the animal tree remains uncertain, Figure 1). Conservation of the neurogenic toolkit between cnidarians and bilaterians extends to all functional gene categories from TFs (see comprehensive lists compiled in Refs 14, 47, 48) and components of signaling pathways (Notch, Wnt, TGF‐β, FGF, Hedgehog, and Jak/Stat) to post‐transcriptional regulators acting at the mRNA level (e.g., Elav and Musashi).6, 49
As a general rule, multigenic families of neurogenic genes diversified before the last common ancestor of cnidarians and bilaterians, but many of them underwent further diversification within the bilaterian lineage(s). As a result, there are examples of well‐known bilaterian neural development genes with no strict orthologue in cnidarians, such as Neurogenin and NeuroD (bHLH TFs),50 SoxD (HMG domain‐containing TFs),46, 51, 52 or Engrailed (Antp‐class homeodomain‐containing TFs).53 These deductions rely on gene phylogenies that are often difficult to interpret, however, such that in most cases it is hard to say if absence of a given orthologue in cnidarians reflects a primitive state (with respect to a genetic novelty of bilaterians) or is the result of gene loss. This difficulty also causes frequent discrepancies with respect to orthology assessments among studies (for example within the Hox, Sox, and Wnt multigenic families). Finally, it must be recognized that our view of the cnidarian neurogenic toolkit is strongly biased toward searching for homologues of known bilaterian genes, with the consequence that the extent of cnidarian‐specific genetic innovations associated with the nervous system remains entirely unevaluated, except for nematocyte‐specific genes.54, 55
THE CELLULAR BASIS OF CNIDARIAN NEUROGENESIS
Interstitial Stem Cells Function in Hydrozoan Neurogenesis
Until recently, neurogenesis in cnidarians had been mainly studied in the context of the adult polyp of the freshwater Hydra, in which production of new nerve cells takes place continuously for tissue homeostasis and is also needed for budding or regeneration of lost body parts.56, 57, 58 In Hydra, the three types of nerve cells derive from a common pool of stem cells known as interstitial stem cells, located in the interspaces between ectodermal epithelial cells of the body column. The cell lineage deriving from interstitial stem cells, which also comprises glandular cells and germ cells, is in Hydra independent from the ectodermal and endodermal epithelio‐muscular cell lineages,59, 60, 61, 62 although in Hydractinia, i‐cells can also generate epithelio‐muscular cells.63, 64 Within the Hydra interstitial cell lineage, there is a subset of i‐cells that is restricted to the generation of nematocytes and other nerve cell types,65 although it is not known whether individual i‐cells in this population can give rise to different neural cell types. In Hydra, stem cells are exclusively found in the body column where they divide, and progenitor cells are then displaced (together with epithelial cells) toward one of the body extremities (either the oral or basal pole).66, 67 They sequentially differentiate and transdifferentiate into particular subtypes of sensory or ganglion cells depending on their position along the body axis68, 69, 70, 71 and are eventually eliminated at the extremities. Production of nematocytes follows a similar process but nematoblasts, arranged in clusters resulting from several synchronous cell cycle divisions in the body column, differentiate (capsule formation) before actively migrating toward their final destination, i.e., mainly to the tentacles.61
There are experimental indications of significant variation concerning cellular aspects of neurogenesis in hydrozoans other than Hydra. Sensory cells (but not ganglion cells or nematocytes) can form in the absence of interstitial cells, and thus probably from epithelial cells, in the planula larvae of Pennaria tiarella and Clytia (formerly Phialidium) gregaria. 72, 73 Isolated striated muscular cells of the medusa of Podocoryne carnea were observed in Petri dishes to transdifferentiate into smooth muscle cells and then into nerve cells.74
During embryonic development in hydrozoans, interstitial cells first appear in the endoderm shortly after gastrulation.75, 76 In the early planula, the endodermal interstitial cells give rise to nematoblasts and neuroblasts, which then migrate to the ectoderm (Figure 2).75 Therefore in hydrozoans, whereas adult interstitial stem cells are located in the ectoderm, the nervous system is of endodermal embryonic origin.
Figure 2.
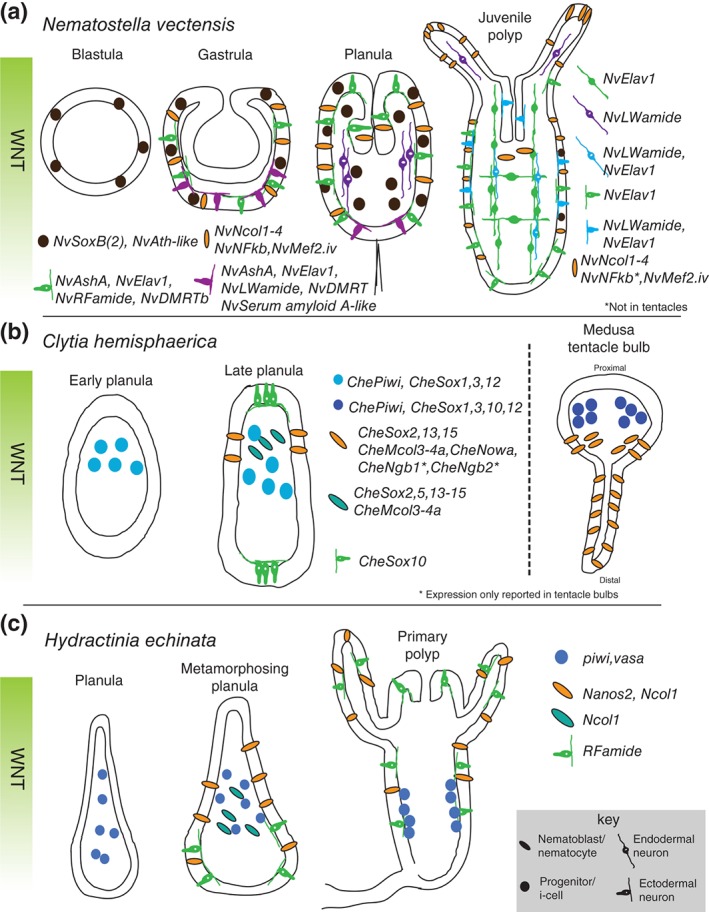
Summary of neural gene expression during embryogenesis. Some of the known neural gene expression patterns for Nematostella vectensis (a), Clytia hemisphaerica (b), and Hydractinia echinata (c) are shown. Developmental time progresses from left to right with depicted stage indicated above each image. In all planulae and polyp stages, the images are oriented with oral pole facing up. The orientation of the Clytia tentacle (b, right side) is proximal up and distal down. Note that the expression patterns of CheSox genes are depicted in a simplified manner.77 CheNgb, neuroglobin. 78
Dedicated Neural Progenitor Cells Contribute to Neurogenesis in Nematostella
Neurogenesis in Nematostella commences at blastula stage with the emergence of neural progenitor cells (NPC); differentiation of neural cells can first be observed in the ectoderm during gastrulation and subsequently also in the endoderm6, 49, 79 (Figure 2). Transplantation experiments have been used to generate chimeric embryos in which the endoderm carries the neuron‐specific NvElav1::mOrange transgene and these embryos revealed that the endoderm itself can generate neurons.6 In Nematostella, no morphological equivalent of i‐cells has been identified; instead, neural cells are derived from epithelial progenitors. A transgenic line expressing mOrange under the control of regulatory elements of the NvSoxB(2) gene revealed that the population of NvSoxB(2)‐expressing cells gives rise to sensory cells, ganglion cells, and nematocytes but not to non‐neural cell types.79
In bilaterian neurogenesis, differences in cell cycle characteristics often reflect functionally asymmetric cell divisions that result in different fates of the daughter cells, generating for example one neuron and one intermediate progenitor cell or two different types of intermediate progenitor cells.80, 81, 82 A similar situation can be observed in Nematostella. Small clusters of NvSoxB(2)::mOrange positive cells (assumed to be clones derived from one NPC) can contain both even and odd numbers of cells, suggesting that there is no strictly synchronous cell division in the progeny of an NPC. This is further supported by EdU pulse labeling experiments, which showed that even after a 2 h pulse, only one cell in pairs of NvSoxB(2)::mOrange positive cells has incorporated EdU and thus been in S‐phase.79 These observations suggest that NPCs in Nematostella can divide more than once and that their developmental program includes asymmetric divisions. The developmental potential of individual NvSoxB(2) expressing cells is currently not known, e.g., whether a single cell can give rise to all three principal classes of neural cells or whether different NvSoxB(2) + NPCs are limited to the generation of either sensory cells, ganglion cells or nematocytes (Figure 3). It is also not clear whether NvSoxB(2) + cells self‐renew, i.e., whether they are stem cells. Alternatively, they might represent progenitor cells that after a series of divisions differentiate into neurons and that are continuously replenished from an as yet unidentified pool of self‐renewing stem cells or directly from epidermal cells (Figure 3).
Figure 3.
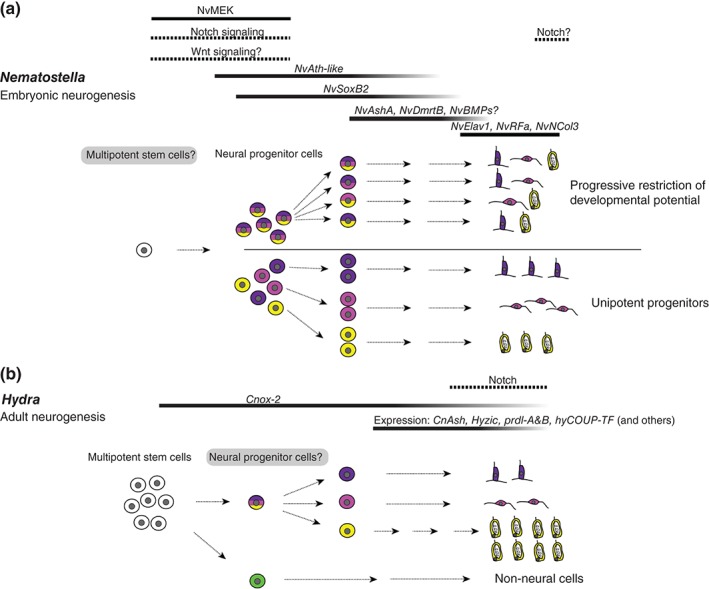
Neurogenesis in Nematostella embryos and in adult Hydra. (a) In Nematostella, a pool of dedicated neural progenitor cells (NPCs) gives rise to the three major classes of neural cells (sensory cells, ganglion cells, and nematocytes) during embryogenesis. Individual NPCs may give rise to different classes (upper part) or to only one class of neural cells (lower part). Note that the existence of these two types of NPCs is not mutually exclusive. NPCs might be derived from multipotent stem cells, but experimental evidence for such stem cells is missing. Bars above the figure depict the stages at which the indicated genes act during the progression of neurogenesis, according to functional data described in the text (except for NvNCol3 and NvRFa). Notch signaling has a role in regulating the number of NPCs and likely in the differentiation of nematocytes. (b) In adult Hydra, multipotent interstitial stem cells (i‐cells) give rise to the different classes of neural cells, but also to non‐neural cells. As for Nematostella NPCs, the developmental potential of individual i‐cells in vivo is not clear. The generation of neural cells may involve a dedicated NPC. Except for cnox‐2, there are no functional data for the indicated genes.
Neurogenesis in Scyphozoa
As in anthozoans, there is no evidence for the existence of an interstitial cell lineage in scyphozoans.83 During embryonic development, differentiating nerve cells are first observed in the ectoderm of the planula and there is no indication that their progenitors would originate in the endoderm.84
THE MOLECULAR REGULATION OF CNIDARIAN NEUROGENESIS
As discussed above, cnidarian genomes contain the bilaterian complement of ‘neurogenic’ genes. A table summarizing expression of known bilaterian neural genes across cnidarian species has been provided in a previous review paper.14 Here, we focus on functional evidence that suggests that neurogenesis in cnidarians and bilaterians is likely conserved beyond the superficial observation that they possess a similar complement of genes.
Sox Action Upstream of bHLH Proneural Gene Transcription Factors Represents a Conserved Neurogenic Cascade
Most functional data has come from disruption of candidate neurogenic genes (Table 2). bHLH proneural genes belonging to the achaete‐scute and atonal families have been the focus of study in multiple cnidarians because of their highly conserved roles in bilaterian neurogenesis (reviewed in Ref 85). Cnash, a cnidarian achaete‐scute homologue (ash) gene identified in Hydra, is expressed in developing nematocytes and sensory neurons.86, 87 The endogenous neurogenic function of cnidarian ash genes was unclear until NvAshA was shown to be both necessary and sufficient for development of a subset of the Nematostella nervous system.88 Although there is a conserved role for ash genes between multiple cnidarian species and bilaterians, some key expression differences exist. Expression of cnidarian ash genes appears restricted to non‐proliferative differentiating neurons,86, 89 whereas in bilaterian species ash genes are expressed in both proliferative progenitor/stem cells and early differentiating neurons.85, 90
Table 2.
Genes Required for Nervous System Development in Cnidarians
|
Nematostella vectensis
Gene |
Expression | Approach | Effect of Manipulation | Ref(s) |
|---|---|---|---|---|
| NvAshA | Scattered cells | lof—MO | Fewer SCs and GCs (ISH, qPCR) | 88, 91 |
| gof—mRNA | More SCs and GCs (ISH, qPCR) | 88, 91 | ||
| NvAth‐like/NvArp3 | Scattered cells | lof—MO | Fewer SCs, GCs, and NCs (ISH, qPCR, NvElav1::mOrange, IHC) | 89, 92 |
| NvSoxB(2) | Scattered cells | lof—MO | Fewer SCs, GCs, and NCs (ISH, qPCR, NvElav1::mOrange, IHC) | 79 |
| NvNotch | Scattered cells (gastrula) | lof—inhibitor (DAPT), MO | More NPCs, SCs, and GCs (ISH, qPCR, NvElav1::mOrange), more immature NCs, fewer mature NCs (ISH, IHC) | 89, 91, 93 |
| gof—NICD mRNA | Fewer NvAshA + neural precursors, fewer SCs and GCs (ISH, qPCR) | 91 | ||
| NvDelta | Scattered cells (gastrula) | lof—MO | Increased NvAshA expression (qPCR) | 91 |
| gof—mRNA | Fewer NvAshA + neural precursors (ISH, qPCR) | 91 | ||
| NvSoxBa/NvSox1 | Broad in oral domain | lof—MO | Fewer NvRFa+ and NvGLWa+ neurons (IHC), no effect on NvElav1::mOrange+ neurons | 92 |
| NvAshB | Broad in oral domain | lof—MO | Fewer NvRFa+ and NvGLWa+ neurons (IHC) | 92 |
| NvArp6 | One sided in endoderm (planula) | lof—MO | Fewer GLW+ neurons (IHC, qPCR) | 92 |
| Nvβ‐catenin | nd | lof—inhibitor (iCRT14) | Fewer NvRFa+, NvGLWa+ (IHC), and NvElav1::mOrange+ neurons | 92 |
| gof—GSK3 inhibitors | More NvRFa+ and NvGLWa+ neurons (IHC), no effect on NvElav1::mOrange | 92, 94 | ||
| NvElav1 | Scattered cells | lof—MO | Fewer NvRFa+, NvGLWa+ (IHC), and NvElav1::mOrange+ neurons | 6 |
| NvDmrtB | Scattered cells | lof—MO | Fewer endodermal NvElav1::mOrange+ neurons | 95 |
| NvBMPs | One sided in oral domain | lof—MO | Fewer NvRFa+ and NvGLWa+ neurons (IHC) | 92, 96 |
| gof—protein | No effect at gastrula, fewer NvRFa+, and NvGLWa+ neurons at planula (IHC) | 92 | ||
| NvMEK | nd | lof—inhibitor (UO126) | Fewer NPCs, SCs, and GCs (ISH) | 97 |
| Hydra | ||||
| Cnox2 | Scattered cells (i‐cells, neurons, nematoblasts) | lof—dsRNA | Fewer SCs and GCs (ISH, IHC) | 65 |
| Hydractinia echinata | ||||
| nanos 2 | Scattered cells (nematoblasts, nematocytes) | lof—MO | Fewer NCs (IHC), more RFa+—neurons (ISH) | 32 |
| gof—plasmid/transgene | More NCs (IHC), fewer RFa+—neurons (ISH) | 32 | ||
GC, ganglion cell; gof, gain‐of‐function; ISH, in situ hybridization; IHC, immunohistochemistry; lof, loss‐of‐function; NC, nematocyte; MO, morpholino antisense oligonucleotide; qPCR, quantitative polymerase chain reaction; SC, sensory cell.
Nematostella has multiple atonal‐like bHLH genes, but exact homology assignments are not clear.50 Regardless, NvAth‐like (also called NvArp3) promotes neural development in Nematostella. 89, 92 Another atonal family gene NvArp6 is necessary for development of GLWamide+ neurons during larval development of Nematostella. Unlike NvAsh genes, NvAth‐like is clearly expressed in proliferating progenitor cells,89 and loss of NvAth‐like function results in a decrease in the expression of NvAsh and other neural markers such as NvElav1.89, 92 It is still unclear if the exact function of NvAth‐like is to promote neurogenesis by regulating the fate of already existing neural progenitors or whether it functions in their initial specification.
sox family TFs are one of the earliest expressed genes in the neural ectoderm of both Drosophila and vertebrates98, 99 and sox function is required for neurogenesis in both groups.98, 100, 101 Expression of sox genes has been characterized in both hydrozoans (Clytia hemisphaerica) and anthozoans (N. vectensis, Acropora millepora) during development51, 79, 102 and in an adult medusa.77 Interestingly, sox genes are expressed in neural progenitor/stem cells in Clytia and Nematostella (Figures 2 and 3). Morpholino mediated knockdown of NvSoxB(2) reduces the number of neurons and the expression of NvAshA and NvAth‐like. 79, 89 Additionally, knockdown of a different soxB gene, NvSoxB2a, reduces expression of NvAshA, NvAshB, and NvAth‐like. 92 The observations that two distinct NvSoxB genes act upstream of proneural gene TFs and that NvSoxB(2) is expressed in progenitor cells suggest that soxB function upstream of proneural genes at early steps in neurogenesis is a conserved feature of cnidarian and bilaterian neural programs.
Notch in Cnidarian Neurogenesis
A highly conserved bilaterian neural regulatory pathway is the Notch signaling pathway. Notch activity in cnidarians has been investigated using both pharmacological and gene specific functional analyses. During Nematostella development, treatment with DAPT, which inhibits Notch activity by disrupting γ‐secretase mediated cleavage of Notch, results in an increased neural marker expression at embryonic and larval stages.89, 91, 93 Specific knockdown of NvNotch also resulted in increased neurogenesis, and hyper‐activation of NvNotch suppresses neural differentiation.91 These data are consistent with Notch having conserved neurogenic function between Nematostella and bilaterian animals. More specifically, NvNotch regulates the number of NvAth‐like+ and NvSoxB(2)+ NPCs,89, 91 which is also in line with the observed role for Notch regulation of neural progenitor fates in multiple bilaterian species.103 Interestingly, Notch effects on neurogenesis in Nematostella are likely mediated by a hes‐ and ‘suppressor of hairless’‐independent pathway,91 which indicates an ancient role for the still poorly understood non‐canonical Notch signaling mechanisms in neurogenesis. It is not yet known if Notch can act at multiple steps of neurogenesis in Nematostella. For example, in Drosophila Notch activity is first required to select neuroblast progenitor cells in the ventral neuroectoderm and then to regulate neuroblast and ganglion mother cell fates after each neuroblast division.104, 105, 106, 107 Currently, there is no experimental evidence in Nematostella in support of or refuting the possibility of Notch acting at multiple levels of the neurogenic cascade. On its own the work in Nematostella suggests that Notch is a conserved neurogenic signaling pathway.
In contrast to the situation in Nematostella, Hydra polyps treated with DAPT generate normal numbers of neurons and nematocytes, but nematocytes fail to fully differentiate.108, 109 Nematocyte maturation defects are also observed in Nematostella after DAPT treatment.89 The lack of a clear neural phenotype in DAPT treated Hydra blurs the comparison between Notch activity in cnidarians and bilaterians. However, it is notable that the Hydra studies were done in adult polyps. Without a better comparison of Notch activity at embryonic stages in other cnidarians, it is difficult to establish that neurogenic phenotypes for Notch in cnidarians and bilaterians represent a deep conservation for this pathway in early neural development.
Does Neurogenesis in Nematostella Require an Inductive Cue?
Neurogenesis in many bilaterians requires molecular cues that confer the competence of a tissue to generate neurons; this process is termed neural induction.110, 111, 112, 113 Whether comparable inductive signals exist in cnidarians is not irrevocably clear, but there is evidence that suggests cells in Nematostella exhibit differential abilities to become neuronal. For example, ubiquitous misexpression of NvAshA is able to upregulate neural marker expression in Nematostella. 88 However, close examination reveals a number of cells that are unresponsive to the ectopic NvAshA. This is reminiscent of unilateral misexpression of ash genes in Xenopus expanding the nervous system, but not neuralizing an entire half embryo.114 Both the Xenopus and Nematostella proneural misexpression phenotypes indicate that cells require a signal to sensitize them toward competence to respond to proneural gene activity. Additional support that not all cells will generate neurons comes from studies investigating Notch signaling. Notch suppresses neurogenesis in Nematostella, but inhibition of Notch by either morpholino or DAPT, does not result in ubiquitous neural marker expression and neither does the simultaneous inhibition of Notch and overexpression of NvAshA. 89, 91 These data argue that Notch regulates the total number of neurons, but that it does not act on embryonic tissue that has a uniform neurogenic potential. Rather, Notch signaling appears to act on a distributed population of cells that has already acquired neurogenic potential. The question then remains whether the differential neurogenic potential is the result of some inductive cue or if it reflects some inherent properties of a subpopulation of embryonic cells.
There are a number of studies of candidate molecules that can provide some insight regarding the potential for neural induction in Nematostella. Inhibition of BMP2/4 signaling is perhaps the best‐known neural induction mechanism in bilaterians.111, 112, 115, 116 In Nematostella embryos, until gastrulation, BMP signaling activity is hardly detectable (as measured by phosphorylation of the signal transducer NvSmad 1/5/8) and the onset of neurogenesis at blastula stage thus occurs at very low levels of BMP signaling.92, 117, 118 Treatment with human BMP2 protein until mid‐blastula stage does not affect the number of NvFMRF+ and NvGLW+ neurons at planula stage; however, it is not clear whether the onset of neurogenesis is delayed until the BMP protein is washed out, which would be expected if inhibition of BMP signaling is required for neurogenesis. Prolonged treatment with human BMP2 (until planula stage) reduces expression of neural markers at larval stages of Nematostella development,92 but injection of the NvBMP2/4 morpholino has the same effect.92, 96 Thus, it remains unclear whether the absence of BMP activity is a prerequisite for the initiation of neural development.
Reports from chicken and zebrafish indicate that FGF signaling promotes neurogenesis by both inducing the expression of the BMP2/4 inhibitors chordin and noggin as well as directly acting to promote expression of neural genes.113, 119 Broad inhibition of FGF signaling with SU5402 does not impact expression of NvashA and thus does not appear to have a neurogenic role.97 However, FGF independent activity of the Mitogen Activated Protein Kinase kinase MEK is necessary for the expression of NvSoxB(2), NvAth‐like, and NvAshA. 97 Additionally, ectopic NvAshA cannot promote neural fates when MEK activity is blocked with the pharmacological inhibitor U0126,97 which taken together suggests that some instruction is necessary to impart neurogenic potential in embryonic cells.
Wnt signaling has also been linked to neural development in Nematostella. Inhibition of Wnt/β‐catenin signaling impairs neural development and reduces the expression of early markers of neurogenesis already at blastula stage.92 While the exact step at which Wnt/β‐catenin signaling regulates neurogenesis remains to be determined, it is interesting to note that the activity of this pathway is highest at the oral pole of the blastula and gastrula, whereas expression of neural markers becomes excluded from the high Wnt activity oral domain just prior to gastrulation.79, 89 Loss of neurogenesis following disruption of Wnt, BMP, and MEK signaling suggests that external cues are able to promote neural fates, and the requirement of MEK activity for NvAshA to promote neural fates suggests that ‘neural’ is not an inherent/default state in some or all of Nematostella cells. Taken together current observations suggest that a process similar to neural induction is required to make cells in Nematostella competent to become neural, but the exact identity of the inductive cue remains elusive.
THE RELATION OF ECTODERMAL AND NEURAL PATTERNING
Gene expression studies have identified distinct ectodermal territories with sharp boundaries along the oral–aboral axis of cnidarian planulae, most prominently in Nematostella. In Nematostella, Clytia, Hydractinia, and Hydra Wnt signaling has been shown to be an important regulator of oral–aboral patterning, with high levels of Wnt/β‐catenin signaling promoting the development of oral identity, and different Wnt genes having nested expression domains starting from the oral pole.31, 94, 120, 121, 122, 123, 124, 125 In bilaterians, the nervous system is patterned along the antero‐posterior axis in register with the ectodermal expression domains of regulatory genes such as otx or the hox genes. Morphological regionalisation along the oral–aboral axis is not much pronounced in the cnidarian planula larva (except for the mouth at the oral pole and the apical organ at the aboral pole), but when looking at the precise distribution of cell types, it appears that there is oral–aboral regionalization of the planula nervous system. In hydrozoan planula larvae, RFamide and GLWamide immunoreactive sensory cells are concentrated in the aboral region, and nematocytes at the oral pole, at least in some species (e.g., Clava multicornis 126; C. hemisphaerica 127 and unpublished observations; Hydractinia echinata 16, 128, 129, 130). There is also regionalized distribution of molecularly defined neural cell types in anthozoan and scyphozoan planulae,6, 49, 131 showing that patterning of the nervous system along the oral–aboral axis is common in cnidarians.
A correspondence of these domains of neural marker expression to those of ectodermal patterning genes remains to be established. Consistent with its function in the patterning of the oral–aboral axis, Wnt/β‐catenin signaling is required for the formation of RFamide and GLWamide immunoreactive neurons in the oral territory of Nematostella. 92 Similarly, NvSix3/6, which regulates aboral development, is required for the expression of NvDmrtB in the aboral domain.132 However, the expression of RFamide, GLWamide and NvDmrtB is not strictly limited to oral and aboral domains, respectively; rather their expression can be detected in different densities along the oral–aboral axis. Thus, while regionalization of the nervous system along the oral–aboral axis is present in cnidarian planulae, the molecular mechanisms that control this regionalization remain to be explored. As a first step, a better molecular definition of distinct neural cell types (by the expression of e.g., receptors, neuropeptides, neurotransmitters, or related biosynthetic enzymes and transporters) will be required.
SUBPOPULATIONS OF NEURONS
Subpopulations of neurons can be characterized by their function (e.g., as chemo‐ or mechanosensory cells), their morphology (e.g., the pattern of neurite projections) and by molecular features (e.g., by neurotransmitter or gene expression) or by combinations of these characters. Each of these three categories of features has been used to describe neural cell types in cnidarians, but there is hardly any information on the developmental programs that control the generation of these cell types. Nematocytes have mechanosensory properties and they contain a sophisticated extrusive organelle, the nematocyst.133, 134 Despite the existence of several different types of nematocysts, the nematocytes might thus be considered a rather well‐defined class of neural cells. In Hydra polyps, nests of nematoblasts derived from up to five rounds of synchronous divisions allow the relation of gene expression patterns to nematocyte development.135 The zinc finger gene HyZic is expressed in proliferating nematoblast nests but not in sensory or ganglion cells, identifying it as a nematocyte‐specific regulator that might be useful to elucidate the molecular basis of nematocyte development.136 Recently, the Hydractinia nanos 2 gene has been shown to promote the formation of nematocytes at the expense of neurons, thus acting as a switch between two classes of neural cells.32 In Nematostella, in situ hybridization combined with counterstaining using neural markers (e.g., neuropeptides) or the use of transgenic reporter lines can tie expression patterns to specific neural cell types. For the nematocyte lineage, NvNF‐κB has been described as a specific regulator,137 whereas NvElav1 is expressed in and required for the development of subsets of sensory and ganglion cells, but not nematocytes.6
For a better understanding of the development of classes of neural cells, it will be necessary to find markers for subpopulations of mature neurons. Subpopulations of sensory cells can potentially be identified by genes related to sensory functions in bilaterians, even though it is often not straightforward to assign particular sensory modalities to these genes. Genes related to vertebrate chemoreceptors and to insect gustatory receptors have been identified in the Nematostella genome,138, 139 but their expression patterns are either not known, or (in the case of the putative gustatory receptor NvGrl1) do not suggest a role in chemoreception. For other candidate sensory cell receptors (e.g., TRP channels, TMCs, and Piezo) expression analyses are so far also limited. A TRPV‐like gene is expressed in the apical organ of Nematostella,140 which contains a tuft of long cilia that are thought to have mechano‐ and/or chemo‐sensory functions; and an antibody against NvTRPA1 labels potentially mechanosensory hair cells in the tentacles.141 The expression of some candidate regulators of neural development in Nematostella (e.g., NvRough and NvEvx 142) is confined to a small number of distributed cells, potentially identifying neural subpopulations defined by a shared developmental program. Functional analyses, however, have so far focused on early and broadly acting regulators of neural development in Nematostella (and other cnidarians), and the knowledge derived from these studies can now be used to test whether a particular candidate gene is indeed expressed in the neural lineage. The generation of new transgenic lines will be essential to describe the composition of the Nematostella nervous system and to refine the cellular and molecular program that generates neural cell type diversity. This more detailed knowledge will in turn provide the basis for comparisons of cell type specific regulatory programs between cnidarians and bilaterians that can inform the reconstruction of the evolution of neural cell types.
IMPLICATIONS CONCERNING THE EVOLUTION OF NEURAL DEVELOPMENT
It is now clear that Nematostella shares several cellular and molecular features of neurogenesis with bilaterian model organisms. Epithelial NPCs divide repeatedly to give rise to different neural cell types; their number is regulated by Notch signaling; soxB, atonal, and achaete‐scute genes are broadly required for neural development and Wnt signaling is involved in the patterning of the nervous system. These features are strong candidates for being shared ancestral traits of cnidarians and bilaterians. A more detailed understanding of the transcriptional regulation of conserved neurogenic genes in cnidarians will likely help to understand how their expression became restricted to defined parts of the ectoderm in many groups of bilaterians and how this relates to the evolution of centralized nervous systems.
Comparison of neurogenesis in hydrozoans and Nematostella reveals similarities at a general level, like the broad neurogenic potential during development and regeneration, but also clear differences. The generation of hydrozoan neurons by interstitial stem cells and the lack of an effect of Notch inhibition on the number of neurons108, 109 indicate substantial differences in the cellular source and the molecular regulation of neurogenesis (Figure 3). Such differences may provide an opportunity to study the evolutionary plasticity of neural development, but such attempts will require a much improved understanding of neurogenesis in hydrozoans, Nematostella and other cnidarians. Current comparisons are to a large extent based on observations in adult Hydra polyps, whereas data for Nematostella is exclusively derived from embryonic neurogenesis. Other important questions that need to be addressed are the origin of the NvSoxB(2) + NPCs in Nematostella and the developmental potential of individual i‐cells during unperturbed hydrozoan development. Hypothetically, the NvSoxB(2) + NPCs could be derived from multipotent epithelial stem cells (functionally resembling i‐cells) and the i‐cells might contain subpopulations that are dedicated to the generation of neural cells (resembling NPCs, Figure 3). New data on these questions will improve the reconstruction of ancestral and derived aspects of cnidarian neurogenesis and in consequence that of shared features of cnidarian and bilaterian neurogenesis.
DEVELOPMENT OF CONNECTIVITY IN CNIDARIAN NERVOUS SYSTEMS
Establishing functional neural circuits requires the outgrowth of neurites and the formation of synaptic connections to other neurons and/or to effector cells, e.g., contractile or secretory cells. In bilaterians, neurites are usually distinguished into dendrites, which receive signals at their postsynaptic sites, and axons, which transmit signals to other cells via presynaptic sites. The mechanisms that control neurite outgrowth differ for dendrites and axons and are better understood for the latter.143, 144 In cnidarians, it is not known whether a clear separation of neurites into axons and dendrites exists. Neurons, in particular ganglion cells, can have multiple neurites, but they usually do not display obvious morphological features (e.g., large synaptic terminals or dendritic spines) that would identify them as dendrites or axons. Cnidarian chemical synapses can be unidirectional or bidirectional (i.e., with synaptic vesicles on both sides of the synaptic cleft), with varying relative abundance,145, 146, 147 but neither the distribution of pre‐ and postsynaptic sites nor the polarity of microtubules (which differs between bilaterian dendrites and axons148) have been mapped systematically.
Cnidarians also differ from bilaterians with respect to the development of neural connectivity. Neurons (including ganglion cells) are generated throughout most of the body column in Nematostella and the distribution of i‐cells in Clytia and Hydractinia planulae suggests that this is also the case in hydrozoans. This widely distributed origin of neurons contrasts with the spatially more restricted generation of neurons (in particular interneurons) in the main bilaterian model organisms. In these animals, neurites are guided by a combination of permissive and instructive cues that are provided by the extracellular matrix and by intermediate ‘signpost cells’ or the eventual targets for innervation.143, 144 The formation of a nerve net starting from distributed neurons is, however, also conceivable without target‐derived guidance cues. Neurites might grow out randomly and the formation of stable synaptic contacts could be determined by the ‘availability’ of target cells, i.e., target cells would accept only a limited number of synaptic contacts. In such a scenario, neurites would compete for the available target sites and the consolidation of synapses would potentially depend on activity that reflects integration into neural circuits. Two observations, however, argue that the outgrowth of neurites in Nematostella is not an entirely random process. At the early planula stage, a dense net of basi‐ectodermal neurites is present but they are almost entirely excluded from a small region at the aboral pole.79 In contrast, at a slightly later timepoint, the neurites of the NvElav1and GLWamide expressing sensory neurons all project in an aboral direction.6 While this is a transient phenomenon (later born NvElav1 and GLWamide neurons project rather in transverse orientation), it suggests that the aboral pole may have a role in regulating the orientation of neurite outgrowth, first negatively and subsequently positively for a subpopulation of neurons.
In anthozoans, bundles of neurites run along the base of the mesenteries, which are endodermal infoldings that structure the gastric cavity and bear the retractor muscles and the gonads.6, 49 While these neurite bundles are the most prominent morphological feature of the nervous system, the neurites within these bundles can project in oral or aboral direction, suggesting that there is no uniform mechanism that regulates their formation.6 Molecularly, genes encoding for several of the major receptor‐ligand pairs involved in bilaterian neurite guidance (Semaphorin and Plexin, Ephrin and Eph, Wnt and Ryk, Netrin and Neogenin/DCC, Unc5, RGM) are present in the Nematostella genome.48 Interestingly, Netrin and RGM are expressed in different subdomains in the aboral territory, consistent with a possible role in the attraction and/or repulsion of neurites in this area.117, 149 Functional characterization of these conserved candidate genes will likely provide interesting first insights into the mechanisms that direct the establishment of the nervous system architecture in cnidarian polyps.
CONCLUSIONS AND FUTURE PERSPECTIVES
Continued study of cnidarian neurogenesis will impact our understanding of nervous system function, evolution of nervous systems, and potentially provide critical clues about mechanisms regarding neural regeneration. Now that tools have been developed (namely transgenesis) in multiple cnidarian species it will be possible to begin to unravel the connectivity of cnidarian nerve nets. Use of neural specific promoters to express calcium sensitive fluorescent proteins, light‐controlled ion channels, and anterograde and retrograde labeling reagents, such as the C‐terminal fragment of the tetanus toxin (TTC) and wheat germ agglutinin (WGA),157 will allow us to assemble wiring maps which in turn can help to understand how neural patterning in neurogenesis might contribute to the formation of specific neural circuits.
Functional studies demonstrate that some cnidarian nervous systems are using the same generic neurogenic programs that bilaterian animals deploy. This links cnidarian nerve nets and bilaterian nervous systems to a common origin. Expression profiling of animals in which neurogenesis has been enhanced or decreased experimentally or of fluorescently labeled neural cells now allows looking at cnidarian neural development at broader scale. Such studies will likely also identify roles of non‐conserved, taxonomically restricted regulators of neural development. More detailed knowledge of the neurogenic program in cnidarians will likely provide insight as to what evolutionary modifications of neural gene regulatory networks gave rise to bilaterian nervous systems and in particular to the brain(s) and central nervous system(s).
While neuronal patterning in both cnidarians and bilaterians is tied to axial patterning, the identity of neurons within distinct domains cannot be easily homologized and there is no adequate description of cnidarian neuronal cell types that would allow specific comparisons between cnidarian nerve nets and bilaterian nervous systems. Thus, there is currently insufficient data to make definitive statements about potential homology of neural cell types or particular regions of the nervous systems of cnidarians and bilaterians.
An exciting aspect of cnidarian neurogenesis is the potential to utilize this highly regenerative group of animals to better understand how nervous systems regenerate. During regeneration, new neurons must reintegrate with existing neurons to reform a functional system. This process is still poorly understood in animals. One of the challenges of neural regeneration is neurite pathfinding and re‐establishing connectivity in an adult environment. Data about how this occurs naturally in animals are relatively limited, but one study in zebrafish suggests that axon pathfinding during regeneration requires molecular programs distinct from those used during development.158 This observation implies that even if research on regenerative neurogenesis can be guided by development, independent studies specifically focused on understanding regeneration must be carried out. Multiple cnidarian species are now accessible to experimental manipulation during development and regeneration and they are poised for studies identifying and comparing developmental and regenerative neurogenic mechanisms.
ACKNOWLEDGMENTS
We thank Chiara Sinigaglia, Thomas Condemine, Uri Frank, and Muriel Jager for providing photographs for Figure 1. All authors contributed equally to the manuscript and FR co‐ordinated the writing.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
FURTHER READING
- Schmidt‐Rhaesa A, Hartzsch S, Purschke G, eds. Structure and Evolution of Invertebrate Nervous Systems. Oxford University Press; 2016. [Google Scholar]
RELATED WIREs ARTICLES
Initial neurogenesis in Drosophila
REFERENCES
- 1. dos Reis M, Thawornwattana Y, Angelis K, Telford MJ, Donoghue PC, Yang Z. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol 2015, 25:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collins AG. Phylogeny of medusozoa and the evolution of cnidarian life cycles. J Evol Biol 2002, 15:418–431. [Google Scholar]
- 3. Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwater B. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol 2006, 55:97–115. [DOI] [PubMed] [Google Scholar]
- 4. Cartwright P, Collins A. Fossils and phylogenies: integrating multiple lines of evidence to investigate the origin of early major metazoan lineages. Integr Comp Biol 2007, 47:744–751. [DOI] [PubMed] [Google Scholar]
- 5. Koizumi O, Hamada S, Minobe S, Hamaguchi‐Hamada K, Kurumata‐Shigeto M, Nakamura M, Namikawa H. The nerve ring in cnidarians: its presence and structure in hydrozoan medusae. Zoology (Jena) 2015, 118:79–88. [DOI] [PubMed] [Google Scholar]
- 6. Nakanishi N, Renfer E, Technau U, Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 2012, 139:347–357. [DOI] [PubMed] [Google Scholar]
- 7. Batham EJ, Pantin CFA, Robson EA. The nerve‐net of the sea‐anemone Metridium senile: the mesenteries and the column. Q J Microsc Sci 1960, 101:487–510. [Google Scholar]
- 8. Satterlie RA. Neuronal control of swimming in jellyfish: a comparative story. Can J Zool 2002, 80:1654–1669. [Google Scholar]
- 9. Satterlie RA. Do jellyfish have central nervous systems? J Exp Biol 2011, 214:1215–1223. [DOI] [PubMed] [Google Scholar]
- 10. Mackie G, Meech R. Central circuitry in the jellyfish Aglantha. II: the ring giant and carrier systems. J Exp Biol 1995, 198:2271–2278. [DOI] [PubMed] [Google Scholar]
- 11. Mackie G, Meech R. Central circuitry in the jellyfish Aglantha. I: the relay system. J Exp Biol 1995, 198:2261–2270. [DOI] [PubMed] [Google Scholar]
- 12. Westfall JA, Elliott CF, Carlin RW. Ultrastructural evidence for two‐cell and three‐cell neural pathways in the tentacle epidermis of the sea anemone Aiptasia pallida . J Morphol 2002, 251:83–92. [DOI] [PubMed] [Google Scholar]
- 13. Thurm U, Brinkmann M, Golz R, Holtmann M, Oliver D, Sieger T. Mechanoreception and synaptic transmission of hydrozoan nematocytes. Hydrobiologia 2004, 530:97–105. [Google Scholar]
- 14. Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic‐Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev Biol 2009, 332:2–24. [DOI] [PubMed] [Google Scholar]
- 15. Koizumi O, Sato N, Goto C. Chemical anatomy of hydra nervous system using antibodies against hydra neuropeptides: a review. Hydrobiologia 2004, 530:41–47. [Google Scholar]
- 16. Martin VJ. Characterization of a RFamide‐positive subset of ganglionic cells in the hydrozoan planular nerve net. Cell Tissue Res 1992, 269:431–438. [DOI] [PubMed] [Google Scholar]
- 17. Moya A, Huisman L, Ball EE, Hayward DC, Grasso LC, Chua CM, Woo HN, Gattuso JP, Foret S, Miller DJ. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO(2)‐driven acidification during the initiation of calcification. Mol Ecol 2012, 21:2440–2454. [DOI] [PubMed] [Google Scholar]
- 18. Meyer E, Aglyamova GV, Wang S, Buchanan‐Carter J, Abrego D, Colbourne JK, Willis BL, Matz MV. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics 2009, 10:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol 2003, 13:2190–2195. [DOI] [PubMed] [Google Scholar]
- 20. Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Bohm AM, Hemmrich G, Sommer F, et al. Regulation of polyp‐to‐jellyfish transition in Aurelia aurita . Curr Biol 2014, 24:263–273. [DOI] [PubMed] [Google Scholar]
- 21. Nakanishi N, Camara AC, Yuan DC, Gold DA, Jacobs DK. Gene expression data from the moon jelly, Aurelia, provide insights into the evolution of the combinatorial code controlling animal sense organ development. PLoS One 2015, 10:e0132544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rachamim T, Morgenstern D, Aharonovich D, Brekhman V, Lotan T, Sher D. The dynamically evolving nematocyst content of an anthozoan, a scyphozoan, and a hydrozoan. Mol Biol Evol 2015, 32:740–753. [DOI] [PubMed] [Google Scholar]
- 23. Momose T, Houliston E. Two oppositely localised frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol 2007, 5:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Houliston E, Momose T, Manuel M. Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet 2010, 26:159–167. [DOI] [PubMed] [Google Scholar]
- 25. Wittlieb J, Khalturin K, Lohmann JU, Anton‐Erxleben F, Bosch TC. Transgenic hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A 2006, 103:6208–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohmann JU, Endl I, Bosch TC. Silencing of developmental genes in hydra. Dev Biol 1999, 214:211–214. [DOI] [PubMed] [Google Scholar]
- 27. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of hydra. Nature 2010, 464:592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chera S, de Rosa R, Miljkovic‐Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J Cell Sci 2006, 119:846–857. [DOI] [PubMed] [Google Scholar]
- 29. Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton‐Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg HH, Domazet‐Loso T, et al. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol 2012, 29:3267–3280. [DOI] [PubMed] [Google Scholar]
- 30. Kunzel T, Heiermann R, Frank U, Muller W, Tilmann W, Bause M, Nonn A, Helling M, Schwarz RS, Plickert G. Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP‐transgenic animals and chimeras. Dev Biol 2010, 348:120–129. [DOI] [PubMed] [Google Scholar]
- 31. Duffy DJ, Plickert G, Kuenzel T, Tilmann W, Frank U. Wnt signaling promotes oral but suppresses aboral structures in Hydractinia metamorphosis and regeneration. Development 2010, 137:3057–3066. [DOI] [PubMed] [Google Scholar]
- 32. Kanska J, Frank U. New roles for Nanos in neural cell fate determination revealed by studies in a cnidarian. J Cell Sci 2013, 126:3192–3203. [DOI] [PubMed] [Google Scholar]
- 33. Soza‐Ried J, Hotz‐Wagenblatt A, Glatting KH, del Val C, Fellenberg K, Bode HR, Frank U, Hoheisel JD, Frohme M. The transcriptome of the colonial marine hydroid Hydractinia echinata . FEBS J 2010, 277:197–209. [DOI] [PubMed] [Google Scholar]
- 34. Hensel K. Wnt signalling in the hydrozoan Hydractinia echinata PhD Thesis, National University of Ireland Galway, 2013. Available at: https://aran.library.nuigalway.ie/xmlui/handle/10379/4374. (Accessed June 2, 2016).
- 35. Renfer E, Amon‐Hassenzahl A, Steinmetz PR, Technau U. A muscle‐specific transgenic reporter line of the sea anemone, Nematostella vectensis . Proc Natl Acad Sci U S A 2009, 107:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis . Development 2008, 135:1761–1769. [DOI] [PubMed] [Google Scholar]
- 37. Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol 2007, 305:483–497. [DOI] [PubMed] [Google Scholar]
- 38. Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear β‐catenin in the evolution of axial polarity and germ layer segregation. Nature 2003, 426:446–450. [DOI] [PubMed] [Google Scholar]
- 39. Ikmi A, McKinney SA, Delventhal KM, Gibson MC. TALEN and CRISPR/Cas9‐mediated genome editing in the early‐branching metazoan Nematostella vectensis . Nat Commun 2014, 5:5486. [DOI] [PubMed] [Google Scholar]
- 40. Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317:86–94. [DOI] [PubMed] [Google Scholar]
- 41. Hudry B, Thomas‐Chollier M, Volovik Y, Duffraisse M, Dard A, Frank D, Technau U, Merabet S. Molecular insights into the origin of the Hox‐TALE patterning system. Elife 2014, 3:e01939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwaiger M, Schonauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 2014, 24:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Layden MJ, Rentzsch F, Rottinger E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. WIREs Dev Biol 2016, 5:408–428. doi: 10.1002/wdev.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 2008, 451:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sebe‐Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz‐Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki . Mol Biol Evol 2011, 28:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 2008, 25:980–996. [DOI] [PubMed] [Google Scholar]
- 47. Galliot B, Quiquand M. A two‐step process in the emergence of neurogenesis. Eur J Neurosci 2011, 34:847–862. [DOI] [PubMed] [Google Scholar]
- 48. Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ 2009, 51:167–183. [DOI] [PubMed] [Google Scholar]
- 49. Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol 2009, 69:235–254. [DOI] [PubMed] [Google Scholar]
- 50. Simionato E, Ledent V, Richards G, Thomas‐Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix‐loop‐helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol 2007, 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shinzato C, Iguchi A, Hayward DC, Technau U, Ball EE, Miller DJ. Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol 2008, 8:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jager M, Queinnec E, Houliston E, Manuel M. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol Phylogenet Evol 2006, 39:468–477. [DOI] [PubMed] [Google Scholar]
- 53. Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian‐bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis . Genome Biol 2006, 7:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Milde S, Hemmrich G, Anton‐Erxleben F, Khalturin K, Wittlieb J, Bosch TCG. Characterization of taxonomically restricted genes in a phylum‐restricted cell type. Genome Biol 2009, 10:R8. doi: 10.1186/gb-2009-10-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hwang JS, Ohyanagi H, Hayakawa S, Osato N, Nishimiya‐Fujisawa C, Ikeo K, David CN, Fujisawa T, Gojobori T. The evolutionary emergence of cell type‐specific genes inferred from the gene expression analysis of hydra. Proc Natl Acad Sci U S A 2007, 104:14735–14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bosch TC, Anton‐Erxleben F, Hemmrich G, Khalturin K. The hydra polyp: nothing but an active stem cell community. Dev Growth Differ 2010, 52:15–25. [DOI] [PubMed] [Google Scholar]
- 57. Watanabe H, Hoang VT, Mattner R, Holstein TW. Immortality and the base of multicellular life: lessons from cnidarian stem cells. Semin Cell Dev Biol 2009, 20:1114–1125. [DOI] [PubMed] [Google Scholar]
- 58. Hobmayer B, Jenewein M, Eder D, Eder MK, Glasauer S, Gufler S, Hartl M, Salvenmoser W. Stemness in hydra – a current perspective. Int J Dev Biol 2012, 56:509–517. [DOI] [PubMed] [Google Scholar]
- 59. Bosch TCG, David CN. Stem cells of hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol 1987, 121:182–191. [Google Scholar]
- 60. David CN, Murphy S. Characterization of interstitial stem cells in hydra by cloning. Dev Biol 1977, 58:372–383. [DOI] [PubMed] [Google Scholar]
- 61. Bode HR. The interstitial cell lineage of hydra: a stem cell system that arose early in evolution. J Cell Sci 1996, 109:1155–1164. [DOI] [PubMed] [Google Scholar]
- 62. Bode HR, David CN. Regulation of a multipotent stem‐cell, interstitial cell of hydra. Prog Biophys Mol Biol 1978, 33:189–206. [DOI] [PubMed] [Google Scholar]
- 63. Muller WA, Teo R, Frank U. Totipotent migratory stem cells in a hydroid. Dev Biol 2004, 275:215–224. [DOI] [PubMed] [Google Scholar]
- 64. Gahan JM, Bradshaw B, Flici H, Frank U. The interstitial stem cells in Hydractinia and their role in regeneration. Curr Opin Genet Dev 2016, 40:65–73. [DOI] [PubMed] [Google Scholar]
- 65. Miljkovic‐Licina M, Chera S, Ghila L, Galliot B. Head regeneration in wild‐type hydra requires de novo neurogenesis. Development 2007, 134:1191–1201. [DOI] [PubMed] [Google Scholar]
- 66. David CN, Hager G. Formation of a primitive nervous‐system – nerve‐cell differentiation in the polyp hydra. Perspect Dev Neurobiol 1994, 2:135–140. [PubMed] [Google Scholar]
- 67. Boehm AM, Bosch TCG. Migration of multipotent interstitial stem cells in hydra. Zoology 2012, 115:275–282. [DOI] [PubMed] [Google Scholar]
- 68. Koizumi O. Developmental neurobiology of hydra, a model animal of cnidarians. Can J Zool 2002, 80:1678–1689. [Google Scholar]
- 69. Koizumi O, Bode HR. Plasticity in the nervous system of adult hydra. I. The position‐dependent expression of FMRFamide‐like immunoreactivity. Dev Biol 1986, 116:407–421. [DOI] [PubMed] [Google Scholar]
- 70. Koizumi O, Bode HR. Plasticity in the nervous system of adult hydra. III. Conversion of neurons to expression of a vasopressin‐like immunoreactivity depends on axial location. J Neurosci 1991, 11:2011–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Koizumi O, Heimfeld S, Bode HR. Plasticity in the nervous system of adult hydra. II. Conversion of ganglion cells of the body column into epidermal sensory cells of the hypostome. Dev Biol 1988, 129:358–371. [DOI] [PubMed] [Google Scholar]
- 72. Martin VJ, Thomas MB. The origin of the nervous system in Pennaria tiarella, as revealed by treatment with colchicine. Biol Bull 1981, 160:303–310. [Google Scholar]
- 73. Thomas MB, Freeman G, Martin VJ. The embryonic origin of neurosensory cells and the role of nerve cells in metamorphosis in Phialidium gregarium (Cnidaria, Hydrozoa). Int J Inver Rep Dev 1987, 11:265–285. [Google Scholar]
- 74. Alder H, Schmid V. Cell cycles and in vitro transdifferentiation and regeneration of isolated, striated muscle of jellyfish. Dev Biol 1987, 124:358–369. [DOI] [PubMed] [Google Scholar]
- 75. Martin VJ. Cnidarians, the jellyfish and hydras In: Gilbert SF, Raunio AM, eds. Embryology. Sunderland, MA: Sinauer Associates; 1997, 57–86. [Google Scholar]
- 76. Leclére L, Jager M, Barreau C, Chang P, Le Guyader H, Manuel M, Houliston E. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev Biol 2012, 364:236–248. [DOI] [PubMed] [Google Scholar]
- 77. Jager M, Queinnec E, Le Guyader H, Manuel M. Multiple Sox genes are expressed in stem cells or in differentiating neuro‐sensory cells in the hydrozoan Clytia hemisphaerica . Evodevo 2011, 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lechauve C, Jager M, Laguerre L, Kiger L, Correc G, Leroux C, Vinogradov S, Czjzek M, Marden MC, Bailly X. Neuroglobins, pivotal proteins associated with emerging neural systems and precursors of metazoan globin diversity. J Biol Chem 2013, 288:6957–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Richards GS, Rentzsch F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis . Development 2014, 141:4681–4689. [DOI] [PubMed] [Google Scholar]
- 80. Cheffer A, Tarnok A, Ulrich H. Cell cycle regulation during neurogenesis in the embryonic and adult brain. Stem Cell Rev 2013, 9:794–805. [DOI] [PubMed] [Google Scholar]
- 81. Homem CC, Repic M, Knoblich JA. Proliferation control in neural stem and progenitor cells. Nat Rev Neurosci 2015, 16:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hardwick LJ, Ali FR, Azzarelli R, Philpott A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res 2015, 359:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gold DA, Jacobs DK. Stem cell dynamics in Cnidaria: are there unifying principles? Dev Genes Evol 2013, 223:53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yuan D, Nakanishi N, Jacobs DK, Hartenstein V. Embryonic development and metamorphosis of the scyphozoan Aurelia. Dev Genes Evol 2008, 218:525–539. [DOI] [PubMed] [Google Scholar]
- 85. Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 2002, 3:517–530. [DOI] [PubMed] [Google Scholar]
- 86. Hayakawa E, Fujisawa C, Fujisawa T. Involvement of hydra achaete‐scute gene CnASH in the differentiation pathway of sensory neurons in the tentacles. Dev Genes Evol 2004, 214:486–492. [DOI] [PubMed] [Google Scholar]
- 87. Grens A, Mason E, Marsh JL, Bode HR. Evolutionary conservation of a cell fate specification gene: the hydra achaete‐scute homolog has proneural activity in Drosophila. Development 1995, 121:4027–4035. [DOI] [PubMed] [Google Scholar]
- 88. Layden MJ, Boekhout M, Martindale MQ. Nematostella vectensis achaete‐scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development 2012, 139:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Richards GS, Rentzsch F. Regulation of Nematostella neural progenitors by SoxB, Notch and bHLH genes. Development 2015, 142:3332–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 2008, 58:52–64. [DOI] [PubMed] [Google Scholar]
- 91. Layden MJ, Martindale MQ. Non‐canonical Notch signaling represents an ancestral mechanism to regulate neural differentiation. Evodevo 2014, 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Watanabe H, Kuhn A, Fushiki M, Agata K, Ozbek S, Fujisawa T, Holstein TW. Sequential actions of β‐catenin and Bmp pattern the oral nerve net in Nematostella vectensis . Nat Commun 2014, 5:5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Marlow H, Roettinger E, Boekhout M, Martindale MQ. Functional roles of Notch signaling in the cnidarian Nematostella vectensis . Dev Biol 2012, 362:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis . Dev Biol 2013, 380:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Parlier D, Moers V, Van Campenhout C, Preillon J, Leclere L, Saulnier A, Sirakov M, Busengdal H, Kricha S, Marine JC, et al. The Xenopus doublesex‐related gene Dmrt5 is required for olfactory placode neurogenesis. Dev Biol 2013, 373:39–52. [DOI] [PubMed] [Google Scholar]
- 96. Saina M, Genikhovich G, Renfer E, Technau U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc Natl Acad Sci U S A 2009, 106:18592–18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Layden MJ, Johnston H, Amiel AR, Havrilak J, Steinworth B, Chock T, Rottinger E, Martindale MQ. MAPK signaling is necessary for neurogenesis in Nematostella vectensis . BMC Biol 2016, 14:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development 2002, 129:4193–4203. [DOI] [PubMed] [Google Scholar]
- 99. Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic‐related‐1 and Sox‐2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 1998, 125:579–587. [DOI] [PubMed] [Google Scholar]
- 100. Overton PM, Meadows LA, Urban J, Russell S. Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila. Development 2002, 129:4219–4228. [DOI] [PubMed] [Google Scholar]
- 101. Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2‐mediated signaling for differentiation of early Xenopus neuroectoderm. Development 2000, 127:791–800. [DOI] [PubMed] [Google Scholar]
- 102. Magie CR, Pang K, Martindale MQ. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis . Dev Genes Evol 2005, 215:618–630. [DOI] [PubMed] [Google Scholar]
- 103. Hartenstein V, Stollewerk A. The evolution of early neurogenesis. Dev Cell 2015, 32:390–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Louvi A, Artavanis‐Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci 2006, 7:93–102. [DOI] [PubMed] [Google Scholar]
- 105. Hartenstein V, Wodarz A. Initial neurogenesis in Drosophila. WIREs Dev Biol 2013, 2:701–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 2011, 69:840–855. [DOI] [PubMed] [Google Scholar]
- 107. Udolph G. Notch signaling and the generation of cell diversity in Drosophila neuroblast lineages. Adv Exp Med Biol 2012, 727:47–60. [DOI] [PubMed] [Google Scholar]
- 108. Kasbauer T, Towb P, Alexandrova O, David CN, Dall'armi E, Staudigl A, Stiening B, Bottger A. The Notch signaling pathway in the cnidarian hydra. Dev Biol 2007, 303:376–390. [DOI] [PubMed] [Google Scholar]
- 109. Khalturin K, Anton‐Erxleben F, Milde S, Plotz C, Wittlieb J, Hemmrich G, Bosch TC. Transgenic stem cells in hydra reveal an early evolutionary origin for key elements controlling self‐renewal and differentiation. Dev Biol 2007, 309:32–44. [DOI] [PubMed] [Google Scholar]
- 110. Bier E. Anti‐neural‐inhibition: a conserved mechanism for neural induction. Cell 1997, 89:681–684. [DOI] [PubMed] [Google Scholar]
- 111. Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today 2009, 87:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ozair MZ, Kintner C, Brivanlou AH. Neural induction and early patterning in vertebrates. WIREs Dev Biol 2013, 2:479–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stern CD. Neural induction: old problem, new findings, yet more questions. Development 2005, 132:2007–2021. [DOI] [PubMed] [Google Scholar]
- 114. Turner DL, Weintraub H. Expression of achaete‐scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev 1994, 8:1434–1447. [DOI] [PubMed] [Google Scholar]
- 115. De Robertis EM, Kuroda H. Dorsal‐ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol 2004, 20:285–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Mizutani CM, Bier E. EvoD/Vo: the origins of BMP signalling in the neuroectoderm. Nat Rev Genet 2008, 9:663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Leclére L, Rentzsch F. RGM regulates BMP‐mediated secondary axis formation in the sea anemone Nematostella vectensis . Cell Rep 2014, 9:1921–1930. [DOI] [PubMed] [Google Scholar]
- 118. Genikhovich G, Fried P, Prunster MM, Schinko JB, Gilles AF, Fredman D, Meier K, Iber D, Technau U. Axis patterning by BMPs: cnidarian network reveals evolutionary constraints. Cell Rep 2015, 10:1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Guillemot F, Zimmer C. From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron 2011, 71:574–588. [DOI] [PubMed] [Google Scholar]
- 120. Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 2005, 132:2907–2916. [DOI] [PubMed] [Google Scholar]
- 121. Momose T, Derelle R, Houliston E. A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica . Development 2008, 135:2105–2113. [DOI] [PubMed] [Google Scholar]
- 122. Plickert G, Jacoby V, Frank U, Muller WA, Mokady O. Wnt signaling in hydroid development: formation of the primary body axis in embryogenesis and its subsequent patterning. Dev Biol 2006, 298:368–378. [DOI] [PubMed] [Google Scholar]
- 123. Rottinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for “endomesoderm” specification: the inputs of ss‐catenin/TCF signaling. PLoS Genet 2012, 8:1‐28. doi:e1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Trevino M, Stefanik DJ, Rodriguez R, Harmon S, Burton PM. Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis . Dev Dyn 2011, 240:2673–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Leclère L, Bause M, Sinigaglia C, Steger J, Rentzsch F. Development of the aboral domain in Nematostella requires β‐catenin and the opposing activities of Six3/6 and Frizzled5/8. Development 2016, 143:1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Piraino S, Zega G, Di Benedetto C, Leone A, Dell'Anna A, Pennati R, Carnevali DC, Schmid V, Reichert H. Complex neural architecture in the diploblastic larva of Clava multicornis (Hydrozoa, Cnidaria). J Comp Neurol 2011, 519:1931–1951. [DOI] [PubMed] [Google Scholar]
- 127. Bodo F, Bouillon J. Etude histologique du développement embryonaire de quelques Hydroméduses de Roscoff: Phialidium hemisphaericum (L.), Obelia sp. Péron et Lesueur, Sarcia eximia (Allman), Podocoryne carnea (Sars), Gonionemus vertens (Agassiz). Cah Biol Mar 1968, 9:69–104. [Google Scholar]
- 128. Leitz T, Lay M. Metamorphosin‐a is a neuropeptide. Rouxs Arch Dev Biol 1995, 204:276–279. [DOI] [PubMed] [Google Scholar]
- 129. Seipp S, Schmich J, Will B, Schetter E, Plickert G, Leitz T. Neuronal cell death during metamorphosis of Hydractinia echinata (Cnidaria, Hydrozoa). Invert Neurosci 2010, 10:77–91. [DOI] [PubMed] [Google Scholar]
- 130. Groger H, Schmid V. Larval development in Cnidaria: a connection to Bilateria? Genesis 2001, 29:110–114. [DOI] [PubMed] [Google Scholar]
- 131. Nakanishi N, Yuan D, Jacobs DK, Hartenstein V. Early development, pattern, and reorganization of the planula nervous system in Aurelia (Cnidaria, Scyphozoa). Dev Genes Evol 2008, 218:511–524. [DOI] [PubMed] [Google Scholar]
- 132. Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol 2013, 11:e1001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Beckmann A, Ozbek S. The nematocyst: a molecular map of the cnidarian stinging organelle. Int J Dev Biol 2012, 56:577–582. [DOI] [PubMed] [Google Scholar]
- 134. Ozbek S. The cnidarian nematocyst: a miniature extracellular matrix within a secretory vesicle. Protoplasma 2011, 248:635–640. [DOI] [PubMed] [Google Scholar]
- 135. David CN, Challone D. Distribution of interstitial‐cells and differentiating nematocytes in nests in hydra‐attenuata. Am Zool 1974, 14:537–542. [Google Scholar]
- 136. Lindgens D, Holstein TW, Technau U. Hyzic, the hydra homolog of the zic/odd‐paired gene, is involved in the early specification of the sensory nematocytes. Development 2004, 131:191–201. [DOI] [PubMed] [Google Scholar]
- 137. Wolenski FS, Bradham CA, Finnerty JR, Gilmore TD. NF‐κB is required for cnidocyte development in the sea anemone Nematostella vectensis . Dev Biol 2013, 373:205–215. [DOI] [PubMed] [Google Scholar]
- 138. Churcher AM, Taylor JS. The antiquity of chordate odorant receptors is revealed by the discovery of orthologs in the cnidarian Nematostella vectensis . Genome Biol Evol 2011, 3:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Saina M, Busengdal H, Sinigaglia C, Petrone L, Oliveri P, Rentzsch F, Benton R. A cnidarian homologue of an insect gustatory receptor functions in developmental body patterning. Nat Commun 2015, 6:6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sinigaglia C, Busengdal H, Lerner A, Oliveri P, Rentzsch F. Molecular characterization of the apical organ of the anthozoan Nematostella vectensis . Dev Biol 2015, 398:120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Mahoney JL, Graugnard EM, Mire P, Watson GM. Evidence for involvement of TRPA1 in the detection of vibrations by hair bundle mechanoreceptors in sea anemones. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 2011, 197:729–742. [DOI] [PubMed] [Google Scholar]
- 142. Ryan JF, Mazza ME, Pang K, Matus DQ, Baxevanis AD, Martindale MQ, Finnerty JR. Pre‐bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis . PLoS One 2007, 2:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chilton JK. Molecular mechanisms of axon guidance. Dev Biol 2006, 292:13–24. [DOI] [PubMed] [Google Scholar]
- 144. Dickson BJ. Molecular mechanisms of axon guidance. Science 2002, 298:1959–1964. [DOI] [PubMed] [Google Scholar]
- 145. Anderson PAV, Schwab WE. The organization and structure of nerve and muscle in the jellyfish cyanea‐capillata (Coelenterata, Scyphozoa). J Morphol 1981, 170:383–399. [DOI] [PubMed] [Google Scholar]
- 146. Horridge GA, Mackay B, Chapman DM. Naked axons and symmetrical synapses in an elementary nervous system. Nature 1962, 193:899–900. [DOI] [PubMed] [Google Scholar]
- 147. Peteya DJ. Light and electron‐microscope study of nervous‐system of Ceriantheopsis americanus (Cnidaria, Ceriantharia). Z Zellforsch Mikrosk Anat 1973, 141:301–317. [DOI] [PubMed] [Google Scholar]
- 148. Rolls MM, Jegla TJ. Neuronal polarity: an evolutionary perspective. J Exp Biol 2015, 218:572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci U S A 2006, 103:11195–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Nakanishi N, Hartenstein V, Jacobs DK. Development of the rhopalial nervous system in Aurelia sp.1 (Cnidaria, Scyphozoa). Dev Genes Evol 2009, 219:301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nakanishi N, Yuan D, Hartenstein V, Jacobs DK. Evolutionary origin of rhopalia: insights from cellular‐level analyses of Otx and POU expression patterns in the developing rhopalial nervous system. Evol Dev 2010, 12:404–415. [DOI] [PubMed] [Google Scholar]
- 152. Chia FS, Koss R. Fine structural studies of the nervous system and the apical organ in the planula larva of the sea anemone Anthopleura elegantissima . J Morph 1979, 160:275–298. [DOI] [PubMed] [Google Scholar]
- 153. Denker E, Manuel M, Leclere L, Le Guyader H, Rabet N. Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev Biol 2008, 315:99–113. [DOI] [PubMed] [Google Scholar]
- 154. Seipel K, Yanze N, Schmid V. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea . Int J Dev Biol 2004, 48:1–7. [DOI] [PubMed] [Google Scholar]
- 155. Deves M, Bourrat F. Transcriptional mechanisms of developmental cell cycle arrest: problems and models. Semin Cell Dev Biol 2012, 23:290–297. [DOI] [PubMed] [Google Scholar]
- 156. Coste A, Jager M, Chambon JP, Manuel M. Comparative study of Hippo pathway genes in cellular conveyor belts of a ctenophore and a cnidarian. Evodevo 2016, 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Huh Y, Oh MS, Leblanc P, Kim KS. Gene transfer in the nervous system and implications for transsynaptic neuronal tracing. Expert Opin Biol Ther 2010, 10:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Binari LA, Lewis GM, Kucenas S. Perineurial glia require Notch signaling during motor nerve development but not regeneration. J Neurosci 2013, 33:4241–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]


