Abstract
Biasing the sex ratio of populations of different organisms, including plants, insects, crustacean, and fish, has been demonstrated by genetic and non‐genetic approaches. However, biasing the sex ratio of mammalian populations has not been demonstrated genetically. Here, we provide a first proof of concept for such a genetic system in mammals by crossing two genetically engineered mouse lines. The maternal line encodes a functional Cas9 protein on an autosomal chromosome, whereas the paternal line encodes guide RNAs on the Y chromosome targeting vital mouse genes. After fertilization, the presence of both the Y‐encoded guide RNAs from the paternal sperm and the Cas9 protein from the maternal egg targets the vital genes in males. We show that these genes are specifically targeted in males and that this breeding consequently self‐destructs solely males. Our results pave the way for a genetic system that allows biased sex production of livestock.
Keywords: CRISPR‐Cas, genetic system, mammals, sex ratio, Y chromosome
Subject Categories: Development & Differentiation; Genetics, Gene Therapy & Genetic Disease; Methods & Resources
Introduction
Some aquatic organisms as well as plants that benefit from single‐sex cultivation have been produced mostly by hormonal feminization of males or by masculinization of females and the subsequent production of a single‐sex progeny. This was demonstrated in crustaceans 1 and fish 2, 3, 4, 5, and is also a common practice in growing Cannabis sativa, where feminized seeds are desired. However, these practices are not feasible for terrestrial livestock.
The sex ratio in a population of mosquitoes and flies was shifted by manipulating specific genes that distort the sex ratio 6, 7, 8. In recent breakthrough studies, researchers have even completely distorted the sex ratio, accompanied by the sterility of females, thus resulting in a collapsed population 9, 10, 11, 12, 13. Such an outcome is desirable for disease‐transferring insects in the wild, but not for domesticated livestock.
Manipulated animals that produce only one sex are impossible to sustain by self‐crossing, because either the male or the female is absent, or due to sub‐fertility or infertility of the manipulated animal. Thus, despite the identification of genetic factors such as Sry, Sox9, Foxl2, and Wnt4, that determine the sex of animals, and the success of reversing animal's sex, fertility and other genetic restrictions preclude a system that reliably produces a single‐sex progeny 14, 15, 16, 17, 18.
Results and Discussion
We chose to provide a proof of concept in mice for an approach that produces mainly single‐sex progeny while retaining a reproductive reservoir of males and females. We used two self‐sustained mouse lines, each producing males and females at an equal ratio. One homozygous line, henceforth termed the “Cas9‐line”, encoded the CRISPR‐Cas9 enzyme from Streptococcus pyogenes, expressed from a CAG promoter on an autosomal chromosome 19. We generated the other hemizygous line, henceforth termed the “Y‐line”, encoding on its Y chromosome three CRISPR guide RNAs (gRNAs) targeting three autosomal genes (see Fig EV1 and the Appendix for details on mouse construction). The selected target genes, Atp5b, Cdc20, and Casp8, were all shown to be essential for mouse early development (Atp5b deficiency in mice results in embryonic lethality prior to organogenesis at embryonic day 9.5 (E9.5) 20; Cdc20 deficiency in mice results in metaphase arrest in the two‐cell stage embryos and consequently in early embryonic death not later than E3.5 21; Casp8 deficiency results in necroptosis and consequently in embryonic death at E10.5 22). We selected targeting three different genes to reduce the probability of simultaneous non‐targeting or in‐frame corrections of three genes, or such combinations that may result in viable males. We hypothesized that crossing these two lines would result in a progeny consisting of mostly female mice, since the three vital genes in male mice are targeted. We further hypothesized that the litter size would be half the normal size, since half of the progeny are eliminated. To test our hypotheses, we crossed Y‐line males with Cas9‐line females, and as a control, we crossed Y‐line males with wild‐type B6J females (Fig 1A). The control cross between the Y‐line males and the B6J females produced 92 pups with an average of 6.57 pups per litter (Figs 1B and EV2). The cross between the Y‐line males and the Cas9‐line females produced 115 pups with an average of 3.71 pups per litter, which is approximately half of the litter size of control cross, as hypothesized (Figs 1B and EV2). At birth, the number of males in the control cross was statistically comparable to the number of females (49 vs. 35; eight infanticides with undetermined sex), whereas in the cross between the Y‐line and Cas9‐line, the number of males significantly decreased (9 vs. 104; two infanticides with undetermined sex; Figs 1C and EV2). At weaning, the female‐to‐male ratio in the latter cross even increased (3 vs. 89; Fig 1C). These results demonstrate, for the first time, a genetic system for significantly biasing the sex ratio in mammals while maintaining a reservoir of fertile parents.
Figure EV1. Schematic summary of Y‐line generation.
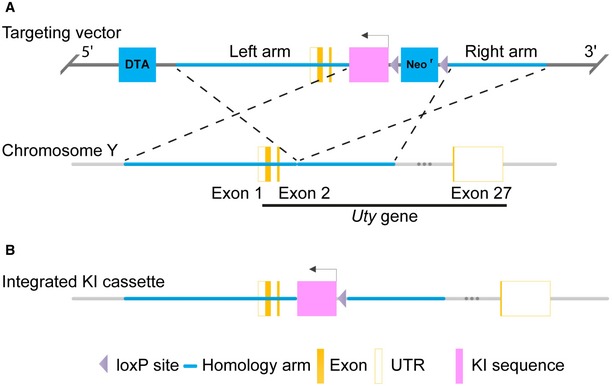
- Mouse genomic fragments containing homology arms to the Uty gene located on mouse chromosome Y were amplified from a BAC clone and sequentially assembled into a targeting vector along with negative and positive selection markers (DTA and Neo, respectively). There are only few mice reported with a transgene on the Y chromosome, one of which is on the Uty gene 28, and thus, we chose to target the KI cassette into this gene. The KI cassette, encoding the gRNAs targeting genes Atp5b, Cdc20, and Casp8, each expressed from a U6 promoter, was inserted into the targeting vector, which was designed to integrate in reverse orientation of the 2nd exon of the Uty gene.
- The KI cassette integrated into the Uty gene and the Neo cassette was self‐deleted due to the loxP sites. Details on the linearization of the targeting vector, its transfection into C57BL/6N ES cells, as well as PCR and Southern blot analyses and ES implantation procedures, are provided in the Appendix.
Figure 1. Biasing the sex ratio in mice.
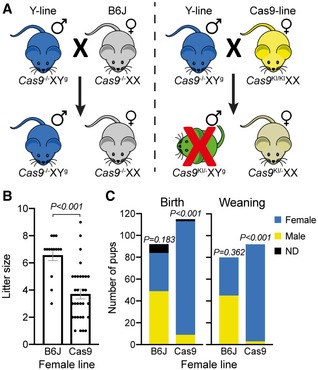
- The crosses between the Y‐line males (blue) with B6J females (gray) are illustrated on the top left and those of the Y‐line males with Cas9‐line females (yellow) on the top right. The presence or absence of the Cas9 allele is annotated as Cas9 KI or Cas9 −, respectively, whereas the Y‐linked gRNA allele is annotated as Yg. The X chromosome is unmodified in all instances.
- Litter size of the crosses of the Y‐line males with the B6J‐ (n = 14) or Cas9‐ (n = 31) line females. Dots represent individual litter size, and bars represent average litter size ± SD. Significance was determined using a two‐tailed unpaired parametric t‐test.
- Pie charts of the sex distribution of the total pups from the crosses of the Y‐line males with the indicated female line at day of birth and at weaning. Significance was determined using a two‐tailed binomial test, assuming equal male‐to‐female ratio.
Figure EV2. Sex of pups from individual Y‐line males.
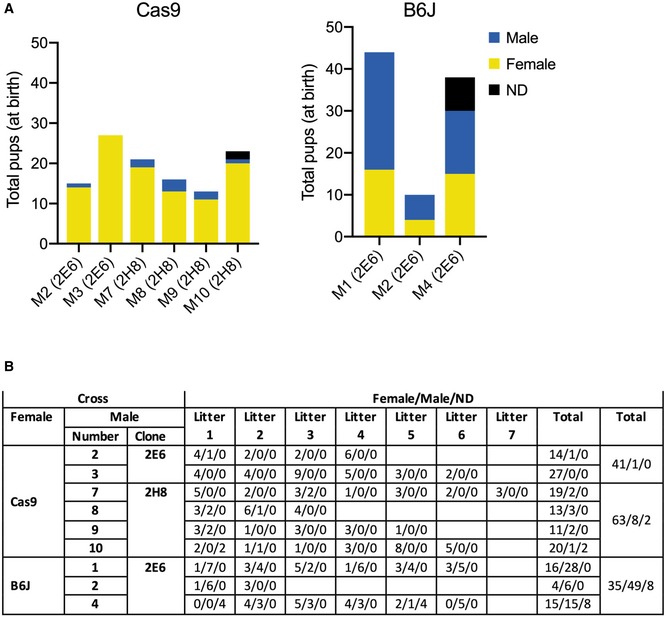
-
A, BTotal pups and their sex resulting from crosses between the indicated males from indicated clones (2E6 and 2H8—detailed in the Appendix) and the indicated females in graphical (A) and numerical (B) representations. ND, not determined due to parental infanticide.
In the control cross, 12 mice did not survive to weaning, whereas in the cross of the Y‐line and the Cas9‐line, 23 mice did not survive to weaning. Six of the dead pups from the latter cross were males, as determined by PCR amplification of the Y chromosome. One of these males was deformed, lacking developed limbs, and most likely died upon birth, suggesting that the genetic system eliminated it (Fig 2). Two appeared smaller compared to their siblings suggesting that their premature death was also caused by the genetic system (Fig 2). Indeed, DNA sequencing of the three target genes demonstrated that they were all disrupted in these three males to different extents (Fig 2). We further analyzed DNA from the three surviving males from the cross between the Y‐line males and Cas9‐line females. Also in these mice, at least two target genes were disrupted to different extents (Fig EV3). Although no sign of illness was observed, one male from the surviving three was found dead after 13.5 weeks. At 4 months of age, the other two males showed no signs of illness. As a control that the targeting is specific for males, we sequenced samples from a female from the cross of the Y‐line and the Cas9‐line that died within 3 days, and as a control that the gRNA by itself does not target these genes, we sequenced samples from a male from the cross of the Y‐line with B6J. Both controls showed that all these target genes were intact (Fig EV3). These results indicate that all males were specifically targeted by the genetic system, but they also indicate that lethality could be delayed or abolished probably due to differences in the type and extent of disruption of the target genes. These sporadic occurrences of male late lethality or even viability could likely be eliminated by simultaneous targeting of more genes using gRNAs that target multiple chromosomal regions 23 or by addition of gRNAs targeting more genes than the current system.
Figure 2. Disruption of male gene targets by the genetic system.
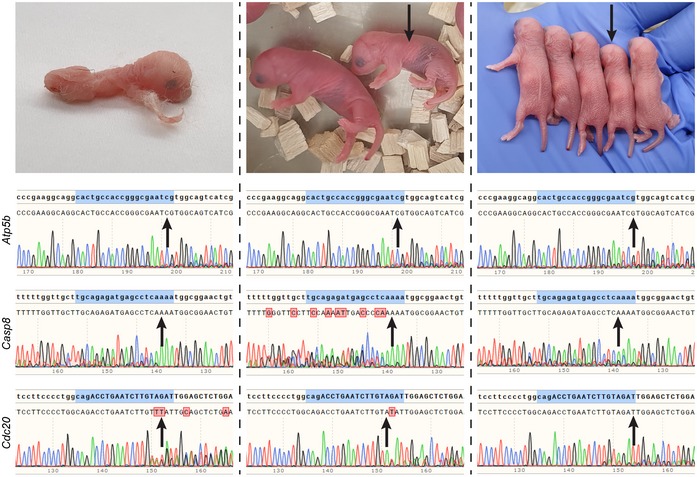
Pictures of a deformed (top left) and of small males (arrowed, top center and right) born from a cross between the Y‐line males and the Cas9‐line females. These males died within 3 days from birth. Below each picture are chromatograms of Sanger DNA sequencing of the indicated PCR amplified genes from samples of these males taken at birth. Targeted regions homologous to the gRNAs are highlighted in blue. Red boxes represent deviations between the expected and observed sequences. Arrows point to indel locations (P < 0.001, two‐tailed t‐test of the variance–covariance matrix of the standard errors), as determined by Tracking of Indels by Decomposition 27.
Figure EV3. Partial disruption of target genes in viable male pups.
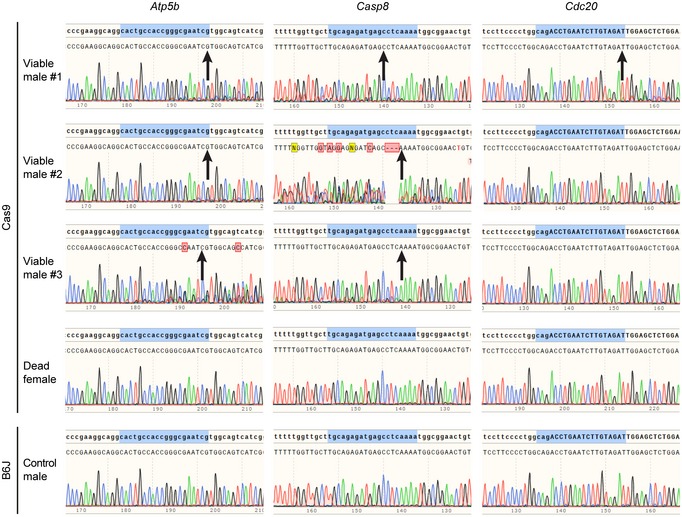
Chromatograms of Sanger DNA sequencing of the indicated PCR amplified genes from samples of mice taken at P0 from the cross of the Y‐line males and the indicated females. Targeted regions homologous to the gRNAs are highlighted in blue. Red boxes represent deviations between the expected and observed sequences. Arrows point to indel locations (P < 0.001, two‐tailed t‐test of the variance–covariance matrix of the standard errors), as determined by Tracking of Indels by Decomposition 27.
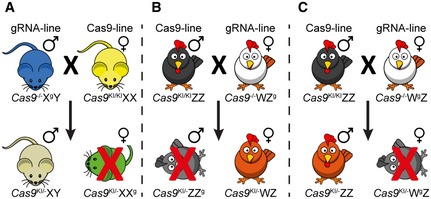
Based on similar principles, one can also establish lines producing only male progeny. For such an outcome, the paternal line should be engineered to encode the gRNAs on its X chromosome and should be crossed with the maternal Cas9‐line (Fig EV4A). Applying this system in animals in which the female is the heterogametic organism, such as chicken, where the female carries the Z and W sex chromosome, and the male carries two copies of the Z chromosome 24 can also be manipulated similarly. In these cases, for female‐only progeny, the Z chromosome of the maternal line should encode the gRNAs, and the paternal line should encode Cas9 (Fig EV4B). For male‐only progeny, the maternal W chromosome should encode the gRNAs and paternal line should encode Cas9 (Fig EV4C). Thus, this system can be manipulated to accommodate changing requirements in different organisms and for both sexes.
Figure EV4. Alternative biasing of sex in mice and birds.
-
A–CSchematic illustrations of hypothetical crosses between (A) mice producing only males, (B) birds producing only females, and (C) birds producing only males.
The females obtained using this approach are genetically modified organisms (GMO) because they inherit a copy of the Cas9 enzyme in their genome. In mice, these are normal‐looking, fertile, and viable animals 19. Nevertheless, from a regulatory point of view, a non‐enzymatic transgene, such as short gRNAs, may be considered more tolerable than the Cas9 nuclease is. Thus, the system can be modified to obtain females that instead encode the gRNAs in their genome, if in that case Cas9 is encoded on the paternal Y chromosome. In this respect, it is noteworthy that GMO crops are approved for consumption by the US Food and Drug Administration (FDA). Furthermore, a GMO salmon, which grows faster than its parental non‐GMO strain, owing to a transgene regulating a growth hormone, has been approved by the FDA for the food industry 25. Thus, GMOs may, in principle, be approved for food production.
A commercial GMO must meet several statutory requirements for safety and effectiveness under the FDA act. First, the GMO products must be safe to consume. Second, the introduced DNA must be safe to the modified organism itself. Third, the modified organism should be superior, at least in one trait, over a non‐GMO. Lastly, the potential environmental impact of the GMO should be non‐significant. Thus, before the proposed genetic system could be used for producing, e.g., cattle, a food safety comparison between the meat and milk from non‐GM and GM cows must first be conducted. The test should compare key hormones, such as estradiol, testosterone, insulin‐like growth factor‐1, and other hormones in the samples. It should also assess the key nutritional constituents such as protein, carbohydrate, and fat levels. No significant difference between the samples should be found in both tests. We expect that animals produced by the proposed genetic system will pass these tests, as the transgenes that are used are not known to have any direct impact on the levels of these factors. The transgenes in the proposed system are harmful to the GM male animal, as few males are born with genetic defects and deformations (Fig 2). Although only a minor percentage of males are born with defects, such a system cannot be approved in its present form. A more robust elimination system should be applied, which halts development of all male zygotes in utero. One way to achieve such elimination is by targeting the essential genes with more than a single gRNA for each target. As shown in Fig 2, all gene targets were disrupted to some extent in the males; however, in some cases, this was insufficient to eliminate them in utero. Using, multiple gRNAs against each gene, should ensure that at least one gene would be deactivated at the early embryo stage, thus resulting in lethality, and consequently in a transgene that does not affect the safety of the male GMO. It is noteworthy that most of the male embryos probably die early during the pregnancy. A thorough study characterizing 5,000 knockout mice lines showed that only 4% of 410 lethal embryonic phenotypes occurred between E12.5 and E18.5, whereas the rest are lethal at earlier stages 20. We speculate that in our experiments too, lethality of most of the male embryos occurred early (i.e., prior to E12.5) and was absorbed during the pregnancy. This speculation is supported by the fact that only one deformed male pup was born (Fig 2) and also with the choice of target genes: Atp5b, Cdc20, and Casp8, all shown to be essential for mouse early development 20, 21, 22. The safety of the transgene to the female GMOs, which are the desired animals in the system, is arguably sound. Each of the transgenes (Cas9 or gRNAs) alone does not cause any DNA damage by itself. The female embryo does not produce at any stage the gRNAs, as they are encoded on the Y chromosome. We have further validated by DNA sequencing of a control female that the target genes are intact (Fig EV3). In addition, female offspring of the Cas9‐ and Y‐line cross showed no sign of illness over a monitoring period of 4 months. Furthermore, these females were examined at 6 weeks of age for fertility by crossing them with Cas9 males and monitoring the pregnancy and sex ratio of their pups. All examined females were fertile, providing normal litter size and normal sex ratio, as expected (average litter size was 6.75 totaling 14 males, 12 females, and one infanticide). Thus, it is probable that the safety and welfare of the GMO females are not affected compared to their non‐GMO counterparts. Regarding efficacy, the system produces significantly higher proportion of desired females compared to the non‐GMO. This ratio will further be increased when a more robust elimination system is used, as indicated above. A system producing solely females without males will completely overcome the requirement for manual separation of the sexes. Lastly, environmental safety should be maintained by multiple redundant containment conditions to prevent their escape into the wild and consequent propagation there. For example, the animals should be fenced at all time and the husbandries should be locked to prevent theft and unauthorized and untraceable distribution of the animals. Small location devices can also be individually applied for large animals such as cows, in which the device's cost is minor compared to the animal's cost. It is important to note that a GM cow inadvertently released to the wild or escaping the husbandry, will unlikely cause a significant long‐term damage, and will most likely be recaptured. This is in contrast to smaller fast‐reproducing animals such as fish in which case the containment methods should be multi‐layered including biological barriers (e.g., infertility) in addition to physical barriers of escape. Thus, it may be assumed that the environmental impact of GM cows would be considered insignificant. Overall, the proposed system at its current form would only partially meet the statutory requirements for safety and effectiveness. Nevertheless, we believe that upon further improvements listed above, it may satisfy these requirements.
The current leading approach for manipulating the sex ratio of domesticated livestock is by using sexed semen, which is sorted for desired gametes by flow cytometry 26. This methodology constitutes only 5% of the artificial insemination market partially because it constitutes a substantial economic burden, shows reduced fertility rates, and its efficacy in biasing the sex ratio is variable 26. Sexing the semen is nevertheless superior to the current proposed genetic system, as the litter size is not reduced, and the offspring is not GMO. There are therefore further obstacles for optimizing the current methodology for biasing the sex and for making it commercially sound. Nevertheless, the proposed proof of principle is a first step in this direction.
Materials and Methods
Ethical statement
All animal experiments conformed with the guidelines of Tel Aviv University's Animal Ethics Committee, which follow the state law of prevention of animal cruelty (1994), the guidelines for preventions of animal cruelty published by the council for animal experimentation (2001), and The NRC Guide for the Care and Use of Laboratory Animals. The animal facilities in Tel Aviv University also work under a permit by the NIH [F16‐00009 (A5010‐01)].
Housing and husbandry
All mice were housed and bred under specific pathogen‐free conditions maintained in the accredited animal facility in the Sackler Faculty of Medicine at Tel Aviv University. Mice were housed with an inverse 12‐h day–night cycle with lights on at 7:00 AM in a temperature (22 ± 1°C)‐ and humidity (55 ± 5%)‐controlled room. All mice were allowed free access to water and food including sunflower seeds. All cages contained wood shavings, bedding, and a cardboard tube for environmental enrichment.
Experimental animals
For all breeding experiments, males and females (> 20 g), 8–52 weeks of age, were used (n = 29). All obtained mice were acclimatized for at least 96 h. Vendor health reports indicated that the mice were free of known viral, bacterial, and parasitic pathogens.
Two independent mice of the Y‐line (generated from two positive ES cloned: 2E6 and 2H8) were constructed by Cyagen Biosciences (California, USA). These C57BL/6N mice encode the following guide RNAs on their Y chromosome: 5′‐CACTGCCACCGGGCGAATCG‐3′; 5′‐CAGACCTGAATCTTGTAGAT‐3′; 5′‐TGCAGAGATGAGCCTCAAAA‐3′ targeting the genes Atp5b, Cdc20, and Casp8, respectively. These guides were cloned into a vector targeting the reverse orientation of the 2nd exon of the Y chromosome Uty gene, which is not part of the pseudoautosomal Y region. Figure EV1 provides a schematic summary, and the Appendix provides detailed description of the Y‐line construction.
Mice of the Cas9‐line were purchased from Jackson Laboratories (Stock No: 026179; Rosa26‐Cas9 knockin on B6J) 19. These mice encode a cassette in the Rosa26 locus on chromosome 6 constitutively expressing the SpCas9 endonuclease from a CAG promoter.
Study design
Ten Cas9‐line and five B6J females were crossed with Y‐line males. Six F1 females from the Cas9‐line and the Y‐line cross were further crossed with Cas9 males to confirm their normal breeding and offspring sex ratio.
Experimental procedures
Pregnancy was monitored daily in the above crosses for any sign of stress. Sex was determined by PCR of the Y chromosome on DNA extracted from the animal's tail. Sex was further confirmed at day 7 and at weaning by observing the genitals. Sanger sequencing of the target regions was carried out following PCR amplification of these regions (see Appendix Tables S1 and S2 for oligonucleotides and PCR set‐ups).
Breeding pairs were monitored daily and pups were monitored twice a day till weaning and twice a week following weaning. Signs of illness (decreased mobility, ruffled fur, pause in weight gaining, or labored breathing) were monitored by researchers and animal technicians and by consultation with a veterinarian. During the entire duration of the experiments, none of the Y‐line males or their pups displayed overt signs of sickness, except the reported deformed male (Fig 2).
Statistics
Data are presented as the mean ± SD. Comparisons were performed using two‐tailed unpaired parametric t‐test or two‐tailed binomial test, assuming normal distribution or two‐tailed t‐test of the variance–covariance matrix of the standard errors.
Author contributions
Conceptualization, IY, UQ; Methodology, IY, LE‐B, RG, IS, AM, MG, and UQ; Investigation, IY, IS, RG, LE‐B, MG, and UQ; Writing—Original Draft, UQ; Writing—Review & Editing, IY, MG, AM, and UQ; Funding Acquisition, MG and UQ; Supervision, MG and UQ.
Conflict of interest
I.Y., M.G., and U.Q. have submitted a provisional patent on the described technology in January 2018. Other authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
The research was funded by the European Research Council StG program (grant 336079), the European Research Council PoC program (grant 811322), the Israel Science Foundation (grants 268/14, 1416/15, 818/18), the Israeli Ministry of Science and Technology (grant 3‐14351), and the Varda and Boaz Dotan Research Center.
EMBO Reports (2019) 20: e48269
See also: MJ Smanski & D Zarkower (August 2019)
Contributor Information
Motti Gerlic, Email: mgerlic@post.tau.ac.il.
Udi Qimron, Email: ehudq@post.tau.ac.il.
References
- 1. Levy T, Rosen O, Eilam B, Azulay D, Aflalo ED, Manor R, Shechter A, Sagi A (2016) A single injection of hypertrophied androgenic gland cells produces all‐female aquaculture. Mar Biotechnol 18: 554–563 [DOI] [PubMed] [Google Scholar]
- 2. Liu H, Guan B, Xu J, Hou C, Tian H, Chen H (2013) Genetic manipulation of sex ratio for the large‐scale breeding of YY super‐male and XY all‐male yellow catfish (Pelteobagrus fulvidraco (Richardson)). Mar Biotechnol 15: 321–328 [DOI] [PubMed] [Google Scholar]
- 3. Yamamoto TO (1975) A YY male goldfish from mating estrone‐induced XY female and normal male. J Hered 66: 2–4 [DOI] [PubMed] [Google Scholar]
- 4. Chevassus B, Devaux A, Chourrout D, Jalabert B (1988) Production of YY rainbow trout males by self‐fertilization of induced hermaphrodites. J Hered 79: 89–92 [DOI] [PubMed] [Google Scholar]
- 5. Thresher R, van de Kamp J, Campbell G, Grewe P, Canning M, Barney M, Bax NJ, Dunham R, Su B, Fulton W (2014) Sex‐ratio‐biasing constructs for the control of invasive lower vertebrates. Nat Biotechnol 32: 424–427 [DOI] [PubMed] [Google Scholar]
- 6. Hickey WA, Craig GB Jr (1966) Genetic distortion of sex ratio in a mosquito, Aedes aegypti . Genetics 53: 1177–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Novitski E, Hanks GD (1961) Analysis of irradiated Drosophila populations for meiotic drive. Nature 190: 989 [DOI] [PubMed] [Google Scholar]
- 8. Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MA, Chen XG et al (2015) SEX DETERMINATION. A male‐determining factor in the mosquito Aedes aegypti. Science 348: 1268–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galizi R, Hammond A, Kyrou K, Taxiarchi C, Bernardini F, O'Loughlin SM, Papathanos PA, Nolan T, Windbichler N, Crisanti A (2016) A CRISPR‐Cas9 sex‐ratio distortion system for genetic control. Sci Rep 6: 31139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, Burt A, Stoddard BL, Windbichler N, Crisanti A (2014) A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun 5: 3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, Nolan T, Crisanti A (2018) A CRISPR‐Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol 36: 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S et al (2016) A CRISPR‐Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae . Nat Biotechnol 34: 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kandul NP, Liu J, Sanchez CH, Wu SL, Marshall JM, Akbari OS (2019) Transforming insect population control with precision guided sterile males with demonstration in flies. Nat Commun 10: 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell‐Badge R (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351: 117–121 [DOI] [PubMed] [Google Scholar]
- 15. Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, Cai C, Qian G, Capel B (2018) The histone demethylase KDM6B regulates temperature‐dependent sex determination in a turtle species. Science 360: 645–648 [DOI] [PubMed] [Google Scholar]
- 16. Harris A, Siggers P, Corrochano S, Warr N, Sagar D, Grimes DT, Suzuki M, Burdine RD, Cong F, Koo BK et al (2018) ZNRF3 functions in mammalian sex determination by inhibiting canonical WNT signaling. Proc Natl Acad Sci USA 115: 5474–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quinn A, Koopman P (2012) The molecular genetics of sex determination and sex reversal in mammals. Semin Reprod Med 30: 351–363 [DOI] [PubMed] [Google Scholar]
- 18. Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev 87: 1–28 [DOI] [PubMed] [Google Scholar]
- 19. Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M et al (2014) CRISPR‐Cas9 knockin mice for genome editing and cancer modeling. Cell 159: 440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H et al (2016) High‐throughput discovery of novel developmental phenotypes. Nature 537: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M, York JP, Zhang P (2007) Loss of Cdc20 causes a securin‐dependent metaphase arrest in two‐cell mouse embryos. Mol Cell Biol 27: 3481–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaiser WJ, Upton JW, Long AB, Livingston‐Rosanoff D, Daley‐Bauer LP, Hakem R, Caspary T, Mocarski ES (2011) RIP3 mediates the embryonic lethality of caspase‐8‐deficient mice. Nature 471: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, He B, Wang X, Shi L, Ping J et al (2017) CRISPR/Cas9‐mediated targeted chromosome elimination. Genome Biol 18: 224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takagi N, Sasaki M (1974) A phylogenetic study of bird karyotypes. Chromosoma 46: 91–120 [DOI] [PubMed] [Google Scholar]
- 25. Waltz E (2017) First genetically engineered salmon sold in Canada. Nature 548: 148 [DOI] [PubMed] [Google Scholar]
- 26. Holden SA, Butler ST (2018) Review: Applications and benefits of sexed semen in dairy and beef herds. Animal 12: s97–s103 [DOI] [PubMed] [Google Scholar]
- 27. Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: e168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One‐step generation of mice carrying mutations in multiple genes by CRISPR/Cas‐mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


