Abstract
Nonsense‐mediated mRNA decay (NMD) was originally coined to define a quality control mechanism that targets mRNAs with truncated open reading frames due to the presence of a premature termination codon. Meanwhile, it became clear that NMD has a much broader impact on gene expression and additional biological functions beyond quality control are continuously being discovered. We review here the current views regarding the molecular mechanisms of NMD, according to which NMD ensues on mRNAs that fail to terminate translation properly, and point out the gaps in our understanding. We further summarize the recent literature on an ever‐rising spectrum of biological processes in which NMD appears to be involved, including homeostatic control of gene expression, development and differentiation, as well as viral defense. WIREs RNA 2016, 7:661–682. doi: 10.1002/wrna.1357
This article is categorized under:
-
1
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
-
2
RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms
-
3
RNA Turnover and Surveillance > Regulation of RNA Stability
INTRODUCTION
More than 30 years ago, it was discovered that mutations terminating the open reading frame (ORF) prematurely reduce the half‐life of the affected messenger RNA (mRNA) in yeast and human cells.1, 2 In 1993, Peltz et al. introduced the term ‘nonsense‐mediated mRNA decay’ (NMD) to describe this phenomenon.3 Thus, in these early days, NMD was essentially defined by the substrates of the process: it stood for the specific degradation of mRNAs harboring premature termination codons (PTCs) and hence NMD was viewed as a quality control mechanism that prevents cells from producing potentially deleterious truncated proteins. While it is clear that quality control of gene expression is indeed one biological function of NMD, the classical definition has changed during the last 10 years, as genome‐wide studies revealed that the stability of many apparently ‘normal’ mRNAs (i.e., encoding a functional, full‐length protein) was also altered by NMD.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 NMD directly or indirectly influences the steady‐state levels of approximately 10% of mRNAs in mammalian cells thereby making a significant contribution to the posttranscriptional regulation of gene expression. We have only just begun to identify the biological functions and pathways that are regulated by NMD and the overall importance of NMD in regulating gene expression is not yet clear. The first insights into different biological functions of NMD are summarized in the second part of this review. Interestingly, there exists a correlation between the complexity of an organism and its dependency on NMD: while NMD is essential in vertebrates,14, 15 NMD‐deficient Caenorhabditis elegans exhibit a vulva malformation phenotype but remain fertile, and NMD‐deficient Saccharomyces cerevisiae strains grow well under laboratory conditions.
Despite the plethora of available biochemical data that have been comprehensively summarized in recent reviews, the molecular mechanism of NMD still remains elusive and we will discuss several models that have been proposed based on data from different organisms. While these models differ in several aspects, they all agree that NMD depends on translation. Translation inhibitors (e.g., cycloheximide, emetine, and puromycin) and extended stem–loops in the 5′ UTR that inhibit translation initiation both effectively inhibit NMD.16, 17 The finding that even PTC‐free mRNAs can be degraded by NMD revitalized a central question that is currently under intensive investigation: what determines whether an mRNA becomes a substrate for NMD? Moreover, the PTC‐centric definition of NMD became obsolete and based on the current mechanistic models, which we discuss in the first part of this review, NMD can currently be defined as an mRNA degradation pathway that requires a number of well‐characterized NMD factors (see below) and targets transcripts that fail to properly terminate translation at their stop codons. But before immersing ourselves in the discussion of different NMD models, we need to first get familiar with the involved NMD effectors.
NMD FACTORS
Genetic screens in S. cerevisiae led to the identification of the first three NMD factors called up‐frameshift (Upf) 1, 2, and 318 and genetic screens in C. elegans for suppressors with morphogenetic effects on genitalia (SMG) revealed seven NMD effectors (SMG‐1 to SMG‐7).19 SMG‐2, SMG‐3, and SMG‐4 turned out to be homologous to the yeast factors Upf1, Upf2, and Upf3, respectively, and they constitute the evolutionarily conserved core set of NMD factors that is present in all late‐branching eukaryotes.20 In contrast, homologs of SMG‐1, SMG‐5, SMG‐6, and SMG‐7 are only found in metazoans (Figure 1).
Figure 1.
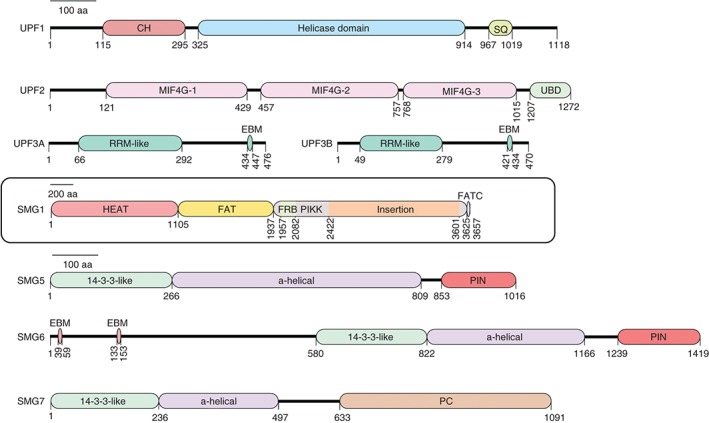
Schematic illustration of domains and motifs of important mammalian NMD factors. Amino acid numbering relates to the human proteins. All proteins are drawn to the same scale except for SMG1. CH: cysteine‐histidine rich domain; SQ: serine‐glutamine rich domain; MIF4G: middle of 4G‐like domains; UBD: UPF1‐binding domain; RRM: RNA recognition motif; EBM: exon junction binding motif; HEAT: Huntingtin, elongation factor 3 (EF3), https://en.wikipedia.org/wiki/Protein_phosphatase_2#Proteinphosphatase2A (PP2A), yeast kinase TOR1 domain; FAT: focal adhesion kinase domain; PIKK: phosphatidylinositol 3‐kinase‐related protein kinase domain; FATC: C‐terminal FAT domain; PIN: PilT N‐terminus domain; PC: C‐terminal proline‐rich region.
UPF1 is a monomeric, highly regulated superfamily 1 (SF1) helicase that is essential for NMD in all eukaryotes. Its central helicase domain is composed of two flexible RecA domains with the ATP‐binding site located in the cleft between the two domains.21 The helicase domain binds single‐stranded RNA and DNA and elegant in vitro experiments using magnetic tweezers recently revealed that UPF1 has the ability to translocate on nucleic acids slowly but very processively and to unwind long double‐stranded structures.22 Its affinity for RNA is lower in the ATP‐bound state23, 24 and the ATPase and helicase activity is essential for NMD in both S. cerevisiae and humans.25, 26 The central helicase of UPF1 is flanked by a conserved N‐terminal domain rich in cysteine and histidine (CH) and a serine‐ and glutamine‐rich (SQ) C‐terminal domain, which in metazoans gets phosphorylated by SMG1 at multiple SQ motifs.27 The CH and SQ domains both suppress UPF1's helicase activity in vitro 28 and UPF2 interaction with the CH domain induces a large conformational change in UPF1, which is a prerequisite for its phosphorylation and promotes the ATPase and helicase activity of UPF1.21, 29, 30, 31, 32 The link between UPF1 phosphorylation and its helicase activity is currently not understood, but likely constitutes a central aspect of NMD activation.
UPF2, the second core NMD factor, functions as a ring‐like scaffold linking UPF1 and UPF3.29, 33, 34 The UPF2 protein consists of three middle portion of eIF4G (MIF4G) domains with the first two domains providing structural support35 and the third domain interacting with UPF3B.36 A highly conserved portion of the first MIF4G domain was recently shown to be essential for NMD in yeast.37 UPF1 interacts with the C‐terminal part (UBD) of UPF2.38
UPF3 is the least conserved of the three core NMD factors.39 Vertebrates contain two UPF3 paralogs, UPF3A and UPF3B, which in humans both encode to alternatively spliced mRNA isoforms. In human cells, UPF3B appears to be the main contributor to NMD and only upon UPF3B depletion, UPF3A is stabilized and substitutes for 3B in NMD.40, 41 UPF3 contains an N‐terminal RNA recognition motif (RRM) that contrary to expectation does not bind RNA but instead is the interaction surface to UPF2.36 Through a short motif in the C‐terminus (called the EJC‐binding motif; EBM), vertebrate UPF3 interacts with the EJC core factors eIF4A3, MAGOH, and Y14.29, 42 UPF3 is a shuttling protein that at steady state is found primarily in the nucleus,38 where it is thought to associate with the EJCs deposited on the newly spliced mRNAs. Like UPF2, UPF3 has also been shown to stimulate the ATPase and helicase activity of UPF1 in vitro.29
The metazoan‐specific SMG1 complex consists of the phosphatidylinositol 3‐kinase‐related kinase SMG1, which is responsible for UPF1 phosphorylation, and the two regulatory factors SMG8 and SMG9, which interact with SMG1's N‐terminal extended stretch of HEAT repeats and confine SMG1 in a kinase inactive conformation.27, 43, 44, 45 The C‐terminus of SMG1 encompasses the catalytic PIKK domain flanked by an FRB (FKBP12‐rapamycin‐binding) domain and two FAT domains and forms the globular ‘head’ region, while the N‐terminal HEAT repeats are included in the ‘arm’ region.32, 43, 46 Upon dissociation of SMG8 and SMG9 from SMG1, the kinase is activated by the third MIF4G domain of UPF2 interacting with the FRB domain of SMG1 and by the helicase domain of UPF1 interacting with a region proximal to the FRB domain.32
SMG5, SMG6, and SMG7 all contain a domain formed by nine antiparallel α helices that fold similarly to 14‐3‐3 proteins.47 The 14‐3‐3‐like domains of SMG5 and SMG7 interact with each other back‐to‐back in a perpendicular orientation48 and as a heterodimer interact with phosphorylated S1096 and additional phosphorylated residues in the C‐terminus of UPF1.49, 50 SMG5 and SMG7 co‐immunoprecipitate protein phosphatase 2A (PP2A) and thus are thought to play a role in the dephosphorylation of UPF1.49, 50 The C‐terminal portion of SMG7 was recently shown to recruit the CCR4‐NOT deadenylase complex by interacting with CNOT8 (POP2),51 providing a molecular link to RNA degradation. In contrast to the SMG5–SMG7 heterodimer, SMG6 appears to function as a monomer. Like SMG5, it contains a C‐terminal PIN domain folded similarly to the RNase H family ribonucleases, but unlike SMG5, which lacks the canonical motif of three aspartic acid residues essential for ribonuclease activity,52 the PIN domain of SMG6 is an active endonuclease.53, 54 Accordingly, SMG6‐dependent endonucleolytic mRNA cleavage in the vicinity of NMD‐triggering termination codons (TCs) has been reported in Drosophila and human cells.53, 55, 56, 57 While these studies did not reveal a preferred cleavage sequence for SMG6, an additional recent genome‐wide analysis of SMG6‐ and UPF1‐dependent 5′ termini of decay intermediates found the pentameric (U/A)‐(G/A)↓(A/C)‐N‐(C/U) motif in >60% of the detected cleavage sites.58 SMG6 associates with UPF1 in a phosphorylation‐dependent and in a phosphorylation‐independent way. The former interaction involves the phospho‐T28 of UPF1,50 and the latter occurs between a low‐complexity region of SMG6 located just N‐terminal to the 14‐3‐3‐like domain and the unique stalk region of the UPF1 helicase domain plus a contribution from the C‐terminal SQ portion.59, 60 At its very N‐terminus, SMG6 has two conserved EBMs through which it interacts with the EJC.61 Notably, not all metazoans possess the full SMG5/6/7 trio: Drosophila melanogaster lacks a SMG7 homolog and plants have two nonredundant SMG7 genes, but no SMG5 and SMG6.
Besides these well‐characterized NMD factors, several additional factors required for NMD have been identified using RNA interference (RNAi) screens in C. elegans.62, 63 Among those, DHX34, NBAS, GNL2, and SEC13 also seem to be required for NMD in human cells. DHX34 and NBAS are highly conserved among metazoans and part of an autoregulatory circuit that regulates endogenous NMD targets.64 Both proteins are required for proper embryonic development of zebrafish and their depletion leads to similar developmental defects as the depletion of UPF1, SMG5, or SMG6.14, 65 The RNA‐binding DExH/D box helicase DHX34 interacts with NMD, decapping, and release factors, and it promotes UPF1 phosphorylation and the interaction of UPF1 with UPF2 and EJC factors.66 Based on these results, DHX34 was proposed to function in the conversion of the SURF to the DECID complex (see below).
NMD TRIGGERING FEATURES
The finding that many different endogenous mRNAs appear to be targets for NMD put the question of substrate recognition into the limelight of current NMD research. What are the features that distinguish an NMD‐targeted transcript from one that remains unaffected by NMD? In mammalian cells, EJCs located sufficiently downstream of the TC that they are not displaced by elongating ribosomes can induce NMD by recruiting NMD factors to such mRNPs.34, 42, 67 In addition, mRNAs with long 3′ UTRs or with upstream ORFs (uORFs) are also frequently targeted by NMD in mammalian as well as in yeast cells.7, 13, 68, 69, 70, 71 However, it is currently not possible to predict which transcripts will be subjected to NMD and which ones remain intact based on sequence features. In fact, many mRNAs with long 3′ UTRs are resistant to NMD13, 56, 72, 73 and not all uORFs or PTCs trigger NMD.73 For a subset of the NMD‐insensitive transcripts with long 3′ UTRs, AU‐rich sequences located within the first 200 nucleotides downstream of the TC have been recently shown to confer NMD resistance.74 Another well‐characterized NMD‐inhibiting sequence is the retroviral RNA stability element (RSE) of the Rous sarcoma virus (RSV), which is located downstream of the TC of the Gag gene and protects the unspliced viral transcript from being degraded by NMD.75 The RSE has recently been shown to interact with the polypyrimidine tract‐binding protein 1 (PTBP1), which suppresses NMD by sterically blocking access of UPF1 to this region.76
CURRENT WORKING MODEL FOR NMD
Despite intensive research over the last two decades that produced a wealth of biochemical and genetic data, our insights into the molecular mechanism of NMD still remain fragmented. Several different NMD models have been proposed and revised over the years and we will describe here a current working model that is based on the pooled data of many different groups and to which we refer as the ‘unified model’ (Figure 2). In its most condensed form, the model posits that NMD results from aberrant translation termination.77, 78, 79, 80 Increased toe‐print signals at NMD‐triggering TCs observed in yeast extracts81 and rabbit reticulocyte lysate82 indicated that ribosomes reside longer at such TCs, which implies mechanistic differences to the faster termination process at normal TCs.
Figure 2.
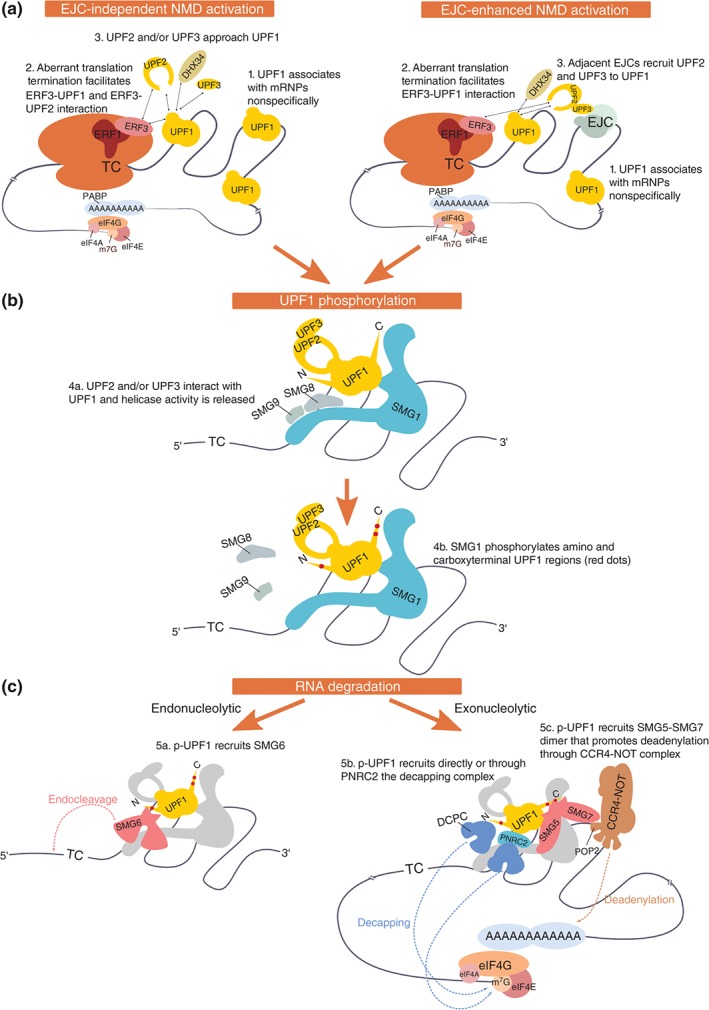
Nonsense‐mediated mRNA decay activation and target degradation. (a) Aberrant translation termination occurs when the termination codon (TC) is distant to the poly(A) tail. NMD licensing includes the interaction of ribosome‐bound ERF3 with UPF1 instead of PABP, and UPF1 activation relies on the recruitment of UPF2 and/or UPF3. In this model, UPF1 is proposed to initially bind mRNA nonspecifically and interact with ERFs on aberrantly terminating ribosomes. UPF1 is activated by UPF2 and/or UPF3 (EJC‐independent NMD activation). Adjacent EJCs enhance UPF2/UPF3 recruitment and therefore facilitate NMD activation (EJC‐enhanced NMD activation). (b) SMG1 kinase is activated when its inhibitory counterparts SMG8 and SMG9 are released, which is facilitated by UPF2 and DHX34. SMG1 phosphorylates UPF1 on N‐ and C‐terminal Ser and Thr residues followed by Gln (S/T‐Q). The helicase activity of UPF1 stimulates downstream NMD events. (c) Phosphorylated UPF1 (p‐UPF1) can promote mRNA degradation by at least three distinct mechanisms: SMG6‐mediated endonucleolytic cleavage (a), recruitment of the decapping complex (DCPC) directly by UPF1 or via PNRC2 (b), or SMG7‐mediated recruitment of the CCR4‐NOT complex (c). NMD‐targeted mRNAs are further degraded by general cellular exonucleolytic activities.
When a ribosome arrives at a TC, the tRNA‐shaped eukaryotic release factor 1 (ERF1) together with GTP‐bound ERF3 binds the A‐site of the ribosome. GTP hydrolysis then leads to the dissociation of ERF3, which is followed by the interaction of the termination factor ABCE1 (Rli1 in yeast) with ERF1.83, 84 An ABCE1‐induced structural rearrangement in ERF1 orients the highly conserved GGQ loop of ERF1 toward the peptidyltransfer center, stimulating the release of the polypeptide chain from the P‐site bound tRNA.85 The ATPase activity of ABCE1 subsequently triggers the dissociation and recycling of the two ribosomal subunits.83, 84, 86 In contrast to bacteria, translation termination and ribosome recycling appear to be mechanistically coupled in eukaryotes.87 Importantly with regard to the NMD model, the C‐terminal part of poly(A)‐binding protein (PABPC1 in mammals, Pab1 in yeast) interacts with ERF388, 89, 90, 91 and PABPC1 can antagonize NMD when tethered downstream of otherwise NMD‐triggering TCs,69, 71, 81, 92, 93, 94 suggesting that PABPC1 promotes correct translation termination.95 However, how exactly PABPC1 contributes to translation termination is not yet known. In addition, UPF1 has also been reported to interact with ERFs31, 93, 96 and there is evidence that in vitro PABPC1 competes with UPF1 for binding ERF3,71 implying that the difference between correct and aberrant, NMD‐eliciting translation termination might ultimately rely on whether PABPC1 or UPF1 interacts with ERF3 at the terminating ribosome.97 Although recent evidence suggests that this ‘UPF1 versus PABPC1 competition model’ might be oversimplified,98, 99, 100 it is consistent with the majority of the available experimental data.
Open Questions Regarding the Role of UPF1 in Determining NMD Targets
Given the evidence for UPF1's presence at terminating ribosomes, the currently most pressing questions are when and how UPF1 is recruited and what exactly are the consequences of this recruitment. Regarding the ‘when and how,’ different studies came to controversial conclusions. On one hand, UPF1 was found to be preferentially associated with transcripts targeted for NMD,101, 102, 103 which suggested that UPF1 recruitment to inefficiently terminating mRNAs was the decisive step to subject such mRNAs to NMD. On the other hand, UPF1 was reported to bind 3′ UTRs in a length‐dependent manner and independently of translation.72 Further support for UPF associating with mRNA independent of translation came from two CLIP studies that detected UPF1 on long noncoding RNAs and along entire mRNAs when cells were treated with translation inhibitors before crosslinking.73, 104 In the absence of translation inhibitors, UPF1 crosslinked primarily to 3′ UTRs, implying that UPF1 binds mRNA before translation begins and elongating ribosomes subsequently strip off UPF1 from the coding sequence.104 The accumulating evidence suggesting that UPF1 associates indiscriminately with NMD targets and nontargets73, 104, 105, 106 begs the question which other step determines whether or not NMD ensues on a given mRNA. Using an antibody that specifically detects UPF1 phosphorylated at serine 1116, it was shown that phospho‐UPF1 preferentially co‐immunoprecipitates NMD‐targeted transcripts,105 suggesting that the SMG1‐mediated phosphorylation of UPF1 might be the NMD‐activating step. However, how aberrant translation termination exactly leads to UPF1 phosphorylation in this scenario remains a crucial unanswered question.
Although a contribution by other kinases cannot be ruled out, SMG1 is the kinase responsible for the phosphorylation of the numerous SQ and TQ motifs located in the N‐ and C‐terminal regions of UPF1.27 Noteworthy, most of the 19 (S/T)Q sites of human UPF1 are conserved in other vertebrates and some of these phosphosites are involved in recruitment of NMD effectors (see below).49, 50 Hypophosphorylated UPF1 was co‐precipitated in a complex with SMG1, ERF1, and ERF3, suggesting that this complex might form on terminating ribosomes.31 Subsequent SMG1‐mediated phosphorylation of UPF1 appears to require a UPF2‐dependent conformational change to activate SMG1.32, 107 Thus, instead of UPF1, it could be that SMG1 or UPF2 preferentially associates with NMD‐susceptible mRNPs.
Alternatively to UPF1 phosphorylation being the step discriminating between NMD targets and nontargets, a recent study indicated that target specificity might be achieved by preferential release of UPF1 from nontargets and that this step requires the ATPase activity of UPF1.108 While wild‐type (WT) UPF1 co‐precipitated preferentially, but not exclusively, NMD‐targeted transcripts, ATPase‐deficient UPF1 mutants co‐precipitated targets and nontargets indiscriminately. Collectively, the results presented in this study suggested that aberrant translation termination might activate NMD by delaying UPF1‐mediated ATP hydrolysis, thereby allowing NMD complexes to form and initiate RNA degradation. However, it is clear that subsequently during NMD, UPF1's ATPase is still required, because ATPase‐inactivating mutations in UPF1 completely abolish NMD.25, 31
SMG1‐Mediated Phosphorylation of UPF1
The so‐called SURF complex (consisting of SMG1 and its regulators SMG8 and SMG9, hypophosphorylated UPF1, ERF1, and ERF3) was shown to interact with DHX34.66 Recent structural and biochemical data indicate that DHX34 might function as a scaffold to recruit UPF1 to SMG1.109 DHX34 further promotes the interaction of UPF2 with UPF166 and UPF2 in turn was shown to induce a large conformational change in UPF1, to stimulate UPF1's helicase activity, and to promote SMG1 kinase activity by dissociating SMG8 from SMG1, finally resulting in the phosphorylation of UPF1.21, 29, 32, 43 The presence of an EJC complex in the 3′ UTR of a SURF‐associated mRNA is thought to greatly enhance the UPF1:UPF2 interaction by serving as an mRNA‐attached anchoring point for the assembly of a decay‐inducing NMD complex on such transcripts.78 Using recombinant proteins, an RNA‐bound EJC core consisting of EIF4A3, Y14, MAGOH, and a fragment of MLN51 could be assembled into a heptameric complex with UPF3B, UPF2, and UPF129 and structural information was obtained by cryo‐electron microscopy combined with modeling of known high‐resolution structures of subcomplexes into the obtained electron density hull.34 Through these interactions, the EJC is thought to enhance NMD activation, but it is important to note that even mRNAs with no predicted EJC in the 3′ UTR can be targeted by NMD, although typically with a lower efficiency.68 It is not known when and how in the EJC‐independent mode of NMD UPF2 and UPF3B are interacting with UPF1, but it is conceivable that in the absence of EJC‐mediated prepositioning of UPF2 and UPF3B on SURF‐associated mRNAs, UPF1 phosphorylation would take longer to occur, thereby increasing the probability for competing interactions (e.g., PABPC1 with ERF3) to promote correct translation termination and inhibit NMD activation during this time window.110 In line with this view, EJC‐independent NMD seems to be more sensitive than EJC‐enhanced NMD to reduced UPF2 and UPF3B concentrations.111 Interestingly, recent findings suggest that UPF2 directly interacts with ERF3 and the ribosome, independently of UPF3B.112
Notably, there is evidence that under certain circumstances or on special transcripts NMD can also occur in the absence of UPF2 or UPF3 in mammalian cells, suggesting the existence of different branches of the NMD pathway with differential co‐factor requirements.113, 114, 115 Based on recent findings, it is conceivable that DHX34 could functionally substitute for UPF2 in promoting UPF1 phosphorylation,109 but further work is needed to clarify this point.
In summary, these findings support the idea that a kinetic competition between efficient translation termination and the assembly of a degradation‐triggering NMD complex determines whether an mRNA survives or not. Interestingly, such a kinetic competition between correct mRNP function and RNA degradation is a recurring concept for many of the posttranscriptional quality control systems in cells.97, 116
NMD Targets CBC‐ and eIF4E‐Bound Transcripts
The above‐described NMD model predicts that every ribosome reaching a stop codon can lead to NMD activation if translation termination fails to take place quick enough. This view however clashed with earlier publications claiming that NMD in mammalian cells could only be elicited during the very first, the so‐called pioneer round of translation or at least that it was restricted to CBC‐bound mRNAs.117 While in yeast it has been long known that NMD targets CBC‐ and eIF4E‐associated mRNAs alike,118, 119 it has recently been demonstrated that the same is also true in human cells,120, 121 consistent with the prediction of the unified NMD model.
Degradation of NMD‐Targeted mRNAs
Irrespectively of the exact mechanisms of NMD target identification, eventually hyperphosphorylated UPF1 will be associated with target RNA,105 and hyperphosphorylated UPF1 serves as the binding platform for the NMD effectors SMG5, SMG6, and SMG7.49, 50 There is evidence that in mammals, rapid degradation of NMD targets can be achieved through at least two different pathways (reviewed in Ref 80) and it is unclear if transcript‐specific or cell type‐specific preferences for one or the other pathway exist, or if they act redundantly.122
SMG6 cleaves mRNA endonucleolytically in the vicinity of PTCs and in human cells, and the majority of NMD targets are apparently degraded through this pathway.53, 54, 56, 57 The decapping‐dependent exonucleolytic pathway may function as a backup, as it is enhanced upon SMG6 depletion.57, 58 After SMG6‐mediated cleavage, for which SMG6 needs to interact with UPF1,60 the 3′ RNA fragment is rapidly degraded by XRN1 and the 5′ fragment seems to be digested by the exosome.53, 54, 55, 56, 57
An alternative RNA decay pathway is promoted by the highly stable heterodimer SMG5–SMG7,48 which interacts with phosphoserines in the C‐terminal region of UPF1. In tethering assays, the C‐terminal portion of SMG7 is sufficient to trigger degradation of the reporter transcript51, 123 and the deadenylase CNOT8 (POP2) interacts with this C‐terminal proline‐rich region of SMG7, indicating that SMG5–SMG7 recruits the CCR4‐NOT complex to NMD targets to induce their deadenylation‐dependent decapping and subsequent XRN1‐mediated degradation.51 Consistently, SMG7‐mediated mRNA decay requires the presence of DCP2 and XRN1 but not of SMG6.123
UPF1 also associates with decapping complex subunits DCP1A, DCP2, and PNRC2, indicating that direct deadenylation‐independent decapping might constitute a third route to degrade aberrantly terminating mRNAs.124, 125, 126, 127, 128, 129, 130 By contacting directly DCP1A and UPF1, PNRC2 is thought to function as the bridge between the NMD and the decapping complexes.127, 130 Recently, PNRC2 was also claimed to induce RNA degradation mainly through an interaction with SMG5,127 but others have not been able to confirm this interaction.51
BIOLOGICAL FUNCTIONS OF NMD
NMD Linked to Alternative Splicing (AS‐NMD) Regulates Gene Expression Levels by Generating Homeostatic Feedback Loops
Classically, NMD has been viewed as a quality control system whose function is to rid mRNAs with prematurely truncated CDS from cells. In mammalian cells, a large portion of such classical PTC‐containing NMD targets arise by unproductive splicing. While a part of these NMD substrates arises by genuine splicing errors due to inherent inaccuracies of the complex underlying biochemical processes, there exist a number of well‐documented cases where proteins involved in regulating splicing regulate the splicing of their cognate pre‐mRNAs.57, 131, 132, 133, 134 This results in homeostatic feedback loops in which splicing of the transcript isoform encoding the correct protein is favored when this protein is at low intracellular concentration, and in which high concentrations of the protein promote the production of an alternatively spliced isoform that contains a PTC and hence is degraded by NMD (Figure 3). Besides most SR proteins, many hnRNPs as well as other proteins involved in mRNA metabolism were found to regulate their own levels in this way. A recent study comparing AS‐NMD events that are conserved between human and mouse brain cortex revealed, besides transcripts involved in mRNA metabolism, a highly significant enrichment of transcripts encoding chromatin modifiers.135
Figure 3.
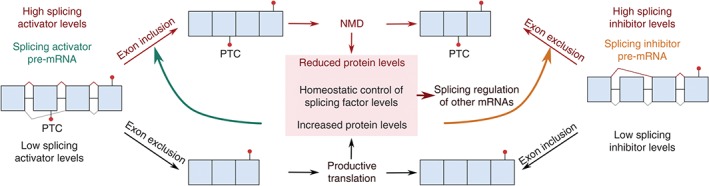
Homeostatic regulation of splicing factor levels by AS‐NMD. Splicing activators and repressors can autoregulate their own protein levels through alternative splicing of their cognate pre‐mRNAs. When the splicing factor is present at high levels in the cell, it inhibits its own expression by stimulating the production of mRNA splice variants that will be targeted for degradation by NMD. Splicing activators stimulate the inclusion of stop codon exons, whereas splicing inhibitors repress the inclusion of a coding exon and hence generate PTC+ mRNAs. The relative levels of splicing activators and inhibitors further impact the alternative splicing of all other pre‐mRNAs in the cell.
An interesting example of an unusual AS‐NMD circuit was recently reported for HPS1, a subunit of a guanine nucleotide exchange factor that is essential for biogenesis of lysosome‐related organelles and mutated in patients with Hermansky–Pudlak syndrome136: PTBP1 promotes utilization of an intrinsically weak 5′ splice site in the HPS1 pre‐mRNA, giving rise to stable mRNA encoding full‐length HPS1 protein. Reduced PTBP1 levels lead to preferential utilization of a stronger 5′ splice site further downstream resulting in an NMD‐susceptible transcript. This regulatory circuit seems to account for the tight correlation of HPS1 and PTBP1 expression levels observed across mammalian tissues. Noteworthy, AS‐NMD is by no means restricted to mammalian cells, and it was also reported in plants,137 zebrafish,64 and even in S. cerevisiae which in general shows very little AS.138 Thus, AS‐NMD represents a widespread autoregulatory gene expression mechanism.
Besides usage of alternative 5′ or 3′ splice sites and exon skipping, intron retention in specific subsets of transcripts has recently been detected to play a role in various cellular differentiation programs. As most transcripts with a retained intron are degraded by NMD, the true extent of intron retention occurring in cells has long been underestimated and became only apparent by recent deep‐sequencing approaches under NMD‐inactivating conditions.139 For example, it was recently shown that normal granulopoiesis requires downregulation of the nuclear protein LMNB1, which occurs by intron retention and NMD of the resulting aberrant mRNAs.140
NMD Factor‐Encoding mRNA Levels Are Regulated by NMD
Another form of feedback regulation was discovered in mRNAs encoding NMD factors, seven of which were found to be targeted by NMD: UPF1, UPF2, UPF3B, SMG1, SMG5, SMG6, and SMG7 mRNAs were upregulated in HeLa cells upon knockdown of other NMD factors.13, 115 All these mRNAs have a longer than average 3′ UTR and some of them also contain uORFs.13 It was shown that the 3′ UTRs of UPF1, SMG5, and SMG7 and the uORF of SMG5 are indeed sufficient to convert a reporter transcript into a substrate for the NMD pathway.13, 71, 115 SMG7 and UPF3 mRNAs in Arabidopsis and SMG5 mRNA in Drosophila have also been reported as NMD targets,9, 141, 142 suggesting that NMD regulation by such a feedback control might be evolutionarily conserved. This autoregulation is most likely important to buffer NMD activity against environmental changes, ensuring robust and tightly controlled NMD activity under different conditions.
NMD Factors Are Required for Mammalian Development
Knockouts of several NMD factors have been attempted in mice but were found to be embryonic lethal, suggesting that NMD might be essential for the normal mammalian embryonic development. For example, homozygous UPF1 knockout mice die shortly after implantation and preimplantation blastocysts cannot be maintained in culture due to massive induction of apoptosis.15 Similarly, SMG1‐deficient mouse embryos die around 8.5 days postcoitum (E8.5) and show severe developmental defects.143 UPF2 is also required for normal embryonic development, because UPF2 knockout mouse embryos die in utero before E9.5.144 Subsequent conditional UPF2 knockout strategies enabled the study of UPF2 functions in different tissues and developmental stages. A conditional UPF2 knockout in the hematopoietic system resulted in a complete loss of hematopoietic stem and progenitor cell populations and subsequent death of the mice within 10 days. In contrast, a specific knockout of UPF2 in the myeloid linage had no gross phenotypic consequences, suggesting that UPF2 might be more important for proliferating than for terminally differentiated cells.144 Specific UPF2 knockout in the liver of mouse embryos at E10 impaired terminal differentiation of liver cells and led to postnatal death, and induced UPF2 knockout in adult mice resulted in extensive liver damage and impaired regeneration.145 Recently, an essential role for UPF2 has also been associated with male fertility. Conditional ablation of UPF2 in mouse embryonic Sertoli cells caused severe testicular atrophy, leading to sterility in adulthood.146
SMG6 knockout mice also show early embryonic lethality, but using a conditional knockout strategy, SMG6−/− mouse embryonic stem cells (ESCs) could be generated that proliferated normally and were morphologically indistinguishable from control ESCs.147 Interestingly, such cultured SMG6 knockout ESCs were unable to undergo differentiation and retained high expression of pluripotency markers, and analysis of chimeric mice showed that the SMG6−/− ESCs failed to differentiate into all three germ layers. Moreover, this study provided compelling evidence that NMD rather than the attributed function of SMG6 in telomere maintenance is responsible for controlling ESC differentiation. c‐MYC mRNA was shown to be an NMD target and NMD inactivation resulted in increased c‐MYC protein levels, which in turn prevented differentiation and kept the ESCs in their pluripotency stage147 (Figure 4(a)).
Figure 4.
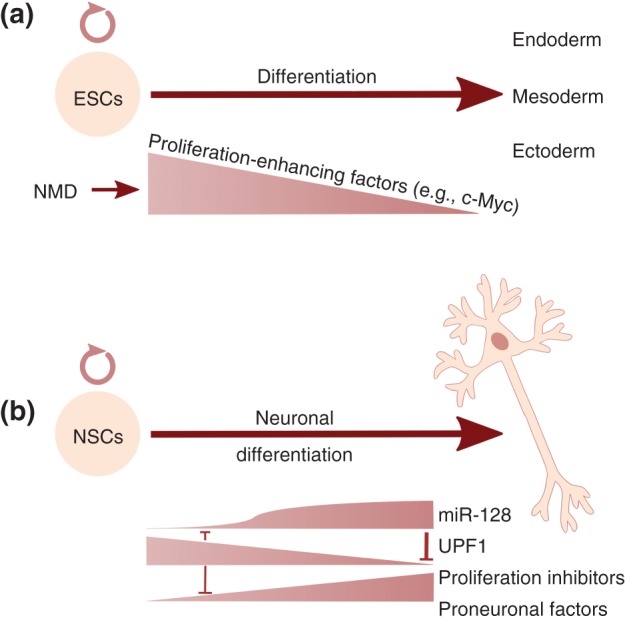
Suggested roles for NMD in stem cell differentiation. (a) NMD promotes the differentiation of ESCs into the three germ layers, at least partially, by downregulating the mRNA levels of pluripotency genes, like c‐Myc.147 (b) UPF1 promotes the proliferative, undifferentiated state of neuronal stem cells, by inducing the decay of mRNAs encoding proneural factors and proliferation inhibitors. Neuronal differentiation is triggered when a neurogenic signal causes a rapid increase in the levels of the neuronally expressed miR‐128, which downregulates UPF1 mRNA by binding to the 3′ UTR of the UPF1 mRNA, and consequently represses NMD. UPF1 and miR‐128, a part of a self‐reinforcing negative feedback control system, can act as a molecular switch to lock in the different cellular states.148
NMD Contributes to Brain Development
Another NMD factor whose depletion has been related to increased self‐renewal and altered cell differentiation is UPF3B.149 UPF3B knockdown in E18.5 mouse cortical neuronal progenitor cells (NPCs) resulted in an increased proliferation capacity of primary NPCs, at the expense of their differentiation capacity. In primary hippocampal neurons, UPF3B downregulation resulted in altered neurite growth, further showing that UPF3B function is also required in postmitotic neurons. UPF3B knockdown altered the expression of SIX3 (a master regulator of cortical development) in NPCs and NRCAM and ROBO1 (involved in axon guidance and growth) in hippocampal neurons, genes whose expression is also deregulated in human patient cells harboring loss‐of‐function mutations in UPF3B.150 A recent study suggests that the neurodevelopmental phenotype of UPF3B missense mutations results from altered neuronal differentiation induced by impaired NMD.151 In rat neural stem cells, the number and complexity of the branching of neurites was significantly reduced in cells expressing UPF3B protein mutants found in patients with neurodevelopmental disorders. In these cells, increased expression of endogenous NMD targets involved in neuronal plasticity and branching was observed (e.g., ATF4 and ARHGAP24 isoform 1). This work shows that despite the observed downregulation of NMD factors during neuronal differentiation, a fully functional NMD machinery appears to be critical for proper differentiation.151
Evidence that downmodulation of NMD activity is an essential step in the differentiation of neural progenitors has recently been reported.148 It was shown that the abundance of several NMD factors (UPF1, UPF2, UPF3B, SMG6, and less pronounced SMG1) was decreased during neuronal differentiation of mouse and human neural stem/progenitor cells, resulting in a significant reduction of NMD activity as exemplified by the increased mRNA levels of several endogenous NMD targets in differentiating P19 cells (GAS5, ATF3, and GADD45B). Prevention of UPF1 downregulation by the expression of modest levels of exogenous UPF1 in differentiating P19 cells resulted in continued expression of proliferation markers and prevented the upregulation of neural differentiation markers. Conversely, knockdown of UPF1 was sufficient to elicit the initial steps of neuronal differentiation in P19 cells. The authors provided evidence that UPF1 promoted the G1/S cell cycle transition by eliciting the decay of mRNAs coding for proliferation inhibitory factors like p21 (CDKN1A), p27 (CDKN1B), p57 (CDKN1C), and ERK3 (MAPK6) and for neural differentiation factors like ASCL1 and POU3F2. In addition, several mRNAs encoding inhibitors of TGF‐β signaling were found to be NMD targets (SMURF1, SMURF2, SMAD6, and SMAD7). Active TGF‐β signaling promotes mesoderm differentiation and its repression leads to neural differentiation of progenitor cells.152 Consistent with this, UPF1 knockdown reduced the ability of P19 cells to differentiate to mesoderm linage and instead promoted neural differentiation. Furthermore, knockdown of SMAD7 prevented the UPF1 knockdown‐induced neuronal differentiation by rescuing TGF‐β signaling, underscoring the view that NMD controls neuronal differentiation by targeting inhibitors of TGF‐β signaling and thus regulating TGF‐β activity.148 A central question in this regulatory circuit is how NMD activity is regulated in the first place. It was shown that UPF1 mRNA has binding sites for the microRNA miR‐128 and that in neuronal precursor cells at the outset of differentiation, induction of miR‐128 reduces UPF1 expression.153 Expression of high levels of a UPF1 mRNA lacking miR‐128‐binding sites inhibited the miR‐128‐induced neural differentiation of P19 cells and vice versa, miR‐128 expression was strongly induced in P19 cells upon UPF1 depletion or inhibition of TGF‐β signaling, revealing mutually reinforcing negative feedback loops that are predicted to form a bistable circuit153 (Figure 4(b)). Interestingly, this UPF1‐miRNA circuitry appears to be highly conserved and was recently documented in Xenopus laevis embryos.148
NMD has also been implicated in commissural axon guidance in the spinal cord.154 When axons cross the ventral midline, they are initially attracted toward the midline by expressing isoform 1 of the guidance cue receptor Robo3 (Robo3.1). After midline crossing, the axons are repelled from the midline due to the loss of Robo3.1 expression and the upregulation of the alternatively spliced Robo3.2 isoform. Robo3.2 mRNA retains an intron that harbors a stop codon and is therefore a predicted NMD target. Colak et al. showed that the Robo3.2 mRNA is selectively trafficked to commissural axons and remains translationally repressed until the axons are exposed to floor plate signals in the spinal cord midline. Upon translation, Robo3.2 transcripts are targeted by NMD, ultimately limiting Robo3.2 protein levels in postcrossing axons. Using UPF2 conditional knockout mice,144 it was shown that NMD‐deficient postcrossing axons exhibit elevated Robo3.2 levels and over‐repulsion from the midline, leading to aberrant postcrossing axonal trajectories.154 Intriguingly, UPF1, UPF2, and SMG1 are also highly enriched in growth cones of different neurons that do not express Robo3, suggesting that localized NMD might play a much broader role in axon physiology of diverse neuronal types.154
Role of NMD in Modulating the Stress Response
Transcriptome‐wide identification of NMD targets revealed many mRNAs coding for stress response factors,7, 10, 57, 58, 144 suggesting that NMD may contribute to the regulation of various stress response pathways (Figure 5). A role of NMD in increasing the threshold for activation of the unfolded protein response (UPR) pathway upon endoplasmic reticulum (ER) stress has recently been reported,155 in agreement with an earlier study documenting an inverse correlation between UPR and NMD.156 Accumulation of unfolded proteins in the ER lumen is sensed by the three transmembrane proteins IRE1, PERK, and ATF6, each of which then activates a network of signaling pathways collectively referred to as the UPR that ultimately leads to reduced global protein synthesis, upregulation of folding‐promoting chaperones, and increased capacity to degrade misfolded proteins.157 Based on knockdowns of UPF1 or UPF3 (A and B forms together), 10 NMD‐targeted transcripts encoding UPR components were identified, among them IRE1α mRNA which was shown to be an NMD target because of its long 3′ UTR.155 The authors suggested that by dampening the levels of these mRNAs, NMD prevents UPR activation by innocuous ER stress, whereas strong ER stress signaling represses NMD to ensure the establishment of a robust adaptive program. Supporting this view, they demonstrated in UPF3B knockout mice that NMD deficiency leads to elevated UPR responses after treatment with low doses of the potent ER stress inducer tunicamycin, whereas high doses of tunicamycin triggered a normal UPR response. Indeed, strong ER stress suppressed NMD by promoting eIFα2 phosphorylation and led to the stabilization of IRE1α mRNA in human and mouse cells. This study moreover provided evidence that NMD plays a role in the timely termination of the UPR and therewith might protect cells from ER stress‐induced apoptosis.155 These findings are supported by a very recent paper showing that inhibition of NMD increases protein synthesis of downstream targets of the IRE1 and PERK pathways, suggesting a positive feedback loop in which ER stress dampens NMD, which in turn promotes the ER stress response.158
Figure 5.
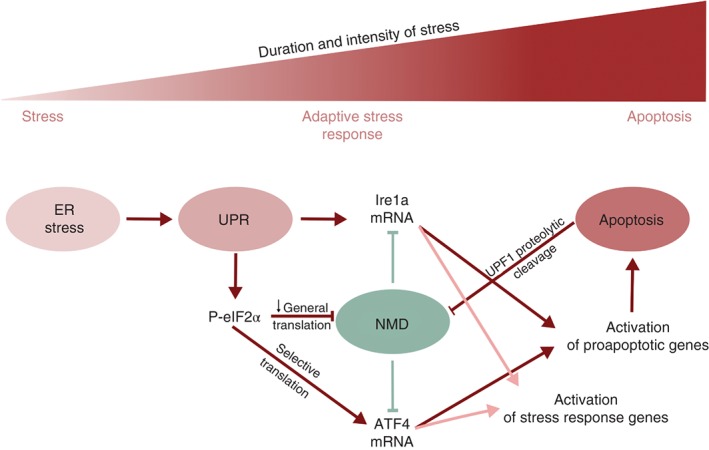
NMD activity is regulated during stress response. Under normal conditions (green) NMD destabilizes IRE1α mRNA, a central UPR component, thereby preventing UPR activation. Under ER stress (red) UPR is activated and inhibits NMD via eIF2α phosphorylation, triggering IRE1α accumulation and hence a robust UPR activation. Stress‐induced inhibition of NMD also results in a higher accumulation of ATF4, a transcriptional activator of the integrated stress response (ISR) with uORFs in its mRNA. Therefore, modulation of NMD activity plays a key role in the initial phase of the stress response. In conditions of prolonged stress, the UPR and ISR pathways activate a proapoptotic program. In cells undergoing apoptosis, NMD activity can be further downregulated by the production of caspase‐derived proteolytic forms of UPF1. This establishes a reinforcing feedback loop that will irreversibly commit the cell to apoptosis.
Other types of stress like serum starvation, hypoxia, and osmotic stress also affect NMD activity, with implications in apoptosis, autophagy, and amino acid transport.159, 160, 161 Although annotated as noncoding RNA, the prosurvival RNA GAS5 is an NMD target with short translated ORFs.162, 163 UPF1 knockdown or serum starvation, which also inhibits NMD due to reduced UPF1 phosphorylation,164 both stabilized GAS5 RNA and so increased its steady‐state levels.160 The elevated GAS5 RNA in turn downregulated apoptosis‐related genes, including the cellular inhibitor of apoptosis 2 (cIAP2) and the serum and glucocorticoid‐regulated kinase 1 (SGK1), and GAS5 knockdown suppressed the effects of serum starvation on the expression of the apoptosis‐related genes.160 NMD inhibition, by UPF1 or UPF2 knockdown or by expressing a dominant‐negative UPF1 mutant, was further found to activate autophagy and augment the intracellular concentration of certain amino acids.161 Vice versa, hyperactivation of NMD repressed the induction of autophagy in response to a variety of cellular stresses, documenting a functional link between an mRNA and a protein surveillance system. Supporting the view that induction of autophagy in response to NMD inhibition represents an adaptive mechanism to rid the cell of misfolded, mutated, aggregated, and otherwise deleterious proteins, cells died when NMD and autophagy were inhibited simultaneously in the colon cancer cell line HCT116.161 The regulation of autophagy by NMD is achieved, at least in part, through the NMD‐sensitive ATF4 transcripts, which directly transactivates the autophagosome membrane component LC3B and ATG5, an E2 ubiquitin ligase necessary for autophagy.161 Based on these results, the authors proposed that co‐administration of pharmacological NMD and autophagy inhibitors could be a selective and effective chemotherapeutic regimen.
Among the endogenous NMD targets, there are also many mRNAs coding for amino acid transporters.7 Amino acid deprivation is a typical stress in fast‐growing tumor cells and the observed inhibition of NMD under such conditions appears to be part of the cell's adaptive response to increase the intracellular levels of amino acids, which are used by tumor cells as energy source and to produce the reduced oxygen species (ROS) scavenger glutathione from cysteine.165 Oxidative stress or depletion of UPF1 or UPF2 upregulated the NMD‐sensitive SLC7A11 mRNA, which codes for a subunit of the xCT cystine/glutamate amino acid transport system.159 This led to an increased intracellular concentration of cysteine and glutathione and ultimately protected cells against oxidative stress.159 This study showed that NMD inhibition contributes to the survival of cells exposed to oxidative stress through a mechanism that completely depends on SLC7A11. Pharmacological attenuation of NMD using the NMD inhibitor NMDI 1166 was recently shown to facilitate the response of cancer cells to the topoisomerase inhibitor doxorubicin, a widely used anticancer drug.167 Doxorubicin induces double‐stranded DNA breaks and severe DNA damage subsequently leads to apoptosis, a controlled cell suicide program that involves caspase‐mediated cleavage of many intracellular proteins. Among those proteins, UPF1 and UPF2 were found to be cleaved by caspases 3 and/or 7.167, 168 The cleavage of UPF1 at aspartate 37 (D37) generates a dominant‐interfering truncated form of UPF1 that inhibits NMD and leads to the upregulation of several mRNAs encoding factors promoting cell cycle arrest and apoptosis.167 Accordingly, expression of a caspase cleavage‐resistant form of UPF1 (D37N) blunted the efficacy of doxorubicin to induce apoptosis in cancer cells.167 These studies suggest that NMD functions in a proapoptotic reinforcing mechanism, in which apoptosis induces NMD inhibition and caspase‐cleaved UPF fragments promote apoptosis168 (Figure 5).
Roles of NMD Factors in Viral Replication
RNA viruses are dependent on mRNA processing, mRNP remodeling, and translation by the respective host cell machineries, begging the question of whether NMD also affects the life cycle of such viruses. With their multicistronic gene organization, the genomic RNA of RNA viruses often have stop codons in places where one would expect them to trigger NMD, for example, in the middle of the RNA leaving a long 3′ UTR. In plants, where the RNAi pathway is well known to restrict RNA virus replication,169 a genetic screen carried out with a recombinant GFP‐expressing potato virus X (PVX) in Arabidopsis attenuated for RNAi to uncover additional host restriction mechanism revealed that NMD targets PVX and restricts its replication.170 The authors showed that two of the subgenomic PVX RNAs, both with long 3′ UTRs, are targeted by NMD. Moreover, overexpression of an NMD suppressing dominant‐negative UPF1 mutant in tobacco plants also led to the accumulation of genomic RNA of the Turnip crinkle virus (TCV), another (+)strand virus, suggesting that restriction of (+)strand RNA viruses might be a general function of NMD.170 Supporting this view, NMD has also been found to restrict the replication of two closely related mammalian alphaviruses, Semliki forest virus (SFV) and Sindbis virus (SINV)171 (Figure 6). For SFV, it was shown that UPF1 knockdown increased the half‐life of the genomic viral RNA. Surprisingly however, shortening of the long 3′ UTR in SFV still resulted in an UPF1‐dependent restriction of the virus‐like particles, indicating that in this particular case the 3′ UTR might not be the NMD‐triggering feature.171 Besides UPF1, SMG5 and SMG7 were documented to also play a role in SFV restriction in this study.
Figure 6.
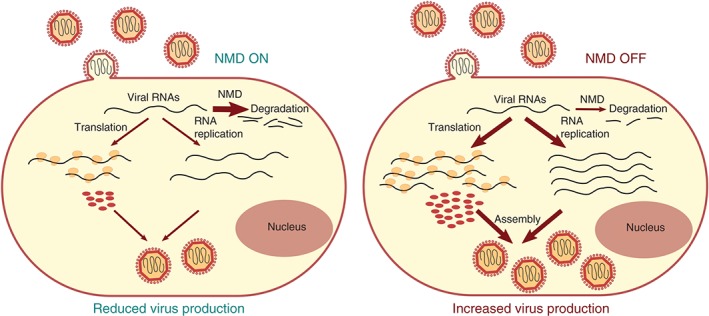
NMD as a cell‐intrinsic viral defense mechanism. NMD can restrict RNA+ alphavirus replication by targeting the viral genome for degradation during early cytosolic steps of the infection. When NMD is impaired (e.g., by depletion of UPF1, SMG5, or SMG7), all steps in the replication cycle are stimulated, leading to the production and release of more virus progeny.
If NMD indeed plays a general role in protecting cells from viruses, one would expect that many viruses have evolved countermeasures to evade or inhibit NMD. As already mentioned, an RSE that protects the unspliced transcript of the avian retrovirus RSV from degradation by the NMD pathway has been described.75, 172 The RSE is located just downstream of the stop codon of the gag ORF and its deletion or mutation rendered the RSV unspliced RNA sensitive to NMD. RSV insertion downstream of the PTC of NMD reporter genes rendered these reporter transcripts immune to NMD in a position‐dependent manner.172 The 400 nucleotides RSV RSE contains 11 CU‐rich clusters, which are binding sites for PTBP1, and they were recently shown to be necessary and sufficient for suppressing NMD.76 The authors showed that PTBP1 binding prevented the interaction of UPF1 with the 3′ UTR regions of otherwise NMD‐sensitive transcripts and revealed a genome‐wide correlation between PTBP1 enrichment near stop codons, 3′ UTR length, and resistance to NMD.76
An alternative viral strategy to evade NMD is to directly inhibit the NMD pathway. Two viruses that appear to have chosen this strategy are human‐T‐cell leukemia virus type I (HTLV‐1)173 and hepatitis C virus (HCV).174 The genomic and full‐length mRNAs of HTLV‐1, a retrovirus causing adult T‐cell leukemia, are sensitive to NMD, but expression of the viral RNA‐binding protein Rex was found to inhibit NMD.173 In the presence of Rex, not only viral transcripts were stabilized but also known NMD reporters and endogenous NMD‐sensitive mRNAs, suggesting that Rex causes a general block of NMD. How exactly Rex inhibits NMD is not yet known. Similarly, a recent study reports that NMD is disrupted in HCV‐infected hepatoma cells, leading to the accumulation of potentially harmful transcripts.174 In an affinity purification coupled to mass spectrometry approach, the authors identified the partner of Y14 and MAGOH (PYM) as an interactor of the viral core protein and showed that expression of HCV core protein reduced PYM interaction with the two EJC factors Y14 and MAGOH. PYM has been shown to promote EJC dissociation from mRNA during translation175 and the authors speculate that HCV core protein‐mediated sequestration of PYM might interfere with NMD by preventing the cytoplasmic release of EJCs and the recycling of the EJC factors back to the nucleus, leading to reduced EJC deposition on newly synthesized mRNAs in HCV‐infected cells.174
Finally, rather than protecting the cell from the virus as in the examples above, UPF1 was also reported to promote export of unspliced genomic RNA in the case of human immunodeficiency virus (HIV).176, 177 HIV‐1 RNPs were found to contain UPF1 along with the double‐stranded RNA‐binding protein Staufen1 and the viral factor pr55Gag. 177 UPF1 depletion resulted in a reduction and its overexpression in an upregulation of HIV‐1 RNA, respectively.177 The finding that UPF1 decrease rather increased HIV‐1 RNA levels and that the UPF1‐mediated effect was independent of UPF2 strongly suggested that the underlying mechanism is unrelated to NMD. Instead, using Rev‐deficient constructs, the authors recently showed that recruitment of UPF1 to these nuclear retained transcripts promotes their CRM1‐dependent export to the cytoplasm.176 UPF1 remains associated with HIV‐1 RNA in the cytoplasm and together with other cellular RNA‐binding proteins, UPF1 is packed into the HIV‐1 virions and was shown to be crucial for the infectivity of the virus.178 Available evidence suggests that UPF1 functions at the step of reverse transcription and that this function requires the helicase activity of UPF1, but the exact mechanism is not yet understood.178
FUTURE DIRECTIONS
There are many central questions regarding mechanistic aspects of NMD that are not yet understood. A particularly burning question concerns the substrate specificity of NMD, i.e., to understand what makes some mRNAs a substrate for NMD and not others. The currently prevailing view that it has to do with kinetic and/or mechanistic differences during translation termination is supported by a fair amount of indirect evidence, but compelling direct evidence is scarce. Mechanistic studies are hampered by the lack of an in vitro system that can recapitulate NMD and would allow systematic manipulation and titration of the involved factors.
With the discovery that besides aberrant PTC‐containing transcripts, the stability of many apparently normal, PTC‐less mRNAs is also controlled at least in part by NMD, the classical and name‐giving definition of NMD became misleading and a new commonly accepted operational definition of NMD based on a consensus set of easily testable criteria is needed. Until we have a better understanding, a new definition should also consider that the phenomenon commonly called NMD might actually result from several different but overlapping decay pathways. A possible operational definition for NMD based on the current literature could be ‘translation‐dependent mRNA degradation that requires UPF1 and in metazoans additionally SMG1 and SMG6 or SMG7.’
The discovery of a large number of mostly PTC‐free endogenous NMD‐targeted mRNAs opened a whole new area of research investigating how NMD is involved in the regulation of which biological processes. This research has just begun and the examples described above presumably only represent the tip of the iceberg. A look into the long lists of mRNAs identified by genome‐wide approaches as putative NMD targets indicates possible roles for NMD‐mediated gene regulation in a wide variety of biological contexts. Therefore, stay tuned for future surprises.
ACKNOWLEDGMENTS
The NMD research in the lab of O.M. is supported by the NCCR RNA & Disease and by grant 31003A‐162986, both funded by the Swiss National Science Foundation.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA 1979, 76:5134–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable β‐globin mRNA in mRNA‐deficient β o thalassemia. Cell 1981, 27:543–553. [DOI] [PubMed] [Google Scholar]
- 3. Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis‐acting sequence elements and one trans‐acting factor. Genes Dev 1993, 7:1737–1754. [DOI] [PubMed] [Google Scholar]
- 4. Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR. Impact of nonsense‐mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet 2006, 2:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome‐wide analysis of mRNAs regulated by the nonsense‐mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 2003, 12:1439–1452. [DOI] [PubMed] [Google Scholar]
- 6. Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 1999, 19:6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendell JT, Sharifi NA, Meyers JL, Martinez‐Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 2004, 36:1073–1078. [DOI] [PubMed] [Google Scholar]
- 8. Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. High resolution transcriptome maps for wild‐type and nonsense‐mediated decay‐defective Caenorhabditis elegans . Genome Biol 2009, 10:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense‐mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 2005, 11:1530–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 2012, 9:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res 2007, 35:4542–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense‐mediated mRNA decay. Mol Cell Biol 2006, 26:1272–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O. Autoregulation of the nonsense‐mediated mRNA decay pathway in human cells. RNA 2011, 17:2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense‐mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 2009, 29:3517–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans‐effector of nonsense‐mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 2001, 10:99–105. [DOI] [PubMed] [Google Scholar]
- 16. Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T‐cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 1995, 270:28995–29003. [DOI] [PubMed] [Google Scholar]
- 17. Thermann R, Neu‐Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 1998, 17:3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 1991, 5:2303–2314. [DOI] [PubMed] [Google Scholar]
- 19. Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 1993, 7:1885–1897. [DOI] [PubMed] [Google Scholar]
- 20. Chen YH, Su LH, Sun CH. Incomplete nonsense‐mediated mRNA decay in Giardia lamblia . Int J Parasitol 2008, 38:1305–1317. [DOI] [PubMed] [Google Scholar]
- 21. Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA‐dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 2011, 41:693–703. [DOI] [PubMed] [Google Scholar]
- 22. Fiorini F, Bagchi D, Le Hir H, Croquette V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun 2015, 6:7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense‐mediated mRNA decay. RNA 2000, 6:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J 2007, 26:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franks TM, Singh G, Lykke‐Andersen J. Upf1 ATPase‐dependent mRNP disassembly is required for completion of nonsense‐mediated mRNA decay. Cell 2010, 143:938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 1996, 16:5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamashita A. Role of SMG‐1‐mediated Upf1 phosphorylation in mammalian nonsense‐mediated mRNA decay. Genes Cells 2013, 18:161–175. [DOI] [PubMed] [Google Scholar]
- 28. Fiorini F, Boudvillain M, Le Hir H. Tight intramolecular regulation of the human Upf1 helicase by its N‐ and C‐terminal domains. Nucleic Acids Res 2013, 41:2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008, 15:85–93. [DOI] [PubMed] [Google Scholar]
- 30. Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense‐mediated mRNA decay in colon cancer cells. Oncogene 2007, 26:2873–2884. [DOI] [PubMed] [Google Scholar]
- 31. Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG‐1‐Upf1‐eRF1‐eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense‐mediated mRNA decay. Genes Dev 2006, 20:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melero R, Uchiyama A, Castano R, Kataoka N, Kurosawa H, Ohno S, Yamashita A, Llorca O. Structures of SMG1‐UPFs complexes: SMG1 contributes to regulate UPF2‐dependent activation of UPF1 in NMD. Structure 2014, 22:1105–1119. [DOI] [PubMed] [Google Scholar]
- 33. Lykke‐Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense‐mediated decay when bound downstream of a termination codon. Cell 2000, 103:1121–1131. [DOI] [PubMed] [Google Scholar]
- 34. Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O. The cryo‐EM structure of the UPF‐EJC complex shows UPF1 poised toward the RNA 3' end. Nat Struct Mol Biol 2012, 19:498–505, S491–S492. [DOI] [PubMed] [Google Scholar]
- 35. Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense‐mediated decay factors, UPF1 and UPF2. EMBO J 2009, 28:2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense‐mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 2004, 11:330–337. [DOI] [PubMed] [Google Scholar]
- 37. Fourati Z, Roy B, Millan C, Coureux PD, Kervestin S, van Tilbeurgh H, He F, Uson I, Jacobson A, Graille M. A highly conserved region essential for NMD in the Upf2 N‐terminal domain. J Mol Biol 2014, 426:3689–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG‐4). Mol Cell Biol 2001, 21:209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 2003, 13:207–214. [DOI] [PubMed] [Google Scholar]
- 40. Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3‐mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 2009, 16:747–753. [DOI] [PubMed] [Google Scholar]
- 41. Kunz JB, Neu‐Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense‐mediated mRNA decay and translation. RNA 2006, 12:1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC‐UPF3b complex. Proc Natl Acad Sci USA 2010, 107:10050–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arias‐Palomo E, Yamashita A, Fernandez IS, Nunez‐Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O. The nonsense‐mediated mRNA decay SMG‐1 kinase is regulated by large‐scale conformational changes controlled by SMG‐8. Genes Dev 2011, 25:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernandez IS, Yamashita A, Arias‐Palomo E, Bamba Y, Bartolome RA, Canales MA, Teixido J, Ohno S, Llorca O. Characterization of SMG‐9, an essential component of the nonsense‐mediated mRNA decay SMG1C complex. Nucleic Acids Res 2011, 39:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG‐8 and SMG‐9, two novel subunits of the SMG‐1 complex, regulate remodeling of the mRNA surveillance complex during nonsense‐mediated mRNA decay. Genes Dev 2009, 23:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase‐related kinase: characterization of the human SMG‐1 RNA surveillance protein. J Biol Chem 2001, 276:22709–22714. [DOI] [PubMed] [Google Scholar]
- 47. Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14‐3‐3‐like adaptor in the nonsense‐mediated mRNA decay pathway. Mol Cell 2005, 17:537–547. [DOI] [PubMed] [Google Scholar]
- 48. Jonas S, Weichenrieder O, Izaurralde E. An unusual arrangement of two 14‐3‐3‐like domains in the SMG5‐SMG7 heterodimer is required for efficient nonsense‐mediated mRNA decay. Genes Dev 2013, 27:211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG‐5 and hSMG‐7. Mol Cell 2003, 12:1187–1200. [DOI] [PubMed] [Google Scholar]
- 50. Okada‐Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N‐ and C‐terminal Upf1 phosphorylations create binding platforms for SMG‐6 and SMG‐5:SMG‐7 during NMD. Nucleic Acids Res 2012, 40:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Loh B, Jonas S, Izaurralde E. The SMG5‐SMG7 heterodimer directly recruits the CCR4‐NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 2013, 27:2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Glavan F, Behm‐Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 2006, 25:5117–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eberle AB, Lykke‐Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 2009, 16:49–55. [DOI] [PubMed] [Google Scholar]
- 54. Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 2008, 14:2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gatfield D, Izaurralde E. Nonsense‐mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 2004, 429:575–578. [DOI] [PubMed] [Google Scholar]
- 56. Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 3' UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep 2014, 9:555–568. [DOI] [PubMed] [Google Scholar]
- 57. Lykke‐Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH. Human nonsense‐mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 2014, 28:2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, Tenenbaum SA, Belasco JG, Green PJ. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res 2014:43:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chakrabarti S, Bonneau F, Schussler S, Eppinger E, Conti E. Phospho‐dependent and phospho‐independent interactions of the helicase UPF1 with the NMD factors SMG5‐SMG7 and SMG6. Nucleic Acids Res 2014, 42:9447–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nicholson P, Josi C, Kurosawa H, Yamashita A, Muhlemann O. A novel phosphorylation‐independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res 2014, 42:9217–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC‐binding motifs (EBMs) required for nonsense‐mediated mRNA decay. Genes Dev 2010, 24:2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Casadio A, Longman D, Hug N, Delavaine L, Vallejos Baier R, Alonso CR, Caceres JF. Identification and characterization of novel factors that act in the nonsense‐mediated mRNA decay pathway in nematodes, flies and mammals. EMBO Rep 2015, 16:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 2007, 21:1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Caceres JF. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans . Nucleic Acids Res 2013, 41:8319–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Anastasaki C, Longman D, Capper A, Patton EE, Caceres JF. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res 2011, 39:3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hug N, Caceres JF. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay‐inducing complex. Cell Rep 2014, 8:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon‐exon junction complex provides a binding platform for factors involved in mRNA export and nonsense‐mediated mRNA decay. EMBO J 2001, 20:4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O. EJC‐independent degradation of nonsense immunoglobulin‐mu mRNA depends on 3' UTR length. Nat Struct Mol Biol 2006, 13:462–464. [DOI] [PubMed] [Google Scholar]
- 69. Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3' untranslated region. PLoS Biol 2008, 6:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. Genome‐wide identification of alternative splice forms down‐regulated by nonsense‐mediated mRNA decay in Drosophila. PLoS Genet 2009, 5:e1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Singh G, Rebbapragada I, Lykke‐Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense‐mediated mRNA decay. PLoS Biol 2008, 6:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hogg JR, Goff SP. Upf1 senses 3'UTR length to potentiate mRNA decay. Cell 2010, 143:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense‐mediated mRNA decay. Genome Res 2013, 23:1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Toma KG, Rebbapragada I, Durand S, Lykke‐Andersen J. Identification of elements in human long 3' UTRs that inhibit nonsense‐mediated decay. RNA 2015, 21:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Withers JB, Beemon KL. The structure and function of the Rous sarcoma virus RNA stability element. J Cell Biochem 2011, 112:3085–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense‐mediated mRNA decay pathway. Elife 2016, 5:e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense‐mediated mRNA decay. Cell Mol Life Sci 2015, 72:4523–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense‐mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829, 2013:612–623. [DOI] [PubMed] [Google Scholar]
- 79. He F, Jacobson A. Nonsense‐mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet 2015, 49:339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lykke‐Andersen S, Jensen TH. Nonsense‐mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 2015, 16:665–677. [DOI] [PubMed] [Google Scholar]
- 81. Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3'‐UTR promotes aberrant termination and triggers nonsense‐mediated mRNA decay. Nature 2004, 432:112–118. [DOI] [PubMed] [Google Scholar]
- 82. Peixeiro I, Inacio A, Barbosa C, Silva AL, Liebhaber SA, Romao L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG‐proximal nonsense mutations. Nucleic Acids Res 2011, 40:1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 2012, 482:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 2010, 37:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Preis A, Heuer A, Barrio‐Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1‐eRF3 or eRF1‐ABCE1. Cell Rep 2014, 8:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin‐ATPase ABCE1. Proc Natl Acad Sci USA 2011, 108:3228–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 2011, 108:E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kononenko AV, Mitkevich VA, Atkinson GC, Tenson T, Dubovaya VI, Frolova LY, Makarov AA, Hauryliuk V. GTP‐dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res 2010, 38:548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. Poly(A)‐binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell 2002, 94:205–216. [DOI] [PubMed] [Google Scholar]
- 90. Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)‐binding protein in Cap/Poly(A)‐dependent translation. J Biol Chem 2002, 277:50286–50292. [DOI] [PubMed] [Google Scholar]
- 91. Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3'‐poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate‐binding protein. J Biol Chem 1999, 274:16677–16680. [DOI] [PubMed] [Google Scholar]
- 92. Behm‐Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)‐binding protein 1 (PABPC1) in nonsense‐mediated mRNA decay. EMBO J 2007, 26:1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 2008, 27:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)‐binding protein to a premature termination codon inhibits mammalian nonsense‐mediated mRNA decay. RNA 2008, 14:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 2006, 7:415–425. [DOI] [PubMed] [Google Scholar]
- 96. Czaplinski K, Ruiz‐Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter‐Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 1998, 12:1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Muhlemann O, Jensen TH. mRNP quality control goes regulatory. Trends Genet 2012, 28:70–77. [DOI] [PubMed] [Google Scholar]
- 98. Fatscher T, Boehm V, Weiche B, Gehring NH. The interaction of cytoplasmic poly(A)‐binding protein with eukaryotic initiation factor 4G suppresses nonsense‐mediated mRNA decay. RNA 2014, 20:1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Joncourt R, Eberle AB, Rufener SC, Muhlemann O. Eukaryotic initiation factor 4G suppresses nonsense‐mediated mRNA decay by two genetically separable mechanisms. PLoS One 2014, 9:e104391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kervestin S, Li C, Buckingham R, Jacobson A. Testing the faux‐UTR model for NMD: analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 2012, 94:1560–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci USA 2007, 104:20872–20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG‐2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol 2007, 27:5630–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3' UTRs. Proc Natl Acad Sci USA 2013, 110:3357–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zund D, Gruber AR, Zavolan M, Muhlemann O. Translation‐dependent displacement of UPF1 from coding sequences causes its enrichment in 3' UTRs. Nat Struct Mol Biol 2013, 20:936–943. [DOI] [PubMed] [Google Scholar]
- 105. Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. A post‐translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 2014, 28:1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 is a 5' to 3' RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3' UTRs. Mol Cell 2014, 54:573–585. [DOI] [PubMed] [Google Scholar]
- 107. Deniaud A, Karuppasamy M, Bock T, Masiulis S, Huard K, Garzoni F, Kerschgens K, Hentze MW, Kulozik AE, Beck M, et al. A network of SMG‐8, SMG‐9 and SMG‐1 C‐terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res 2015, 43:7600–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke‐Andersen J. Target discrimination in nonsense‐mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 2015, 59:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Melero R, Hug N, Lopez‐Perrote A, Yamashita A, Caceres JF, Llorca O. The RNA helicase DHX34 functions as a scaffold for SMG1‐mediated UPF1 phosphorylation. Nat Commun 2016, 7:10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol 2008, 18:315–321. [DOI] [PubMed] [Google Scholar]
- 111. Metze S, Herzog VA, Ruepp MD, Muhlemann O. Comparison of EJC‐enhanced and EJC‐independent NMD in human cells reveals two partially redundant degradation pathways. RNA 2013, 19:1432–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lopez‐Perrote A, Castano R, Melero R, Zamarro T, Kurosawa H, Ohnishi T, Uchiyama A, Aoyagi K, Buchwald G, Kataoka N, et al. Human nonsense‐mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 2016, 44:1909–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gehring NH, Kunz JB, Neu‐Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon‐junction complex components specify distinct routes of nonsense‐mediated mRNA decay with differential cofactor requirements. Mol Cell 2005, 20:65–75. [DOI] [PubMed] [Google Scholar]
- 114. Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA 2nd, Wilkinson MF. An alternative branch of the nonsense‐mediated decay pathway. EMBO J 2007, 26:1820–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. RNA homeostasis governed by cell type‐specific and branched feedback loops acting on NMD. Mol Cell 2011, 43:950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Doma MK, Parker R. RNA quality control in eukaryotes. Cell 2007, 131:660–668. [DOI] [PubMed] [Google Scholar]
- 117. Maquat LE. Nonsense‐mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004, 5:89–99. [DOI] [PubMed] [Google Scholar]
- 118. Gao Q, Das B, Sherman F, Maquat LE. Cap‐binding protein 1‐mediated and eukaryotic translation initiation factor 4E‐mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci USA 2005, 102:4258–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Maderazo AB, Belk JP, He F, Jacobson A. Nonsense‐containing mRNAs that accumulate in the absence of a functional nonsense‐mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol 2003, 23:842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Durand S, Lykke‐Andersen J. Nonsense‐mediated mRNA decay occurs during eIF4F‐dependent translation in human cells. Nat Struct Mol Biol 2013, 20:702–709. [DOI] [PubMed] [Google Scholar]
- 121. Rufener SC, Muhlemann O. eIF4E‐bound mRNPs are substrates for nonsense‐mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 2013, 20:710–717. [DOI] [PubMed] [Google Scholar]
- 122. Muhlemann O, Lykke‐Andersen J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 2010, 7:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 2004, 16:587–596. [DOI] [PubMed] [Google Scholar]
- 124. Fenger‐Gron M, Fillman C, Norrild B, Lykke‐Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 2005, 20:905–915. [DOI] [PubMed] [Google Scholar]
- 125. Lejeune F, Li X, Maquat LE. Nonsense‐mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 2003, 12:675–687. [DOI] [PubMed] [Google Scholar]
- 126. Lykke‐Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense‐mediated decay. Mol Cell Biol 2002, 22:8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. SMG5‐PNRC2 is functionally dominant compared with SMG5‐SMG7 in mammalian nonsense‐mediated mRNA decay. Nucleic Acids Res 2013, 41:1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Cho H, Kim KM, Kim YK. Human proline‐rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 2009, 33:75–86. [DOI] [PubMed] [Google Scholar]
- 129. Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense‐mediated mRNA decay. Cell 2008, 133:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H. Structural basis of the PNRC2‐mediated link between mRNA surveillance and decapping. Structure 2012, 20:2025–2037. [DOI] [PubMed] [Google Scholar]
- 131. Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446:926–929. [DOI] [PubMed] [Google Scholar]
- 132. Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense‐mediated decay. Genes Dev 2007, 21:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing‐coupled nonsense‐mediated mRNA decay. Mol Cell Biol 2008, 28:4320–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wollerton MC, Gooding C, Wagner EJ, Garcia‐Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense‐mediated decay. Mol Cell 2004, 13:91–100. [DOI] [PubMed] [Google Scholar]
- 135. Yan Q, Weyn‐Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci USA 2015, 112:3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hamid FM, Makeyev EV. Regulation of mRNA abundance by polypyrimidine tract‐binding protein‐controlled alternate 5' splice site choice. PLoS Genet 2014, 10:e1004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Ratsch G, Wachter A. Nonsense‐mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25:3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kawashima T, Douglass S, Gabunilas J, Pellegrini M, Chanfreau GF. Widespread use of non‐productive alternative splice sites in Saccharomyces cerevisiae . PLoS Genet 2014, 10:e1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Braunschweig U, Barbosa‐Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos‐Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 2014, 24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 2013, 154:583–595. [DOI] [PubMed] [Google Scholar]
- 141. Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter‐kingdom conservation of mechanism of nonsense‐mediated mRNA decay. EMBO J 2008, 27:1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Saul H, Elharrar E, Gaash R, Eliaz D, Valenci M, Akua T, Avramov M, Frankel N, Berezin I, Gottlieb D, et al. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense‐mediated mRNA decay pathway. Plant J 2009, 60:1031–1042. [DOI] [PubMed] [Google Scholar]
- 143. McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie‐Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense‐mediated mRNA decay. Proc Natl Acad Sci USA 2010, 107:12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Weischenfeldt J, Damgaard I, Bryder D, Theilgaard‐Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by‐products of programmed DNA rearrangements. Genes Dev 2008, 22:1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Thoren LA, Norgaard GA, Weischenfeldt J, Waage J, Jakobsen JS, Damgaard I, Bergstrom FC, Blom AM, Borup R, Bisgaard HC, et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS One 2010, 5:e11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bao J, Tang C, Yuan S, Porse BT, Yan W. UPF2, a nonsense‐mediated mRNA decay factor, is required for prepubertal Sertoli cell development and male fertility by ensuring fidelity of the transcriptome. Development 2014, 142:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M, et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense‐mediated mRNA decay. EMBO J 2015, 34:1630–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense‐mediated RNA decay pathway. Cell Rep 2014, 6:748–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet 2013, 22:4673–4687. [DOI] [PubMed] [Google Scholar]
- 150. Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, Raynaud M, Hackett A, Field M, Rodriguez J, et al. Transcriptome profiling of UPF3B/NMD‐deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry 2012, 17:1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Alrahbeni T, Sartor F, Anderson J, Miedzybrodzka Z, McCaig C, Muller B. Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain 2015, 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Seuntjens E, Umans L, Zwijsen A, Sampaolesi M, Verfaillie CM, Huylebroeck D. Transforming growth factor type β and Smad family signaling in stem cell function. Cytokine Growth Factor Rev 2009, 20:449–458. [DOI] [PubMed] [Google Scholar]
- 153. Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. Identification of a microRNA that activates gene expression by repressing nonsense‐mediated RNA decay. Mol Cell 2011, 42:500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense‐mediated mRNA decay. Cell 2013, 153:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep 2015, 16:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Oren YS, McClure ML, Rowe SM, Sorscher EJ, Bester AC, Manor M, Kerem E, Rivlin J, Zahdeh F, Mann M, et al. The unfolded protein response affects readthrough of premature termination codons. EMBO Mol Med 2014, 6:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Moore KA, Hollien J. The unfolded protein response in secretory cell function. Annu Rev Genet 2012, 46:165–183. [DOI] [PubMed] [Google Scholar]
- 158. Sieber J, Hauer C, Bhuvanagiri M, Leicht S, Krijgsveld J, Neu‐Yilik G, Hentze MW, Kulozik AE. Proteomic analysis reveals branch‐specific regulation of the unfolded protein response by nonsense‐mediated mRNA decay. Mol Cell Proteomics 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Martin L, Gardner LB. Stress‐induced inhibition of nonsense‐mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene 2015, 34:4211–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non‐coding RNA in mammalian cells. PLoS One 2013, 8:e55684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wengrod J, Martin L, Wang D, Frischmeyer‐Guerrerio P, Dietz HC, Gardner LB. Inhibition of nonsense‐mediated RNA decay activates autophagy. Mol Cell Biol 2013, 33:2128–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Ingolia NT, Brar GA, Stern‐Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein‐coding genes. Cell Rep 2014, 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Smith CM, Steitz JA. Classification of gas5 as a multi‐small‐nucleolar‐RNA (snoRNA) host gene and a member of the 5'‐terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 1998, 18:6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin‐sensitive and rapamycin‐sensitive PI 3‐kinase‐related kinase signaling pathway. RNA 2001, 7:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2012, 2:881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Durand S, Cougot N, Mahuteau‐Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. Inhibition of nonsense‐mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P‐bodies. J Cell Biol 2007, 178:1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Popp MW, Maquat LE. Attenuation of nonsense‐mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun 2015, 6:6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Jia J, Furlan A, Gonzalez‐Hilarion S, Leroy C, Gruenert DC, Tulasne D, Lejeune F. Caspases shutdown nonsense‐mediated mRNA decay during apoptosis. Cell Death Differ 2015, 22:1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell 2007, 130:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Garcia D, Garcia S, Voinnet O. Nonsense‐mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 2014, 16:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, Merits A, Muhlemann O, Azzalin C, Helenius A. The host nonsense‐mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 2014, 16:403–411. [DOI] [PubMed] [Google Scholar]
- 172. Weil JE, Beemon KL. A 3' UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 2006, 12:102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, Tanaka Y, Brighty DW, Watanabe T. Viral interference with host mRNA surveillance, the nonsense‐mediated mRNA decay (NMD) pathway, through a new function of HTLV‐1 Rex: implications for retroviral replication. Microbes Infect 2013, 15:491–505. [DOI] [PubMed] [Google Scholar]
- 174. Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense‐mediated mRNA decay. Mol Cell 2015, 57:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell 2009, 137:536–548. [DOI] [PubMed] [Google Scholar]
- 176. Ajamian L, Abel K, Rao S, Vyboh K, Garcia‐de‐Gracia F, Soto‐Rifo R, Kulozik AE, Gehring NH, Mouland AJ. HIV‐1 recruits UPF1 but excludes UPF2 to promote nucleocytoplasmic export of the genomic RNA. Biomolecules 2015, 5:2808–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ. Unexpected roles for UPF1 in HIV‐1 RNA metabolism and translation. RNA 2008, 14:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Serquina AK, Das SR, Popova E, Ojelabi OA, Roy CK, Gottlinger HG. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol 2013, 87:8853–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]


