Abstract
Background & Aims
The toll‐like receptor‐interferon (TLR–IFN) signalling pathway plays a crucial role in HBV infection. Human leucocyte antigen (HLA) polymorphisms are associated with chronic HBV infection by genome wide association study (GWAS). We aimed to explore interaction between TLR–IFN and HLA gene polymorphisms in susceptibility of chronic HBV infection.
Methods
In the Chinese Southwest Han population, 1191 chronic HBV infection patients and 273 HBV clearance were selected. A total of 39 single nucleotide polymorphism loci in 23 genes of the TLR–IFN pathway and four HLA polymorphism loci associated with chronic HBV infection identified by GWAS were selected for genotyping. SNPStats, QVALUE, and multifactor dimensionality reduction were used for statistical analysis.
Results
A significant association was seen in several of the TLR–IFN pathway genes, TLR9 rs352140 (OR = 0.70, P = 0.0088), IL1B rs16944 (OR = 0.67, P = 0.016), IL12B rs3212227 (OR = 1.38, P = 0.021), IFNGR1 rs3799488 (OR = 1.48, P = 0.0048), IFNGR2 rs1059293 (OR = 0.27, P = 0.011), MX1 rs467960 (OR = 0.68, P = 0.022), as well as four loci in HLA, rs3077 (OR = 0.55, P < 0.0001), rs2856718 (OR = 0.60, P = 4e‐04), rs9277535 (OR = 0.54, P < 0.0001) and rs7453920 (OR = 0.43, P < 0.0001). A synergistic relationship was seen between rs9277535 and rs16944 (0.13%), rs1143623 and rs6613 (0.10%). The combination of rs9277535 in HLA and rs16944 in IL1B was the best model to predict chronic HBV infection (testing accuracy = 0.6040, P = 0.0010, cross‐validation consistency = 10/10).
Conclusions
TLR–IFN pathway gene polymorphisms are associated with chronic HBV infection. Interactions with polymorphisms in these genes may be one mechanism by which HLA polymorphisms influence susceptibility to chronic HBV infection, as specific single nucleotide polymorphism combinations are highly predictive of chronic HBV infection.
Keywords: gene–gene interaction, hepatitis B virus, innate immune, MDR, polymorphism
Abbreviations
- AIC
Akaike information
- ALT
alanine aminotransferase
- CI
confidence interval
- CVC
cross‐validation consistency
- CXCL
chemokine motif (C‐X‐C) ligand
- FDR
false discovery rate
- GMDR
generalized multifactor dimensionality reduction
- GWAS
genome‐wide association studies
- HBcAb
hepatitis B core antibody
- HBeAb
hepatitis B e antibody
- HBeAg
hepatitis B e antigen
- HBsAb
hepatitis B surface antibody
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HLA
human leucocyte antigen
- IFN
interferon
- IL
interleukin
- iMLDR
improved multiplex ligation detection reaction
- MDR
multifactor dimensionality reduction
- OR
odds ratio
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
- TA
testing accuracy
- TLR
Toll‐like receptor
Key Points.
Polymorphisms within the TLR‐IFN pathway key gene are associated with chronic HBV infection.
A synergistic relationship was seen between rs9277535 and rs16944, and rs1143623 and rs6613.
Interactions with TLR‐IFN polymorphisms may be involved in the mechanism by which HLA polymorphisms influence susceptibility to chronic HBV infection.
The combination of HLA‐DPB1 rs9277535 and IL1B rs16944 better predicts chronic HBV infection than rs9277535.
More than 350 million people are chronic hepatitis B virus (HBV) infection 1. HBV‐related end stage liver disease or hepatocellular carcinoma (HCC) are responsible for over 0.5–1 million deaths per year 2. HBV is transmitted from person to person through percutaneous/mucous membrane exposure to blood or other body fluids of an infected person. The mechanism of chronic HBV infection is an important research topic that has attracted extensive attention.
Genetic background is an important factor in susceptibility to disease. Genome wide association studies (GWAS) in the Asian population identified a strong association between chronic HBV infection and variants in the human leucocyte antigen (HLA)‐DP and HLA‐DQ genes 3, 4. Some studies have confirmed the association between chronic HBV infection and gene single nucleotide polymorphisms (SNPs), such as HLA, toll‐like receptor (TLR), interleukin (IL) 10, chemokine motif (C‐X‐C) ligand (CXCL) 10, interferon (IFN) and IL28B 5, 6, 7, 8, 9, 10. However, the mechanism by which such SNPs influence disease susceptibility remains incompletely understood.
The immune system within the liver is characterized mainly by the innate immunity 11. Innate immune response plays an important role in HBV control and expression of immune response genes might occur below the level of detection of the microarray analysis that has been carried out 12. Important in the innate immune response to HBV infection is the TLR–IFN pathway, including TLRs themselves, adaptins and effector molecules for TLRs, IFNs and their cognate receptors, and proteins induced downstream by IFN signalling 12, 13, 14, 15.
Since the identification of SNPs by GWAS, the mechanisms of significant SNP have been attracting the concern of researchers. However, these SNPs appeared to be only genetic markers associated with disease rather than involved in the disease mechanism per se. An example of this type of association is characterized by the association between IL28B and hepatitis C virus (HCV) clearance. Although IL28B rs12979860 is a good predictor for HCV clearance 16, the level of IFN lambda 3, coded for by the IL28B gene, does not crucially influence the outcome in HCV infection 16, 17. However, abnormal innate immune responses such as high levels of IL‐10 and low IL‐12p40 were found in patients with the poor‐response genotype 18, 19, 20, 21.
After considering the published work regarding gene–gene interaction and the role of innate immunity in the control of HBV, we hypothesized that there may be some interaction between polymorphisms in the HLA and TLR–IFN pathway genes. Therefore, we designed this case–control study to identify any SNPs associated with the risk of chronic HBV infection and also to investigate the interaction between HLA and innate immunity gene variations. This study contributes to the broader understanding of the mechanisms behind chronic HBV infection.
Patients and methods
Patients
This study was designed as an analytical observational case–control study, which included 1464 Han people from Chongqing, China. Patients with chronic HBV infection were enrolled in this study (n = 1191, female, 424; male, 767; median age, 36; range, 26–42), and the control population demonstrated previous HBV clearance (n = 273; female, 111; male, 162; median age, 42; range, 33–49). A significant difference was found between HBV clearance and chronic HBV infection for age (P < 0.0001). This was taken into account in the multivariate analysis. No difference was seen between males and females (P = 0.177).
The inclusion criteria for the patient cohort were chronic HBV infection as evidenced by a history of HBV surface antigen (HBsAg) positive for over 6 months, regardless of alanine aminotransferase (ALT) levels, HBV DNA levels or hepatitis B e antigen (HBeAg) status. The inclusion criteria for the control, cleared‐virus group were the following: (a) HBsAg negative; (b) hepatitis B surface antibody (HBsAb), hepatitis B e antibody (HBeAb), and hepatitis B core antibody (HBcAb) positive; (c) undetectable HBV DNA levels; (d) normal serum ALT levels.
Exclusion criteria consisted of the following: (a) co‐infectionwith HCV, hepatitis D virus or human immunodeficiency virus (HIV); (b) coexisting serious medical or psychiatric illness; (c) organ or bone marrow transplantation; (d) recent treatment with systemic corticosteroids, immunosuppressants or chemotherapeutic agents.
Written informed consent was obtained from each patient included in this study. This study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by Ethic Committee of Southwest Hospital, Third Military Medical University.
Serological assays
Routine biochemical tests were performed using automated techniques. Serum HBV DNA was quantified using the TaqMan polymerase chain reaction (PCR) assay on a LightCycler 480 Real‐Time PCR System (Roche, Basel, Switzerland). HBsAg, HBeAg, HBeAb and HBcAb were detected by the Architect i2000SR System (Abbott Laboratories, Abbott Park, IL, USA).
Target genes and SNP selection
A total of 23 genes belonging to the TLR–IFN pathway were selected as target genes. These genes included TLRs (TLR3, TLR4, TLR5 and TLR9), the TLR adaptin MYD88, the TLR effector molecule IRAK1, receptors for IFNs (IFNGR1, IFNGR2, IL27B and IL12B), ligands for IFN receptors (IFNG and IL15), other genes related to IFNs (IL28A, IL29 and IL6), proteins induced by IFN (MX1, CXCL10), and genes of the NFκB pathway (NFKB1, NFKB2, IL1B, IL2, IL10 and IL12A).
A total of 39 SNPs located in the 23 genes were identified from the SNP database in NCBI (http://www.ncbi.nlm.nih.gov/SNP) or selected according to the literature. These were: TLR3 rs3775296, TLR4 rs11536889 and rs1927907, TLR5 rs5744174, TLR9 rs352140, MYD88 rs7744, IRAK1 rs1059702 and rs1059703, IFNG rs2069705, IFNGR1 rs3799488 and rs10457655, IFNGR2 rs1059293 and rs2834211, IL15 rs10519613 and rs10833, IL28B rs12979860 and rs8099917, IL29 rs30461, IL6 rs1800796, IL27B rs4905 and rs6613, IL12B rs3212227, NFKB1 rs4648068, NFKB2 rs7897947, IL1B rs1143623, rs1143627, and rs16944, IL2 rs2069772, IL10 rs1800872 and rs1800896, IL12A rs2243115 and rs568408, CXCL10 rs4256246, rs4508917, and rs8878, MX1 rs1557370, rs2070229, rs467960, and rs469390. Moreover, HLA‐DPA1 rs3077, HLA‐DQA2 rs2856718, HLA‐DPB1 rs9277535 and HLA‐DQB2 rs7453920 were selected to investigate gene–gene interactions according to GWAS on susceptibility to chronic HBV infection. Basic information regarding the SNPs is demonstrated in Table S1.
SNP genotyping
Genomic DNA was isolated from 200 μL of EDTA non‐coagulated blood samples using QuickGene DNA whole blood kit S (QuickGene, FujiFilm, Japan). The SNP genotyping was performed using an improved multiplex ligation detection reaction (iMLDR) technique (Genesky Biotechnologies Inc., Shanghai, China). The basic principle for this technique is illustrated in Figure S1.
We genotyped the selected SNP loci in one ligation reaction. Two multiplex PCR reactions were designed to amplify fragments covering all SNP loci. The primer information of the two reaction mixtures is described in Tables S2 and S3. The PCR programme for both reactions was 95°C, 2 min; 11 cycles × (94°C, 20 s; 65°C–0.5°C/cycle, 40 s; 72°C, 1 min 30 s); 24 cycles × (94°C, 20 s; 59°C, 30 s; 72°C, 1 min 30 s); 72°C, 2 min; hold at 4°C. The ligation cycling programme was 95°C, 2 min; 38 cycles × (94°C, 1 min; 56°C, 4 min); hold at 4°C. Half a microlitre of ligation product was loaded into the ABI 3730XL and the raw data were analysed by GeneMapper 4.1. All primers, probes and labelling oligos were designed by and ordered from Genesky Biotechnologies Inc.
Analysis of gene–gene interaction
In this study, there are four steps to analyse gene–gene interaction by multifactor dimensionality reduction (MDR; v 3.0.2) 22 and generalized multifactor dimensionality reduction (GMDR; v 0.7) 23. The first step is to select SNPs that are most likely to interact by GMDR. The second step is to analyse information gain – that is, how much information is gained about case–control status from knowledge about genotypes at one or more SNPs – gene–gene interaction dendrogram and entropy algorithms were carried out by MDR. The third step is to analyse the best predictive model by GMDR, which includes multiple processor threading by the permutation testing software compared with the MDR method. The fourth step is to construct a new attribute including all SNPs selected in the best model using MDR attribute construction. The fifth and final step is to evaluate a classification model using the newly constructed attribute(s) by MDR.
Statistical analysis
SNPStats (http://bioinfo.iconcologia.net/snpstats/start.htm) and spss 18.0 (SPSS Inc., Chicago, IL, USA) were used to obtain odds ratios (ORs), 95% confidence intervals (CIs), and P‐values. The Hardy–Weinberg equilibrium of each SNP was tested using SNPStats. Multiple logistic regression models were applied in the analysis of genotypes. Each component of the model was: co‐dominant model (major allele homozygotes vs. heterozygotes vs. minor allele homozygotes), dominant model (major allele homozygotes vs. heterozygotes + minor allele homozygotes), recessive model (major allele homozygotes vs. minor allele homozygotes), overdominant model (major allele homozygotes + heterozygotes vs. minor allele homozygotes) and log‐additive model. Akaike information (AIC) was used to choose the inheritance model that best fits the data. Age and gender were included as covariates and adjusted to obtain statistical significance. A multiple comparison was carried out by QVALUE with a false discovery rate level of 0.05. The missing genotypes were addressed by MDR Data Tool (v 0.4.3). In the statistical analysis, P < 0.05 was considered significant.
Results
Association between SNP and susceptibility to chronic HBV infection
Analysis of allele, genotype frequencies, and Hardy–Weinberg equilibrium of the selected SNPs is demonstrated in Tables S4–S6 respectively. Notably, rs1059702 and rs1059703 are located on the X chromosome and therefore the Hardy–Weinberg equilibrium cannot be accurately calculated. As a result, rs1059702 and rs1059703 were not included in the response analysis.
After adjustment by age, a significant association between SNP and susceptibility to chronic HBV infection was seen in 10 SNPs. For the four control SNPs in the HLA gene, the results were consistent with previous GWAS, rs3077 (OR = 0.55; 95% CI, 0.42–0.72; P < 0.0001), rs2856718 (OR = 0.60; 95% CI, 0.45–0.80; P = 4e‐04), rs9277535 (OR = 0.54; 95% CI, 0.44–0.66; P < 0.0001) and rs7453920 (OR = 0.43; 95% CI, 0.30–0.62; P < 0.0001). The most significant difference was seen in rs7453920 (dominant model) and rs9277535 (log‐additive model). Similarly, this result indicated that the selection of case and control in this study was reasonable.
For the TLR–IFN pathway genes, a significant association between SNP and the risk of chronic HBV infection was seen in TLR9 rs352140, IL1B rs16944, IL12B rs3212227, IFNGR1 rs3799488, IFNGR2 rs1059293 and MX1 rs467960. For rs352140, the relative risk of chronic HBV infection for genotype CT was lower than for genotype CC‐TT (overdominant model; OR = 0.70; 95% CI, 0.53–0.91; P = 0.0088). For rs16944, the relative risk of chronic HBV infection for genotype AA was lower than for genotype GG‐GA (recessive model; OR = 0.67; 95% CI, 0.49–0.92; P = 0.016). For rs3212227, the relative risk of chronic HBV infection for genotype GT was higher than for genotype TT‐GG (overdominant model; OR = 1.38; 95% CI, 1.05–1.81; P = 0.021). For rs3799488, the relative risk of chronic HBV infection for genotype CT‐CC was higher than for genotype TT (dominant model; OR = 1.48; 95% CI, 1.13–1.95; P = 0.0048. For rs1059293, the relative risk of chronic HBV infection for genotype CC was lower than for genotype TT‐CT (recessive model; OR = 0.27; 95% CI, 0.10–0.71; P = 0.011. For rs467960, the relative risk of chronic HBV infection for genotype CT–TT was lower than for genotype CC (dominant model; OR = 0.68; 95% CI, 0.49–0.94; P = 0.022). For the remaining 33 SNPs, there was no significant association between SNP and susceptibility to chronic HBV infection. Five models of SNP association with chronic HBV infection are presented in Table S7 (10 significant SNPs) and Table S8 (33 non‐significant SNPs).
Given the total of 43 SNPs in this study, we carried out a multiple comparison by QVALUE to reduce the chance of false positives. In addition to the 10 significant SNPs mentioned above, a significant association was seen in three new SNPs, IL1B rs1143627 (OR = 0.74; 95% CI, 0.54–1.01; P = 0.057, q = 0.0467), IL10 rs1800872 (OR = 0.77; 95% CI, 0.59–1.01; P = 0.069, q = 0.0467) and CXCL10 rs4256246 (OR = 1.21; 95% CI, 0.98–1.49; P = 0.069, q = 0.0495). The results are shown in Table 1 (significant) and Table S9 (non‐significant). Therefore, there was a total of 13 SNP loci with a significant association after multiple comparisons. Nevertheless, the new three SNPs demonstrated only borderline significance.
Table 1.
Multiple comparisons of SNP association with chronic HBV infection by QVALUE (significance only)
| Gene | SNP | Model | Genotype | Control | Case | OR (95% CI) | P‐value | q‐value |
|---|---|---|---|---|---|---|---|---|
| HLA‐DPA1 | rs3077 | Dominant | G/G | 124 (45.6%) | 724 (61%) | 1.00 | <1e‐04 | <0.0002 |
| G/A‐A/A | 148 (54.4%) | 462 (39%) | 0.55 (0.42–0.72) | |||||
| HLA‐DPB1 | rs9277535 | Log‐additive | – | – | – | 0.54 (0.44–0.66) | <1e‐04 | <0.0002 |
| HLA‐DQA2 | rs2856718 | Dominant | T/T | 82 (30.3%) | 504 (42.7%) | 1.00 | 4e‐04 | 0.0009 |
| C/T‐C/C | 189 (69.7%) | 677 (57.3%) | 0.60 (0.45–0.80) | |||||
| HLA‐DQB2 | rs7453920 | Dominant | G/G | 213 (78.9%) | 1061 (89.8%) | 1.00 | <1e‐04 | <0.0002 |
| G/A‐A/A | 57 (21.1%) | 121 (10.2%) | 0.43 (0.30–0.62) | |||||
| TLR9 | rs352140 | Overdominant | C/C‐T/T | 122 (44.7%) | 639 (53.6%) | 1.00 | 0.0088 | 0.0137 |
| C/T | 151 (55.3%) | 552 (46.4%) | 0.70 (0.53–0.91) | |||||
| IFNGR1 | rs3799488 | Dominant | T/T | 163 (59.7%) | 608 (51.1%) | 1.00 | 0.0048 | 0.0089 |
| C/T‐C/C | 110 (40.3%) | 582 (48.9%) | 1.48 (1.13–1.95) | |||||
| IFNGR2 | rs1059293 | Recessive | T/T‐C/T | 265 (97.1%) | 1180 (99.2%) | 1.00 | 0.011 | 0.0146 |
| C/C | 8 (2.9%) | 10 (0.8%) | 0.27 (0.10–0.71) | |||||
| IL12B | rs3212227 | Overdominant | T/T‐G/G | 151 (55.9%) | 582 (49%) | 1.00 | 0.021 | 0.0205 |
| G/T | 119 (44.1%) | 606 (51%) | 1.38 (1.05–1.81) | |||||
| IL1B | rs1143627 | Recessive | A/A‐G/A | 197 (72.4%) | 921 (77.6%) | 1.00 | 0.057 | 0.0467 |
| G/G | 75 (27.6%) | 266 (22.4%) | 0.74 (0.54–1.01) | |||||
| IL1B | rs16944 | Recessive | G/G‐G/A | 172 (70.5%) | 882 (77.5%) | 1.00 | 0.016 | 0.0186 |
| A/A | 72 (29.5%) | 256 (22.5%) | 0.67 (0.49–0.92) | |||||
| IL10 | rs1800872 | Overdominant | T/T‐G/G | 138 (51.1%) | 698 (58.8%) | 1.00 | 0.06 | 0.0467 |
| G/T | 132 (48.9%) | 490 (41.2%) | 0.77 (0.59–1.01) | |||||
| CXCL10 | rs4256246 | Log‐additive | – | – | – | 1.21 (0.98–1.49) | 0.069 | 0.0495 |
| MX1 | rs467960 | Dominant | C/C | 207 (75.8%) | 982 (82.5%) | 1.00 | 0.022 | 0.0205 |
| C/T‐T/T | 66 (24.2%) | 209 (17.6%) | 0.68 (0.49–0.94) |
Analysis of gene–gene interaction
To observe gene–gene interactions, we first filtered the data to reduce the number of attributes considered in the analysis by GMDR. Among the 43 SNPs investigated, five were filtered for further analysis, including HLA‐DPB1 rs9277535, IL1B rs16944, IL1B rs1143627, IL1B rs1143623 and IL27B rs6613. We applied interaction entropy algorithms to facilitate interpretation of the relationship between these five SNPs.
As shown in Figure 1A, the interaction dendrogram placed rs16944 and rs1143627, rs9277535 and rs6613 on the same branch. Their position in the diagram indicates the interaction between these SNPs, but the evidence showed that the interaction is not strong between the five SNPs. As shown in Figure 1B, the more percentages of the entropy in case–control status were explained by rs9277535 (1.65%) and rs16944 (0.22%), while the strongest positive percentage entropy indicated an interaction between these two loci (0.13%). Furthermore, a positive marginal effect was seen between rs1143623 and rs6613 (0.10%). This suggested that a synergistic relationship exists between rs9277535 and rs16944, rs1143623 and rs6613.
Figure 1.
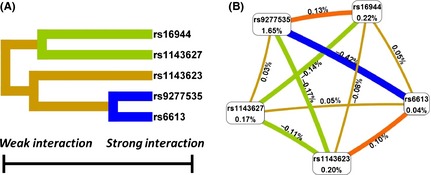
The interaction between the five‐filtered SNPs. (A) Gene–gene interaction dendrogram. The shorter the line connecting two attributes, the stronger the interaction. The colour of the line indicates the type of interaction. Yellow indicates independence. Green and blue indicate redundancy or correlation. Red and orange indicate that there is a synergistic relationship. (B) Entropy algorithms with marginal effects. These interaction models describe the per cent entropy in case–control status, that is, explained by two‐way interaction. Each gene is shown in a box with the per cent entropy below the label. Two‐way interactions between SNPs are depicted as a colour curve accompanied by a per cent of entropy explained by that interaction. Orange with a positive per cent entropy suggests there is a synergistic relationship. Yellow with a positive or negative per cent entropy suggests independence. Green and blue with a negative per cent entropy suggest redundancy or correlation.
Analysis of best models in evaluating the risk of chronic HBV infection
According to the data filtered by GMDR, we further evaluated the risk of chronic HBV infection associated with genetic variation in more than one gene. The GMDR model analysis is shown in Table 2. Rs9277535 was the best single factor for predicting chronic HBV infection risk [testing balance accuracy (TA) = 0.5842; P = 0.0107; cross‐validation consistency (CVC) = 10/10]. The combination of rs9277535 and rs16944 was the best two‐factor model (TA = 0.6040; P = 0.0010; CVC = 10/10). The three‐factor model added rs6613 to the rs9277535 and rs16944 model (TA = 0.5600; P = 0.0107; CVC = 3/10). The four‐factor model included rs9277535, rs16944, rs6613 and rs1143623 (TA = 0.5532; P = 0.0547; CVC = 10/10). The five‐factor model added rs1143627 to the four‐factor model (TA = 0.5487; P = 0.1719; CVC = 10/10). Therefore, the best model is the two‐factor model, rs9277535 plus rs16944. Figure 2A and Table S10 summarize the MDR analysis for the best model. Of all nine possible genotype combinations, nine were observed in the samples, four genotypes were predicted to be high risk (dark‐grey cells), and five genotypes were predicted to be low risk (light‐grey cells). Figure 2B illustrates the distribution of cases and controls when the two functional SNPs were considered jointly. A new single attribute was constructed by pooling the “high‐risk” genotype combinations into a single group (1) and the ‘low‐risk’ into another (0).
Table 2.
The best models to predict chronic HBV infection by GMDR
| Factor | Modela | Training Bal. Acc.b | Testing Bal. Acc.b | Sign Test (P)c | CV Consistencyd |
|---|---|---|---|---|---|
| 1 | rs9277535 | 0.5847 | 0.5842 | 9 (0.0107) | 10/10 |
| 2 | rs9277535 rs16944e | 0.6045 | 0.6040 | 10 (0.0010) | 10/10 |
| 3 | rs9277535 rs16944 rs6613 | 0.6104 | 0.5600 | 9 (0.0107) | 3/10 |
| 4 | rs9277535 rs16944 rs6613 rs1143623 | 0.6267 | 0.5532 | 8 (0.0547) | 10/10 |
| 5 | rs9277535 rs16944 rs1143627 rs6613 rs1143623 | 0.6312 | 0.5487 | 7 (0.1719) | 10/10 |
The best combination of attributes for each order model.
Ratio of correct classifications to the total number of instances classified within the training or testing set. This excludes instances that could not be classified. If cross‐validation is used, this value is estimated from the average of all accuracies across the n subsets.
The number of testing accuracies greater than 0.5 and, in parentheses, the P‐value computed using the nonparametric sign test.
Number of times in a particular cross‐validated run that a given attribute combination was selected as the best model.
Whole dataset statistics: Training Balanced Accuracy, 0.6045; Training Accuracy, 0.6045; Training Sensitivity, 0.6742; Training Specificity, 0.5348; Training Odds ratio, 2.3792 (1.6188, 3.4968); Training χ² (P), 19.7912 (P < 0.0001); Training Precision, 0.5917; Training Kappa, 0.2090; Training F‐Measure, 0.6303.
Figure 2.
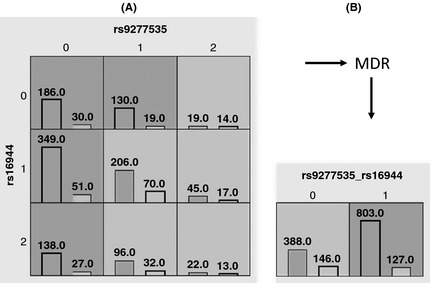
Summary of the best MDR model (rs9277535 + rs16944). (A) A two‐locus model has nine multifactorial cells, each of which is filled with the distribution of cases (left bars) and controls (right bars) for the corresponding genotypes. Each cell is labelled either ‘high risk’ (dark‐grey) or ‘low risk’ (light‐grey) based on its case–control ratio. For rs9277535, 0, 1, and 2 represent the genotypes GG, GA and AA respectively. This has also been adopted for rs16944. (B) A new single attribute is constructed by pooling the ‘high‐risk’ genotype combinations into a single group (1) and the ‘low‐risk’ into another group (0).
Discussion
Chronic HBV infection most frequently occurs via mother‐to‐child transmission or following childhood infection in China 24. Innate immunity plays a crucial role in HBV control 12. Susceptibility to chronic infection rather than virus clearance after HBV exposure, presentation of various clinical phenotypes and differing clinical outcomes in chronic HBV infection, as well as differing responses to antivirus treatment demonstrate that the genetic background of the innate immune system is a crucial factor in determining the outcome of HBV infection.
In terms of genetics, HBV infection is a complex disease. Several SNPs located in the HLA gene cluster, namely rs2856718, rs7453920, rs3077 and rs9277535, were identified by GWAS as being associated with the risk of chronic infection with HBV in Asian individuals 3, 4, 25, 26. Genetic variants in the HLA‐DP locus are strongly associated with persistent HBV infection in the northern Han Chinese population and in individuals of both European‐American and African‐American ancestry 27, 28. Unlike the IL28B SNP, rs12979860, which is associated with HCV clearance, there are few studies focusing on the mechanisms by which HLA gene variations may influence the development of HBV infection. Furthermore, studies investigating the mechanism between IL28B variation and HCV clearance have shown that IL28B variation is associated with other immune factors 19, 21, 29, 30. Therefore, we attempted to define the association between genes of the innate immunity and HLA variation.
In this study, as expected, a significant association was seen in all four of the HLA SNPs. The two most significantly associated SNPs, rs7453920 and rs9277535, are located in HLA‐DQB1 and HLA‐DPB1, which encode β‐chain paralogs of HLA‐DQ and HLA‐DP respectively. Class II molecules are expressed in antigen‐presenting cells such as B lymphocytes, dendritic cells, and macrophages and antigen polymorphisms in these molecules depend on β‐chain paralogs. Therefore, the two SNP loci may affect the presentation of peptides derived from extracellular proteins by influencing the class II molecules. Furthermore, in addition to an association with HBV infection, rs7453920 and rs9277535 were also found to be associated with cervical cancer in Chinese females, and with multiple sclerosis 31, 32.
Although 39 SNPs within 23 genes of the TLR–IFN pathway were considered, significant associations were only seen in six SNP loci. Heterozygosity at TLR9 rs352140 and MX1 rs467960 was protective against the risk of chronic hepatitis B, while heterozygosity at IL12B rs3212227 was associated with susceptibility to chronic HBV infection. The minor alleles of IL1B rs16944 and IFNGR2 rs1059293 were protective against the risk of chronic HBV infection, while the minor allele of IFNGR1 rs3799488 conferred susceptibility. Notably, a significant association was not seen for IL28B rs12979860 and rs8099917, two SNP loci associated with HCV clearance.
TLRs play a key role in innate immunity by recognizing invading pathogens. In a study of HBV transgenic mice, a single intravenous injection of ligands specific for TLR3, TLR4, TLR5, TLR7 and TLR9 can inhibit HBV replication 33. TLR9 rs352140 was found to be associated with various diseases, such as systemic lupus erythematosus, cancer, glomerulonephritis and malaria 34, 35, 36, 37. Notably, in our study, no significant correlation was seen in four SNP loci of TLR3, TLR4 and TLR5. This suggested that TLR9 may play a more influential role in anti‐HBV immunity than other TLRs involved in the antiviral response.
The IL12B gene encodes the beta subunit (p40) of IL‐12 and IL‐23, which can activate the transcription activator STAT4 and stimulate the production of IFN‐γ. IL12B rs3212227, located in the 3'UTR of exon 6, is associated with various diseases, especially those mediated by the immune system 38, 39. Moreover, serum IL‐12p40 levels of rs3212227 CC/AC genotypes were less than AA genotypes in osteosarcoma patients 40. The mechanism by which rs3212227 influences HBV infection may be associated with influencing cytokine protein expression.
Interleukin‐1 beta (IL‐1β), encoded by the IL1B gene, is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation, differentiation and apoptosis. IL1B rs16944 has been associated with several diseases, including stroke, HIV infection and chronic obstructive pulmonary disease 41, 42, 43. Although the mechanism by which rs16944 is involved in such diseases is not clear, such a wide association with so many conditions indicates that rs16944 plays an important role in the immune system.
In this study, five SNP loci in IFNG and IFNGR were also considered. However, a significant association was seen in IFNGR1 and IFNGR2, which together encode the heterodimeric interferon gamma receptor. IFNGR1 rs3799488 and IFNGR2 rs1059293 were reportedly associated with cancer 44, 45, 46. Mx1 is an interferon‐inducible intracytoplasmic protein that mediates antiviral activity against invading viruses. Studies have shown that overexpression of the HBV capsid protein led to selective downregulation of Mx1 gene expression and also indicated that Mx1 protein exerts an antiviral effect on HBV 47, 48, 49. Among four SNP loci within Mx1, a significant association was only seen in rs467960. MX1 rs467960 C to T in exons encodes a synonymous mutation of isoleucine. Although synonymous SNPs do not produce altered coding sequences and are not expected to change the function of the protein, a study has shown that a synonymous SNP in the Multidrug Resistance 1 gene altered not only drug and inhibitor interactions but also the structure of substrate and inhibitor interaction sites 50.
To investigate potential gene–gene interactions, we further observed the gene–gene interactions among all 43 SNPs. Among five filtered SNPs, three SNP loci were located in IL1B, but only rs9277535 and rs16944 were significant SNP loci. Gene–gene interactions with synergistic relationships were only seen in two combinations in all 10 combinations. This result suggests that the main mechanism of these significant SNP loci may be independent of one another.
Furthermore, we evaluated the risk of chronic HBV infection associated with genetic variation in more than one gene. The best GMDR model was the combination of rs9277535 and rs16944, which demonstrated the highest testing accuracy and CVC. A weak positive percentage entropy in case–control status was observed. Therefore, this suggested that the rs9277535 and rs16944 model should be used for evaluating the risk of chronic HBV infection, and IL1B rs16944 plays an important role in the mechanism of chronic HBV infection when in combination with HLA‐DPB1 rs9277535.
This study has several important features. First, our focus on the mechanism of chronic HBV infection was concentrated around the association between the innate immune pathway and previously published GWAS findings. Second, the selected target genes cover the key factors in the TLR–IFN pathway and most proteins encoded by these genes are soluble and readily detectable. Third, MDR was applied to analyse the observed gene–gene interactions.
In conclusion, in the Chinese Southwest Han population, key gene polymorphisms within the TLR–IFN pathway are associated with susceptibility to chronic HBV infection. Interactions with such TLR–IFN SNPs may be involved in the mechanism behind the observed association between HLA polymorphisms and chronic HBV infection. Finally, the combination of HLA‐DPB1 rs9277535 and IL1B rs16944 is a more reliable predictor than rs9277535 of susceptibility to chronic HBV infection.
Supporting information
Figure S1. Illustration for Multiplex SNP Genotyping Using iMLDRTM technique.
Table S1. Basic information of the selected SNP loci in this study.
Table S2. The primer information of the selected SNP loci.
Table S3. Primer information of double reaction.
Table S4. Analysis of allele of the selected SNPs in this study.
Table S5. Genotype frequencies of the selected SNPs in this study.
Table S6. SNP loci exact test for Hardy–Weinberg equilibrium.
Table S7. Significant SNPs association with chronic HBV infection (adjusted by age).
Table S8. Non‐significant SNPs association with chronic HBV infection (adjusted by age).
Table S9. Multiple comparisons by QVALUE (only non‐significance).
Table S10. Detail of the best model (rs9277535+rs16944).
Acknowledgments
We acknowledge helpful suggestions, discussions and excellent technical assistance from Prof. Qing Mao, Prof. Guohong Deng and Prof. Xiaohong Wang. We acknowledge SNP genotyping by the Center for Genetic & Genomic Analysis, Genesky Biotechnologies Inc., Shanghai 201203, China. This study was supported by grants from the National Natural Science Foundation of China (81270563) and the State Key Project specialized for HBV‐related severe hepatitis of China (2012ZX10002004).
Conflict of interests: The authors do not have any disclosures to report.
Liver Int. 2015; 35: 1941–1949
Handling Editor: Vincent wong
References
- 1. European Association For The Study Of The Liver . EASL clinical practice guidelines: management of chronic hepatitis B virus infection. J Hepatol 2012; 57: 167–85. [DOI] [PubMed] [Google Scholar]
- 2. Ganem D, Prince AM. Hepatitis B virus infection–natural history and clinical consequences. N Engl J Med 2004; 350: 1118–29. [DOI] [PubMed] [Google Scholar]
- 3. Kamatani Y, Wattanapokayakit S, Ochi H, et al A genome‐wide association study identifies variants in the HLA‐DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009; 41: 591–5. [DOI] [PubMed] [Google Scholar]
- 4. Mbarek H, Ochi H, Urabe Y, et al A genome‐wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 2011; 20: 3884–92. [DOI] [PubMed] [Google Scholar]
- 5. Deng G, Zhou G, Zhang R, et al Regulatory polymorphisms in the promoter of CXCL10 gene and disease progression in male hepatitis B virus carriers. Gastroenterology 2008; 134: 716–26. [DOI] [PubMed] [Google Scholar]
- 6. Sawhney R, Visvanathan K. Polymorphisms of toll‐like receptors and their pathways in viral hepatitis. Antivir Ther 2011; 16: 443–58. [DOI] [PubMed] [Google Scholar]
- 7. Wu H, Zhao G, Qian F, et al Association of IL28B polymorphisms with peginterferon treatment response in Chinese Han patients with HBeAg‐positive chronic hepatitis B. Liver Int 2014; DOI: 10.1111/liv.12491 [DOI] [PubMed] [Google Scholar]
- 8. Cheong JY, Cho SW, Chung SG, et al Genetic polymorphism of interferon‐gamma, interferon‐gamma receptor, and interferon regulatory factor‐1 genes in patients with hepatitis B virus infection. Biochem Genet 2006; 44: 246–55. [DOI] [PubMed] [Google Scholar]
- 9. Gong QM, Kong XF, Yang ZT, et al Association study of IFNAR2 and IL10RB genes with the susceptibility and interferon response in HBV infection. J Viral Hepat 2009; 16: 674–80. [DOI] [PubMed] [Google Scholar]
- 10. Wang S, Huang D, Sun S, Ma W, Zhen Q. Interleukin‐10 promoter polymorphism predicts initial response of chronic hepatitis B to interferon alfa. Virol J 2011; 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology 2008; 47: 729–36. [DOI] [PubMed] [Google Scholar]
- 12. Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol 2005; 5: 215–29. [DOI] [PubMed] [Google Scholar]
- 13. Khoo JJ, Forster S, Mansell A. Toll‐like receptors as interferon‐regulated genes and their role in disease. J Interferon Cytokine Res 2011; 31: 13–25. [DOI] [PubMed] [Google Scholar]
- 14. Jung MC, Pape GR. Immunology of hepatitis B infection. Lancet Infect Dis 2002; 2: 43–50. [DOI] [PubMed] [Google Scholar]
- 15. Li J, Liu K, Liu Y, et al Exosomes mediate the cell‐to‐cell transmission of IFN‐alpha‐induced antiviral activity. Nat Immunol 2013; 14: 793–803. [DOI] [PubMed] [Google Scholar]
- 16. Ge D, Fellay J, Thompson AJ, et al Genetic variation in IL28B predicts hepatitis C treatment‐induced viral clearance. Nature 2009; 461: 399–401. [DOI] [PubMed] [Google Scholar]
- 17. Suppiah V, Moldovan M, Ahlenstiel G, et al IL28B is associated with response to chronic hepatitis C interferon‐alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–4. [DOI] [PubMed] [Google Scholar]
- 18. Rallon NI, Soriano V, Naggie S, et al Impact of IL28B gene polymorphisms on interferon‐lambda3 plasma levels during pegylated interferon‐alpha/ribavirin therapy for chronic hepatitis C in patients coinfected with HIV. J Antimicrob Chemother 2012; 67: 1246–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Asahina Y, Tsuchiya K, Muraoka M, et al Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology 2012; 55: 20–9. [DOI] [PubMed] [Google Scholar]
- 20. Naggie S, Osinusi A, Katsounas A, et al Dysregulation of innate immunity in HCV genotype 1 IL28B unfavorable genotype patients: Impaired viral kinetics and therapeutic response. Hepatology 2012; 56: 444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Umemura T, Joshita S, Yoneda S, et al Serum interleukin (IL)‐10 and IL‐12 levels and IL28B gene polymorphisms: pretreatment prediction of treatment failure in chronic hepatitis C. Antivir Ther 2011; 16: 1073–80. [DOI] [PubMed] [Google Scholar]
- 22. Moore JH, Gilbert JC, Tsai CT, et al A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol 2006; 241: 252–61. [DOI] [PubMed] [Google Scholar]
- 23. Lou XY, Chen GB, Yan L, et al A generalized combinatorial approach for detecting gene‐by‐gene and gene‐by‐environment interactions with application to nicotine dependence. Am J Hum Genet 2007; 80: 1125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu FM, Li T, Liu S, Zhuang H. Epidemiology and prevention of hepatitis B virus infection in China. J Viral Hepat 2010; 17(Suppl 1): 4–9. [DOI] [PubMed] [Google Scholar]
- 25. Matsuura K, Tanaka Y, Nishida N, et al A genome‐wide association study identifies genetic variants in the HLA‐DP locus associated with chronic hepatitis B. Hepatology 2009; 50: 1457. [DOI] [PubMed] [Google Scholar]
- 26. Nishida N, Sawai H, Matsuura K, et al Genome‐wide association study confirming association of HLA‐DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS ONE 2012; 7: e39175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo X, Zhang Y, Li J, et al Strong influence of human leukocyte antigen (HLA)‐DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 2011; 53: 422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas R, Thio CL, Apps R, et al A novel variant marking HLA‐DP expression levels predicts recovery from hepatitis B virus infection. J Virol 2012; 86: 6979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pasha HF, Radwan MI, Hagrass HA, Tantawy EA, Emara MH. Cytokines genes polymorphisms in chronic hepatitis C: impact on susceptibility to infection and response to therapy. Cytokine 2013; 61: 478–84. [DOI] [PubMed] [Google Scholar]
- 30. Bes M, Sauleda S, Campos‐Varela I, et al IL28B genetic variation and hepatitis C virus‐specific CD4(+) T‐cell responses in anti‐HCV‐positive blood donors. J Viral Hepat 2012; 19: 867–71. [DOI] [PubMed] [Google Scholar]
- 31. Jiang J, Li N, Shen Y, et al Genetic variants in HLA‐DP/DQ contribute to risk of cervical cancer: a two‐stage study in Chinese women. Gynecol Oncol 2013; 129: 401–5. [DOI] [PubMed] [Google Scholar]
- 32. Field J, Browning SR, Johnson LJ, et al A polymorphism in the HLA‐DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS ONE 2010; 5: e13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll‐like receptor signaling inhibits hepatitis B virus replication in vivo . J Virol 2005; 79: 7269–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J, Zhu Q, Meng F, Lei H, Zhao Y. Association study of TLR‐9 polymorphisms and systemic lupus erythematosus in northern Chinese Han population. Gene 2014; 533: 385–8. [DOI] [PubMed] [Google Scholar]
- 35. Chen YT, Wei CC, Ng KL, et al Toll‐like receptor 9 SNPs are susceptible to the development and progression of membranous glomerulonephritis: 27 years follow‐up in Taiwan. Ren Fail 2013; 35: 1370–5. [DOI] [PubMed] [Google Scholar]
- 36. Omar AH, Yasunami M, Yamazaki A, et al Toll‐like receptor 9 (TLR9) polymorphism associated with symptomatic malaria: a cohort study. Malar J 2012; 11: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L, Qin H, Guan X, Zhang K, Liu Z. The TLR9 gene polymorphisms and the risk of cancer: evidence from a meta‐analysis. PLoS ONE 2013; 8: e71785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou L, Yao F, Luan H, et al Functional polymorphisms in the interleukin‐12 gene contribute to cancer risk: evidence from a meta‐analysis of 18 case‐control studies. Gene 2012; 510: 71–7. [DOI] [PubMed] [Google Scholar]
- 39. Van Wanrooij RL, Zwiers A, Kraal G, Bouma G. Genetic variations in interleukin‐12 related genes in immune‐mediated diseases. J Autoimmun 2012; 39: 359–68. [DOI] [PubMed] [Google Scholar]
- 40. Wang J, Nong L, Wei Y, et al Association of interleukin‐12 polymorphisms and serum IL‐12p40 levels with osteosarcoma risk. DNA Cell Biol 2013; 32: 605–10. [DOI] [PubMed] [Google Scholar]
- 41. Fernandez‐Cadenas I, Del Rio‐Espinola A, Giralt D, et al IL1B and VWF variants are associated with fibrinolytic early recanalization in patients with ischemic stroke. Stroke 2012; 43: 2659–65. [DOI] [PubMed] [Google Scholar]
- 42. Ahir S, Chaudhari D, Chavan V, et al Polymorphisms in IL‐1 gene cluster and its association with the risk of perinatal HIV transmission, in an Indian cohort. Immunol Lett 2013; 153: 1–8. [DOI] [PubMed] [Google Scholar]
- 43. Issac MS, Ashur W, Mousa H. Genetic Polymorphisms of Surfactant Protein D rs2243639, Interleukin (IL)‐1beta rs16944 and IL‐1RN rs2234663 in Chronic Obstructive Pulmonary Disease, Healthy Smokers, and Non‐Smokers. Mol Diagn Ther 2014; 18: 343–54. [DOI] [PubMed] [Google Scholar]
- 44. Slattery ML, Lundgreen A, Bondurant KL, Wolff RK. Interferon‐signaling pathway: associations with colon and rectal cancer risk and subsequent survival. Carcinogenesis 2011; 32: 1660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu G, Pan D, Zheng T, et al Polymorphisms in Th1/Th2 cytokine genes, hormone replacement therapy, and risk of non‐Hodgkin lymphoma. Front Oncol 2011; pii: 00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quan L, Gong Z, Yao S, et al Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int J Cancer 2014; 134: 1408–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gordien E, Rosmorduc O, Peltekian C, et al Inhibition of hepatitis B virus replication by the interferon‐inducible MxA. J Virol 2001; 75: 2684–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosmorduc O, Sirma H, Soussan P, et al Inhibition of interferon‐inducible MxA protein expression by hepatitis B virus capsid protein. J Gen Virol 1999; 80: 1253–62. [DOI] [PubMed] [Google Scholar]
- 49. Peltekian C, Gordien E, Garreau F, et al Human MxA protein participates to the interferon‐related inhibition of hepatitis B virus replication in female transgenic mice. J Hepatol 2005; 43: 965–72. [DOI] [PubMed] [Google Scholar]
- 50. Kimchi‐Sarfaty C, Oh JM, Kim IW, et al A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007; 315: 525–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Illustration for Multiplex SNP Genotyping Using iMLDRTM technique.
Table S1. Basic information of the selected SNP loci in this study.
Table S2. The primer information of the selected SNP loci.
Table S3. Primer information of double reaction.
Table S4. Analysis of allele of the selected SNPs in this study.
Table S5. Genotype frequencies of the selected SNPs in this study.
Table S6. SNP loci exact test for Hardy–Weinberg equilibrium.
Table S7. Significant SNPs association with chronic HBV infection (adjusted by age).
Table S8. Non‐significant SNPs association with chronic HBV infection (adjusted by age).
Table S9. Multiple comparisons by QVALUE (only non‐significance).
Table S10. Detail of the best model (rs9277535+rs16944).


