Abstract
Detergents are often used to investigate the structure and dynamics of membrane proteins. Whereas the structural integrity seems to be preserved in detergents for many membrane proteins, their functional activity is frequently compromised, but can be restored in a lipid environment. Herein we show with per‐residue resolution that while OmpX forms a stable β‐barrel in DPC detergent micelles, DHPC/DMPC bicelles, and DMPC nanodiscs, the pico‐ to nanosecond and micro‐ to millisecond motions differ substantially between the detergent and lipid environment. In particular for the β‐strands, there is pronounced dynamic variability in the lipid environment, which appears to be suppressed in micelles. This unexpected complex and membrane‐mimetic‐dependent dynamic behavior indicates that the frequent loss of membrane protein activity in detergents might be related to reduced internal dynamics and that membrane protein activity correlates with lipid flexibility.
Keywords: membrane proteins, NMR spectroscopy, OmpX, protein dynamics, relaxation
As cellular gatekeepers, integral membrane proteins are essential in all branches of life. As such, their structural plasticity appears to be critical for functionality and selectivity. G‐protein coupled receptors (GPCRs), for example, seem to sample several inactive and active conformations whose equilibrium can be shifted by ligand binding.1 Ion‐conducting channels, such as VDAC (voltage‐dependent anion channel) and KcsA (potassium channel), selectively regulate ion fluxes across cell membranes through their open and closed states.2 Functional studies of integral membrane proteins have traditionally used detergent micelles,3 but recently, lipid‐detergent bicelles,4 amphipathic polymers (amphipols),5 and detergent‐free lipid‐bilayer encircling nanodiscs6 have gained attention as mimetics of hydrophobic membrane environments. However, owing to the capabilities of detergents in terms of protein extraction, refolding, and crystallization, detergent micelles remain the most common membrane mimetic for biophysical studies.7 Unfortunately, detergents frequently reduce or abolish the functionality of membrane proteins,4a, 8 or interfere with functional assays at higher concentration.9 Furthermore, detergents also seem to influence membrane protein dynamics. A recent NMR study revealed that the exchange rate between the active and inactive conformation of the β2 adrenergic receptor is on the millisecond timescale in lipid‐bilayer nanodiscs, which is remarkably different from those measured in detergent micelles.10 In a single‐molecule force spectroscopy study, the human voltage dependent anion channel 1 (hVDAC1) showed lower mechanical rigidity and a higher number of conformational states in liposomes than in detergent micelles.11 In another recent NMR study, the dynamic behavior of BamA was globally characterized in micelles, bicelles, and nanodiscs.12 However, studies investigating and comparing fast and slow motions in detergent and lipid environments on a residue‐specific level have not been reported to date. Hence, we herein compare in detail on a per‐residue level the fast and slow timescale dynamics of the outer membrane protein X (OmpX) upon incorporation into dodecylphosphocholine (DPC) micelles, dihexanoylphosphocholine/dimyristoylphosphocholine (DHPC/DMPC) bicelles, and DMPC nanodiscs. The fast and slow motions for OmpX in DPC micelles have low dynamic variability, similar to what has been reported previously for OmpA, PagP, and hVDAC.13 In a lipid environment, however, the dynamic variability in the β‐strands of OmpX increases significantly, especially on the micro‐ to millisecond timescale, suggesting that the inherent flexibility of membrane proteins is seriously compromised in detergent micelles.
The virulence‐related membrane protein OmpX is an eight‐stranded antiparallel β‐barrel.14 1H–15N TROSY HSQC 2D spectra of OmpX reconstituted in micelles, bicelles, and nanodiscs show the characteristic broad dispersion of HN chemical shifts (Figure 1), indicative of a β‐structure, in accordance with previous NMR investigations.4b, 15 The broad HN dispersion allowed for the unambiguous assignment of 125 (84 %), 135 (91 %), and 131 (89 %) out of 148 residues for OmpX in micelles, bicelles, and nanodiscs, respectively (see the Supporting Information and Figure S1 for details regarding the incorporation of OmpX in the different membrane mimetics).4b, 15a, 16 A closer inspection of the 2D 1H–15N TROSY HSQC spectra revealed chemical shift differences between the DPC detergent environment and the DMPC lipid environment for most resonances (Figure S2). The formation of DHPC/DMPC bicelles from DHPC micelles induced chemical shift changes for OmpX that agree well with the chemical shifts of OmpX in DMPC nanodiscs (Figure 1 d and Figure S2), indicating that OmpX in DHPC/DMPC bicelles is indeed surrounded by DMPC and not by DHPC.
Figure 1.
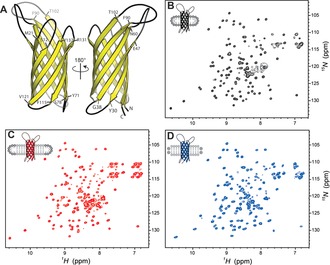
Spectral overview of OmpX. A) Cartoon representation of OmpX. Residues flanking the loops are specified. 2D 1H–15N TROSY HSQC spectra of OmpX in B) DPC micelles, C) DHPC/DMPC bicelles, and D) DMPC‐containing nanodiscs.
Next, we determined the dynamics of OmpX over a broad range of timescales using NMR relaxation.17 The measurement of [1H]–15N hetNOE values (Figure 2 A) and 15N R 1 relaxation rates (Figure 2 B) probes pico‐ to nanosecond motions of the H−N bonds. High [1H]–15N hetNOE values and small R 1 rates indicate a rigid, stably folded region whereas low [1H]–15N hetNOE values and high R 1 rates point to flexible or unstructured regions. The relaxation rates of OmpX in DPC micelles allowed for clear identification of all eight β‐strands, which are characterized by high hetNOE values of approximately 0.8 and low R 1 rates of about 0.6 s−1. The strands are interrupted by flexible loops with low hetNOE values of about 0.4 and higher R 1 rates compared to the β‐strands, similar to findings for outer membrane protein A (OmpA) in DPC micelles.13b For OmpX in DPC micelles, the β‐strand regions show uniform fast‐timescale dynamics as indicated by very similar hetNOE values and R 1 rates. For OmpX in nanodiscs and bicelles, we observed a similar general behavior of overall more rigid β‐strands that are interrupted by flexible loop regions, which appear to be even more dynamic in bicelles and nanodiscs than in detergent micelles, with hetNOE values between 0.2–0.4. Furthermore, as indicated by substantial variations in the hetNOE values and R 1 rates, we observed less uniform fast‐timescale (ps to ns) dynamics within the rigid β‐strands and a more gradual transition of mobility between the loops and β‐strands in the lipid environment. One example is the β3‐strand where, according to the hetNOE values, the most rigid residues are located centrally. Furthermore, the β6‐strand shows a gradual increase in rigidity from the N‐ to the C‐terminal; this effect is most pronounced in lipid nanodiscs and also observable in bicelles but absent in detergent micelles. For the β5‐strand, however, the pattern of gradually decreasing flexibility from the loop regions to the β‐strands is most pronounced in bicelles. The gradual transition between rigid and flexible parts of OmpX is also reflected by the R 1 rates. Notably, the R 1 rates of around 0.3–0.4 s−1 for OmpX in nanodiscs and bicelles are due to the slower tumbling times of approximately 40 ns and about 35 ns, respectively, which are much slower than the about 22 ns for OmpX in micelles, estimated from the transverse 15N dipolar/CSA cross‐correlated relaxation rates, ηxy , of the most rigid residues (Figure S3). To investigate the conformational plasticity of OmpX on the micro‐ to millisecond time scale, we measured the 15N transverse relaxation rates of the slowly relaxing 15N[1H] doublet component (R 2β) in a Hahn echo experiment.18 Whereas R 1 is dominated by fast‐timescale motions, exchange contributions, R ex, that are due to dynamics on the micro‐ to millisecond timescale will contribute to R 2β.19
Figure 2.
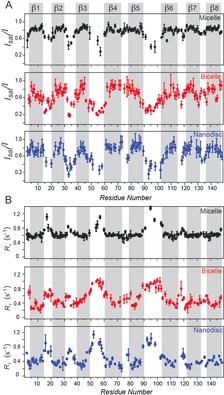
Fast‐timescale (pico‐ to nanosecond) motions of OmpX. A) [1H]–15N hetNOE values for the residues of OmpX in DPC micelles (black circles), DHPC/DMPC bicelles (red circles), and DMPC nanodiscs (blue circles). The β‐strands exhibit high hetNOE values, indicating a rigid, stable secondary structure, whereas the connecting loops show low hetNOE values, indicating flexibility. In contrast to the striking separation between β‐strands and loops for OmpX in detergent environments, a gradual transition between rigid β‐strands and flexible loops was observed for OmpX in the lipid environment. B) A similar pattern was found for the 15N R 1 relaxation rates. Low rates indicate a rigid secondary structure while high rates suggest flexibility on the pico‐ to nanosecond timescale.
Although elevated R 2β rates can be an indicator for conformational exchange on the micro‐ to millisecond timescale, they can also be the result of a large anisotropic diffusion tensor.20 Notably, whereas the R 2β/R 1 ratios are also influenced by the anisotropy of the diffusion tensor and the resulting effective correlation time, these contributions cancel out for the product R 1×R 2β.21 We therefore plotted the R 2β rates along with the R 1×R 2β values (green bars in the upper panels in Figure 3 A–C). For OmpX in detergent micelles, elevated R 2β rates were only found for residues located within the loops and N‐terminal Thr2 (Figure 3 A, black circles in the upper panel).
Figure 3.
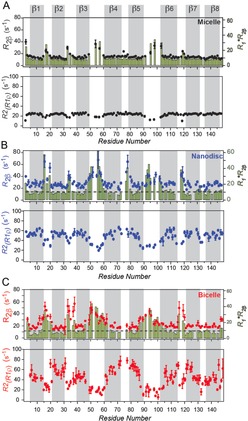
Slow‐timescale (micro‐ to millisecond) motions of OmpX. A) OmpX in DPC micelles shows exchange contributions on the micro‐ to millisecond timescale for loop residues, indicated by increased R 2β rates (black circles) and consistent R 1×R 2β values (green bars). The exchange contributions were refocused with a spin‐lock field of 2000 Hz (lower panel). B) R 2β (red circles), R 1×R 2β (green bars), and R2R1ρ (lower panel) values for OmpX in nanodiscs. C) R 2β (blue circles), R 1×R 2β (green bars), and R2R1ρ (lower panel) values for OmpX in bicelles.
Otherwise, a planar profile of R 2 rates was observed, similar to the hetNOE and R 1 rates. Furthermore, the R 1×R 2β values revealed the same trend. However, for R 2 rates derived from R 1ρ measurements (R2R1ρ), which refocus R ex contributions that are slower than about 80 μs, an opposite behavior with lower rates for the (more flexible) loop region is apparent. This comparison indicates that elevated R 2 values are not caused by diffusional anisotropy but result from conformational exchange on the micro‐ to millisecond timescale (Figure 3 A, green bars in the upper panel) that is slower than about 80 μs. For OmpX in nanodiscs, a similar picture emerged. Whereas the loop regions and Thr2 undergo exchange on the micro‐ to millisecond timescale, the β‐strand regions appear flat and free of exchange (Figure 3 B, upper panel), but with slightly more pronounced dynamic variability. In bicelles, however, Thr2, Ser3, and several residues of the β‐strands 1, 3, 5, and 6 show conformational exchange (Figure 3 C, upper panel). As R 1×R 2β shows a similar pattern, this behavior cannot be explained by rotational anisotropy but indeed also points to conformational exchange on the micro‐ to millisecond timescale.
The conformational variability and the number of residues showing conformational exchange are higher in bicelles than in nanodiscs. These findings correlate strikingly well with the higher flexibility and reduced order of lipids in liposomes when compared to lipids in nanodiscs,22 showing that lipid flexibility directly impacts protein dynamics. In contrast, deuterated lipids do not influence the relaxation rates (Figure S1).
In conclusion, we have compared the dynamics of OmpX incorporated in DPC micelles, DHPC/DMPC bicelles, and DMPC nanodiscs. Although OmpX forms a stable β‐barrel with a similar 3D structure in all three membrane mimetics,4b, 15, 23 the pico‐ to nanosecond and micro‐ to millisecond motions differ between the detergent and the lipid environment. In detergent micelles, all eight β‐strands of OmpX display similar dynamic behavior, resulting in flat profiles for the hetNOE, R 1, and R2R1ρ values, which are sharply interrupted by flexible loop regions, similar to what is found for OmpA, PagP, and hVDAC.13 In a lipid environment, a more gradual transition from rigid β‐strands to flexible loops was observed. In all three environments, the loops show high flexibility on the pico‐ to nanosecond timescale and conformational exchange on the micro‐ to millisecond timescale. The relaxation data suggest that the inherent flexibility of membrane proteins is seriously compromised in detergent micelles, which may explain the often observed decrease in functional activity.8b Furthermore, lipid flexibility seems to have a substantial influence on membrane protein dynamics.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank Sebastian Hiller for the OmpX plasmid and Oliver Zerbe for the MSPΔH5 plasmid (with permission from Gerhard Wagner). This project was funded by the Swiss National Science Foundation (grant number 154461) and the EU Marie Curie program (grant number 627767).
L. Frey, N.-A. Lakomek, R. Riek, S. Bibow, Angew. Chem. Int. Ed. 2017, 56, 380.
References
- 1.
- 1a. Kobilka B. K., Deupi X., Trends Pharmacol. Sci. 2007, 28, 397–406; [DOI] [PubMed] [Google Scholar]
- 1b. Nygaard R., Zou Y. Z., Dror R. O., Mildorf T. J., Arlow D. H., Manglik A., Pan A. C., Liu C. W., Fung J. J., Bokoch M. P., Thian F. S., Kobilka T. S., Shaw D. E., Mueller L., Prosser R. S., Kobilka B. K., Cell 2013, 152, 532–542; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Liu J. J., Horst R., Katritch V., Stevens R. C., Wüthrich K., Science 2012, 335, 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Peng S., Blachlydyson E., Forte M., Colombini M., Biophys. J. 1992, 62, 123–135; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Baker K. A., Tzitzilonis C., Kwiatkowski W., Choe S., Riek R., Nat. Struct. Mol. Biol. 2007, 14, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Tanford C., Reynolds J. A., Biochim. Biophys. Acta Rev. Biomembr. 1976, 457, 133–170; [DOI] [PubMed] [Google Scholar]
- 3b. Helenius A., Simons K., Biochim. Biophys. Acta Rev. Biomembr. 1975, 415, 29–79. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Sanders C. R., Landis G. C., Biochemistry 1995, 34, 4030–4040; [DOI] [PubMed] [Google Scholar]
- 4b. Lee D., Walter K. F. A., Bruckner A. K., Hilty C., Becker S., Griesinger C., J. Am. Chem. Soc. 2008, 130, 13822–13823. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Tribet C., Audebert R., Popot J. L., Proc. Natl. Acad. Sci. USA 1996, 93, 15047–15050; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Chae P. S., Rasmussen S. G., Rana R. R., Gotfryd K., Kruse A. C., Manglik A., Cho K. H., Nurva S., Gether U., Guan L., Loland C. J., Byrne B., Kobilka B. K., Gellman S. H., Chem. Eur. J. 2012, 18, 9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Bayburt T. H., Carlson J. W., Sligar S. G., J. Struct. Biol. 1998, 123, 37–44; [DOI] [PubMed] [Google Scholar]
- 6b. Glück J. M., Wittlich M., Feuerstein S., Hoffmann S., Willbold D., Koenig B. W., J. Am. Chem. Soc. 2009, 131, 12060–12061; [DOI] [PubMed] [Google Scholar]
- 6c. Lyukmanova E. N., Shenkarev Z. O., Paramonov A. S., Sobol A. G., Ovchinnikova T. V., Chupin V. V., Kirpichnikov M. P., Blommers M. J., Arseniev A. S., J. Am. Chem. Soc. 2008, 130, 2140–2141. [DOI] [PubMed] [Google Scholar]
- 7. Linke D., Methods Enzymol. 2009, 463, 603–617. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Poget S. F., Girvin M. E., Biochim. Biophys. Acta Biomembr. 2007, 1768, 3098–3106; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Hwang P. M., Bishop R. E., Kay L. E., Proc. Natl. Acad. Sci. USA 2004, 101, 9618–9623; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8c. Catoire L. J., Warnet X. L., Warschawski D. E., Membrane Proteins Production for Structural Analysis, Springer, Heidelberg, 2014, pp. 315–345. [Google Scholar]
- 9. Ding Y., Fujimoto L. M., Yao Y., Plano G. V., Marassi F. M., Biochim. Biophys. Acta Biomembr. 2015, 1848, 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kofuku Y., Ueda T., Okude J., Shiraishi Y., Kondo K., Mizumura T., Suzuki S., Shimada I., Angew. Chem. Int. Ed. 2014, 53, 13376–13379; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 13594–13597. [Google Scholar]
- 11. Ge L., Villinger S., Mari S. A., Giller K., Griesinger C., Becker S., Muller D. J., Zweckstetter M., Structure 2016, 24, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morgado L., Zeth K., Burmann B. M., Maier T., Hiller S., J. Biomol. NMR 2015, 61, 333–345. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Hwang P. M., Choy W. Y., Lo E. I., Chen L., Forman-Kay J. D., Raetz C. R., Prive G. G., Bishop R. E., Kay L. E., Proc. Natl. Acad. Sci. USA 2002, 99, 13560–13565; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Liang B., Arora A., Tamm L. K., Biochim. Biophys. Acta Biomembr. 2010, 1798, 68–76; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Jaremko M., Jaremko L., Villinger S., Schmidt C. D., Griesinger C., Becker S., Zweckstetter M., Angew. Chem. Int. Ed. 2016, 55, 10518; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 10674. [Google Scholar]
- 14. Vogt J., Schulz G. E., Structure 1999, 7, 1301–1309. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. Hagn F., Etzkorn M., Raschle T., Wagner G., J. Am. Chem. Soc. 2013, 135, 1919–1925; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15b. Fernandez C., Adeishvili K., Wüthrich K., Proc. Natl. Acad. Sci. USA 2001, 98, 2358–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. F. A. Walter, Georg-August-Universität Göttingen, 2011.
- 17.
- 17a. Kleckner I. R., Foster M. P., Biochim. Biophys. Acta Proteins Proteomics 2011, 1814, 942–968; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17b. Jarymowycz V. A., Stone M. J., Chem. Rev. 2006, 106, 1624–1671; [DOI] [PubMed] [Google Scholar]
- 17c. Morin S., Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 245–262. [DOI] [PubMed] [Google Scholar]
- 18. Lakomek N. A., Kaufman J. D., Stahl S. J., Louis J. M., Grishaev A., Wingfield P. T., Bax A., Angew. Chem. Int. Ed. 2013, 52, 3911–3915; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 4003–4007. [Google Scholar]
- 19. Lakomek N. A., Ying J., Bax A., J. Biomol. NMR 2012, 53, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kroenke C. D., Loria J. P., Lee L. K., Rance M., Palmer A. G., J. Am. Chem. Soc. 1998, 120, 7905–7915. [Google Scholar]
- 21. Kneller J. M., Lu M., Bracken C., J. Am. Chem. Soc. 2002, 124, 1852–1853. [DOI] [PubMed] [Google Scholar]
- 22.
- 22a. Nakano M., Fukuda M., Kudo T., Miyazaki M., Wada Y., Matsuzaki N., Endo H., Handa T., J. Am. Chem. Soc. 2009, 131, 8308–8312; [DOI] [PubMed] [Google Scholar]
- 22b. Mörs K., Roos C., Scholz F., Wachtveitl J., Dotsch V., Bernhard F., Glaubitz C., Biochim. Biophys. Acta Biomembr. 2013, 1828, 1222–1229. [DOI] [PubMed] [Google Scholar]
- 23. Bibow S., Carneiro M. G., Sabo T. M., Schwiegk C., Becker S., Riek R., Lee D., Protein Sci. 2014, 23, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


