ABSTRACT
Objective
Accurate gestational‐age (GA) estimation, preferably by ultrasound measurement of fetal crown–rump length before 14 weeks' gestation, is an important component of high‐quality antenatal care. The objective of this study was to determine how GA can best be estimated by fetal ultrasound for women who present for the first time late in pregnancy with uncertain or unknown menstrual dates.
Methods
INTERGROWTH‐21st was a large, prospective, multicenter, population‐based project performed in eight geographically defined urban populations. One of its principal components, the Fetal Growth Longitudinal Study, aimed to develop international fetal growth standards. Each participant had their certain menstrual dates confirmed by first‐trimester ultrasound examination. Fetal head circumference (HC), biparietal diameter (BPD), occipitofrontal diameter (OFD), abdominal circumference (AC) and femur length (FL) were measured every 5 weeks from 14 weeks' gestation until delivery. For each participant, a single, randomly selected ultrasound examination was used to explore all candidate biometric variables and permutations to build models to predict GA. Regression equations were ranked based upon minimization of the mean prediction error, goodness of fit and model complexity. An automated machine learning algorithm, the Genetic Algorithm, was adapted to evaluate > 64 000 potential polynomial equations as predictors.
Results
Of the 4607 eligible women, 4321 (94%) had a pregnancy without major complications and delivered a live singleton without congenital malformations. After other exclusions (missing measurements in GA window and outliers), the final sample comprised 4229 women. Two skeletal measures, HC and FL, produced the best GA prediction, given by the equation loge(GA) = 0.03243 × (loge(HC))2 + 0.001644 × FL × loge(HC) + 3.813. When FL was not available, the best equation based on HC alone was loge(GA) = 0.05970 × (loge(HC))2 + 0.000000006409 × (HC)3 + 3.3258. The estimated uncertainty of GA prediction (half width 95% interval) was 6–7 days at 14 weeks' gestation, 12–14 days at 26 weeks' gestation and > 14 days in the third trimester. The addition of FL to the HC model led to improved prediction intervals compared with using HC alone, but no further improvement in prediction was afforded by adding AC, BPD or OFD. Equations that included other measurements (BPD, OFD and AC) did not perform better.
Conclusions
Among women initiating antenatal care late in pregnancy, a single set of ultrasound measurements combining HC and FL in the second trimester can be used to estimate GA with reasonable accuracy. We recommend this tool for underserved populations but considerable efforts should be implemented to improve early initiation of antenatal care worldwide. © 2016 Authors. Ultrasound in Obstetrics & Gynecology published by John Wiley & Sons Ltd on behalf of International Society of Ultrasound in Obstetrics and Gynecology.
Keywords: dating, fetal growth, gestational age
Short abstract
INTRODUCTION
Reliable estimation of gestational age (GA) is essential as it allows appropriate scheduling of a woman's antenatal care, informs obstetric management decisions and facilitates the correct interpretation of fetal growth assessment1. Abnormal fetal growth patterns such as growth restriction or macrosomia may be missed or diagnosed incorrectly if GA is unknown or incorrect. Reliable GA estimation is also important at a population level to calculate rates of preterm delivery and small‐for‐gestational‐age neonates at delivery. The lack of accurate GA estimation, particularly in geographical regions at greatest risk of these conditions, means that preterm delivery and small‐for‐gestational‐age rates are mere approximations in many parts of the world2, 3.
Traditionally, GA is estimated using the first day of the last menstrual period (LMP), which assumes that ovulation occurs on day 14 of the menstrual cycle. Irregular menses, unknown or uncertain dates, oral contraceptive use or recent pregnancy or breastfeeding, issues that occur in a large proportion of women, may all influence the accuracy of this method4, 5, 6. In such cases, early (< 14 weeks' gestation) ultrasound measurement of fetal crown–rump length (CRL) is recommended7, 8. First‐trimester GA assessment is more accurate than is dating in late pregnancy because, with advancing gestation, fetal ultrasound measurements have a larger absolute error9 and growth disturbances become more noticeable, resulting in potential underestimation of GA for an abnormally small fetus and overestimation for a macrosomic fetus.
Unfortunately, in many settings in which high‐risk pregnancies are prevalent, women attend their first antenatal care visit late in pregnancy or even at the time of delivery. This makes it difficult to manage complications, evaluate fetal growth and implement evidence‐based interventions, such as the administration of corticosteroids for fetal lung maturation in cases of threatened preterm labor.
The present analysis of the Fetal Growth Longitudinal Study (FGLS), one of the main components of the INTERGROWTH‐21st Project, aimed to complement our previous work of early GA estimation by ultrasound measurement of CRL10. We explored a set of equations to estimate GA using fetal biometric measurements acquired during a single ultrasound scan performed between 14 and 34 weeks' gestation.
SUBJECTS AND METHODS
INTERGROWTH‐21st was a multicenter, multiethnic, population‐based project, conducted between 2009 and 2014 in eight countries11. Its primary aim was to study growth, health, nutrition and neurodevelopment from < 14 weeks' gestation to 2 years of age, using the same conceptual framework as that of the WHO Multicentre Growth Reference Study12, 13.
Eight urban areas located at low altitude (≤ 1600 m) were chosen as study sites, within which we selected all institutions that provided pregnancy and intrapartum care and at which > 80% of deliveries occurred. Women receiving antenatal care had to plan to deliver in these institutions or in a similar hospital located in the same geographical area and there had to be an absence or low levels of major, known, non‐microbiological contamination such as pollution, domestic smoke, radiation or any other toxic substances14.
Women from these populations with a singleton pregnancy that was conceived naturally and who met the individual inclusion criteria were recruited prospectively and consecutively into the FGLS. The study methods have been described in detail elsewhere11, 15.
The true GA (GAtrue) was defined by the woman's LMP determined at the first visit at < 14 + 0 weeks' gestation, provided that: (i) the date was certain; (ii) she had a regular 24–32‐day menstrual cycle; (iii) she had not been using hormonal contraception or breastfeeding in the preceding 2 months; and (iv) it was in agreement (within 7 days) with the measurement of fetal CRL at 9 + 0 to 13 + 6 weeks' gestation15.
The same type of ultrasound machine (HD‐9; Philips Ultrasound, Bothell, WA, USA) with curvilinear abdominal transducers (C5‐2, C6‐3, V7‐3) was used for all fetal measurements at ≥ 14 + 0 weeks' gestation. To reduce expected value bias, the ultrasound machines were specially adapted so that the measurements were not visible on the screen. However, as women presented for their first visit at different clinics within the geographical area, for those ultrasound scans performed at < 14 + 0 weeks' gestation (CRL measurements only), it was considered acceptable to use other, locally available machines, provided that they were evaluated and approved by the study team. All sonographers (n = 39) at the eight study sites underwent rigorous training and standardization. In accordance with the protocol, CRL and other fetal biometry measures were assessed for quality; the former were also reviewed blindly by our collaborators at the Société Française pour l'Amélioration des Pratiques Echographiques16, 17.
Women were invited for follow‐up ultrasound scans every 5 weeks (within 1 week either side) after the initial dating scan, so that the possible ranges after the dating scan were: 14–18, 19–23, 24–28, 29–33, 34–38 and 39–42 weeks' gestation. At each visit, fetal head circumference (HC), biparietal diameter (BPD), occipitofrontal diameter (OFD), abdominal circumference (AC) and femur length (FL) were measured three times from three separately obtained ultrasound images of each structure18.
Head measurements were taken in an axial view at the level of the thalami, with an angle of insonation as close as possible to 90°. The head had to be oval in shape, symmetrical, centrally positioned and filling at least 30% of the monitor. The midline echo (representing the falx cerebri) had to be broken anteriorly, at one‐third of its length, by the cavum septi pellucidi. The thalami had to be located symmetrically on either side of the midline. Calipers were then placed on the outer border (‘outer to outer’) of the parietal bones at the widest or longest part of the skull for the BPD and OFD, respectively; the HC was measured using the ellipse facility of the ultrasound machine on the outer border of the skull.
The measurements of the fetal abdomen were taken in a cross‐sectional view (as close as possible to a circle), with the umbilical vein in the anterior third of the abdomen (at the level of the portal sinus) and the stomach bubble visible. The operator was instructed to avoid applying too much pressure with the transducer as this can distort the circular shape of the fetal abdomen. The abdomen had to fill at least 30% of the monitor screen; the spine preferably had to be positioned at either 3 or 9 o'clock to avoid internal shadowing; and the kidneys and bladder did not have to be visible. For the measurements, the contour of the ellipse was placed on the outer border of the abdomen.
Finally, the FL was measured using a longitudinal view of the fetal thigh closest to the probe and with the femur as close as possible to the horizontal plane. The angle of insonation of the ultrasound beam was approximately 90° with the full length of the bone visualized, unobscured by shadowing from adjacent bony parts, and the femur had to fill at least 30% of the monitor screen. The intersection of the calipers was placed on the outer borders of the edges of the femoral diaphysis (outer to outer) ensuring clear femoral edges.
Detailed measurement protocols, standardization procedures and quality‐control methods employed across all sites are described in detail elsewhere15, 19, 20, 21.
Statistical analysis
For each woman included in the study, a single ultrasound scan between 14 + 0 and 40 + 0 weeks' gestation was selected randomly using the ‘sample’ function in Stata (version 13). At each scan, the routinely measured fetal biometric variables were recorded. To overcome the problem of data truncation at the lower end of gestation (<14 + 0 weeks), we followed the approach described previously and applied to CRL data22. Using the international fetal growth equations for HC, AC, FL, BPD and OFD18, we simulated 20 observations for each day between 12 + 0 and 13 + 6 weeks' gestation (n = 280), which is approximately the same number of observations for each day of GA in the untruncated dataset. After simulation, we restricted the data based on HC by excluding values < 85 mm or > 330 mm and visually inspecting a plot of the data to assess whether the truncation problem had been overcome. Using the augmented dataset, fractional polynomial regression analyses were employed, using the Xrigls function in Stata, to model the mean and SD of GA for each biometric variable22.
In order to establish the relationship of fetal biometric variables and GA we used an automated machine learning ‘Genetic Algorithm’ (Appendix S1). This method was chosen because a more traditional fractional polynomial approach, which is well‐suited to modeling a single variable, has limited scope when used with multiple biometric variables that are highly correlated. By virtue of the automated approach, the Genetic Algorithm is able to evaluate large numbers (in this case > 64 000) of potential combinations of biometric variables that are used to build polynomial equations as predictors of GA, which would not be feasible using conventional approaches. By specifying a mathematical definition of optimal performance, based upon minimization of the mean prediction error (root mean squared error (RMSE)), the first stage of model development was entirely automated with the capacity to assemble, evaluate and modify equations. We were therefore able to use the data themselves to generate preliminary models in an entirely objective manner.
Briefly, a large number of preliminary candidate equations were developed using combinations of all candidate biometric variables (including powers (0.5, 1, 2 and 3), their logs and their products). Each of the candidate equations was used to obtain for each fetus a predicted GA (GApredicted) as an estimate of their GAtrue. After preliminary analysis it was clear that the GApredicted values were not normally distributed; this was addressed by predicting the natural logarithm of GApredicted. The equations were then ranked to assess which had the lowest uncertainty based on the 95% prediction interval.
We used a four‐step approach to determine our final equation (see Appendix S1 for more detailed explanation).
Equation discovery using the Genetic Algorithm. The automated Genetic Algorithm was used to determine the equation providing the best prediction of GA using combinations of fetal biometric variables. Briefly, the model initiated itself by assigning polynomial equations linking fetal biometry within the dataset and GAtrue. Model terms, coefficients and powers were selected randomly within specified limits. Once defined, the individual equations were each used to predict GAtrue using the observed fetal biometry data. The performance of individual equations was measured by calculating the RMSE between the GAtrue and GApredicted at each iteration of the Genetic Algorithm. For each combination of biometric variables, the equation with the lowest RMSE was selected automatically and modified by methods that mimic the genetic principles of mutation and crossover. Thus, a second generation of equations was developed with the ‘positive predictive qualities’ of the first generation preserved in their structure. Furthermore, random variation was introduced as ‘mutations’ into the best‐performing equations in order to assess whether such mutations conferred an advantage in prediction, evaluated using the RMSE. By repeating the process over many iterations, the structure of equations was continuously refined until there was convergence upon the equation, or series of equations, that most accurately predicted GAtrue. All data processing at this stage was performed using the ‘GAPolyfitn’ function in MATLAB version R2014b.
Goodness of fit. Visual inspection of scatter plots was used to compare GAtrue with GApredicted for each candidate equation obtained from the Genetic Algorithm. Quantile–quantile (QQ) plots were used to compare the distributions. Well‐fitting models were identified by a QQ plot with minimal deviation from the line of equality. For each equation, absolute residuals between GAtrue − GApredicted were regressed on GA using fractional polynomial methods (powers ± 0.5, 1, 2 and 3) to provide an equation that approximated the SD, and multiplied by a constant to estimate the 2.5th and 97.5th centiles using the Xrigls function in Stata23. The goodness of fit of these estimated SDs was assessed by calculating the proportion of GApredicted that were outside the 95% prediction intervals (± 1.96 SD), which should be 5%.
Evaluation of model complexity. To facilitate a suitable balance between parsimony and model performance, estimates of Akaike's information criterion (AIC)24 were calculated and compared. The AIC combines an estimate of the goodness of fit of a model with a penalty for increasing model complexity. In addition, candidate equations with similar indices were compared in terms of number of terms and complexity, defined as the sum of the powers of each variable. Where two equations demonstrated similar performance, the equation with a less complex structure was preferred.
Postproduction model refinement. After examining the model complexity, it appeared that most of the contribution to the prediction of GA was based on HC. Therefore, simplified models were constructed, restricted to biometry of the fetal head (HC, OFD and BPD).
The INTERGROWTH‐21st Project was approved by the Oxfordshire Research Ethics Committee ‘C’ (ref: 08/H0606/139), the research ethics committees of the individual participating institutions, as well as the corresponding regional health authorities in which the project was implemented.
RESULTS
Of the 4607 women recruited into the FGLS, 4321 delivered live singletons without congenital malformations (Figure 1). Women with missing fetal measurements (n = 84), and outliers defined as fetal measurements > 5 SD above/below the mean (n = 7) were excluded. We further restricted the actual data at the top end by excluding HC values > 330 mm (n = 1), resulting in a total of 4229 women who contributed a single, randomly selected ultrasound scan between 14 + 0 and 40 + 0 weeks' gestation (Figure 1). Of the 280 observations simulated and added to actual data, we similarly excluded HC values < 85 mm (n = 148) and obtained a final analysis sample of 4361 observations. The baseline characteristics and perinatal events of the study population (n = 4229 excluding simulated observations) are shown in Table 1.
Figure 1.

Flowchart of recruitment of women with singleton pregnancy to the Fetal Growth Longitudinal Study (FGLS). *> 5 SD above or below mean fetal measurement. HC, head circumference.
Table 1.
Maternal and pregnancy characteristics of 4229 women enrolled in Fetal Growth Longitudinal Study of the INTERGROWTH‐21st Project who had a singleton live delivery
| Characteristic | Value |
|---|---|
| Maternal age (years) | 27.8 ± 3.8 |
| Maternal height (cm) | 162.2 ± 5.8 |
| Maternal weight (kg) | 61.5 ± 9.2 |
| Maternal BMI (kg/m2) | 23.2 ± 3.0 |
| GA at first visit (weeks) | 11.3 ± 1.4 |
| Nulliparous | 2815 (66.6) |
| Pre‐eclampsia | 31 (< 1) |
| Preterm delivery (< 37 weeks' gestation) | 189 (4.5) |
| Birth weight (kg)* | 3.3 ± 0.4 |
| Birth weight < 2500 g* | 127 (3.1) |
| Newborn sex male | 2101 (49.7) |
Data are given as mean ± SD or n (%). Maternal baseline characteristics were measured at < 14 weeks' gestation.
≥ 37 weeks' gestation only.
BMI, body mass index; GA, gestational age.
The equations that best estimated GAtrue based on lowest RMSE, best fit and optimal AIC are shown in Table 2. The equations selected were based on HC alone and a combination of HC and FL; despite including multiple measures (HC, BPD, OFD, AC and FL) in the models, only HC and FL were retained after the selection process.
Table 2.
Equations for estimating gestational age (GA) and its SD in late pregnancy, derived from biometric data of 4229 singleton pregnancies
| Equation | Variables in equation (mm) | Equation to estimate logeGApredicted (days) | Equation to estimate SD of GApredicted (days) |
|---|---|---|---|
| 1 | HC | 0.05970 × (loge(HC))2 + 0.000000006409 × (HC)3 + 3.3258 | 0.6492 × (GA × 0.01)3 + 2.991 |
| 2 | HC, FL | 0.03243 × (loge(HC))2 + 0.001644 × FL × loge(HC) + 3.813 | 0.04009 × GA – 1.149 |
FL, femur length (in mm); HC, head circumference (in mm); loge, natural logarithm.
As an example, to calculate GA using Equation 1, if HC is 250 mm, median GA =exp [0.05970 × (loge(250))2 + 0.000000006409 × (250)3 +3.3258] = exp [5.245986506] = 189.8 days (equivalent to 27.1 weeks). To calculate GA using Equation 2, if HC is 250 mm and FL is 55 mm, median GA = exp [0.03243 × (loge(250))2 + 0.001644 × 55 × loge(250) + 3.813] = exp [5.300929] = 200.5 days (equivalent to 28.6 weeks).
Using equations of the median and SD one can easily compute any desired centiles using the relation Pth centile = median + KSD, where K is the normal equivalent deviate (z‐score) corresponding to a particular centile, e.g. K = 1.88 for the 97th centile and −1.88 for the 3rd centile. The SDs in this equation are the predicted estimates from the regression analysis. For example, the 3rd centile for GA = exp (0.05970 × (loge(HC))2 + 0.000000006409 ×(HC)3 + 3.3258) + (−1.88 × (0.6492 × (median GA × 0.01)3 + 2.991)).
Overall, based on a model using HC only, the uncertainty of estimated GA gradually increased with advancing GA, from 6–7 days in either direction at 14 + 0 weeks' gestation to 15–20 days at 32 + 0 weeks' gestation (Table 3). Inclusion of FL led to an improvement in the accuracy of prediction throughout gestation of about 1–6 days. Inclusion of the other parameters led to no further improvement and therefore these were not included in the equations resulting from the Genetic Algorithm search.
Table 3.
Characteristics of goodness of fit of an equation based on fetal head circumference (HC) (Equation 1) and an equation based on fetal HC and femur length (FL) (Equation 2) for predicting gestational age (GA) in late pregnancy
| Equation | ||
|---|---|---|
| Characteristic | 1 | 2 |
| Variables in equation | HC | HC, FL |
| RMSE (log days) | 0.0423 | 0.0352 |
| R 2 | 0.98 | 0.99 |
| Goodness of fit (%)* | 5.21 | 6.20 |
| AIC | 16.33 | 14.69 |
| Variation (days) around mean GA estimate† at: | ||
| 14 weeks | 7.1 | 5.4 |
| 16 weeks | 7.7 | 6.5 |
| 18 weeks | 8.4 | 7.6 |
| 20 weeks | 9.4 | 8.7 |
| 22 weeks | 10.5 | 9.8 |
| 24 weeks | 11.9 | 10.9 |
| 26 weeks | 13.5 | 12.0 |
| 28 weeks | 15.4 | 13.2 |
| 30 weeks | 17.6 | 14.3 |
| 32 weeks | 20.1 | 15.4 |
| 34 weeks | 23.0 | 16.5 |
Percentage of predicted estimates of GA outside 95% prediction interval across all GA.
Half width of 95% prediction interval.
AIC, Akaike's information criterion; RMSE, root mean squared error.
The plots of GApredicted vs GAtrue between 14 + 0 and 34 + 0 weeks' gestation demonstrated good model fitting (Figure 2 for the scatterplots and Figures S1 and S2 for the QQ plots).
Figure 2.
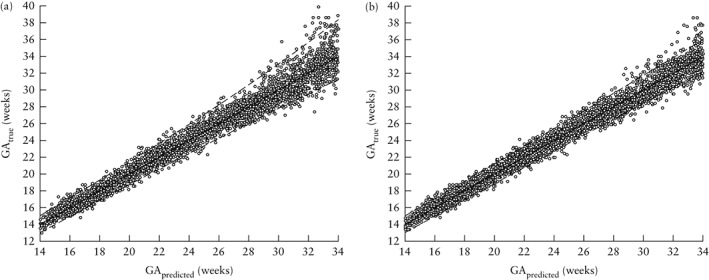
Scatterplots showing predicted gestational age (GA) based on fetal head circumference (a) and on fetal head circumference and femur length (b) at 14 to 34 weeks of true GA. Solid line is line of equality and dashed lines are ± 2 SD.
Apart from estimating the most likely GA (by using GApredicted for a set of measurements), we also present the lower and upper bounds of the estimation of GA (Table S1). The lower bound can be used in clinical management for women who present in late pregnancy; it is an estimate of the likely ‘least GA’, e.g. they are 97.5% likely to be at least X weeks.
DISCUSSION
Main findings
We have shown that a single set of basic ultrasound measurements of HC and FL in the second trimester can be used to estimate GA with reasonable accuracy. The estimation is best at lower GAs where the 95% prediction interval is within 6 days, whereas it is just over 12 days at 26 + 0 weeks' gestation. The addition of FL to HC leads to considerable improvement over just using HC, but no further improvement in prediction is afforded by adding AC, BPD or OFD.
Strengths and weaknesses
We have produced equations for GA assessment that are more precise than those used currently in routine clinical practice (Table 4). This may be due to the prospective nature of the study, a large sample size, accurately dated pregnancies, a clearly defined measurement protocol, quality control measures and a statistical approach that searched for the optimal combination of factors iteratively, rather than relying upon a user‐controlled search. The multicenter, international setting of the study with measurements taken by a large group of sonographers provides external validity.
Table 4.
Commonly used pregnancy‐dating equations and their imprecision in estimating gestational age (GA) (half width of 95% prediction intervals)
| Imprecision in GA estimation (days) for assessment at: | |||||
|---|---|---|---|---|---|
| Reference | R 2 | 12–18 weeks | 18–24 weeks | 24–30 weeks | 30–36 weeks |
| Hadlock et al. (1984)29 | |||||
| BPD | 0.967 | 8.3 | 12.1 | 15.3 | 21.6 |
| HC | 0.973 | 8.3 | 10.4 | 14.4 | 20.9 |
| AC | 0.969 | 11.6 | 14.4 | 15.3 | 20.7 |
| FL | 0.971 | 9.7 | 12.6 | 14.6 | 20.7 |
| HC + BPD | 0.974 | 7.6 | 10.4 | 13.9 | 20.0 |
| HC + FL | 0.976 | 8.4 | 10.6 | 13.9 | 18.8 |
| HC + AC | 0.98 | 7.6 | 9.4 | 13.0 | 17.6 |
| HC + FL + BPD | 0.981 | 7.3 | 9.5 | 12.7 | 17.6 |
| HC + AC + FL | 0.981 | 8.0 | 10.2 | 13.2 | 17.6 |
| HC + BPD + AC + FL | 0.981 | 7.6 | 9.8 | 12.6 | 17.1 |
| Altman and Chitty (1997)30 | |||||
| HC | NR | 8.0 | 13.0 | 17.0 | 22.0 |
AC, abdominal circumference; BPD, biparietal diameter; FL, femur length; HC, head circumference; NR, not reported.
There is an intrinsic limitation when estimating GA by fetal anthropometric‐based equations, i.e. that the measurement is of fetal size not GA. Fetal size may vary for reasons other than differences in GA, especially as factors conditioning abnormal fetal growth are more prevalent in the populations among which the equation is most likely to be used. In other words, it is important to take into account the impact of pathology (fetal growth restriction and overgrowth) on GA estimation. This is true for any equation estimating GA; the accuracy at an individual level will depend on the ‘normality’ of the fetal size and, at the population level, on the prevalence of abnormal growth patterns. Thus, efforts should focus on modifying health systems and referral pathways to prevent late presentation in pregnancy, rather than simply achieving technological advances in fetal size‐based dating.
Interpretation
Ultrasound assessment of GA is performed assuming that fetal size can be used as a proxy for GA. This assumption depends on: (i) the GA at which biometry is performed (at earlier GAs growth is more uniform and there is less measurable growth impairment); (ii) the choice of biometric variable; (iii) accuracy of the measurement, which is affected by technical aspects of imaging and operator skill; and (iv) absence of pathology that could affect growth.
The most accurate way to estimate GA is by measuring fetal CRL between 8 + 0 and 14 + 0 weeks' gestation, which is associated with a 95% prediction interval of 2.7 days10, 25. This method is the basis of recommended pregnancy‐dating policies throughout much of the developed world26. Beyond 14 + 0 weeks' gestation, fetal flexion limits the accuracy of CRL measurements for dating purposes and GA estimates are based on measurement of the HC, BPD, AC and FL or a combination of these8, 27.
Our results demonstrate the relative inaccuracy of late GA assessment, which is due to the increasing biological variability in fetal size as well as the increasing absolute error of fetal measurements with advancing GA9. Therefore, all information (clinical and imaging) should be considered when dating pregnancies and providing obstetric care, particularly after late pregnancy dating. Thus, we recommend that the following principles should be applied in clinical practice.
Assessment of fetal age should be based on the earliest available ultrasound measurement after 8 + 0 weeks' gestation, provided that the measurements are technically adequate. CRL should be used before 14 + 0 weeks' gestation and Equation 2 (HC and FL) after 14 + 0 but before 26 + 0 weeks' gestation. Equation 1 can be used in settings in which only HC is available.
If menstrual dates are reliable and within the prediction limits of the fetal measurement, ultrasound examination should merely confirm the GA assessed by LMP.
When menstrual dates are reliable and fall outside the prediction interval of ultrasound assessment there are two interpretations: the menstrual dates are in fact incorrect and GA should be based on ultrasound measurement, or the GA is correct as assessed by LMP and the fetus is an abnormal size for that GA (or both). Clinical features of growth restriction, e.g. reduced amniotic fluid or abnormal uterine or umbilical artery blood flow, should be taken into consideration, as should factors that may lead to overgrowth, e.g. maternal diabetes. An interval ultrasound scan should then be carried out to confirm GA.
When menstrual dates are unknown, GA estimation should be based on ultrasound examination, which has reasonable accuracy until 26 + 0 weeks' gestation, and a further ultrasound scan should be carried out.
Finally, when GA is estimated in the third trimester, the possible error is large and must be taken into account to ensure safe obstetric practice. The use of the concept of a ‘minimum’ GA, by using the lower limit of the prediction interval from the equation can be useful in this instance (Table S1). For example, if a woman presents with threatened preterm labor and the ultrasound examination suggests a median GA of 34 + 6 weeks based on HC and FL, it should be appreciated that the GA could be as low as 32 + 6 weeks, i.e. it is most likely that the GA is 34 + 6 weeks, but we are 95% certain that the GA is at least 32 + 6 weeks. However, if the fetus is growth restricted, the GA could be as much as 36 + 5 weeks. This analysis is very relevant to clinical decision making, e.g. when administering prophylactic corticosteroids or transferring a neonate to a higher level of care. In contrast, labor induction may be considered at 40 + 0 weeks' gestation based on late assessment, as the GA could be more advanced. Such a clinically cautious approach is particularly important as it is known that unreliable reporting of LMP and late antenatal care are both associated with adverse pregnancy outcome5, 28.
Conclusion
We have shown that a single set of ultrasound measurements in the second trimester can be used to estimate GA with relative accuracy. We recommend these tools for the management of women who present late in pregnancy. However, we strongly encourage, as a priority, the promotion of early antenatal care in regions and subpopulations that are not yet benefiting from this practice.
Supporting information
Appendix S1 Details of statistical analytical strategy
Appendix S2 Members of the International Fetal and Newborn Growth Consortium for the 21st Century (INTERGROWTH‐21st) and its Committees
Figures S1‐S2: Q‐Q plots to assess the goodness of fit of the models for head circumference (S1); and head circumference and femur length (S2).
Table S1 Estimation of gestational age in late pregnancy using an equation based on fetal head circumference (HC) (Equation 1) and an equation based on fetal HC and femur length (FL) (Equation 2)
ACKNOWLEDGMENTS
This project was supported by a generous grant from the Bill & Melinda Gates Foundation to the University of Oxford, for which we are very grateful. We would also like to thank the Health Authorities in Pelotas, Brazil; Beijing, China; Nagpur, India; Turin, Italy; Nairobi, Kenya; Muscat, Oman; Oxford, UK and Seattle, USA, which facilitated the project by allowing participation of these study sites as collaborating centers. We are extremely grateful to Philips Medical Systems, which provided the ultrasound equipment and technical assistance throughout the project. We also thank MedSciNet U.K. Ltd for setting up the INTERGROWTH‐21st website and for the development, maintenance and support of the online data management system. We thank the parents and infants who participated in the studies and the more than 200 members of the research teams who made the implementation of this project possible. The participating hospitals included: Pelotas, Brazil (Hospital Miguel Piltcher, Hospital São Francisco de Paula, Santa Casa de Misericórdia de Pelotas and Hospital Escola da Universidade Federal de Pelotas), Beijing, China (Beijing Obstetrics & Gynecology Hospital, Shunyi Maternal & Child Health Centre and Shunyi General Hospital), Nagpur, India (Ketkar Hospital, Avanti Institute of Cardiology Private Limited, Avantika Hospital, Gurukrupa Maternity Hospital, Mulik Hospital & Research Centre, Nandlok Hospital, Om Women's Hospital, Renuka Hospital & Maternity Home, Saboo Hospital, Brajmonhan Taori Memorial Hospital and Somani Nursing Home), Nairobi, Kenya (Aga Khan University Hospital, MP Shah Hospital and Avenue Hospital), Turin, Italy (Ospedale Infantile Regina Margherita Sant' Anna and Azienda Ospedaliera Ordine Mauriziano), Muscat, Oman (Khoula Hospital, Royal Hospital, Wattayah Obstetrics & Gynaecology Poly Clinic, Wattayah Health Centre, Ruwi Health Centre, Al‐Ghoubra Health Centre and Al‐Khuwair Health Centre), Oxford, UK (John Radcliffe Hospital) and Seattle, USA (University of Washington Hospital, Swedish Hospital and Providence Everett Hospital). Members of INTERGROWTH‐21st and its committees are listed in Appendix S2. Full acknowledgement of all those who contributed to the development of the INTERGROWTH‐21st Project protocol appears at www.intergrowth21.org.uk
REFERENCES
- 1. Ioannou C, Talbot K, Ohuma E, Sarris I, Villar J, Conde‐Agudelo A, Papageorghiou AT. Systematic review of methodology used in ultrasound studies aimed at creating charts of fetal size. BJOG 2012; 119 : 1425–1439. [DOI] [PubMed] [Google Scholar]
- 2. Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller A, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 2012; 379 : 2162–2172. [DOI] [PubMed] [Google Scholar]
- 3. de Onis, Blössner M , Villar J. Levels and patterns of intrauterine growth retardation in developing countries. Eur J Clin Nutr 1998; 52 Suppl 1 : S5–15. [PubMed] [Google Scholar]
- 4. Savitz DA, Terry JW, Dole N, Thorp JM, Siega‐Riz AM, Herring AH. Comparison of pregnancy dating by last menstrual period, ultrasound scanning, and their combination. Am J Obstet Gynecol 2002; 187 : 1660–1666. [DOI] [PubMed] [Google Scholar]
- 5. Thorsell M, Kaijser M, Almström H, Andolf E. Expected day of delivery from ultrasound dating versus last menstrual period – obstetric outcome when date mismatch. BJOG 2008; 115 : 585–589. [DOI] [PubMed] [Google Scholar]
- 6. Nguyen TH, Larsen T, Engholm G, Moller H. Evaluation of ultrasound estimated date of delivery in 17,450 spontaneous singleton births: do we need to modify Naegele's rule? Ultrasound Obstet Gynecol 1999; 14 : 23–28. [DOI] [PubMed] [Google Scholar]
- 7. Hughes R, Aitken E, Anderson J, Barry C, Benton M, Elliot J. Antenatal Care. Routine Care for the Healthy Pregnant Woman. National Institute for Health and Clinical Excellence. NICE clinical guideline 62. RCOG Press: London, 2008. [Google Scholar]
- 8. Butt K, Lim K; Society of Obstetricians and Gynaecologists of Canada . Determination of gestational age by ultrasound. J Obstet Gynaecol Can 2014; 36 : 171–183. [DOI] [PubMed] [Google Scholar]
- 9. Sarris I, Ioannou C, Chamberlain P, Ohuma E, Roseman F, Hoch L, Altman DG, Papageorghiou AT; International Fetal and Newborn Growth Consortium for the 21st Century (INTERGROWTH‐21st). Intra‐ and interobserver variability in fetal ultrasound measurements. Ultrasound Obstet Gynecol 2012; 39 : 266–273. [DOI] [PubMed] [Google Scholar]
- 10. Papageorghiou AT, Kennedy SH, Salomon LJ, Ohuma EO, Cheikh Ismail L, Barros FC, Lambert A, Carvalho M, Jaffer YA, Bertino E, Gravett MG, Altman DG, Purwar M, Noble JA, Pang R, Victora CG, Bhutta ZA, Villar J; International Fetal and Newborn Growth Consortium for the 21st Century (INTERGROWTH‐21st). International standards for early fetal size and pregnancy dating based on ultrasound measurement of crown–rump length in the first trimester of pregnancy. Ultrasound Obstet Gynecol 2014; 44 : 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villar J, Altman DG, Purwar M, Noble JA, Knight HE, Ruyan P, Cheikh Ismail L, Barros FC, Lambert A, Papageorghiou AT, Carvalho M, Jaffer YA, Bertino E, Gravett MG, Bhutta ZA, Kennedy SH; International Fetal and Newborn Growth Consortium for the 21st Century . The objectives, design and implementation of the INTERGROWTH‐21st Project. BJOG 2013; 120 (Suppl 2): 9–26, v. [DOI] [PubMed] [Google Scholar]
- 12. Garza C, de Onis M. Rationale for developing a new international growth reference. Food Nutr Bull 2004; 25 ( Suppl ): S5–14. [DOI] [PubMed] [Google Scholar]
- 13. de Onis M, Garza C, Onyango AW, Martorell R. WHO child growth standards. Acta Paediatr Suppl 2006; 450 : 1–101. [Google Scholar]
- 14. Eskenazi B, Bradman A, Finkton D, Purwar M, Noble JA, Pang R, Burnham O, Cheikh Ismail L, Farhi F, Barros FC, Lambert A, Papageorghiou AT, Carvalho M, Jaffer YA, Bertino E, Gravett MG, Altman DG, Ohuma EO, Kennedy SH, Bhutta ZA, Villar J; International Fetal and Newborn Growth Consortium for the 21st Century . A rapid questionnaire assessment of environmental exposures to pregnant women in the INTERGROWTH‐21st Project. BJOG 2013; 120 ( Suppl 2 ): 129–138. [DOI] [PubMed] [Google Scholar]
- 15. Papageorghiou AT, Sarris I, Ioannou C, Todros T, Carvalho M, Pilu G, Salomon LJ; International Fetal and Newborn Growth Consortium for the 21st Century . Ultrasound methodology used to construct the fetal growth standards in the INTERGROWTH‐21st Project. BJOG 2013; 120 ( Suppl 2 ): 27–32. [DOI] [PubMed] [Google Scholar]
- 16. Ioannou C, Sarris I, Hoch L, Salomon LJ, Papageorghiou AT; International Fetal and Newborn Growth Consortium for the 21st Century . Standardisation of crown‐rump length measurement. BJOG 2013; 120 ( Suppl 2 ): 38–41. [DOI] [PubMed] [Google Scholar]
- 17. Wanyonyi SZ, Napolitano R, Ohuma EO, Salomon LJ, Papageorghiou AT. Image‐scoring system for crown‐rump length measurements. Ultrasound Obstet Gynecol 2014; 44 : 649–654. [DOI] [PubMed] [Google Scholar]
- 18. Papageorghiou AT, Ohuma EO, Altman DG, Todros T, Cheikh Ismail L, Lambert A, Jaffer YA, Bertino E, Gravett MG, Purwar M, Noble JA, Pang R, Victora CG, Barros FC, Carvalho M, Salomon LJ, Bhutta ZA, Kennedy SH, Villar J; International Fetal and Newborn Growth Consortium for the 21 Century (INTERGROWTH‐21st) . International standards for fetal growth based on serial ultrasound measurements: the Fetal Growth Longitudinal Study of the INTERGROWTH‐21st Project. Lancet 2014; 384 : 869–879. [DOI] [PubMed] [Google Scholar]
- 19. Sarris I, Ioannou C, Ohuma EO, Altman DG, Hoch L, Cosgrove C, Fathima S, Salomon LJ, Papageorghiou AT; International Fetal and Newborn Growth Consortium for the 21st Century . Standardisation and quality control of ultrasound measurements taken in the INTERGROWTH‐21st Project. BJOG 2013; 120 ( Suppl 2 ): 33–37. [DOI] [PubMed] [Google Scholar]
- 20. Sarris I, Ioannou C, Dighe M, Mitidieri A, Oberto M, Qingqing W, Shah J, Sohoni S, Al Zidjali W, Hoch L, Altman DG, Papageorghiou AT; International Fetal and Newborn Growth Consortium for the 21st Century . Standardization of fetal ultrasound biometry measurements: improving the quality and consistency of measurements. Ultrasound Obstet Gynecol 2011; 38 : 681–687. [DOI] [PubMed] [Google Scholar]
- 21.INTERGROWTH‐21st: The International Fetal and Newborn Growth Consortium. http://www.intergrowth21.org.uk [Accessed 10 September 2016].
- 22. Ohuma EO, Papageorghiou AT, Villar J, Altman DG. Estimation of gestational age in early pregnancy from crown‐rump length when gestational age range is truncated: the case study of the INTERGROWTH‐21st Project. BMC Med Res Methodol 2013; 13 : 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Altman DG. Construction of age‐related reference centiles using absolute residuals. Stat Med 1993; 12: 917–924. [DOI] [PubMed] [Google Scholar]
- 24. Akaike H. A new look at the statistical model identification. IEEE Trans Autom Cont 1974; 19 : 716–723. [Google Scholar]
- 25. Robinson HP. Sonar measurement of fetal crown‐rump length as means of assessing maturity in first trimester of pregnancy. BMJ 1973; 4 : 28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Napolitano R, Dhami J, Ohuma EO, Ioannou C, Conde‐Agudelo A, Kennedy SH, Villar J, Papageorghiou AT. Pregnancy dating by fetal crown‐rump length: a systematic review of charts. BJOG 2014; 121 : 556–565. [DOI] [PubMed] [Google Scholar]
- 27. Salomon LJ, Alfirevic Z, Bilardo CM, Chalouhi GE, Ghi T, Kagan KO, Lau TK, Papageorghiou AT, Raine‐Fenning NJ, Stirnemann J, Suresh S, Tabor A, Timor‐Tritsch IE, Toi A, Yeo G. ISUOG practice guidelines: performance of first‐trimester fetal ultrasound scan. Ultrasound Obstet Gynecol 2013; 41 : 102–113. [DOI] [PubMed] [Google Scholar]
- 28. Nguyen TH, Larsen T, Engholm G, Møller H. Increased adverse pregnancy outcomes with unreliable last menstruation. Obstet Gynecol 2000; 95 : 867–873. [DOI] [PubMed] [Google Scholar]
- 29. Hadlock FP, Deter RL, Harrist RB, Park SK. Estimating fetal age: computer‐assisted analysis of multiple fetal growth parameters. Radiology 1984; 152 : 497–501. [DOI] [PubMed] [Google Scholar]
- 30. Altman DG, Chitty LS. New charts for ultrasound dating of pregnancy. Ultrasound Obstet Gynecol 1997; 10 : 174–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Details of statistical analytical strategy
Appendix S2 Members of the International Fetal and Newborn Growth Consortium for the 21st Century (INTERGROWTH‐21st) and its Committees
Figures S1‐S2: Q‐Q plots to assess the goodness of fit of the models for head circumference (S1); and head circumference and femur length (S2).
Table S1 Estimation of gestational age in late pregnancy using an equation based on fetal head circumference (HC) (Equation 1) and an equation based on fetal HC and femur length (FL) (Equation 2)


