Abstract
Hepatocellular carcinoma (HCC) is the third most lethal cancer worldwide. The lack of effective biomarkers for the early detection of HCC results in unsatisfactory curative treatments. Here, metabolite biomarkers were identified and validated for HCC diagnosis. A total of 1,448 subjects, including healthy controls and patients with chronic hepatitis B virus infection, liver cirrhosis, and HCC, were recruited from multiple centers in China. Liquid chromatography–mass spectrometry–based metabolomics methods were used to characterize the subjects' serum metabolic profiles and to screen and validate the HCC biomarkers. A serum metabolite biomarker panel including phenylalanyl‐tryptophan and glycocholate was defined. This panel had a higher diagnostic performance than did α‐fetoprotein (AFP) in differentiating HCC from a high‐risk population of cirrhosis, such as an area under the receiver‐operating characteristic curve of 0.930, 0.892, and 0.807 for the panel versus 0.657, 0.725, and 0.650 for AFP in the discovery set, test set, and cohort 1 of the validation set, respectively. In the nested case–control study, this panel had high sensitivity (range 80.0%‐70.3%) to detect preclinical HCC, and its combination with AFP provided better risk prediction of preclinical HCC before clinical diagnosis. Besides, this panel showed a larger area under the receiver‐operating characteristic curve than did AFP (0.866 versus 0.682) to distinguish small HCC, and 80.6% of the AFP false‐negative patients with HCC were correctly diagnosed using this panel in the test set, which was corroborated by the validation set. The specificity and biological relevance of the identified biomarkers were further evaluated using sera from another two cancers and HCC tissue specimens, respectively. Conclusion: The discovered and validated serum metabolite biomarker panel exhibits good diagnostic performance for the early detection of HCC from at‐risk populations. (Hepatology 2018;67:662‐675).
Abbreviations
- AFP
α‐fetoprotein
- AUC
area under the receiver‐operating characteristic curve
- CHB
chronic hepatitis B
- Cir
cirrhosis group
- ESI
electrospray ionization
- GCA
glycocholate
- GSC
gastric cancer
- HCC
hepatocellular carcinoma
- HCT
HCC tissue
- ICC
intrahepatic cholangiocarcinoma
- LC‐MS
liquid chromatography‐mass spectrometry
- NC
normal controls
- Phe‐Trp
phenylalanyl‐tryptophan
- QC
quality control
- S‐HCC
small HCC
- VIP
variable important in the projection
Hepatocellular carcinoma (HCC) is one of the most fatal malignancies, causes approximately 7 million deaths worldwide, and exhibits a significant decrease in the 1‐year to 5‐year survival rate from 47% to 10%.1, 2 About 50% of all new cases and deaths related to liver cancer worldwide occur in China.2 Although risk factors (e.g., liver cirrhosis) are well recognized, the early diagnosis of HCC in high‐risk populations remains challenging due to the early recessive clinical symptoms and the difficulties in differential diagnosis with cirrhosis.3 Imaging methods (such as computed tomography and ultrasonography) and serum biomarkers (e.g., α‐fetoprotein [AFP]) are commonly used to screen and diagnose HCC in the clinic. However, it is still challenging to distinguish small HCC (S‐HCC) from cirrhosis nodules solely by imaging. On the other hand, the sensitivity of AFP is limited to 65% for clinical HCC diagnosis and <40% for preclinical prediction.4 Therefore, it is still urgent to discover novel biomarkers for the screening of HCC.
Circulation biomarkers can facilitate the screening of cancer, understanding of tumor biology, and early discovery of recurrence with minimum invasion.5 The study of circulating biomarkers is of great concern due to the identification of more and more serum molecules, such as proteins, metabolites, and microRNAs recently.6 Liquid chromatography–mass spectrometry (LC‐MS)–based metabolomics is a powerful tool that has been used to discover novel circulating biomarkers for many diseases.7, 8, 9 An increasing number of studies have screened candidate biomarkers from body fluids (plasma, serum, and urine) or liver tissue that may be used to diagnose HCC.9, 10, 11 For example, a urinary biomarker panel consisting of four metabolites has been reported to classify HCC and cirrhosis with an area under the receiver‐operating characteristic curve (AUC) of 0.9.12 However, most metabolic markers were discovered based on small pilot studies discriminating HCC from liver disease subjects.13, 14 We have also established several biomarker panels for HCC in some pilot and cross‐sectional studies.15, 16 These studies may reflect different metabolic aspects of HCC, but the limited study cohort or lack of effective validation restricts further clinic applications of these biomarkers.17 Due to the heterogeneity and variety of genetic backgrounds of patients, the lack of data from large‐scale samples for the validation of HCC biomarkers is a main deterrent to the development of these biomarkers.12
A large number of samples from multiple centers is required to validate the reliability of these biomarkers. A commonly encountered problem during LC‐MS‐based metabolic profiling analysis in a large‐scale study is the limited repeatability of the measurement. To address the challenge, we developed a new technology known as a pseudotargeted metabolomics method,18 which acquires profiling data in the targeted multiple reaction monitoring mode but contains the metabolites defined from a nontargeted full‐scan mode; it can provide high‐quality and information‐rich data for a large‐scale metabolomics study.19
The goal of the current LC‐MS‐based metabolomics study is to define more reliable serum biomarkers for diagnosing HCC using metabolome data from a large number of samples from multiple centers. Therefore, a total of 1,448 participants from six clinic centers in China were enrolled, and a three‐step analysis strategy, including discovery, test, and validation, was employed to identify and validate the clinical practicability of these novel biomarkers. Additionally, patients with S‐HCC were specifically recruited to assess the performance of the biomarkers in diagnosing early‐stage HCC. Furthermore, a nested case–control study, prospectively collecting sera from patients with preclinical HCC and at‐risk controls before clinical diagnosis, was used to evaluate the predictive capacity of these identified biomarkers for preclinical HCC. Two other cancers, gastric cancer (GSC) and intrahepatic cholangiocarcinoma (ICC), were used to assess the specificity of these potential biomarkers for HCC. The biological relevance of these biomarkers was further evaluated using HCC tissue specimens.
Patients and Methods
STUDY DESIGN AND PARTICIPANTS
In the present study, a total of 1,448 participants, including normal controls (healthy volunteers, NC) and patients with chronic hepatitis B (CHB) infections, liver cirrhosis (Cir), HCC, GSC, or ICC, from six clinic centers were enrolled between September 2008 and May 2014. The exclusion criteria were abnormal liver biochemistry, a history of liver disease or other systematic diseases for the healthy controls, and a history of acute diseases or other types of malignant diseases for patients with liver disease. In the discovery stage, all 108 fasting serum samples were collected at the First Hospital of Jilin University (Changchun, China). The test cohort of 684 participants was from First Hospital of Jilin University, the Eastern Hepatobiliary Surgery Institute of the Second Military Medical University (Shanghai, China). The validation cohort 1 of 572 participants was from First Hospital of Jilin University, the Eastern Hepatobiliary Surgery Institute of the Second Military Medical University, Zhongshan Hospital of Xiamen University (Xiamen, China), Shandong Provincial Hospital Affiliated to Shandong University (Ji'nan, China), and Peking University People's Hospital (Beijing, China). The validation cohort 2 of 84 participants came from the Dongfeng–Tongji cohort including 27,009 participants20 in the nested case–control study. Sera of these 42 new cases of HCC were collected between September 2008 and March 2009 at an average of 1.7 (range 0.2‐3.0) years before the clinical diagnosis, and sera of these 42 well‐matched at‐risk controls, who had hepatitis B virus or liver cirrhosis but did not have HCC during the follow‐up period, were collected at a similar time (Fig. 1). Written informed consent was obtained from each participant. The study was approved by the ethics committee of each center and conformed to the ethical guidelines of the 1975 Declaration of Helsinki. The HCC diagnosis was confirmed with ultrasound, computed tomography, or magnetic resonance imaging; and most cases were further diagnosed by histopathology according to the guidelines of the American Association for the Study of Liver Diseases.21 Cirrhosis was diagnosed based on clinical evidence of portal hypertension or hepatic decompensation according to the same guidelines. CHB was defined as the presence of hepatitis B surface antigen for >6 months, concentrations of hepatitis B virus DNA >105 copies/mL, and elevated aspartate aminotransferase or alanine aminotransferase levels.22 The distributions of gender and age among groups were matched as much as possible, and detailed information is listed in Table 1.
Figure 1.
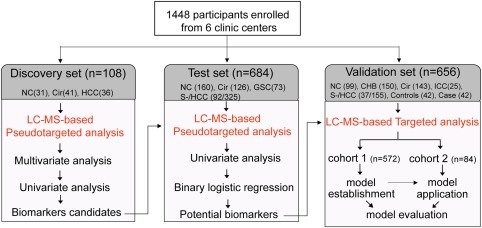
Design of the study.
Table 1.
Clinic Characteristics of the Subjects in Each Set
| Discovery Set (n = 108) | Test Set (n = 684) | Validation Set (n = 656) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort 1 (n = 572) | Cohort 2 (n = 84) | |||||||||||||
| Characteristics | NC | Cir | HCC | NC | Cir | HCC | GSC | NC | CHB | Cir | HCC | ICC | Controls | Cases |
| n | 31 | 41 | 36 | 160 | 126 | 325 | 73 | 99 | 150 | 143 | 155 | 25 | 42 | 42 |
| Age (years) | 50.2 ± 10.8 | 53.0 ± 9.9 | 58.4 ± 10.6 | 47.5 ± 6.5 | 51.9 ± 9.0 | 54.3 ± 10.5 | 62.2 ± 12.9 | 52.0 ± 4.5 | 46.2 ± 11.9 | 53.6 ± 10.4 | 53.6 ± 10.4 | 58.5 ± 11.4 | 66.5 ± 7.9 | 66.2 ± 8.1 |
| Sex (male/female) | 24/7 | 31/10 | 29/7 | 89/71 | 94/32 | 272/53 | 52/21 | 79/20 | 92/58 | 103/40 | 130/25 | 17/7 | 32/10 | 32/10 |
| AFP (ng/mL) | – | 107.1 ± 282.8 | 487.5 ± 575.7 | – | 56.6 ± 202 | 372.8 ± 507.6 | – | – | 5.1 ± 6.4 | 79.2 ± 226 | 292.6 ± 452.2 | – | 3.5 ± 4.5 | 82.2 ± 162.1 |
| AST (U/L) | 21.4 ± 6.4 | 80.5 ± 98.6 | 110 ± 186.7 | 23.4 ± 5.6 | 72.5 ± 85.3 | 75.4 ± 68.8 | 25.3 ± 17.0 | 20.9 ± 4.6 | 37.8 ± 25.5 | 166.3 ± 997.8 | 67.6 ± 79.3 | – | 24.3 ± 6.8 | 46.4 ± 42.8 |
| ALT (U/L) | 20.1 ± 8.5 | 73.4 ± 117.4 | 76.9 ± 73.7 | 20.8 ± 9.4 | 71.1 ± 109.6 | 62.2 ± 54.3 | 18.8 ± 2.9 | 19.8 ± 7.3 | 47.8 ± 66.4 | 134.9 ± 696.2 | 76.5 ± 104.5 | – | 22.0 ± 10.5 | 40.5 ± 38.3 |
| TBIL (μmol/L) | 13.4 ± 5.4 | 46.3 ± 56.3 | 68.3 ± 67.5 | 13.1 ± 5.6 | 51.7 ± 70.8 | 68.4 ± 174.2 | – | 15.2 ± 3.5 | 21.1 ± 19.4 | 55.2 ± 102 | 65.5 ± 79.4 | 28.0 ± 23.1 | 13.1 ± 5 | 19.5 ± 9.1 |
| DBIL (μmol/L) | 3.6 ± 1.1 | 20.7 ± 34.7 | 33.6 ± 45.6 | 3.8 ± 1.5 | 24.6 ± 41.5 | 64.2 ± 171.8 | – | 3.1 ± 0.9 | 7.1 ± 12.9 | 25.6 ± 51.6 | 37.6 ± 54.3 | 17.9 ± 20.0 | 3.9 ± 1.5 | 7.1 ± 5.1 |
| IBIL (μmol/L) | 9.8 ± 4.4 | 25.6 ± 24.4 | 42.5 ± 28.9 | 9.3 ± 4.5 | 27.1 ± 30.2 | 50.5 ± 62.8 | – | 12.0 ± 2.6 | 14.0 ± 9.0 | 29.7 ± 52.6 | 38.8 ± 32.7 | 10.1 ± 4.5 | 9.4 ± 3.9 | 12.5 ± 5.1 |
| TP (g/L) | 74.9 ± 5 | 63 ± 6.4 | – | 75.3 ± 5.6 | 62.9 ± 8.5 | – | – | 75.0 ± 5.1 | 71.0 ± 5.5 | 63.2 ± 8.4 | – | 65.0 ± 8.6 | – | – |
| ALB (g/L) | 46.6 ± 2.8 | 31.4 ± 5.4 | – | 47.8 ± 4.1 | 31.0 ± 7 | – | – | 48.4 ± 3.7 | 45.1 ± 5.6 | 31.5 ± 7.1 | – | 36.6 ± 5.2 | – | – |
| Tumor diameter (cm) | ||||||||||||||
| ≤3 (%) | – | – | 0 (0) | – | – | 92 (28) | – | – | – | – | 37 (24) | – | – | – |
| >3 (%) | – | – | 36 (100) | – | – | 195 (60) | – | – | – | – | 42 (27) | – | – | – |
| NA (%) | – | – | 0 (0) | – | – | 38 (12) | – | – | – | – | 76 (49) | – | – | – |
Data are presented as mean as the mean ± SD. In the cohort 2 of validation set, the cases were the pre‐HCC subjects before clinical diagnosis.
Abbreviations: ALB, albumin; ALT, alanine aminotransferase; AST, aspartate transaminase; DBIL, direct bilirubin; IBIL, indirect bilirubin; NA, not available; TBIL, total bilirubin; TP, total protein.
SERUM SAMPLE COLLECTION AND PRETREATMENT
Serum samples were collected at 6:00‐8:00 am after an overnight fast to eliminate the disturbance of diet.23 Then all samples were immediately stored in a –80°C refrigerator and thawed on ice before analysis.24 Eighty microliters of sample were mixed with 320 μL of acetonitrile by vortexing for 60 seconds. Then, the sample was centrifuged at 18,920 g for 10 minutes (4°C) to precipitate the protein. Two 150‐μL aliquots of supernatant were transferred and lyophilized for the analysis in positive and negative electrospray ionization (ESI+ and ESI−) mode, respectively. Fifty microliters of a 25% (by volume) acetonitrile aqueous solution were used to reconstitute the sample before the LC‐MS analysis. A quality control (QC) sample was prepared by pooling a large number of serum samples from the biobank in our laboratory. Pretreatment of the QC samples paralleled and was the same as that of the study samples. The QC samples were evenly inserted in each set of the analysis running sequence to monitor the stability of the large‐scale analysis.19
LC‐MS ANALYSIS
A modified pseudotargeted method18 was used to acquire the LC‐MS spectra in the discovery and test sets. The acquisition was performed with an ACQUITY UPLC system (Waters, Milford, MA) coupled to a Q‐Trap 5500 mass spectrometer system (AB SCIEX, Framingham, MA). A Waters ACQUITY BEH C8 column (100 × 2.1 mm, 1.7 μm) and an HSS T3 column (100 × 2.1 mm, 1.8 μm) were used in ESI+ and ESI– mode, respectively. A targeted analysis was used to quantify the identified biomarkers in the validation set. Separation and acquisition were performed using the LC (Nexera x2)‐MS (TQ8050) system (Shimadzu, Kyoto, Japan). Detailed experimental conditions of LC separation and MS detection are described in the Supporting Information.
TISSUE PRETREATMENT AND LC‐MS ANALYSIS
HCC tissues (HCT), paired adjacent noncancerous tissue (2 cm from the tumor), and distal noncancerous tissues (>5 cm from the tumor) were collected from 50 HCC patients. The metabolic profiles of these tissues were detected using LC‐MS. Detailed information regarding the pretreatment and LC‐MS analysis was published in our previous report.25
DATA ANALYSIS
A multivariate analysis was performed using the software SIMCA‐P 11.0 (Umetrics AB, Umea, Sweden). An unsupervised model of principal component analysis with unit variance scaling was applied to assess the holistic metabolome alterations among groups and monitor the stability of the study. A supervised model of partial least squares discriminant analysis with unit variance scaling was performed to maximize the distance between groups and identify important variables with an important contribution to the classification according to its variable important in the projection (VIP).26 A permutation test was performed 200 times to assess the risk of overfitting for the model.
A univariate analysis was performed using the Multiple Experiment Viewer 4.7.4 software (http://www.tm4.org). A Wilcoxon Mann‐Whitney test with Benjamini‐Hochberg‐based false discovery rate was used for the statistical analysis, setting P < 0.05 and the false discovery rate <0.05 as the significance levels. A heat map was also obtained to express the results of correlation analysis. The biomarker model was established and assessed using SPSS 18.0 software (SPSS, Inc.). Binary logistic regression was used to build the model based on the potential biomarkers. A receiver‐operating characteristic curve was used to evaluate the results of the regression analysis. Correlation analysis of the metabolites and clinical parameters was also performed using SPSS. The pathway analysis based on the differential metabolites was applied by the software MetaboAnalyst (http://www.metaboanalyst.ca) to reveal the important disturbed metabolic pathways.
Results
DEMOGRAPHICS OF THE STUDY COHORT
The workflow of the study is presented in Fig. 1. To define biomarker candidates, 108 serum samples in the discovery set were collected. A total of 684 participants, including 325 patients with HCC, were recruited to test these biomarker candidates and define potential biomarkers in the test set. Patients with a solitary HCC nodule or at most two nodules <3 cm in diameter, which were primarily S‐HCC, were used to assess the diagnostic potential of the metabolite biomarkers for early‐stage HCC in this set. Among the 325 subjects with HCC, 92 (28%) were diagnosed with S‐HCC. Additionally, another independent cohort of 572 subjects (cohort 1 of validation set), containing 99 NC, 143 patients with CHB, 150 patients with Cir, and 155 patients with HCC, and 25 patients with ICC, was used to establish the metabolite panel model and evaluate its diagnostic performance. Finally, this established model was applied to cohort 2, which were from 42 new cases of HCC and 42 well‐matched at‐risk controls, to evaluate the predictive capacity of this identified biomarker panel for preclinical HCC. The clinical information for all subjects is listed in Table 1.
METABOLIC PROFILING OF SERUM
Typical extracted ion chromatograms from two ESI modes are displayed in Supporting Fig. S1. Coefficients of variation of the distribution of peaks in the QC samples showed that the present analysis was stable and repeatable (Supporting Fig. S2). Additionally, in the principal component analysis score plots of both the discovery and test sets (Fig. 2), the QC samples clustered tightly together, which further confirmed the reliability of the present study. Furthermore, the discrimination trends among samples from the NC, Cir, and HCC groups revealed significant systematic metabolic differences among these groups. Finally, 239 were identified based on our previous study,27, 28 searches of our home‐developed database (containing more than 2,000 metabolite standards), and online databases (HMDB and Metlin) or confirmed with authentic standards. These variables were used for the subsequent multivariate and univariate analyses.
Figure 2.
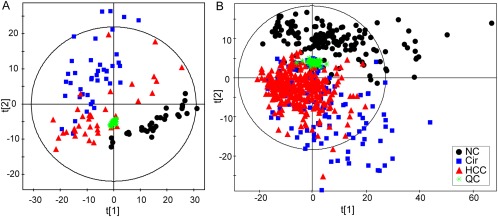
Score plots of principal component analysis based on the combinational data of ESI+ and ESI− modes from the discovery set (A) and the test set (B). Colors and shapes display the subjects from different groups ( , normal controls;
, normal controls;  , patients with cirrhosis;
, patients with cirrhosis;  , patients with HCC; and
, patients with HCC; and  , QC samples).
, QC samples).
DEFINING OF POTENTIAL METABOLIC BIOMARKERS FOR HCC
First, the partial least squares discriminant analysis score plot (Fig. 3A) revealed apparent separations among these groups without overfitting (Supporting Fig. S3) in the discovery set. Fifty‐seven metabolites with VIP >1.0 on two principal components were identified as important variables to contribute the classifications (Fig. 3B). Subsequently, a univariate analysis was used to determine whether these 57 metabolites were significantly altered in the HCC group compared with the NC and Cir groups. Finally, 17 of these metabolites exhibited P < 0.05 and a false discovery rate <0.05 in the two comparisons (Fig. 3C).
Figure 3.
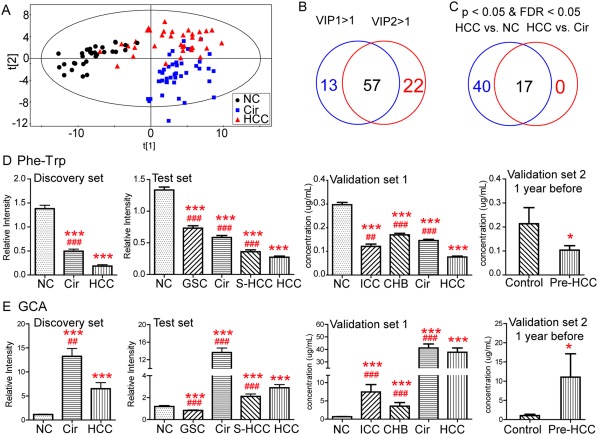
Identification of potential metabolic biomarkers for the diagnosis of HCC. (A) Partial least squares discriminant analysis score plot based on NC, Cir, and HCC groups in the discovery set. (B) Venn diagram displays variables with VIP values >1 on two principal components (VIP1 and VIP2). (C) Venn diagram displays the differential metabolites when the HCC group was compared with the NC and Cir groups in the discovery set, respectively. Serum relative concentrations of defined potential biomarkers of Phe‐Trp (D) and GCA (E) in the discovery, test, and validation sets, respectively. * P < 0.05, ** P < 0.01, and *** P < 0.001 when compared with NC/controls, respectively; # P < 0.05, ## P < 0.01, and ### P < 0.001 when compared with HCC, respectively. All data are presented as mean ± SE. Abbreviation: FDR, false discovery rate.
An independent test cohort of 684 individuals (Fig. 1) was used to evaluate the reliability of 17 biomarker candidates and define the useful biomarkers. These potential biomarkers must satisfy the following criteria: (1) significant differences in the four comparisons (HCC versus NC, HCC versus Cir, S‐HCC versus NC, and S‐HCC versus Cir) and (2) simultaneously maintaining the same change trend as the discovery set for these comparisons. Ultimately, eight metabolites were retained: phenylalanyl‐tryptophan (Phe‐Trp); glycocholate (GCA); taurocholate; lysophosphatidylethanolamines 18:2, 20:5, and 22:4; choline; and taurine. Subsequently, a binary logistic regression analysis and an optimized algorithm of the forward stepwise (Wald) method were employed to construct the best model using these eight potential metabolite biomarkers.29 Finally, the combination of Phe‐Trp and GCA (Fig. 3D,E) was defined as the ideal biomarker panel to distinguish patients with HCC from subjects without HCC.
The diagnostic performance of this metabolic panel was notably higher than that of AFP (with a cutoff of 20 ng/mL), such as AUC values of 0.930 and 0.892 versus 0.657 and 0.725 in the discrimination of HCC from Cir in the discovery and test sets, respectively (Table 2). Furthermore, the serum metabolite panel had a higher sensitivity and similar specificity when compared with AFP (80.2% versus 45.3% and 78.4% versus 78.4%) to identify patients with S‐HCC from the high‐risk population of Cir in the test set (Table 2). Additionally, for the AFP false‐negative (AFP <20 ng/mL) patients with HCC and S‐HCC, the diagnostic accuracy of this panel was 80.6% and 74.5%, respectively (Fig. 4B). Based on these results, the stepwise identification provides reliable metabolite biomarkers for HCC diagnosis.
Table 2.
Results of Measurement of the Serum Metabolite Panel, AFP, or Both in the Diagnosis of HCC
| Discovery Set | Test Set | Validation Set (Cohort 1) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
AUC (95% CI) |
Sensitivity (%) |
Specificity (%) |
AUC (95% CI) |
Sensitivity (%) |
Specificity (%) |
AUC (95% CI) |
Sensitivity (%) |
Specificity (%) |
|
| HCC versus non‐HCC | |||||||||
| 2‐Meta | 0.951 (0.914‐0.989) | 88.9 | 88.9 | 0.936 (0.917‐0.955) | 88.6 | 85.7 | 0.875 (0.846‐0.905) | 91.6 | 72.2 |
| HCC versus Cir | |||||||||
| 2‐Meta | 0.930 (0.871‐0.988) | 88.9 | 82.9 | 0.892 (0.856‐0.929) | 86 | 78.4 | 0.807 (0.753‐0.861) | 92.1 | 52.8 |
| AFP | 0.657 (0.528‐0.786) | 61.8 | 56.1 | 0.725 (0.671‐0.779) | 56.4 | 78.4 | 0.650 (0.582‐0.718) | 50.4 | 73.2 |
| 2‐Meta+AFP | 1.000 (1.000‐1.000) | 100 | 100 | 0.905 (0.872‐0.938) | 81.4 | 87.4 | 0.826 (0.774‐0.877) | 77.9 | 76.4 |
| HCC versus (CHB and Cir) | |||||||||
| 2‐Meta | — | — | — | — | — | — | 0.826 (0.779‐0.873) | 92.1 | 63.8 |
| AFP | — | — | — | — | — | — | 0.682 (0.620‐0.745) | 50.4 | 78.4 |
| 2‐Meta+AFP | — | — | — | — | — | — | 0.847 (0.803‐0.891) | 77.9 | 78.4 |
| S‐HCC versus Cir | |||||||||
| 2‐Meta | — | — | — | 0.866 (0.816‐0.916) | 80.2 | 78.4 | 0.753 (0.664‐0.843) | 80.6 | 52.8 |
| AFP | — | — | — | 0.682 (0.607‐0.757) | 45.3 | 78.4 | 0.676 (0.575‐0.778) | 55.6 | 73.2 |
| 2‐Meta+AFP | — | — | — | 0.870 (0.820‐0.919) | 74.4 | 87.4 | 0.774 (0.688‐0.861) | 66.7 | 76.4 |
| S‐HCC versus (CHB and Cir) | |||||||||
| 2‐Meta | — | — | — | — | — | — | 0.771 (0.689‐ 0.854) | 80.6 | 63.8 |
| AFP | — | — | — | — | — | — | 0.711 (0.614‐0.808) | 55.6 | 78.4 |
| 2‐Meta+AFP | — | — | — | — | — | — | 0.801 (0.722‐0.878) | 66.7 | 78.4 |
Abbreviations: CI, confidence interval; 2‐Meta, serum metabolite panel.
Figure 4.
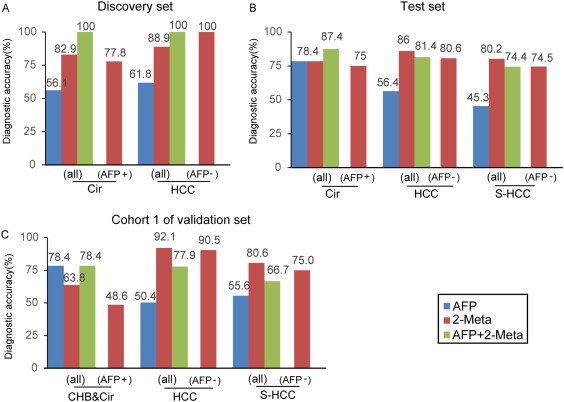
Diagnostic performances of serum metabolite panel (2‐Meta), AFP, and both in the diagnosis of HCC and S‐HCC. (A‐C) Diagnostic accuracy of 2‐Meta in the individuals from groups CHB, Cir, HCC, and S‐HCC or falsely diagnosed patients by AFP in the discovery, test, and validation (cohort 1) sets, respectively. AFP+, false‐positive AFP (CHB and Cir patients with AFP >20 ng/mL), AFP–, false‐negative AFP (HCC and S‐HCC patients with AFP <20 ng/mL).
VALIDATION OF THE BIOMARKER PANEL FOR HCC
To validate the diagnostic potential of this serum metabolite panel for HCC detection, 572 samples from another independent cohort (validation cohort 1) were used. Notably, 150 CHB patients were enrolled in this cohort to enrich the clinical background of the at‐risk controls. The serum concentrations of Phe‐Trp and GCA determined by the isotope‐labeled quantification method were used to establish this metabolic panel by a logistical regression model. The methodology validation of this measurement indicated that the quantification was robust enough to detect Phe‐Trp and GCA in serum (Supporting Table S1). Finally, this metabolite panel for the detection of HCC was constructed as follows: logit[P = HCC] = –23.323 × [Phe‐Trp] + 0.007 × [GCA] + 1.727. In this equation, [P = HCC] is the predicted probability of HCC by this panel, [Phe‐Trp] and [GCA] mean serum concentrations of Phe‐Trp and GCA, respectively. The cutoff value of [P = HCC] was 0.218. Similar to the results of the discovery and test sets, the metabolic panel showed a higher diagnostic performance than that of AFP not only for HCC but also for S‐HCC from the high‐risk population (Cir) (AUC 0.807 versus 0.650, 0.753 versus 0.676, respectively) (Table 2). Moreover, 90.5% and 75.0% of the AFP false‐negative patients with HCC and S‐HCC (AFP < 20 ng/mL) were accurately diagnosed using the metabolite panel (Fig. 4C).
Validation cohort 2 of 84 samples was used to further evaluate the above constructed metabolite panel. This panel had higher sensitivity (range 80.0%‐70.3%) than that of AFP (range 40.0%‐26.2%) in the discrimination of subjects who developed HCC from those who did not before the clinical diagnosis (Table 3). Besides, the sensitivity of this panel for preclinical HCC diagnosis maintained the increased trend with the time interval from 3 years to 1 year (from 71.4% to 80.0%), whereas its specificity (range 47.6%‐80.0%) was lower than that of AFP (range 100%‐93.8%) at every time point before the clinical diagnosis. The combination with AFP resulted in a slight increase of specificity but similar sensitivity when compared with this panel alone (Table 3). Additionally, in the 1 year before clinical diagnosis of samples, the levels of Phe‐Trp and GCA were significantly changed in preclinical HCC individuals when compared with risk controls, whose alterations were consistent with the changes in HCC subjects when compared with controls in the discovery and test sets (Fig. 3D,E). These results indicated that this serum metabolic panel had the potential to screen preclinical HCC from high‐risk populations before clinical diagnosis.
Table 3.
Performance of Serum Metabolite Panel, AFP, or Both in Prediction of HCC by Year Before Clinical Diagnosis in Cohort 2 of the Validation Set
| 1 Year Before (n = 10 × 2) | 1.5 Years Before (n = 16 × 2) | 2 Years Before (n = 28 × 2) | 2.5 Years Before (n = 37 × 2) | 3 Years Before (n = 42 × 2) | |
|---|---|---|---|---|---|
| 2‐Meta | |||||
| AUC | 0.790 | 0.738 | 0.689 | 0.647 | 0.635 |
| (95% CI) | (0.581‐0.999) | (0.557‐0.920) | (0.550‐0.828) | (0.522‐0.773) | (0.513‐0.751) |
| Sensitivity (%) | 80.0 | 75.0 | 75.0 | 70.3 | 71.4 |
| Specificity (%) | 80.0 | 68.8 | 57.1 | 51.4 | 47.6 |
| AFP | |||||
| AUC | 0.740 | 0.674 | 0.633 | 0.619 | 0.622 |
| (95% CI) | (0.518‐0.962) | (0.482‐0.866) | (0.482 ‐0.783) | (0.489‐0.750) | (0.500‐0.745) |
| Sensitivity (%) | 40.0 | 37.5 | 28.6 | 27.0 | 26.2 |
| Specificity (%) | 100.0 | 93.8 | 96.4 | 97.3 | 97.6 |
| 2‐Meta&AFP | |||||
| AUC | 0.880 | 0.789 | 0.742 | 0.726 | 0.721 |
| (95% CI) | (0.723‐1.037) | (0.618‐0.961) | (0.610‐0.874) | (0.608‐0.844) | (0.610‐0.833) |
| Sensitivity (%) | 60.0 | 68.8 | 64.3 | 64.9 | 64.3 |
| Specificity (%) | 100.0 | 87.5 | 78.6 | 78.4 | 78.6 |
Abbreviations: CI, confidence interval; 2‐Meta, serum metabolite panel.
The other two cancers, GSC and ICC, were specifically included in this study to assess the specificity of these potential biomarkers for HCC. Compared to the GSC or ICC group, the serum level of Phe‐Trp significantly declined in the HCC group, whereas the level of GCA was remarkably raised in HCC subjects (Fig. 3D,E). Furthermore, the combination of these two metabolites distinguished patients with HCC from patients with GSC or ICC with AUC of 0.946 or 0.829, respectively (Supporting Fig. S4), suggesting that the metabolic panel is specific for HCC.
ASSOCIATIONS OF THE BIOMARKER PANEL WITH CLINICAL CHARACTERISTICS AND HCC
A pathway analysis of all differential metabolites (Supporting Table S2) revealed that HCC‐induced metabolic perturbations were mainly related to primary bile acid biosynthesis; glycerophospholipid metabolism; glycine, serine, and threonine metabolism; tryptophan metabolism; and taurine and hypotaurine metabolism (Supporting Fig. S6).
A correlation analysis of the content of these differential metabolites (Supporting Table S2) and clinical characteristics was performed to obtain a better understanding of the relationship between the serum metabolite profiles and the HCC phenotype. The levels of several bile acids were positively associated with the levels of aspartate aminotransferase, alanine aminotransferase, total bilirubin, direct bilirubin, and indirect bilirubin and negatively associated with total protein and albumin levels. In contrast, the levels of Phe‐Trp, serotonin, inositol, choline, taurine, etc., showed inverse relationships. Apparently, the levels of these different metabolites were not associated with body mass index, AFP levels, age, and sex (Fig. 5A). We also found that these two biomarkers were not different between hepatitis B and C virus infections.
Figure 5.
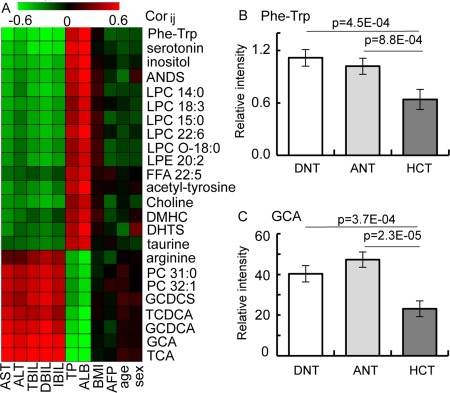
Understanding the biological relevance of the biomarkers for the HCC diagnosis. (A) Heat map of the Pearson correlation coefficients between differential metabolite contents and clinical parameters. Only metabolites with absolute values of correlation coefficient >0.5 and P values <0.05 were left, and the shades of the color represent the strength of the relationship (red, black, and green represent the positive, no, and negative correlations, respectively). (B,C) Histograms of potential biomarkers in distal noncancerous tissue, adjacent noncancerous tissue, and HCT specimens. All data are presented as mean ± SE. Abbreviations: ALB, albumin; ALT, alanine aminotransferase; ANDS, androsterone sulfate; ANT, adjacent noncancerous tissue; AST, aspartate aminotransferase; BMI, body mass index; DBIL, direct bilirubin; DHTS, dihydrotestosterone sulfate; DMHC, dimethylheptanoylcarnitine; DNT, distal noncancerous tissue; FFA, free fatty acid; GCDCA, glycochenodeoxycholate; GCDCS, glycoursodeoxycholate sulfate; IBIL, indirect bilirubin; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; TBIL, total bilirubin; TCDCA, taurochenodeoxycholate; TCA, taurocholate; TP, total protein.
To further reveal the biological relevance of these markers in patients with HCC, we investigated the levels of Phe‐Trp and GCA in paired HCT, adjacent noncancerous tissue, and distal noncancerous tissue specimens from 50 patients with HCC in our previous study.25 Phe‐Trp level was significantly decreased in the HCT compared to the distal noncancerous tissue (Fig. 5B) and accompanied by elevated levels of tryptophan and phenylalanine (Supporting Fig. S5). The levels of several bile acids were significantly decreased in the tumor tissues (Fig. 5C; Supporting Fig. S5).
Discussion
Currently, the identification of novel potential serum biomarkers for the detection of HCC remains a vital goal, particularly for the diagnosis of early‐stage HCC. However, only a few biomarker candidates have been translated to clinical applications due to the limited study cohorts or diagnostic performance. For example, only 9 of 1,261 protein candidates have been approved by the Food and Drug Administration as “tumor‐associated antigens” in nearly 160 years.30
In the present study, several cohorts with 1,448 subjects, including groups of NC, CHB, Cir, and HCC, were recruited from multiple centers (Fig. 1). We employed the “pseudotargeted” metabolomics method to screen biomarkers.18 Some of the biomarkers may be among the unidentified metabolites, but for clinical application, they should be identified. Therefore, in our study we did our best to identify as many metabolites as possible (see Supporting Information). After systematic selection using multivariate and univariate statistical analyses, a biomarker panel consisting of Phe‐Trp and GCA was identified and validated. The serum biomarker panel separated HCC from the high‐risk population (Cir) with higher performance than did AFP (AUC 0.930, 0.892, and 0.807 versus 0.657, 0.725, and 0.650) (Table 2). The diagnostic accuracy range of HCC was 86.0%‐92.5%, and that of cirrhosis was 63.8%‐82.9% by this panel in three sets. Furthermore, among the AFP false‐negative (HCC with AFP < 20 ng/mL) and false‐positive (cirrhosis with AFP > 20 ng/mL) patients, the diagnostic accuracy of the novel metabolite biomarkers was also satisfactory (Fig. 4), which indicates the complementarity of the metabolite biomarker panel and AFP. Additionally, in the comparisons with other two cancers, this metabolite panel showed high selectivity for HCC (Fig. 3D,E; Supporting Fig. S4).
The screening of early‐stage HCC remains a challenge. In the present study, the early diagnostic performance of the serum metabolite panel was also tested in patients with S‐HCC. This metabolite panel effectively discriminated patients with S‐HCC from the high‐risk population in the test cohort with a higher AUC than that of AFP (0.866 versus 0.682), which was further confirmed in cohort 1 of the validation set (Table 2). These results highlight the early diagnostic potential of this metabolite biomarker panel.
Another unique aspect of the present study is the validation of the nested case–control study. In the preclinical HCC screening, the metabolic panel had higher sensitivity than that of AFP, whereas its specificity was lower (Table 3). Additionally, the increased trend of sensitivity of this panel along with the time before clinical diagnosis conformed to the clinical progression of HCC. Furthermore, the combination with AFP could provide a better prediction of pre‐HCC before clinical diagnosis (Table 3), which indicates the complementarity of this panel and AFP. These results demonstrated that this serum metabolic panel had the potential to screen the preclinical HCC from high‐risk populations before clinical diagnosis.
Differential pathway analysis revealed that the metabolic reprogramming was mainly related to metabolism of bile acids, amino acids, and lipids during the progression of HCC (Supporting Fig. S6). What is more, the pathways related to the identified biomarkers will provide insights into the underlying pathogenesis of HCC, such as the perturbations of energy metabolism, oxidative stress, and inflammation during the development of HCC.
The levels of most bile acids were significantly increased in patients with liver disease compared with healthy controls (Supporting Table S2). These results are consistent with many other reports.31, 32 According to recent evidence, bile acids play important roles in regulating cell apoptosis, energy metabolism, oxidative stress, and inflammation by activating the G protein–coupled receptor TGR5 and/or nuclear receptor farnesoid X receptor.33, 34, 35 High levels of bile acids have been shown to induce reactive oxygen species production and cell apoptosis in rat and human hepatocytes,36 thereby leading to liver injury and impaired hepatic function.16 Additionally, bile acids and the gut microbiota have profound influences on each other.37 Consistent with these findings, the levels of circulating bile acids were significantly and positively associated with the levels of alanine aminotransferase, aspartate transaminase, and bilirubin (Figs. 3E and 5A). Increased levels of bile acids in the circulation are usually caused by elevated synthesis of bile acids or liver injury.32, 38 Many studies have revealed that the synthesis of bile acids in malignant cancer cell is down‐regulated by fibroblast growth factor 19, which can promote carcinogenesis of the liver.39, 40, 41 We also found a lowered level of bile acids in tumorous tissues (Fig. 5C). Consistent with many other studies,42, 43 we think the liver injury caused by the development of liver tumor plays an important role in the elevation of bile acids in the circulation. Therefore, bile acids may be used to indicate liver injury and hepatic function during the progression of HCC.
Phe‐Trp is a dipeptide, which is the product of protein breakdown.44 Many studies have revealed that the perturbed dipeptide metabolism plays an important role in disease development.45, 46 In our study, circulating Phe‐Trp levels progressively decreased in NC and patients with Cir and HCC and were positively associated with the total protein and albumin levels. This finding is consistent with the results of a small pilot study by Fitian et al.13 We hypothesized that the decrease in Phe‐Trp levels may be caused by the elevated hydrolysis in the gut. The side chain of tryptophan may be cleaved by gut bacteria, which may decrease the de novo synthesis of nicotinamide adenine dinucleotide, leading to the development of HCC.47, 48 Because aromatic amino acids are mainly metabolized in the gut,49 Phe‐Trp, as a donor of tryptophan, may be altered by gut microbiota during the development of HCC, which may lead to a significant decrease of the dipeptide in the circulation. However, the detailed mechanism should be further studied.
This multicenter metabolomics study provides a practical strategy for screening HCC using a small aliquot of serum. The results may be used as an index for further clinical examinations of patients. However, there are some limitations of this study. Firstly, the metabolites measured in the study were still limited due to the trade‐off among coverage, throughput, and cost. To gain a relatively wide metabolite coverage, we selected an effective and robust reversed‐phase LC‐MS‐based metabolomics method. Secondly, although 27,009 participants constituted the Dongfeng–Tongji cohort, only 42 new cases of HCC were found in the nested case–control study during the 3‐year follow‐up period. Because most cases of HCC were developed from hepatitis B virus infection, we believe that the study will be more powerful when more new HCC cases are recruited in a case–control study. Finally, to improve the outcome of HCC is the most important task of the clinic. We are still collecting prognostic information on these patients, and the outcome results will be reported in the future.
In summary, a biomarker panel consisting of Phe‐Trp and GCA was defined and validated as an effective tool for the detection of HCC by a multicenter cross‐sectional study. The biomarker panel could identify AFP false‐negative HCC patients and discriminate patients with S‐HCC from a high‐risk population, whose diagnostic performance was better than that of AFP. Additionally, this metabolite panel showed high selectivity for HCC among other cancers. Moreover, this panel might predict the risk of HCC development in high‐risk populations before clinical diagnosis, which is meaningful for the surveillance of patients with preclinical HCC. Therefore, we believe this metabolic panel has potential in clinical practice for HCC diagnosis.
Author names in bold designate shared co‐first authorship.
Supporting information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.29561/suppinfo.
Supporting Information
Acknowledgment
We thank Qiang Huang for the measurement of metabolites in liver tissue samples by LC‐MS.
Potential conflict of interest: Nothing to report.
Supported by the National Key Research and Development Program of China (2017YFC0906900), the projects (No. 21375127) and key project (No. 21435006) from the National Natural Science Foundation of China and the National Grand Project (2012ZX10002‐011) of Science and Technology of China.
Contributor Information
Tangchun Wu, Email: wut@mails.tjmu.edu.cn.
Hongyang Wang, Email: hywangk@vip.sina.com.
Junqi Niu, Email: junqiniu@aliyun.com.
Guowang Xu, Email: xugw@dicp.ac.cn.
REFERENCES
- 1. Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol 2009;27:1485‐1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 3. El‐Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557‐2576. [DOI] [PubMed] [Google Scholar]
- 4. Marrero JA, Lok ASF. Newer markers for hepatocellular carcinoma. Gastroenterology 2004;127(5 Suppl. 1):S113‐S119. [DOI] [PubMed] [Google Scholar]
- 5. Nandagopal L, Sonpavde G. Circulating biomarkers in bladder cancer. Bladder Cancer 2016;2:369‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oberg K, Modlin IM, De Herder W, Pavel M, Klimstra D, Frilling A, et al. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncol 2015;16:e435‐e446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan Y, Li Y, Chen Y, Zhao YJ, Liu LW, Li J, et al. Comprehensive metabolomic characterization of coronary artery diseases. J Am Coll Cardiol 2016;68:1281‐1293. [DOI] [PubMed] [Google Scholar]
- 8. Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 2014;20:415‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology 2013;57:2072‐2077. [DOI] [PubMed] [Google Scholar]
- 10. Kimhofer T, Fye H, Taylor‐Robinson S, Thursz M, Holmes E. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: a comprehensive review. Br J Cancer 2015;112:1141‐1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang Q, Liu H, Wang C, Li B. Phenotypic characterization analysis of human hepatocarcinoma by urine metabolomics approach. Sci Rep 2016;6:19763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ladep NG, Dona AC, Lewis MR, Crossey MM, Lemoine M, Okeke E, et al. Discovery and validation of urinary metabotypes for the diagnosis of hepatocellular carcinoma in West Africans. Hepatology 2014;60:1291‐1301. [DOI] [PubMed] [Google Scholar]
- 13. Fitian AI, Nelson DR, Liu C, Xu Y, Ararat M, Cabrera R. Integrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC/MS and UPLC/MS‐MS. Liver Int 2014;34:1428‐1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Hong Z, Tan G, Dong X, Yang G, Zhao L, et al. NMR and LC/MS‐based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer 2014;135:658‐668. [DOI] [PubMed] [Google Scholar]
- 15. Zeng J, Yin P, Tan Y, Dong L, Hu C, Huang Q, et al. Metabolomics study of hepatocellular carcinoma: discovery and validation of serum potential biomarkers by using capillary electrophoresis–mass spectrometry. J Proteome Res 2014;13:3420‐3431. [DOI] [PubMed] [Google Scholar]
- 16. Tan Y, Yin P, Tang L, Xing W, Huang Q, Cao D, et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteomics 2012;11:M111.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavlou MP, Diamandis EP, Blasutig IM. The long journey of cancer biomarkers from the bench to the clinic. Clin Chem 2013;59:147‐157. [DOI] [PubMed] [Google Scholar]
- 18. Chen S, Kong H, Lu X, Li Y, Yin P, Zeng Z, et al. Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high‐performance liquid chromatography/triple quadrupole mass spectrometry. Anal Chem 2013;85:8326‐8333. [DOI] [PubMed] [Google Scholar]
- 19. Luo P, Yin P, Zhang W, Zhou L, Lu X, Lin X, et al. Optimization of large‐scale pseudotargeted metabolomics method based on liquid chromatography–mass spectrometry. J Chromatogr A 2016;1437:127‐136. [DOI] [PubMed] [Google Scholar]
- 20. Wang F, Zhu J, Yao P, Li X, He M, Liu Y, et al. Cohort profile: the Dongfeng–Tongji cohort study of retired workers. Int J Epidemiol 2013;42:731‐740. [DOI] [PubMed] [Google Scholar]
- 21. Bruix J, Sherman M; Practice Guidelines Committee, American Association for the Study of Liver Diseases . Management of hepatocellular carcinoma. Hepatology 2005;42:1208‐1236. [DOI] [PubMed] [Google Scholar]
- 22. Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology 2009;50:661‐662. [DOI] [PubMed] [Google Scholar]
- 23. Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am J Clin Nutr 2006;84:531‐539. [DOI] [PubMed] [Google Scholar]
- 24. Yin P, Peter A, Franken H, Zhao X, Neukamm SS, Rosenbaum L, et al. Preanalytical aspects and sample quality assessment in metabolomics studies of human blood. Clin Chem 2013;59:833‐845. [DOI] [PubMed] [Google Scholar]
- 25. Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, et al. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res 2013;73:4992‐5002. [DOI] [PubMed] [Google Scholar]
- 26. Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res 2007;6:469‐479. [DOI] [PubMed] [Google Scholar]
- 27. Luo P, Dai W, Yin P, Zeng Z, Kong H, Zhou L, et al. Multiple reaction monitoring‐ion pair finder: a systematic approach to transform nontargeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography‐mass spectrometry. Anal Chem 2015;87:5050‐5055. [DOI] [PubMed] [Google Scholar]
- 28. Shao Y, Zhu B, Zheng R, Zhao X, Yin P, Lu X, et al. Development of urinary pseudotargeted LC‐MS‐based metabolomics method and its application in hepatocellular carcinoma biomarker discovery. J Proteome Res 2014;14:906‐916. [DOI] [PubMed] [Google Scholar]
- 29. Nuamah IF, Qu Y, Amini SB. A SAS macro for stepwise correlated binary regression. Comput Methods Programs Biomed 1996;49:199‐210. [DOI] [PubMed] [Google Scholar]
- 30. Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights 2007;1:1‐48. [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X, Xie G, Zhao A, Zheng X, Huang F, Wang Y, et al. Serum bile acids are associated with pathological progression of hepatitis B–induced cirrhosis. J Proteome Res 2016;15:1126‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie G, Wang X, Huang F, Zhao A, Chen W, Yan J, et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int J Cancer 2016;139:1764‐1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pols TWH, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol 2011;54:1263‐1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5‐mediated bile acid sensing controls glucose homeostasis. Cell Metab 2009;10:167‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fiorucci S, Mencarelli A, Palladino G, Cipriani S. Bile‐acid‐activated receptors: targeting TGR5 and farnesoid‐X‐receptor in lipid and glucose disorders. Trends Pharmacol Sci 2009;30:570‐580. [DOI] [PubMed] [Google Scholar]
- 36. Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, et al. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin–sensitive mechanism in murine and human hepatocytes. Hepatology 2005;42:1291‐1299. [DOI] [PubMed] [Google Scholar]
- 37. Xie G, Wang X, Liu P, Wei R, Chen W, Rajani C, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016;7:19355‐19366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 2016;30:909‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha‐hydroxylase gene expression. Hepatology 2009;49:297‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, et al. Definition of a novel growth factor–dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev 2003;17:1581‐1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou M, Wang X, Phung V, Lindhout DA, Mondal K, Hsu JY, et al. Separating tumorigenicity from bile acid regulatory activity for endocrine hormone FGF19. Cancer Res 2014;74:3306‐3316. [DOI] [PubMed] [Google Scholar]
- 42. Chen Y, Song X, Valanejad L, Vasilenko A, More V, Qiu X, et al. Bile salt export pump is dysregulated with altered farnesoid X receptor isoform expression in patients with hepatocellular carcinoma. Hepatology 2013;57:1530‐1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics 2011;10:M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilbert ER, Wong EA, Webb KE Jr. Board‐invited review. Peptide absorption and utilization: implications for animal nutrition and health. J Anim Sci 2008;86:2135‐2155. [DOI] [PubMed] [Google Scholar]
- 45. Soga T, Sugimoto M, Honma M, Mori M, Igarashi K, Kashikura K, et al. Serum metabolomics reveals gamma‐glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J Hepatol 2011;55:896‐905. [DOI] [PubMed] [Google Scholar]
- 46. Fonteh AN, Harrington RJ, Tsai A, Liao P, Harrington MG. Free amino acid and dipeptide changes in the body fluids from Alzheimer's disease subjects. Amino Acids 2007;32:213‐224. [DOI] [PubMed] [Google Scholar]
- 47. Tummala KS, Gomes AL, Yilmaz M, Graña O, Bakiri L, Ruppen I, et al. Inhibition of de novo NAD+ synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 2014;26:826‐839. [DOI] [PubMed] [Google Scholar]
- 48. Surjana D, Halliday GM, Damian DL. Role of nicotinamide in DNA damage, mutagenesis, and DNA repair. J Nucleic Acids 2010;2010. doi: 10.4061/2010/157591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krishnan S, Alden N, Lee K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr Opin Biotechnol 2015;36:137‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.29561/suppinfo.
Supporting Information


