Abstract
Lager beer fermentations rely on specific polyploid hybrids between Saccharomyces cerevisiae and Saccharomyces eubayanus falling into the two groups of S. carlsbergensis/Saaz-type and S. pastorianus/Frohberg-type. These strains provide a terroir to lager beer as they have long traditional associations and local selection histories with specific breweries. Lager yeasts share, based on their common origin, several phenotypes. One of them is low transformability, hampering the gene function analyses required for proof-of-concept strain improvements. PCR-based gene targeting is a standard tool for manipulating S. cerevisiae and other ascomycetes. However, low transformability paired with the low efficiency of homologous recombination practically disable targeted gene function analyses in lager yeast strains. For genetic manipulations in lager yeasts, we employed a yeast transformation protocol based on lithium-acetate/PEG incubation combined with electroporation. We first introduced freely replicating CEN/ARS plasmids carrying ScRAD51 driven by a strong heterologous promoter into lager yeast. RAD51 overexpression in the Weihenstephan 34/70 lager yeast was necessary and sufficient in our hands for gene targeting using short-flanking homology regions of 50 bp added to a selection marker by PCR. We successfully targeted two independent loci, ScADE2/YOR128C and ScHSP104/YLL026W, and confirmed correct integration by diagnostic PCR. With these modifications, genetic alterations of lager yeasts can be achieved efficiently and the RAD51-containing episomal plasmid can be removed after successful strain construction.
Keywords: homologous recombination, gene function analysis, hybrid yeast, fermentation
1. Introduction
Lager beer is the most popular alcoholic beverage worldwide. It is distinguished from top-fermented ale beers by the use of dedicated bottom fermenting lager yeasts. Pure culture lager yeasts were introduced in the late 19th century and the first lager yeast strain became known as Saccharomyces carlsbergensis [1,2]. Lager yeasts are particularly suited for cold fermentations below 15 °C, providing a specific and clean taste [3]. Lager yeast strains are hybrids of two Saccharomyces species, namely S. cerevisiae and S. eubayanus [4,5,6,7,8]. The S. cerevisiae parent is related to ale yeasts while the S. eubayanus parent has recently been isolated from South America and Asia [9,10]. The low temperature fermentation capabilities of lager yeast hybrids may have originated from the parental S. eubayanus mitochondrial DNA that is invariably present in today’s lager yeasts [11].
Genetic manipulations in lager yeast are hampered by the allopolyploid nature of these hybrids but also by their low transformation and homologous recombination (HR) efficiency [12]. This is demonstrated by the very few reports on targeted gene alterations in lager yeast [13,14,15]. These shortcomings of lager yeasts are in sharp contrast to the high efficiency and ease with which S. cerevisiae can be manipulated [16]. A large array of modules for PCR-based gene targeting have been developed for S. cerevisiae and the gene deletion collection was one of its outputs that spurred large scale functional and synthetic genetic array analyses [17,18,19,20,21,22]. PCR-based gene targeting tools were also developed for other ascomycetous fungi with efficient homologous recombination machineries, including Ashbya gossypii, Candida albicans and Schizosaccharomyces pombe [23,24,25,26,27,28,29].
Deletion of Ku70 or Ku80 of the non-homologous end joining (NHEJ) pathway has been shown to be a very useful tool to improve gene targeting in a large variety of fungi, including e.g., Kluyveromyces lactis and Yarrowia lipolytica [30,31,32]. The Ku70 deletion approach has been particularly successful in filamentous ascomycetes in which non-homologous end joining is otherwise predominant, e.g., in Neurospora crassa or Zymoseptoria tritici [33,34]. However, in Cryptococcus neoformans it was found that deletion of Ku70 also alters virulence, which then requires sexual crosses to reinstate wildtype Ku70 in a mutant background [35].
An alternative to impairing NHEJ is improving HR efficiency. Overexpression of RAD51, coding for a protein involved in the recombinational repair of DNA-double strand breaks, in S. cerevisiae enhances gene repair, a feature conserved in human cells that can also be achieved by using RecA from Escherichia coli [36,37,38]. We thought to employ the apparently highly conserved nature of HR improvement upon RAD51 overexpression in lager yeast. Here we show that plasmid-based overexpression of ScRAD51 under the control of a strong heterologous promoter, allows for simplified targeted gene function analyses in lager yeast. This genetic improvement of lager yeast transformation was combined with a lithium acetate/electroporation transformation protocol similar as has previously been described for S. cerevisiae and S. pombe [39,40]. Together, these improvements should aid proof-of-concept analyses in lager yeasts prior to embarking on laborious mutagenesis and selection schemes for strains not genetically modified (non-GM) used in the production of fermented beverages. Additionally, this provides a tool to investigate hybrid biology at the molecular level.
2. Material and Methods
2.1. Strains and Culture Conditions
Yeast strains, used and generated (see Table 1), were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) at 30 °C. Solid media were prepared by adding 2% agar and YPD plates were supplemented with either hygromycin or G418/geneticin. For lager yeasts the antibiotic concentrations used were 100 µg/mL, while S. cerevisiae lab strains were selected at 200 µg/mL final concentration. Minimal media lacking leucine contained 6.7 g/L yeast nitrogen base with NH4SO4 and without amino acids and 0.69 g/L completed synthetic medium leucine drop-out mixture. Plasmids were propagated in Escherichia coli DH5alpha in 2xYT (1.6 % tryptone, 1 % yeast extract, 0.5 % NaCl) containing 100 µg/mL ampicillin.
Table 1.
Strains used in this study.
| Strain Number | Feature/Genotype | Source |
|---|---|---|
| B003 | Saccharomyces cerevisiae BY4741 MATa his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| B237 | Weihenstephan WS34/70 lager yeast | Lab collection |
| B256 | WS34/70; pRAD51 (HYG3, CEN6/ARSH4) | This study |
| B257 | WS34/70; pHYG3 (CEN6/ARSH4) | This study |
| G001 | WS34/70; pRAD51; ade2::YES1 | This study |
| G002 | WS34/70; pRAD51; hsp104::YES1 | This study |
2.2. Plasmid Design and Constructions
The plasmids used in this study are listed in Table 2. The primers required for plasmid constructions or for PCR-based gene targeting are listed in Table 3. Plasmid DNAs were amplified in E. coli and prepared using a Plasmid Midi DNA Purification Kit (Genaxxon, Ulm, Germany). PCR fragments were cloned into pGEM (Promega, Madison, USA). The ScRAD51/YER095W open reading frame was amplified from BY4741 genomic DNA using primers 260 and 261 and the Arthroascus (=Saccharomycopsis) schoenii TEF1 promoter was amplified from A. schoenii genomic DNA using primers 106 and 107, and both were cloned into pGEM. To place LacZ under the control of the AsTEF1-promoter, AsTEF1p was amplified form pGEM-AsTEF1p (E026) with primers 108/109, which added 45 bp of flanking homology region to E025-pRS417-AgTEFp-LacZ (here LacZ is under the control of the Ashbya gossypii TEF-promoter). The AgTEF-promoter was removed from E025 by restriction digestion with KpnI/XhoI, and the vector and AsTEF1p were cotransformed into yeast to combine them via in vivo recombination, yielding plasmid E065. All restriction enzymes were obtained from Thermo Fisher (Asse, Belgium). All primers were obtained from Sigma Aldrich (Overijse, Belgium).
Table 2.
Plasmids used in this study.
| Strain Number | Feature/Genotype | Source |
|---|---|---|
| E008 | pRS415 | [41] |
| E025 | pRS417-AgTEF1p-LacZ-GEN3 | [42] |
| E026 | pGEM-AsTEF1p | This study |
| E054 | pUC57-HYG3 | GenScript |
| E065 | pRS417-AsTEF1p-LacZ | This study |
| E066 | pYES1 | This study |
| E068 | pYES2 | This study |
| E088 | pGEM-YES1 | This study |
| E120 | pGEM-ScRAD51 | This study |
| E150 | pRS-AsTEF1p-ScRAD51-HYG3-GEN3 | This study |
| E160 | pRS-AsoTEF1p-ScRAD51-HYG3-LEU2 | This study |
Table 3.
Primers used in this study.
| Primer Number | Primer Name | Sequence 5′→3′ * |
|---|---|---|
| 106 | 5′-AsTEF1p | GTCCAGAATAACATCAAATC |
| 107 | 3′-AsTEF1p | CTATAAAAAATGTTAGTATGGAG |
| 108 | 5′-AsTEFp-pRS | CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGCTCGAGTCCAGAATAACATCAAATC |
| 109 | 3′-AsTEFp-lacZ | CAATCTTTGGATCGTTTAAATAAGTTTGAATTTTTTCAGTCATGTTCTATAAAAAATGTTAGTATGGAG |
| 226 | P3-pRS415-AsTEF1p | TGTAAAACGACGGCCAGTGAGCGCGCGTAATACGACTCACTATAGGAAGCTTCGTACGCTGCAGGTCGGATCCCCCGGGGGCGCGCCGTCCAGAATAACATCAAATC |
| 227 | P4-AsTEF1p-kanR | GTTGGAGTTCAAACGTGGTCTGGAAACGTGAGTCTTTTCCTTACCCTATAAAAAATGTTAGTATGGAG |
| 228 | P5-kan-ORF | GGTAAGGAAAAGACTCACGTTTCCA |
| 229 | P6-kanR-pRS415 | GGAAACAGCTATGACCATGATTACGCCAAGCGCGCAATTAACCCTTCTGATATCATCGATGAATTCGAGCTCGTTTAAACATTGGTAATAG |
| 230 | P7-5′-HYG3 | CTGACTTTTGTCTTGTTATGGACTCCATACTAACATTTTTTATAGAAAAAACCAGAATTGACTGCTACTTC |
| 231 | P8-3′-HYG3 | CTGATATCATCGATGAATTCGAGCTCGTTTAAACATTGGTAATAGGACCACCTTTGATTGTAAATAG |
| 253 | 5-HYG3+AD | GGCGCGCCAGATCTAGCCTCCTCAGAGAAAATTGCACAAAAAAAAGGAAGCTTCGTACGCTGCAGGTC |
| 254 | 3-HYG3+AD | TTACGCCAAGCGCGCAATTAACCCTCACTAAAGGGAACAAAAGCTGACCACCTTTGATTGTAAATAG |
| 258 | 5-ScRAD51+AD | CTGACTTTTGTCTTGTTATGGACTCCATACTAACATTTTTTATAGTCTCAAGTTCAAGAACAACATATATCAG |
| 259 | 3-ScRAD51+AD | CTTTTTTTTGTGCAATTTTCTCTGAGGAGGCTAGATCTGGCGCGCCGAAAAATACATATATTTCATGGGTGACAG |
| 260 | 5-ScRAD51 | TCTCAAGTTCAAGAACAACATATATCAG |
| 261 | 3-ScRAD51 | GAAAAATACATATATTTCATGGGTGACAG |
| 344 | S1-LEU2 | GGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTAAAGTGCAATTCTTTTTCC |
| 345 | S2-LEU2 | CTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCGGTCGAGGAGAACTTC |
| 355 | G2-YES1 | GAATGAATCTACTGGTTTGG |
| 356 | G3-YES1 | GTGTCGGTATCGCAGAC |
| 369 | S1-ADE2 | CCTACTATAACATTCAAGAAAAACAAGAAAACCGGACAAAACAATCAAGTGTCCAGAATAACATCAAATC |
| 371 | S2-ADE2 | TATATCATTTTATATTATTTGCTGTGCAAGTATATCAATAAACTTATATATAATAAATTATTTTTATTGTTG |
| 373 | G1-ADE2 | GACTCTTGTTGCAGGGCT |
| 389 | G4-ADE2 | GTGATGCATTGAGCCGCC |
| 390 | S1-HSP104 | TATATTACTGATTCTTGTTCGAAAGTTTTTAAAAATCACACTATATTAAAGTCCAGAATAACATCAAATC |
| 391 | S2-HSP104 | AACAAAGAAAAAAGAAATCAACTACACGTACCATAAAATATACAGAATATTAATAAATTATTTTTATTGTTG |
| 392 | G1-HSP104 | CCCGTATTCTAATAATGGACC |
| 393 | G4-HSP104 | CAAACTTATGCAACCTGCCAG |
* Underlined sequences correspond to restriction sites.
E150/pRAD51 (GEN3) was constructed by in vivo recombination of E065 vector backbone with fragments carrying ScRAD51-ORF and HYG3 marker. To this end, E065 was linearized by digesting with EcoRV/SacI, which removed most of the LacZ-ORF, and then the band was gel purified. Flanking adaptor regions, to guide in vivo recombination, were added to ScRAD51-ORF and HYG3 marker fragments using primers 258/259 and 253/254, respectively. Then BY4741 was transformed with the two PCR products and the cut vector. In order to use a G418 resistance marker for PCR-based gene targeting in lager yeast, the GEN3 marker in E150 was replaced by LEU2 in a PCR-based gene targeting approach using BY4741 as a host. To this end, ScLEU2 was amplified from pRS415 with primer 344/345 by PCR and used for transformation, generating E160.
Novel synthetic marker genes YES1 and YES2 were also constructed using in vivo recombination in S. cerevisiae using plasmid pRS415 as the vector backbone. YES1 and YES2 contain the AsTEF1-promoter that controls kanamycin or hygromycin resistance gene-ORFs, respectively. For construction, pRS415 was linearized with SmaI, the AsTEF1p was amplified from E026 using primers 226/227, the kanR-ORF was amplified with primers 228/229 and the hygR-ORF with primers 230/231 to add suitable adaptors for homologous recombination. Vector, promoter and resistance gene ORFs were then cotransformed in S. cerevisiae and the resulting transformants were selected on G418 or hygromycin, respectively. All plasmids obtained by in vivo recombination in S. cerevisiae were verified and shuttled into E. coli for propagation. A. schoenii promoters were used as heterologous promoters in lager yeast and S. cerevisiae.
HYG3 is a synthetic selectable marker that consists of the 694 bp A. schoenii PGK1 promoter, the 1026 bp hygromycin resistance gene ORF and the Candida albicans URA3 terminator, which was synthesized by GenScript (GenScript, Piscataway, NJ, USA). Promoter sequences were derived from the A. schoenii CBS 7425 genome, which can be accessed via GenBank under accession JNFU00000000 CBS 7425 [43].
2.3. Yeast Transformation
For the transformation of S. cerevisiae, the lithium acetate/single strand DNA/polyethylene glycol 4000 protocol was used as described [44]. For lager yeast transformation, we adapted a protocol from Thompson et al. [39]. WS34/70 was grown overnight to stationary phase in 50 mL YPD. Then the culture was diluted to OD600 = 0.3 in 100 mL fresh YPD. Cells were harvested at mid-log growth with OD600 = 0.7–0.8 (after approximately 3 h of incubation). The culture was centrifuged and washed once with sterile water and resuspended in 25 mL of 0.1 M lithium acetate/10 mM dithiothreitol/10 mM TE solution (Tris HCl: EDTA = 10:1) and incubated for 1 h at room temperature. The cells were washed in 25 mL ice-cold distilled sterile water twice and once with 10 ml of ice-cold 1 M sorbitol, then resuspended in 100 µL ice-cold sorbitol. A 100 µL aliquot of the cell suspension was used for transformation. Transforming DNA (15 µL) was mixed with the cells, incubated for 5 min on ice and electroporated with 1.8 kV in 0.2 cm cuvettes. Then, cells were resuspended in 1 ml cold sorbitol and transferred to a tube with 300 µL YPD. This suspension was incubated for 3 h at 30 °C prior to plating on selective plates that did not contain sorbitol.
2.4. Verification of Lager Yeast Transformants
The Weihenstephan strain WS34/70 was used for gene targeting experiments. Target genes were derived from the S. cerevisiae parental subgenome. Sequences for verification primers (and also for the S1 and S2 primers) were deduced from the genome sequence that is available at DDBJ/EMBL/GenBank under the accession AZAA00000000. PCR-based gene targeting and diagnostic PCR were performed as described [45].
3. Results
3.1. Design and Testing of Synthetic Marker and Reporter Genes
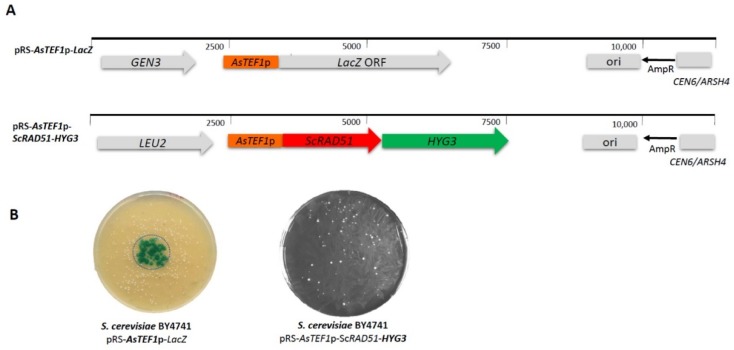
We were interested in developing novel synthetic marker and reporter genes for dual use in non-conventional yeasts, including lager yeasts and S. cerevisiae. To this end, we tested heterologous promoters derived from Arthroascus schoenii (Saccharomycopsis schoenii). Saccharomycopsis species are non-conventional yeasts used in diverse brewing settings, e.g., to generate nuruk [46,47]. We recently obtained several Saccharomycopsis genome sequences including that of A. schoenii [43]. Here we fused the AsTEF1 promoter with the LacZ reporter gene derived from Streptococcus thermophilus [48]. Additionally, we generated a completely new synthetic marker gene based on the AsPGK1 promoter controlling a hygromycin resistance gene ORF. Both genes were introduced into BY4741 on episomal CEN6/ARSH4 plasmids (Figure 1). The functionality of both the synthetic marker and the reporter were demonstrated in vivo in S. cerevisiae: HYG3 was functional as we obtained hygromycin-resistant transformants that carried the plasmids, and the LacZ reporter was active as transformants were able to convert colorless X-Gal into a blue dye (Figure 1B). This indicates that both A. schoenii promoters derived from AsTEF1 and AsPGK1 were functional in the heterologous host S. cerevisiae and we thus went on to employ these genes in lager yeast.
Figure 1.
Generating heterologous synthetic marker and reporter genes. (A) Linear maps of pRS-AsTEF1p-LacZ and pRS-AsTEF1p-ScRAD51-HYG3 plasmids. (B) The AsTEF1p-LacZ reporter gene and the HYG3 resistance marker were tested in Saccharomyces cerevisiae BY4741. The presence of an active β-galactosidase was detected by adding X-Gal to the centre of the transformation plate (left panel, circle indicates the area of X-Gal application). The presence and function of the HYG3 marker was tested by transforming S. cerevisiae with pRS-AsTEF1p-ScRAD51-HYG3 and selecting the transformants in the presence of 100 µg/mL of hygromycin (right).
3.2. Transformation of Lager Yeast
Previous analyses already indicated that lager yeasts’ transformation efficiency is drastically lower than that of S. cerevisiae. In S. cerevisiae lithium acetate/ss DNA/PEG protocols are commonly used [44]. We compared this standard protocol with modified versions, which were developed to deal with yeast strains that are non-responsive to standard lithium acetate or electroporation protocols [39,40]. Our modified protocol included parts of both protocols: a lithium acetate treatment (without DMSO) and an electroporation step (Li-Ac/Ep). To allow time for the expression of the antibiotic resistance genes used in this study, we incubated the cell suspensions after electroporation in an osmotically stabilized medium before plating on selective plates. To compare both protocols, we employed a freely replicating plasmid (E160) with HYG3 as the selectable marker.
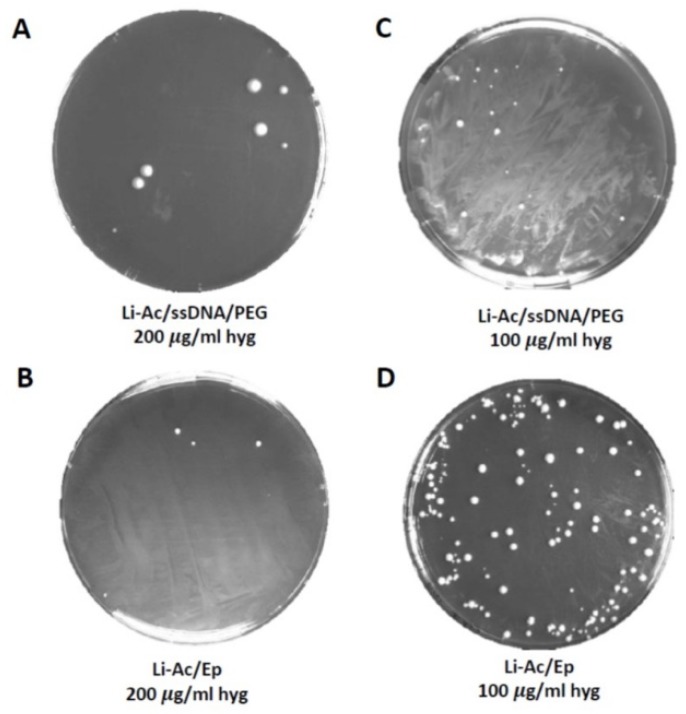
In our hands, the lithium acetate/ss DNA/PEG protocol (with DMSO) yielded only a few transformants per µg of DNA, demonstrating the poor transformation efficiency of lager yeast with this method, even after lowering the temperature of the heat shock step at 40 °C, which we tested as lager yeasts are known to be temperature sensitive and do not grow at elevated temperatures above 34 °C. The Li-Ac/Ep protocol, however, resulted in much higher transformation efficiencies (1–2 orders of magnitude, Figure 2).
Figure 2.
Comparison of two transformation methods in the Weihenstephan lager yeast strain WS 34/70. (A,C) A lithium-acetate/single strand DNA/polyethylene glycol (Li-Ac/ssDNA/PEG) and (B,D) a LiAc/PEG incubation combined with electroporation (Li-Ac/Ep) were used with two different antibiotic concentrations for the selection of transformants. The transformation of WS 34/70 with a freely replicative plasmid carrying a hygromycin resistance marker (pRS-AsTEF1p-ScRAD51-HYG3) yielded few transformants when using the Li-ac/ssDNA/PEG method (A,C). By contrast, the Li-Ac/Ep method together with antibiotic selection at 100 µg/mL final concentration (D), resulted in a higher transformation efficiency compared to the other combinations (A–C).
3.3. PCR-Based Gene Targeting is Enhanced by RAD51 Overexpression
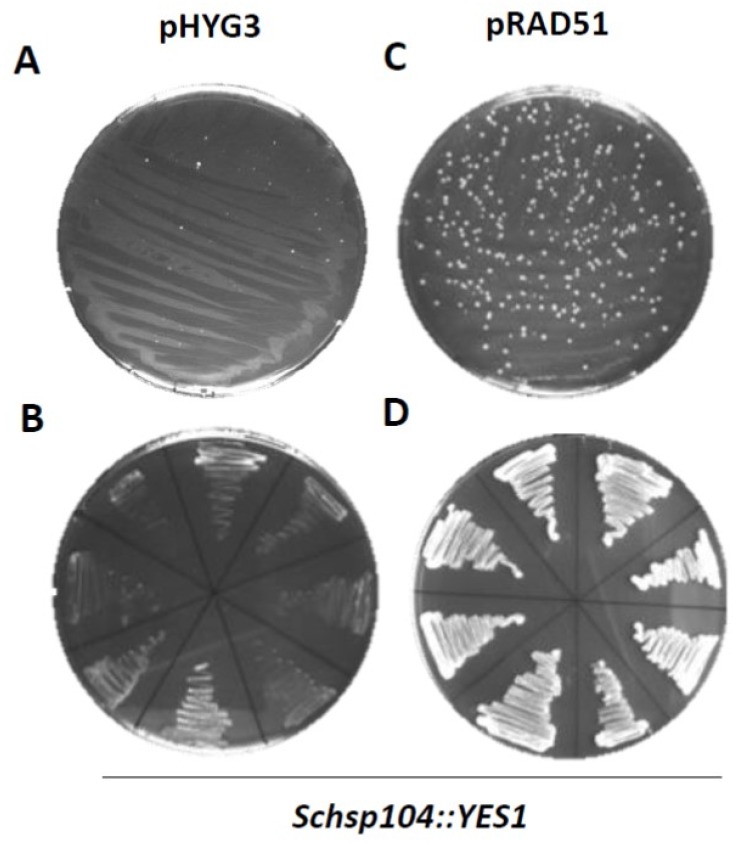
Plasmid transformation is regularly far more efficient than integrative transformation. PCR-based gene targeting approaches are convenient because in one PCR reaction short-flanking homology regions can be added to a selection marker. We used standard S1 and S2 primers that added 50 bases of flanking homology region. However, in our hands, we failed to obtain stable lager yeast transformants with these PCR-based disruption cassettes, even when using the Li-Ac/Epo protocol (Figure 3A). After prolonged incubation on selective plates (>2d) small colonies may appear. However, these colonies did not continue to grow upon restreaking on new selective plates or in selective liquid media and represent background growth (Figure 3B). In contrast, the transformation of a Weihenstephan lager yeast strain that harbors a plasmid-encoded ScRAD51 expressed from a strong TEF promoter yielded a large number of transformants on primary selective plates using disruption cassettes for ScHSP104 (Figure 3C). A randomly selected set of colonies continued to grow upon restreaking and also grew well in liquid YPD supplemented with hygromycin (Figure 3D and data not shown).
Figure 3.
PCR-based gene targeting in lager yeast. The Weihenstephan strain WS34/70 carrying either an empty vector (pHYG3; A) or a RAD51 plasmid (pRAD51, C) was transformed with disruption cassettes containing the YES1 marker harboring 50 bp of flanking homology regions for targeting to the ScHSP104 locus. We used a Li-Ac/Ep transformation protocol and selected transformants on YPD plates supplemented with hygromycin and G418 (100 µg/mL final concentration each). Restreaking of putative transformant colonies on new selective plates indicated stable transformants were only obtained in the lager yeast strain overexpressing pRAD51 (B,D).
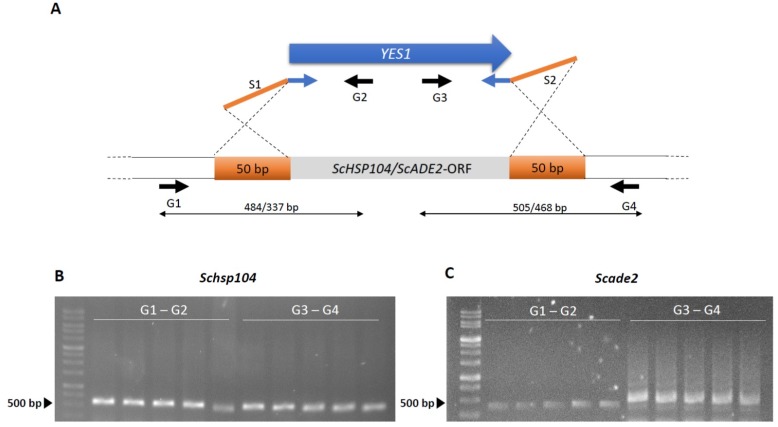
Verification of these transformants that grew upon restreaking and growth in liquid culture was done by standard diagnostic PCR, amplifying the novel joints at the borders of marker integration [45]. This confirmed targeted gene disruption (Figure 4). Overall, between 60–75% of primary transformants could be cultivated further in a liquid culture, and all for all of those diagnostic PCRs indicated correct integration of the marker cassette at the target locus (n > 20). However, we did not obtain any transformants in the same strain without the overexpression of ScRAD51, demonstrating the usefulness of this approach. To verify that the marker integration and PCR-based gene targeting success was not solely locus dependent, we targeted a second gene, ScADE2, in the same manner. Transformation efficiencies were found to be similar for both loci and diagnostic PCR was successfully employed for the verification of the deletion of an ADE2 allele (Figure 4C).
Figure 4.
Diagnostic PCR for verification of correct marker integration. (A) Schematic presentation of PCR-based gene targeting of YES1 amplified with gene specific S1/S2-primers adding 50 bp of target homology region (orange bars) to ScHSP104 or ScADE2, respectively. Diagnostic G1/G2 and G3/G4 primers were used for verification of the correct insertion of YES1 and removal of the Sc HSP104 (B) and Sc ADE2 locus (C) ORFs.
4. Discussion
Lager yeasts are the workhorses of the beer industry. Detailed strain characterizations and molecular genetics-assisted yeast breeding, however, are hampered by the lack of proof-of-concept technologies. A major obstacle in lager yeast research is the surprisingly inefficient gene targeting in lager yeast. Thus, there are only a few reports of the successful molecular genetic manipulation of lager yeast relying on HR [13,14,15]. There are other issues beyond the mere technical difficulties. Primarily, these are complicated by the hybrid nature of lager yeast. Lager yeasts are allopolyploid hybrids between S. cerevisiae and S. eubayanus parents [9]. Group I/Saaz strains are triploid, while Group II/Frohberg strains, to which Weihenstephan 34/70 belongs, are tetraploid [2]. Additionally, aneuploidies could further change allele frequencies, complicating gene knockout experiments. Secondly, due to the hybrid nature, sporulation is severely crippled due to failure to proceed through meiotic divisions, resulting in hybrid sterility, which is more pronounced in triploid Group I strains than in tetraploid Group II strains [49,50]. Thirdly, even though lager yeasts are the workhorses of the beer industry, there is a reluctance to employ genetically modified yeast strains, a notion that has rather been strengthened over recent years.
In addition to their industrial importance, lager yeast hybrids present excellent model systems to study hybrid biology, including hybrid vigor, hybrid sterility, adaptive evolution and the analysis of hybrid protein complexes [51,52]. Major advances in hybrid yeast breeding resolved the F1-sterility problem represented in lager yeast [53].
Our study indicates that RAD51 overexpression in lager yeast opens the tool-box for all genetic manipulations previously only available in S. cerevisiae. Although transformation efficiencies in lager yeasts are far lower than in S. cerevisiae, rational strain design based on targeted gene replacements has become feasible. We have successfully employed this strategy already to other loci, suggesting that potential locus dependent variations in gene targeting are not inhibitory to successful gene targeting. Furthermore, by selecting specific homology regions, even closely related alleles of the S. cerevisiae and S. eubayanus parental genomes can be distinguished. The overexpression of RAD51 has been shown to improve HR in other systems, so it may also be advantageous in other non-conventional yeasts in which molecular genetic studies are hampered by the preferential ectopic integration of gene targeting cassettes.
Two additional advances to improve HR in resilient strains in recent years include the use of Ku70 mutant strains and the establishment of CRISPR/Cas9 methodologies [12]. Deletion of Ku70 inactivates the NHEJ pathway, thus favouring HR and targeted gene alterations [33]. However, in S. cerevisiae, the deletion of Ku70 has been shown to generate defects in telomere maintenance and cell cycle regulation [54,55]. Thus, this may not be favorable in lager yeast as breeding efforts to restore a wildtype Ku70 status after genetic engineering are also quite laborious.
Recent reports established CRISPR/Cas9 in lager yeast, making this a promising tool for strain engineering [12,56]. CRISPR/Cas9 will be particularly useful for simultaneous alterations of multiple alleles in a single step, even more so when the number of alleles may vary due to aneuploidies. In a recent report, simultaneous single and double deletions of SeATF1 and SeATF2 were performed in lager yeast, demonstrating the power of this tool [12]. It will be interesting to see if a combined approach, overexpressing RAD51 and the use of CRISPR/Cas9, will further improve gene replacement efficiencies.
Author Contributions
The authors of this paper contributed in the following way: Conceptualization, J.W. and B.B.; methodology, B.B., Y.K., M.A. and J.W.; validation, B.B., Y.K. and M.A.; investigation, B.B., Y.K., M.A., and J.W.; data curation, B.B. and Y.K.; writing—original draft preparation, B.B. and J.W.; writing—review and editing, B.B., Y.K., M.A. and J.W.
Funding
This research was supported by the European Union Marie Skłodowska-Curie Actions Innovative Training Network Aromagenesis (764364) (https://www.aromagenesis.eu/).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hansen E.C. Grundlagen zur Systematik der Saccharomyceten. Zentralbl. Bakteriol. II Natur. 1904;12:529–538. [Google Scholar]
- 2.Walther A., Hesselbart A., Wendland J. Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 Genes Genomes Genet. 2014;4:783–793. doi: 10.1534/g3.113.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendland J. Lager yeast comes of age. Eukaryot. Cell. 2014;13:1256–1265. doi: 10.1128/EC.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker E., Wang B., Bellora N., Peris D., Hulfachor A.B., Koshalek J.A., Adams M., Libkind D., Hittinger C.T. The Genome Sequence of Saccharomyces eubayanus and the Domestication of Lager-Brewing Yeasts. Mol. Biol. Evol. 2015;32:2818–2831. doi: 10.1093/molbev/msv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson B., Liti G. Saccharomyces pastorianus: Genomic insights inspiring innovation for industry. Yeast. 2015;32:17–27. doi: 10.1002/yea.3033. [DOI] [PubMed] [Google Scholar]
- 6.Okuno M., Kajitani R., Ryusui R., Morimoto H., Kodama Y., Itoh T. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 2016;23:67–80. doi: 10.1093/dnares/dsv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monerawela C., Bond U. Brewing up a storm: The genomes of lager yeasts and how they evolved. Biotechnol. Adv. 2017;35:512–519. doi: 10.1016/j.biotechadv.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Monerawela C., Bond U. The hybrid genomes of Saccharomyces pastorianus: A current perspective. Yeast. 2018;35:39–50. doi: 10.1002/yea.3250. [DOI] [PubMed] [Google Scholar]
- 9.Libkind D., Hittinger C.T., Valerio E., Goncalves C., Dover J., Johnston M., Goncalves P., Sampaio J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bing J., Han P.J., Liu W.Q., Wang Q.M., Bai F.Y. Evidence for a Far East Asian origin of lager beer yeast. Curr. Biol. 2014;24:R380–R381. doi: 10.1016/j.cub.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Baker E.P., Peris D., Moriarty R.V., Li X.C., Fay J.C., Hittinger C.T. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 2019;5:eaav1869. doi: 10.1126/sciadv.aav1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorter de Vries A.R., de Groot P.A., van den Broek M., Daran J.G. CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb. Cell Fact. 2017;16:222. doi: 10.1186/s12934-017-0835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duong C.T., Strack L., Futschik M., Katou Y., Nakao Y., Fujimura T., Shirahige K., Kodama Y., Nevoigt E. Identification of Sc-type ILV6 as a target to reduce diacetyl formation in lager brewers’ yeast. Metab. Eng. 2011;13:638–647. doi: 10.1016/j.ymben.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Murakami N., Miyoshi S., Yokoyama R., Hoshida H., Akada R., Ogata T. Construction of a URA3 deletion strain from the allotetraploid bottom-fermenting yeast Saccharomyces pastorianus. Yeast. 2012;29:155–165. doi: 10.1002/yea.2897. [DOI] [PubMed] [Google Scholar]
- 15.Bolat I., Romagnoli G., Zhu F., Pronk J.T., Daran J.M. Functional analysis and transcriptional regulation of two orthologs of ARO10, encoding broad-substrate-specificity 2-oxo-acid decarboxylases, in the brewing yeast Saccharomyces pastorianus CBS1483. FEMS Yeast Res. 2013;13:505–517. doi: 10.1111/1567-1364.12051. [DOI] [PubMed] [Google Scholar]
- 16.Wendland J. PCR-based methods facilitate targeted gene manipulations and cloning procedures. Curr. Genet. 2003;44:115–123. doi: 10.1007/s00294-003-0436-x. [DOI] [PubMed] [Google Scholar]
- 17.Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 18.Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 20.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D., et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzmin E., VanderSluis B., Wang W., Tan G., Deshpande R., Chen Y., Usaj M., Balint A., Mattiazzi Usaj M., van Leeuwen J., et al. Systematic analysis of complex genetic interactions. Science. 2018;360:eaao1729. doi: 10.1126/science.aao1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Gerami-Nejad M., Berman J., Gale C.A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- 25.Gerami-Nejad M., Hausauer D., McClellan M., Berman J., Gale C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast. 2004;21:429–436. doi: 10.1002/yea.1080. [DOI] [PubMed] [Google Scholar]
- 26.Gerami-Nejad M., Dulmage K., Berman J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast. 2009;26:399–406. doi: 10.1002/yea.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gola S., Martin R., Walther A., Dunkler A., Wendland J. New modules for PCR-based gene targeting in Candida albicans: Rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- 28.Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 2000;242:381–391. doi: 10.1016/S0378-1119(99)00509-0. [DOI] [PubMed] [Google Scholar]
- 29.Amelina H., Moiseeva V., Collopy L.C., Pearson S.R., Armstrong C.A., Tomita K. Sequential and counter-selectable cassettes for fission yeast. BMC Biotechnol. 2016;16:76. doi: 10.1186/s12896-016-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kooistra R., Hooykaas P.J., Steensma H.Y. Efficient gene targeting in Kluyveromyces lactis. Yeast. 2004;21:781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- 31.Wesolowski-Louvel M. An efficient method to optimize Kluyveromyces lactis gene targeting. FEMS Yeast Res. 2011;11:509–513. doi: 10.1111/j.1567-1364.2011.00741.x. [DOI] [PubMed] [Google Scholar]
- 32.Kretzschmar A., Otto C., Holz M., Werner S., Hubner L., Barth G. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 2013;59:63–72. doi: 10.1007/s00294-013-0389-7. [DOI] [PubMed] [Google Scholar]
- 33.Ninomiya Y., Suzuki K., Ishii C., Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidhu Y.S., Cairns T.C., Chaudhari Y.K., Usher J., Talbot N.J., Studholme D.J., Csukai M., Haynes K. Exploitation of sulfonylurea resistance marker and non-homologous end joining mutants for functional analysis in Zymoseptoria tritici. Fungal Genet. Biol. 2015;79:102–109. doi: 10.1016/j.fgb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arras S.D., Fraser J.A. Chemical Inhibitors of Non-Homologous End Joining Increase Targeted Construct Integration in Cryptococcus neoformans. PLoS ONE. 2016;11:e0163049. doi: 10.1371/journal.pone.0163049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanez R.J., Porter A.C. Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 1999;6:1282–1290. doi: 10.1038/sj.gt.3300945. [DOI] [PubMed] [Google Scholar]
- 37.Shcherbakova O.G., Lanzov V.A., Ogawa H., Filatov M.V. Overexpression of bacterial RecA protein stimulates homologous recombination in somatic mammalian cells. Mutat. Res. 2000;459:65–71. doi: 10.1016/S0921-8777(99)00059-2. [DOI] [PubMed] [Google Scholar]
- 38.Liu L., Maguire K.K., Kmiec E.B. Genetic re-engineering of Saccharomyces cerevisiae RAD51 leads to a significant increase in the frequency of gene repair in vivo. Nucleic Acids Res. 2004;32:2093–2101. doi: 10.1093/nar/gkh506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J.R., Register E., Curotto J., Kurtz M., Kelly R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast. 1998;14:565–571. doi: 10.1002/(SICI)1097-0061(19980430)14:6<565::AID-YEA251>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Suga M., Hatakeyama T. High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast. 2001;18:1015–1021. doi: 10.1002/yea.753. [DOI] [PubMed] [Google Scholar]
- 41.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunkler A., Wendland J. Use of MET3 promoters for regulated gene expression in Ashbya gossypii. Curr. Genet. 2007;52:1–10. doi: 10.1007/s00294-007-0134-1. [DOI] [PubMed] [Google Scholar]
- 43.Junker K., Chailyan A., Hesselbart A., Forster J., Wendland J. Multi-omics characterization of the necrotrophic mycoparasite Saccharomycopsis schoenii. PLoS Pathog. 2019 doi: 10.1371/journal.ppat.1007692. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 45.Walther A., Wendland J. PCR-based gene targeting in Candida albicans. Nat. Protoc. 2008;3:1414–1421. doi: 10.1038/nprot.2008.137. [DOI] [PubMed] [Google Scholar]
- 46.Carroll E., Trinh T.N., Son H., Lee Y.W., Seo J.A. Comprehensive analysis of fungal diversity and enzyme activity in nuruk, a Korean fermenting starter, for acquiring useful fungi. J. Microbiol. 2017;55:357–365. doi: 10.1007/s12275-017-7114-z. [DOI] [PubMed] [Google Scholar]
- 47.Farh M.E., Cho Y., Lim J.Y., Seo J.A. A diversity study of Saccharomycopsis fibuligera in rice wine starter nuruk, reveals the evolutionary process associated with its interspecies hybrid. J. Microbiol. 2017;55:337–343. doi: 10.1007/s12275-017-7115-y. [DOI] [PubMed] [Google Scholar]
- 48.Uhl M.A., Johnson A.D. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- 49.Greig D. Reproductive isolation in Saccharomyces. Heredity. 2009;102:39–44. doi: 10.1038/hdy.2008.73. [DOI] [PubMed] [Google Scholar]
- 50.Sipiczki M. Yeast two- and three-species hybrids and high-sugar fermentation. Microb. Biotechnol. 2019 doi: 10.1111/1751-7915.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leducq J.B., Charron G., Diss G., Gagnon-Arsenault I., Dube A.K., Landry C.R. Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genet. 2012;8:e1003161. doi: 10.1371/journal.pgen.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piatkowska E.M., Naseeb S., Knight D., Delneri D. Chimeric protein complexes in hybrid species generate novel phenotypes. PLoS Genet. 2013;9:e1003836. doi: 10.1371/journal.pgen.1003836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfliegler W.P., Antunovics Z., Sipiczki M. Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res. 2012;12:703–718. doi: 10.1111/j.1567-1364.2012.00820.x. [DOI] [PubMed] [Google Scholar]
- 54.Barnes G., Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polotnianka R.M., Li J., Lustig A.J. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 1998;8:831–834. doi: 10.1016/S0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 56.Mertens S., Gallone B., Steensels J., Herrera-Malaver B., Cortebeek J., Nolmans R., Saels V., Vyas V.K., Verstrepen K.J. Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts. PLoS ONE. 2019;14:e0209124. doi: 10.1371/journal.pone.0209124. [DOI] [PMC free article] [PubMed] [Google Scholar]