Summary
Hydroxylation of steroids has acquired special relevance for the pharmaceutical industries. Particularly, the 11β‐hydroxylation of steroids is a reaction of biotechnological importance currently carried out at industrial scale by the fungus Cochliobolus lunatus. In this work, we have identified the genes encoding the cytochrome CYP103168 and the reductase CPR64795 of C. lunatus responsible for the 11β‐hydroxylase activity in this fungus, which is the key step for the preparative synthesis of cortisol in industry. A recombinant Corynebacterium glutamicum strain harbouring a plasmid expressing both genes forming a synthetic bacterial operon was able to 11β‐hydroxylate several steroids as substrates. This is a new example to show that the industrial strain C. glutamicum can be used as a suitable chassis to perform steroid biotransformation expressing eukaryotic cytochromes.
Introduction
Microbial steroid transformation has been used for years as a powerful tool to generate novel steroidal drugs and key synthons of pharmaceutical interest (Donova and Egorova, 2012). Many of these microbial transformations are carried out by P450 cytochromes (CYP) (Bernhardt, 2006). The reactions carried out by CYPs are extremely diverse and contribute to the biotransformation of drugs, the bioconversion of xenobiotics and the biosynthesis of physiologically important compounds such as steroids among others (Ortiz de Montellano and De Voss, 2002; Hannemann et al., 2006). Cytochromes are hemoproteins encoded by a superfamily of genes nearly ubiquitously distributed in many different organisms from all biological kingdoms but especially in fungi, where some fungal genomes have over 150 CYPs, representing over 1% of all genes (Doddapaneni et al., 2005). Although fungal CYPs have been used for different steroid biotransformations, hydroxylation is one of the most studied and used processes at industrial scale (Zakelj‐Mavric and Belic, 1987; Vita et al., 1994; Fernandes et al., 2003; Kristan and Rižner, 2012). Fungal steroid hydroxylation is usually carried out by two‐component systems consisting of a NAD(P)H‐cytochrome P450 reductase (CPR) and a cytochrome P450 monooxygenase which are frequently attached to the membrane of the endoplasmic reticulum (Kristan and Rižner, 2012). Hydroxylation performed by fungi has acquired special relevance for the pharmaceutical industries. Among them, the hydroxyl group in position 11β of the cortisol molecule and its synthetic derivatives are the key functionalization that provides their glucocorticoid effects (Mahato and Garai, 1997; Schiffer et al., 2015). Currently, the 11β‐hydroxylation in the industrial production of corticosteroid precursors is catalysed by fungal cultures of Cochliobolus lunatus (Paraszkiewicz and Długon, 1998; Lu et al., 2007). Moreover, 11β‐hydroxylation of progesterone is another reaction of biotechnological importance carried out by C. lunatus (anamorph Curvularia lunata) (Vita et al., 1994). Although there are many data concerning the CYP encoding genes responsible for 11β‐hydroxylation in mammals (Estabrook, 2005), so far, none of the fungal genes encoding the CYPs having the 11β‐hydroxylating activity have been identified and cloned. Only the 11β‐hydroxylating CYP from C. lunatus has been biochemically characterized (Zuidweg, 1968; Janig et al., 1992; Suzuki et al., 1993), but all attempts to identify the C. lunatus 11β‐hydroxylase coding gene have failed (Berne et al., 2008). The identification and cloning of this gene are fundamental to develop more efficient biotransformation processes since, apart from its limited regioselectivity, 11β‐hydroxylation is generally accompanied by 14α‐ and 7α‐hydroxylations (Janig et al., 1992; Vita et al., 1994), and the fungus produces other steroid derivatives. The recombinant expression of these CYP systems in heterologous hosts appears as a promising alternative for the development of cleaner and more efficient introduction of the 11β‐hydroxyl group. However, only few examples of steroid hydroxylation have been described using recombinant heterologous hosts. For example, Schizosaccharomyces pombe has been used as host in the bioconversion of 11‐deoxycorticosterone to aldosterone and deoxycorticosterone (DOC) to hydroxycortisone using the human mitochondrial cytochromes CYP11B2 and CYP11B1 respectively (Bureik et al., 2002; Drǎgan et al., 2005; Hakki et al., 2008). The 11α‐hydroxylase CYPs from Aspergillus ochraceus and Rhizopus oryzae together with their corresponding oxidoreductases have been expressed in Spodoptera frugiperda (Sf‐9) insect cells (Bolten et al., 2006) and in S. pombe (Bernhardt et al., 2009) respectively. Recently, the 11α‐hydroxylase from the fungus Absidia coerulea has been identified and expressed in Pichia cells (Wang et al., 2017). Although the human 11α‐ or 11β‐hydroxylating CYPs have been expressed in bacteria (Durairaj et al., 2016), to the best of our knowledge, the fungal 11α‐ or 11β‐hydroxylating CYPs have not been expressed in bacteria yet.
In this sense, we propose here to use the bacterium Corynebacterium glutamicum as a host, because it been widely used for industrial purposes, and the publication of its complete genome, (Ikeda and Nakagawa, 2003; Kalinowski et al., 2003) has provided the basis for an enormous progress in the use of this microorganism for other biotechnological applications placing it as an ideal chassis for cell factories (De Lorenzo, 2015). Moreover, we have demonstrated that a recombinant strain of C. glutamicum carrying the 3β‐hydroxysteroid dehydrogenase of Mycobacterium smegmatis was able to catalyse the biotransformation of short chain (C19 and C21) steroids indicating that these compounds are efficiently transported across the cytoplasmic membrane by C. glutamicum (Garcia‐Fernandez et al., 2017). In addition, this work showed that C. glutamicum can be used as a clean host for steroid biotransformations, because it does not introduce additional undesired side reactions on the steroids, thus reducing the contamination of the final products.
In this work, we have identified the gene encoding the CYP responsible for the 11β‐hydroxylase activity of C. lunatus among the 112 putative CYP encoding genes present in this fungus. The gene encoding its auxiliary CPR was also identified. We have cloned and expressed both genes forming a synthetic operon in C. glutamicum R31. The resulting recombinant strain was able to synthesize 11β‐hydroxysteroids from different steroid substrates.
Results
In silico analyses of the CYPome and CPRome from C. lunatus
The genes from C. lunatus that code for putative P450 cytochromes (CYPome) were extracted from its genome annotation at JGI, and 112 non‐redundant putative CYP sequences were identified containing the Pfam PF00067 and InterPro IPR002238 domains (Table S1). The 11β‐hydroxylase was purified and biochemically characterized proposing a molecular weight (MW) of 60 kDa for the CYP protein (Zuidweg, 1968; Janig et al., 1992). Therefore, the sequences in the CYPome rendering MWs lower than 50 kDa or higher than 70 kDa were excluded as 11β‐hydroxylase candidates. These criteria eliminated 20 CYPs from the list (Table S1). The other 92 CYPs candidates were compared using local Blast with 11α‐hydroxylase CYP from A. ochraceus (USP7033807). This comparison rendered eight putative CYPs with scores higher than 100, named CYP116182 (score 335, 37% identity), CYP51519 (score 215, 29% identity), CYP115117 (score 207, 27% identity), CYP103168 (score 170, 26% identity), CYP135200 (score 152, 24% identity), CYP34615 (score 114, 24% identity), CYP31052 (score 113, 23% identity) and CYP56034 (score 105, 24% identity).
Analogously, the genes from C. lunatus encoding putative CPRs (CPRome) were extracted from the genome annotation at JGI and five putative non‐redundant CPR sequences were identified containing the PF00258 (flavodoxin domain), PF00667 (FAD‐binding domain) and PF00175 (NAD binding domain) Pfam domains. Two of the selected sequences, i.e. CPR112637 and CPR152579, were type III CYPs and were discarded. The three remaining CPRs, i.e. CPR59830, CPR64795 and CPR128465, were selected for further analysis.
Identification of CYP and CPR transcripts induced by steroids in C. lunatus
Fungal CYPs are usually induced by their substrates (Undisz et al., 1992), and thus we decided to analyse by RT‐PCR the relative mRNA levels of the selected CYP and CPR encoding genes under induction conditions. C. lunatus mycelium was induced in the presence of DOC, progesterone and androst‐4‐ene‐3,17‐dione (AD) while cholic acid and β‐sitosterol were used as negative control for non‐induced conditions (Undisz et al., 1992). The genes encoding three of the selected eight putative CYPs (CYP115117, CYP34615 and CYP56034) were not expressed neither under induced or non‐induced conditions (Fig. 1A). The genes encoding the CYP116182, CYP51519 or CYP135200 could not be amplified using three different pairs of oligonucleotides neither by using gDNA or cDNA as templates, suggesting the existence of genome differences between the JGI‐sequenced strain and the strain used in our experiments (see below). However, the CYP103168 transcript and to a lesser extent the CYP31052 transcript were detected in the tested conditions (Fig. 1A). The expression of both genes was induced upon exposure to DOC, progesterone and AD compared to the transcript levels in the non‐induced conditions (no inductor or non‐inducer steroids like β‐sitosterol or cholic acid). However, the gene expression data suggest that upon induction conditions the gene coding CYP103168 is being transcribed at a higher level than CYP31052 (Fig. 1A), so we selected it as the probable candidate to perform the 11β‐hydroxylation. The induction fold was calculated by qRT‐PCR in the conditions tested. The expression of CYP103168 in the presence of DOC, progesterone and AD was upregulated 156‐, 9‐ and 7‐fold respectively (Fig. 1B). Based on this result, the CYP103168 was selected for further experiments as the most likely responsible for the 11β‐hydroxylase of C. lunatus.
Figure 1.
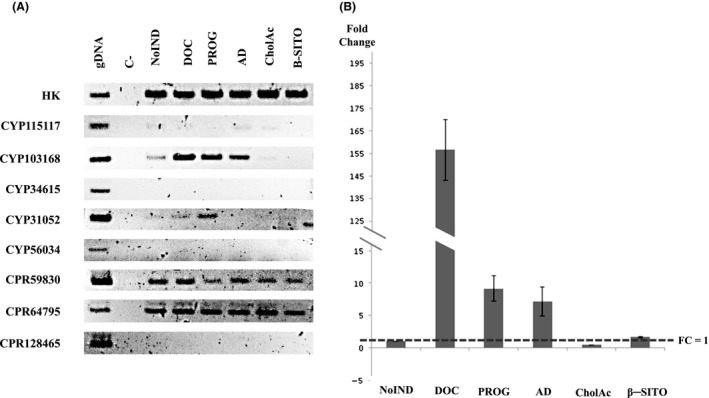
Expression profiles of selected putative CYP genes of C. lunatus exposed to various steroids: deoxycorticosterone (DOC), androstenedione (AD), progesterone (PROG), cholic acid (ChoAc) and β‐sitosterol (Β‐SITO) and C corresponds to the negative control without DNA, NoIND corresponds to the no induced condition (fungal cultures not exposed to steroid, but exposed to the solvent Tyloxapol at same concentration) and gDNA corresponds to the genomic DNA as a positive control. The glyceraldehyde 3‐phosphate dehydrogenase gene has been used as housekeeping, an internal control. A. sqRT‐PCR for CYP transcript determination. B. Fold change of CYP130168 mRNAs determined by qRT‐PCR. Error bars represent the standard deviation of three independent experiments.
Analogously, the expressions of the reductases CPR59830, CPR64795 and CPR128465 were analysed by RT‐PCR under the same conditions. The gene encoding the CPR128465 was not detected, whereas the genes encoding the CPR59830 and CPR64795 were expressed constitutively and selected for further analysis (Fig. 1A).
Gene isolation and phylogenetic studies
Using the oligonucleotides listed in Table S2, we obtained by RT‐PCR the cDNA fragments encoding the CYP103168, CPR64795 from C. lunatus. The three amplicons were sequenced, and their sequences were compared to the annotated genome of C. lunatus at JGI. The cDNA sequences were not identical to the sequences of the database, and we determine a 99.4% identity for CPR64795 and a 98.8% identity for CYP103168. Such discrepancies suggest that the genome of C. lunatus m118 strain sequenced at JGI is not identical to the genome of the C. lunatus CECT2130 strain used in this work. But more important, the protein sequence of CYP103168 lacks 11 residues (ASLKEKNLSHS) when compared to the CYP sequence annotated at JGI. Remarkably, these 11 residues correspond to an intron that was erroneously annotated as an exon in the database (Fig. S1).
The CYP103168 sequence was initially compared with the 11β‐hydroxylases from A. ochraceus, R. oryzae and Homo sapiens showing identity values of 28% (96% coverage), 25% (54% coverage) and 26% (21% coverage) respectively. In addition, to establish a phylogenetic tree, we extracted 100 homologous enzymes from the GenBank database having identity values ranging from 41% to 70% (Fig. S2). The principal clade containing CYP103168 (β) shows 30 homologous sequences, 22 of them belong to the anamorph Cochliobolus genus Bipolaris. In some cases, several homologous enzymes can be detected in a single species, observing up to five homologous proteins in the case of Bipolaris zeicola 26‐R‐13.
Steroid biotransformation
To check the 11β‐hydroxylase activity of the selected CYP and CPR genes, we constructed two synthetic bacterial operons FIN (CYP103168 + CPR59830) and FAN (CYP103168 + CPR64795) and cloned them into the C. glutamicum replicating plasmid pECXK‐99E delivering pXKFIN and pXKFAN respectively (Fig. 2) (see Experimental procedures section). The ability of these recombinant strains to achieve steroid 11β‐hydroxylation was tested using resting cells in the presence of deoxycortisone (DOC) as a substrate during 96 h at 30°C, and the products were analysed by HPLC‐MS. Figure 3 shows that DOC was 11β‐hydroxylated only by the strain harbouring the pXKFAN plasmid generating two hydroxylated compounds: the 11β‐hydroxylated derivative cortisone and another unidentified monohydroxylated side product. According to its molecular mass and characteristic ion fragmentation this peak could be ascribed to the previously described 14α‐hydroxylated (Vita et al., 1994; Andryushina et al., 2013) (Fig. 3A). Interestingly, the same monohydroxylated side product was detected also when DOC hydroxylation was performed with C. lunatus mycelia (Fig 3B). The C. glutamicum (pXKFIN) recombinant strain did not render any 11β‐hydroxylated product (Fig. 3A), but a less polar monohydroxylated compound, according to its molecular mass and characteristic ion fragmentation, was detected. This compound was not detected when DOC hydroxylation was performed with C. lunatus mycelia, suggesting that the reductase CPR31502 is not responsible for 11β‐hydroxylation, and it is not the biological partner of CPR103168. The control strain C. glutamicum (pECXK‐99E) did not modify the DOC substrate (Fig 3A). These results suggested that the operon FAN contains the genes encoding the 11β‐hydroxylase activity of C. lunatus.
Figure 2.
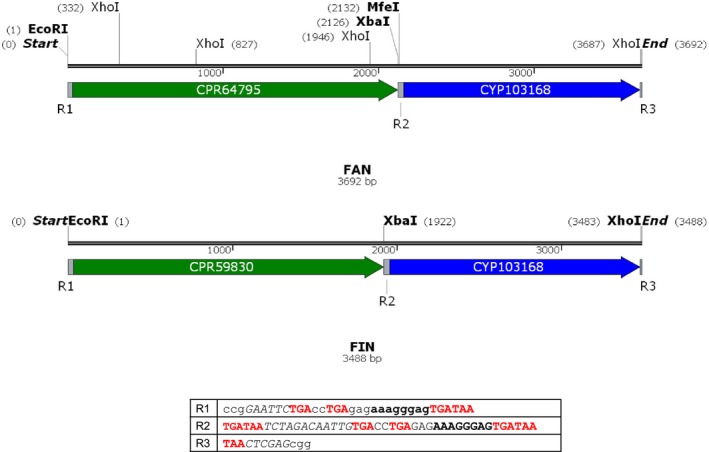
Schematic representation of the genes contained in the FAN operon and FIN operons. The sequences of the intergenic regions (R1‐R3) are indicated in the table. The sequences of the restriction sites are in cursive, and the corresponding restriction enzymes are annotated. The RBS sequences are indicated in bold.
Figure 3.
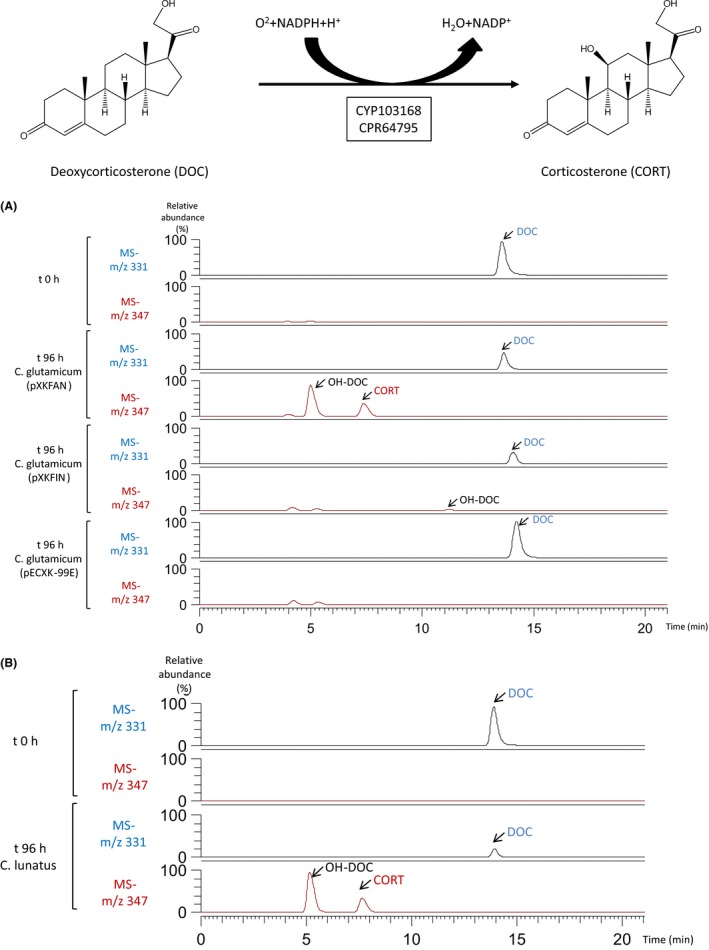
A. Deoxycorticosterone (DOC) biotransformation by C. glutamicum (pXKFAN) and C. glutamicum (pXKFIN). Mass spectra obtained from ion 331 (characteristic ion of DOC) (blue line) and mass spectra obtained from ion 347 (characteristic ion of corticosterone (CORT)) (red line). B. Deoxycorticosterone biotransformation by C. lunatus. Mass spectra obtained from ion 331 (characteristic ion of DOC) (blue line) and mass spectra obtained from ion 347 (characteristic ion of corticosterone (CORT)) (red line). All the experiments were repeated three times and a representative one was chosen.
Once the capability of C. glutamicum (pXKFAN) to hydroxylate DOC was demonstrated, we tested the ability of resting cells to hydroxylate cortexolone (RSS) finding significant amounts of the 11β‐hydroxylated product hydrocorticosterone (HC) and a small proportion of another polar compound previously described Zuidweg (1968), Kollerov et al. (2010) and Andryushina et al. (2013) (Fig. 4) . Remarkably, the recombinant C. glutamicum (pXKFAN) cells performed the transformation of RSS into hydrocortisone without the formation of significant side products. Depending on the operational conditions and the substrates used in the resting cell assays, we have obtained biotransformation yields of 6–49% that are in the range of 10–35% yield described for biotransformations carried out with C. lunatus (Manosroi et al., 2006).
Figure 4.
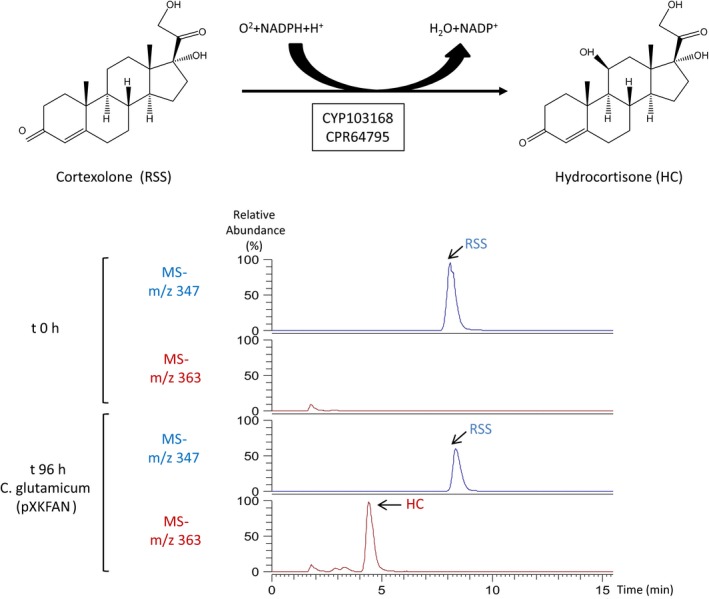
Cortexolone biotransformation by C. glutamicum (pXKFAN). Mass spectra obtained from ion 347 (characteristic ion of cortexolone (Reichstein's Substance S, RSS) (blue line) and mass spectra obtained from ion 363 (characteristic ion of hydrocortisone (HC)) (red line).
Discussion
Biochemical studies of the 11β‐hydroxylase of C. lunatus described it as a microsomal two‐component monooxygenase system which is composed of a CYP and a NADPH‐CPR (Janig et al., 1992; Suzuki et al., 1993). The 11β‐hydroxylase can be specifically induced by some steroids (Undisz et al., 1992). On the other hand, two CPRs named CPR1 (CPR64795) and CPR2 (CPR128465) had been previously characterized in C. lunatus (Lah et al., 2008, 2011) and both enzymes have been expressed in Escherichia coli and reconstituted in vitro with the CYP53A15 of C. lunatus having a benzoate 4‐hydroxylase activity. In spite of having this information, previous attempts to identify the CYP responsible gene for 11β‐hydroxylase in C. lunatus by differential expression were unsuccessful (Berne et al., 2008). Therefore, we started a different approach taking advantage of the genome sequence of C. lunatus published at JGI (Gao et al., 2014).
An in silico analysis of the CYPome from C. lunatus rendered 8 CYPs as the best candidates to represent the 11β‐hydroxylase of this fungus. The transcriptomic analyses suggested that only two out of the eight genes selected were inducible by DOC, progesterone and AD and probably CYP103168 that is expressed at higher level could encode the true 11β‐hydroxylase protein. A similar analysis carried out with the CYPome of R. oryzae showed that only three out of the 48 putative CYPs of R. oryzae were induced in the presence of progesterone, and only one of them turned to be the CYP with 11α‐hydroxylase activity (Petric et al., 2010).
In silico analysis of the CPRome of C. lunatus rendered three CPR candidates (CPR64795, CPR128465 and CPR59830). Two of them corresponding to the CPR1 (CPR64795) and CPR2 (CPR128465) have been previously identified (Lah et al., 2008, 2011), but the CPR3 (CPR59830) was a novel candidate. The transcriptomic analyses showed that CPR2 was not expressed in the tested conditions, whereas CPR1 and CPR3 were expressed constitutively.
The NAD(P)H‐cytochrome P450 reductase from C. lunatus has been purified and biochemically characterized proposing a molecular weight (MW) of 80 kDa for the CPR protein (Janig et al., 1992; Suzuki et al., 1993). The annotated sequence of the two expressed CPRs from C. lunatus (i.e. CPR3 and CPR1) rendered a 71.3 kDa protein for CPR59830 and 77.3 kDa for CPR64795. The latter was shown to be functional in vitro (Lah et al., 2011).
The in silico analysis pointed out that the most probable candidates to encode the 11β‐hydroxylase system of C. lunatus were the CYP103168, in combination with CPR64795 or CPR59830 as the redox partner.
The phylogenetic analysis classifies CYP103168 as a member of the β group (Fig. S1). The 30 sequences of this group might potentially possess 11‐β‐hydroxylase activity; however, the other proteins in the tree, included those contained in the α cluster, are grouped too far from CYP103168 and their biochemical characteristics cannot be anticipated. The presence of 22 sequences belonging to the genus Bipolaris in the β group could indicate that 11‐β‐hydroxylase activity is a specific acquisition of the Cochliobolus genus including their anamorphs.
In the last decade, significant progress was made in the field of heterologous expression of steroidogenic CYP systems in microorganisms allowing generation of strains capable of hydroxylating steroids at specific positions. Successful heterologous expression of mammalian and fungal CYPs have been accomplished using both yeasts and bacteria as hosts (Ikeda et al., 2003; Szczebara et al., 2003; Kolar et al., 2007; Hakki et al., 2008; Petric et al., 2010; Ichinose and Wariishi, 2013). Heterologous expression of membrane‐bound eukaryotic CYPs in bacteria is difficult since eukaryotic and prokaryotic membranes have not the same composition, and the appropriate insertion of the protein in the membrane and its expression requires to perform different strategies, such as for instance a precise modification of the amino‐terminal region of the enzyme to achieve maximal protein expression and optimal catalytic activity (Gonzalez and Korzekwa, 1995; Shet et al., 1997; Sakaki et al., 1999; Ichinose and Wariishi, 2013). On the other hand, there are several studies in which yeasts have been used as hosts for the biotransformation of steroids (Szczebara et al., 2003), but heterologous expression of fungal or mammalian genes in yeasts present some disadvantages like the unexpected formation of by‐products by unexpected side reactions, different glycosylation of the proteins, the difficulty to predict the plasma membrane permeability for a given compound, and the unpredictable targeting of recombinant proteins (Dumas et al., 2006).
Actinobacteria are widely used in steroid industry but to the best of our knowledge, these bacteria have not been used as hosts for CYPs expression. We selected C. glutamicum as a bacterial chassis to express the 11β‐hydroxylase system of C. lunatus for several reasons: (i) this bacterium contains only two CYPs within its genome (Kabus et al., 2007); (ii) the biochemical pathways for sterol/steroid biodegradation are missing (Kalinowski et al., 2003); (iii) it is a robust strain well tested at industrial scale (Becker and Wittmann, 2012; De Lorenzo, 2015; Unthan et al., 2015); (iv) a recombinant C. glutamicum strain has been successfully tested for steroid biotransformation with satisfactory results in a recent publication (Garcia‐Fernandez et al., 2017). Recombinant C. glutamicum (pXKFAN) and C. glutamicum (pXKFIN) strains were constructed and only C. glutamicum (pXKFAN) resting cells containing CYP103168 and CPR64795 genes rendered corticosterone (11β‐hydroxylated DOC) when DOC was used as a substrate. This result suggests that CPR59830 is not part of the 11β‐hydroxylase system of C. lunatus. In addition to the peak corresponding to corticosterone, we have identified product with 347 as characteristic ion more polar than corticosterone that can be ascribed to 14α‐hydroxydeoxycortisone. In this sense, it is known that the purified 11β‐hydroxylase from C. lunatus has bifunctional properties rendering in vitro two hydroxylated derivatives in the presence of different substrates identified as 14α‐hydroxysteroids and 11β‐hydroxysteroids (Suzuki et al., 1993). However, C. glutamicum (pXKFAN) resting cells in the presence of RSS substrate render mainly the 11β‐hydroxylated derivative hydrocortisone. This result is interesting since the differences in the ratio of the 11β‐ and 14α‐hydroxylated compounds observed in vitro varies from 0.5 to 2 depending on the substrate used (Suzuki et al., 1993). In this sense, biotransformations of RSS performed by C. lunatus rendered multiple hydroxylated compounds (e.g. at position 6β, 11α, 14α and other unidentified monohydroxylated compounds (Lu et al., 2007). This result suggests that the enzyme has a different behaviour in vitro than in vivo, at least in C. glutamicum, most probably due to the specific conditions provided by the surrounding cytoplasmic environment. This could be fundamental for the industrial purposes since the efficiency of the 11β‐hydroxylation process and the purity of the final 11β‐hydroxylated steroid might depend on the host used to express the CYP‐CRP system.
Here, we showed a successful heterologous expression of CYP103168 of C. lunatus in C. glutamicum with a minimal N‐terminal sequence modification. Further, modifications carried out by protein engineering can now be made to improve the specificity of the enzyme and the process yield. Our results demonstrated that C. glutamicum constitutes an excellent chassis to achieve steroid biotransformations (Garcia‐Fernandez et al., 2017).
Experimental procedures
Chemicals
Deoxycortisone (DOC) and corticosterone were purchased from Sigma‐Aldrich (Darmstadt, Germany). Cortexolone (RSS), and hydrocortisone was provided from Gadea BioPharma (León, Spain).
Strains, oligonucleotides and culture growth
The strains and plasmids used in this study are listed in Table 1. E. coli was cultured in LB medium at 37°C. C. glutamicum R31 was cultured on Tryptic Soy Agar (TSA) at 30°C in agitation at 250 rpm. The filamentous fungus C. lunatus CECT 2130 strain was obtained from the Spanish Collection of Type Culture and was cultured on Potato Dextrose Agar (PDA). C. lunatus strain was also cultured in agitation at 250 rpm and 30°C in M7 broth containing yeast extract (1 g l−1), glucose (10 g l−1), (NH4)2C4H4O6 (2 g l−1), KH2PO4 (1 g l−1), MgSO4 7H20 (0.50 g l−1), KCl (0.50 g l−1), Na2B4O7 10H2O (0.10 mg l−1), CuSO4 5H2O (0.01 mg l−1), FeSO4 7H2O (0.05 mg l−1), MnSO4 4H2O (0.01 mg l−1), ZnSO4 7H2O (0.07 mg l−1) and (NH4)2MoO4 4H2O (0.01 mg l−1) at pH 5.1. Antibiotics were used where indicated at the following concentrations: kanamycin (50 μg ml−1 for E. coli strains and 25 μg ml−1 for C. glutamicum strains). The oligonucleotides used in this work are listed in Table 2.
Table 1.
Bacterial and fungal strains and plasmids used in this study
| Strains | Genotype | References |
|---|---|---|
| Escherichia coli DH10B | F−, mcrA, Δ(mrr‐hsdRMS‐mcrBC), f80ΔlacZDM15 ΔlacX74, deoR, recA1, endA1, araD139, Δ(ara,leu)7697, galU, galK, rpsL, nupG, λ − | Invitrogen |
| Corynebacterium glutamicum R31 | MeLisR, AecR transformation efficient | Santamaria et al. (1985) |
| Cochliobolus lunatus | Type strain | CECT 2130 |
| Plasmids | ||
| pECXK‐99E | KmR, Escherichia and Corynebacterium replication | Kirchner and Tauch (2003) |
| pXKFAN | KmR, FAN operon into pECXK‐99E | This work |
| pXKFIN | KmR, FIN operon into pECXK‐99E | This work |
Table 2.
Oligonucleotides used in this study
| Primer name | Sequence |
|---|---|
| GAPDH F (HK F) | GACGGCAACAACCTGACT |
| GAPDH R (HK R) | CAGTGCTGCTGGGAATGA |
| 116182 F | GAGACCTTGAAACCTTCAACTGG |
| 116182 R | GCATTCACACAGCGTGATGG |
| 51519 F | CAACTCAATTCCCCATCTTCC |
| 51519 R | AGTCCTCCATAGAGGATCTCTCG |
| 115117 F | ATTCACTTATGGACGGCTCTAGC |
| 115117 R | GAAATCTTGTCGAACTAGCTCTCG |
| 103168 F | GGACCGAAGTCAACATCAACG |
| 103168 R | GTGCTTCTCGCGTGCACG |
| 135200 F | CCAATTGTGAAGACTGGACACC |
| 135200 R | CGTCTCTCTTCTCGCCTTGG |
| 34615 F | GTTGTCATACCGCCAAGTCG |
| 34615 R | GCTTAATCCAATTCTCTGTGTCG |
| 31052 F | CAACGCAGAGCGAGACTATCC |
| 31052 R | CACAGAAGGCTCCATTACTTGC |
| 56034 F | GAGCGAGCTTCATCATCTTACC |
| 56034 R | TTCGTTCAATGCGGAGAGC |
| 64795 F | GCACAAGCTCGAAGAGAACG |
| 64795 R | TTCCTGGTATTGGTTCGAAGC |
| 128465 F | GAGGAATTGGAAATAGTGACAGC |
| 128465 R | GACATCAGCCCTCCACTTCC |
| 59830 F | GTCCAAGATCTCCTTCGACAGC |
| 59830 R | CCATGTTTCTTGTCTATACCGTCC |
| 116182 F2 | GCATTCGGTTCCTCGTTCC |
| 116182 R2 | GCAATGAGGCAGGATCATAGC |
| 116182 F3 | CACTTTGATATTGCTTGCCACC |
| 116182 R3 | ACCTTCTTCGTTCCGGATAGC |
| 51519 F2 | GTCATCGATCCAATCGTACAGG |
| 51519 R2 | TTCTCATGATCGCAGATATCAGC |
| 51519 F3 | TGATATGATTAGCTGGGTTGACG |
| 51519 R3 | CCTTCATCGCACTATCAAGTAGC |
| 135200 F2 | GTCATCGCTCAGATTATTCAAGC |
| 135200 R2 | CGTATCCTTCACGATAGAGTGC |
| 135200 F3 | TGAATTGCTCACGACTATTGTGG |
| 135200 R3 | GGAGGCGAGCTTCTACAACC |
| 128465 F2 | GCAAGAAAGTCGGTCGCATT |
| 128465 R2 | CCAACATCTCCAATACTTGATCC |
| 128465 F3 | CACTGTTACCGTTCTCGTGG |
| 128465 R3 | GCAGCAATGAGAATGAGTGG |
| 103168 XhoR | CCGCTCGAGTTACTACACTACCACTCTCTTGAAAGC |
| 103168 BglIIXbaIMunIF | gaAGATCTTCTAGACAATTGTGACCTGAGAGAAAGGGAGTGATAAATGGATCCCCAGACTGTCG |
| 64795 EcoRIF | ccgGAATTCTgacctgagagaaagggagtgataaATGGCACAACTCGACACGC |
| 64795 XbaIR | GCTCTAGATTATCATGACCAGACGTCTTCCTG |
| 64795 F2 | AATCAGCATTGCTGGCTCC |
| 64795 F3 | CTCCAACTTCAAGCTTCCTTCG |
| 59830 F2 | GGTATTGATGGCTCGTTCCTCC |
| 59830 F3 | CTCTACGACTACACAACACGTCC |
| 64795 F4 | AATACGTCGCTTTCGGTCTCG |
| pXK6906 F | CGACATCATAACGGTTCTGG |
| pXK118 R | TTTATCAGACCGCTTCTGC |
Bioinformatic tools
Putative CYP and CPR sequences from C. lunatus genome were extracted from MycoCosm resource at JGI Genome Portal (Gao et al., 2014; Nordberg et al., 2014). Annotation provided by JGI database was used to extract those cytochrome protein sequences containing the corresponding Pfam and InterPro families (PF00067 and IPR002238 respectively) from C. lunatus genome (JGI Project ID: 403758). Annotation provided by JGI database was used to extract those CPR protein sequences containing PF00258 (flavodoxin domain), PF00667 (FAD‐binding domain) and PF00175 (NAD binding domain) Pfam domains.
Multiple alignments of protein were carried out with the MUSCLE server program at the European Bioinformatics Institute (EBI) (http://www.ebi.ac.uk/Tools/msa/muscle/). The theoretical molecular masses of the proteins were calculated using the program Compute pI/Mw of ExPASy server (http://www.expasy.org/tools/pi_tool.html). Phylogenetic studies were completed using Phylogeny Fr web tool (Kumar et al., 2016). Workflow settings were established performing a multiple alignments using MUSCLE and an alignment curation with Gblocks. Maximum likelihood phylogenetic tree was constructed using a bootstrapping procedure (100 bootstrap replicates). Appropriated substitution model of the alignment was obtained using ProtTest (Darriba et al., 2011) being LG+I+G the best result with a gamma shape of 1.12 and a proportion of 0.01 invariable sites. The resultant phylogenetic tree was visualized using mega 7.0 (Kumar et al., 2016) and figtree 1.4.2 (Rambaut) software.
Isolation of DNA from C. lunatus
The fungal spores were harvested and resuspended in breaking buffer containing 10 mM Tris‐HCl, 10% (v/v) Triton X‐100, 1% (v/v) SDS, 10 mM NaCl and 1 mM EDTA at pH 8. The mixture was shaken at 350 rpm at 70°C with 0.4 mm glass beads. The solution was gently mixed with a volume of phenol:chloroform:isoamyl alcohol (25:24:1), and the aqueous phase was used as template.
Isolation of RNA from C. lunatus. RT‐PCR and qRT‐PCR analyses
Four‐day‐old C. lunatus mycelium cultured in M7 medium was induced with 300 μM of the selected steroid (DOC, AD, progesterone, cholic acid or β‐sitosterol) previously solubilized in 10% (v/v) Tyloxapol. The mycelium was harvested after 40 min by filtration and washed with a solution containing 0.85% (v/v) of NaCl containing 0.05% (v/v) Tween 80 and stored at −80°C until RNA extraction. The frozen mycelium was harvested and homogenized by mechanic disruption using liquid nitrogen. RNA purification was carried out using TRIzol Reagent DNA‐freeTM DNA Removal Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Extracted RNA was treated with RNAse‐free DNAse and the Removal Treatment kit (Invitrogen) following manufacturer's instructions. RNA integrity was checked by agarose gel electrophoresis. The absence of contaminating DNA was analysed by RT‐PCR using specific primers for amplifying the housekeeping gene of C. lunatus encoding the glyceraldehyde 3‐phosphate dehydrogenase as described below.
For RT‐PCR and qRT‐PCR experiments, cDNA was synthesized from 1 μg of purified RNA in poly‐A primers reactions with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basilea, Switzerland). RT‐PCR amplification of the putative CYP and CPR genes was carried out in a total volume of 50 μl using 1 μl of the reverse transcription reactions and 1 U of Taq polymerase (Biotools, Madrid, Spain) and 0.5 μM of gene‐specific primers (Table 2) under the following conditions: denaturation (94°C, 4 min), 25 cycles consisting of denaturation (94°C, 30 s), annealing (55°C, 30 s) and extension (72°C, 30 s), with a final extended extension (72°C, 7 min). The amounts of PCR amplicons were visualized by electrophoresis in a 0.7% (v/v) agarose gel.
Gene expressions analyses were performed by a two‐step RT‐qPCR approach using SYBR Green I dye in a LightCycler 480 II Roche®. After retrotranscription, PCR reactions were carried out in 96‐well plates in a final volume of 20 μl containing: 1 μl of transcribed cDNA, 0.25 μM of each forward and reverse primer and 10 μl of SYBR Green Master Mix (FastStart Taq DNA Polymerase, reaction buffer, dNTP mix, SYBER Green I dye and 8 mM MgCl2). Cycling was performed as follows: pre‐incubation at 95°C for 5 min followed by 55 cycles of 10 s at 95°C, 10 s at 60°C and 30 s at 72°C. After thermo cycling, a melting curve was made to verify the specificity of the amplified PCR product. The sequences of the primers used for this study are listed in Table 2. The analysis was performed in three technical replicates from three biological samples. The results were analysed using the method (Livak and Schmittgen, 2001) with a reference gene previously identified (Liu et al., 2014). Fold change is expressed as a range, which is a result of incorporating the standard deviation of the ΔΔC T value into the fold change calculation, using mRNA levels of the gene encoding the glyceraldehyde 3‐phosphate dehydrogenase of C. lunatus used as housekeeping control gene (Liu et al., 2014).
Construction of the bacterial synthetic FAN (CYP103168‐CPR64765) and FIN (CYP103168‐CPR59830) operons
The full‐length cDNA of CYP103168, CPR64795 and CPR59830 encoding genes was PCR amplified from the total cDNA of C. lunatus obtained after deoxycorticosterone induction. To achieve optimal translation in the host bacterium, the PCR primers contained a Shine Dalgarno sequence (AAAGGGAG) upstream of each gene at 6 bp from the respective start codons. Restriction enzyme sites were also added to the primers to clone the genes into the plasmid vector. Primers used to perform these amplifications are listed in Table S2. The amplicons were sequenced to determine the amino acid compositions of the enzymes. The isolated DNA fragments containing the CPR64795 and CPR59830 encoding gene were further digested with EcoRI and XbaI, whereas the fragment containing the CYP103168 encoding gene was digested with XbaI and XhoI. These fragments were isolated and ligated to the pECXK‐99E shuttle E. coli/C. glutamicum plasmid previously digested with EcoRI and SalI. The ligation delivers the pXKFAN and pXKFIN plasmids carrying the CPR64795 + CYP103168 and CPR59830 + CYP103168 encoding genes forming the synthetic operons which were named as FAN and FIN respectively. The resulting plasmids were transformed into E. coli for its isolation and characterization (Fig. 2). The genes cloned into the pXKFAN and pXKFIN plasmids were sequenced to confirm the accuracy of the construction. Each isolated pXKFAN and pXKFIN plasmids were further electroporated into C. glutamicum R31 competent cells to generate the recombinant C. glutamicum (pXKFAN) and C. glutamicum (pXKFIN) strains respectively.
Steroid biotransformation assay
Recombinant C. glutamicum (pXKFAN) and C. glutamicum (pXKFIN) cells were grown in 200 ml of TSB containing kanamycin and 0.5 mM δ‐aminolevulinic acid at 30°C and 250 rpm. The FAN and FIN operons were induced with 1 mM IPTG when OD600 reached 1.5, during 25 h, and then the cells were harvested by centrifugation (10 min at 4000 g) and washed twice with 0.85% (v/v) NaCl. For the biotransformation assay, C. glutamicum cells were resuspended in 50 mM potassium phosphate buffer (pH 7.4) and the steroid was added at a final concentration of 0.5 mM, from a 5 mM stock prepared in 10% (v/v) of Tyloxapol. Aliquots of 200 μl were taken to analyse the biotransformation of steroids at 96 h.
HPLC‐DAD‐MS analysis
Aliquots of 10 μl of 5 mM testosterone in 10% (v/v) Tyloxapol were added to each 0.2 ml sample taken from the biotransformation experiments prior to extraction with chloroform as an internal standard (ISTD). The samples were extracted using two volumes of chloroform. The aqueous fraction was discarded, and the chloroform fraction was dried at 60°C using a Thermoblock and then dissolved in 0.5 ml of acetonitrile. Each sample (25 μl) was subjected to chromatographic analysis by HPLC‐DAD‐MS. Experiments were carried out using a DAD detector and a LXQ Ion Trap Mass Spectrometer, equipped with an atmospheric pressure chemical ionization source and interfaced to a Surveyor Plus LC system (all from Thermo Electron, San Jose, CA, USA). Data were acquired with a Surveyor Autosampler and MS Pump and analysed with the Xcalibur Software (from Thermo‐Fisher Scientific, San Jose, CA, USA). All the experiments were carried out with the following interface parameters: capillary temperature 350°C, 60°C for gas temperature in the vaporizer, capillary voltage 9 V, amplifier 400 Vp and power source 100 μA. High‐purity nitrogen was used as nebulizer, sheath and auxiliary gas. MS analysis was performed both in full scan and in selected ion monitoring (SIM) mode by scanning all the daughter ions of the products in positive ionization mode. The quantification was performed from parent mass of compounds, and the specificity was obtained by following the specific fragmentations of all compounds. Chromatographic separation was performed on a Mediterranea Sea C18 column (4.6 × 150 mm, particle size 5 mm) (Teknokroma, Barcelona, Spain). The chromatography was performed using water containing 0.1% (v/v) of formic acid and acetonitrile containing 0.1% (v/v) of formic acid as mobile phases A and B respectively (flow 1 ml min−1). Gradient was as follows: 40% B for 5 min, reaching to 80% B in 30 min, hold for 5 min and return 40% B in 5 min. The HPLC column was re‐equilibrated for 15 min in initial conditions. The valve was set to direct LC flow to the mass spectrometer from 1 to 45 min, with the remaining LC eluent diverted to waste.
Conflict of interest
None declared.
Supporting information
Fig. S1. Alignment of CYP103168 predict protein annotated in JGI database and the protein translate from the sequenced gDNA.
Fig. S2. Phylogenetic tree of CYP103168 homologous sequences. Neighbour‐joining tree shows the distances between the 100 nearest homologous sequences to CYP103168 contained in the GenBank database. H. sapiens homologous cytochrome (NP_001021384.1) was used as an outgroup (real distance to root = 7.51). Main bootstrap values (N = 100) are shown in their corresponding nodes. The two main sets of sequences have been called α and β.
Table S1. Oligonucleotides used in this study.
Table S2. CYPome of C. lunatus.
Acknowledgements
We want to thank A. Valencia for technical support. We are deeply indebted to the Protein Chemistry service of CIB, especially to E. Aporta for chromatographic technical and data interpretation support and to the Aspergillus Molecular Genetics Unit of CIB, especially to Dr. E. Espeso and Dr. M. Villarino Villarino too for fungal technical support. We want to thank to Dr. R. Santamaría for providing the plasmid pECXK‐99E and Gadea Pharma S.L. for generously providing the RSS and hydrocortisone. This work was supported by grants from the Ministry of Science and Innovation (BFU2006‐15214‐C03‐01, BFU2009‐11545‐C03‐03) and Ministry of Economy and Competitiveness (BIO2012‐39695‐C02‐01).
Microbial Biotechnology (2019) 12(5), 856–868
Funding Information This work was supported by grants from the Ministry of Science and Innovation (BFU2006‐15214‐C03‐01, BFU2009‐11545‐C03‐03) and Ministry of Economy and Competitiveness (BIO2012‐39695‐C02‐01).
References
- Andryushina, V.A. , Voishvillo, N.E. , Druzhinina, A.V. , Stytsenko, T.S. , Yaderets, V.V. , Petrosyan, M.A. , et al. (2013) 14α-Hydroxylation of steroids by mycelium of the mold fungus Curvularia lunata (VKPM F-981) to produce precursors for synthesizing new steroidal drugs. Pharm Chem J 47: 103–108. [Google Scholar]
- Becker, J. , and Wittmann, C. (2012) Bio‐based production of chemicals, materials and fuels – Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23: 631–640. [DOI] [PubMed] [Google Scholar]
- Berne, S. , Lah, L. , Korošec, B. , Kraševec, N. , and Komel, R. (2008) Progesterone‐induced gene expression profile of the filamentous fungus Cochliobolus lunatus . Acta Chim Slov 55: 93–100. [Google Scholar]
- Bernhardt, R. (2006) Cytochromes P450 as versatile biocatalysts. J Biotechnol 124: 128–145. [DOI] [PubMed] [Google Scholar]
- Bernhardt, R. , Cresnar, B. , Hakki, T. and Petric, S. (2009) Cytochrome P450 from Rhizopus oryzae and uses thereof. WO2011042143.
- Bolten, I.S.L. , Easton, A.M. , Pere, D. , Us, M.O. , Messing, D.M. , Louis, S. , et al (2006) Aspergillus ochraceus 11 alpha hydroxylase and oxidoreductase. US 70033807B2 2.
- Bureik, M. , Lisurek, M. , and Bernhardt, R. (2002) The human steroid hydroxylase CYP11B1 and CYP11B2. Biol Chem 383: 1537–1551. [DOI] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G.L. , Doallo, R. , and Posada, D. (2011) ProtTest‐HPC: fast selection of best‐fit models of protein evolution. Lect Notes Comput Sci (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) LNCS 6586: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo, V. (2015) Chassis organism from Corynebacterium glutamicum: the way towards biotechnological domestication of Corynebacteria. Biotechnol J 10: 244–245. [DOI] [PubMed] [Google Scholar]
- Doddapaneni, H. , Chakraborty, R. , and Yadav, J.S. (2005) Genome‐wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genom 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donova, M.V. , and Egorova, O.V. (2012) Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol 94: 1423–1447. [DOI] [PubMed] [Google Scholar]
- Drǎgan, C.A. , Zearo, S. , Hannemann, F. , Bernhardt, R. , and Bureik, M. (2005) Efficient conversion of 11‐deoxycortisol to cortisol (hydrocortisone) by recombinant fission yeast Schizosaccharomyces pombe . FEMS Yeast Res 5: 621–625. [DOI] [PubMed] [Google Scholar]
- Dumas, B. , Brocard‐Masson, C. , Assemat‐Lebrun, K. , and Achstetter, T. (2006) Hydrocortisone made in yeast: metabolic engineering turns a unicellular microorganism into a drug‐synthesizing factory. Biotechnol J 1: 299–307. [DOI] [PubMed] [Google Scholar]
- Durairaj, P. , Hur, J.S. , and Yun, H. (2016) Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb Cell Fact 15: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook, R.W. (2005) Steroid hydroxylations: a paradigm for cytochrome P450 catalyzed mammalian monooxygenation reactions. Biochem Biophys Res Commun 338: 290–298. [DOI] [PubMed] [Google Scholar]
- Fernandes, P. , Cruz, A. , Angelova, B. , Pinheiro, H.M. , and Cabral, J.M.S. (2003) Microbial conversion of steroid compounds: recent developments. Enzyme Microb Technol 32: 688–705. [Google Scholar]
- Gao, S. , Li, Y. , Gao, J. , Suo, Y. , Fu, K. , Li, Y. , and Chen, J. (2014) Genome sequence and virulence variation‐related transcriptome profiles of Curvularia lunata, an important maize pathogenic fungus. BMC Genom 15: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Fernandez, J. , Galan, B. , Felpeto‐Santero, C. , Barredo, J. , and Garcia, J. (2017) Production of 4‐Ene‐3‐ketosteroids in Corynebacterium glutamicum . Catalysts 316: 1–12. [Google Scholar]
- Gonzalez, F.J. , and Korzekwa, K.R. (1995) Cytochromes P450 expression systems. Annu Rev Pharmacol Toxicol 35: 369–390. [DOI] [PubMed] [Google Scholar]
- Hakki, T. , Zearo, S. , Drǎgan, C.A. , Bureik, M. , and Bernhardt, R. (2008) Coexpression of redox partners increases the hydrocortisone (cortisol) production efficiency in CYP11B1 expressing fission yeast Schizosaccharomyces pombe . J Biotechnol 133: 351–359. [DOI] [PubMed] [Google Scholar]
- Hannemann, F. , Virus, C. , and Bernhardt, R. (2006) Design of an Escherichia coli system for whole cell mediated steroid synthesis and molecular evolution of steroid hydroxylases. J Biotechnol 124: 172–181. [DOI] [PubMed] [Google Scholar]
- Ichinose, H. , and Wariishi, H. (2013) High‐level heterologous expression of fungal cytochrome P450s in Escherichia coli . Biochem Biophys Res Commun 438: 289–294. [DOI] [PubMed] [Google Scholar]
- Ikeda, M. , and Nakagawa, S. (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62: 99–109. [DOI] [PubMed] [Google Scholar]
- Ikeda, M. , Arai, M. , Okuno, T. , and Shimizu, T. (2003) TMPDB: a database of experimentally‐characterized transmembrane topologies. Nucleic Acids Res 31: 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janig, G. , Pfeil, D. , Muller‐Frohne, M. , Riemer, H. , Henning, M. , Schwarze, W. , and Ruckpaul, K. (1992) Steroid 11 beta‐hydroxylationby a fungal microsomal cytochrome P450. J Steroid Biochem Molec Biol 43: 1117–1123. [DOI] [PubMed] [Google Scholar]
- Kabus, A. , Niebisch, A. , and Bott, M. (2007) Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl Environ Microbiol 73: 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski, J. , Bathe, B. , Bartels, D. , Bischoff, N. , Bott, M. , Burkovski, A. , et al (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L‐aspartate‐derived amino acids and vitamins. J Biotechnol 104: 5–25. [DOI] [PubMed] [Google Scholar]
- Kirchner, O. , and Tauch, A. (2003) Tools for genetic engineering in the amino acid‐producing bacterium Corynebacterium glutamicum . J Biotechnol 104: 287–299. [DOI] [PubMed] [Google Scholar]
- Kolar, N.W. , Swart, A.C. , Mason, J.I. , and Swart, P. (2007) Functional expression and characterisation of human cytochrome P45017α in Pichia pastoris . J Biotechnol 129: 635–644. [DOI] [PubMed] [Google Scholar]
- Kollerov, V.V. , Shutov, A.A. , Fokina, V.V. , Sukhodol’skaya, G.V. , Gulevskaya, S.A. , and Donova, M.V. (2010) Bioconversion of C 19-and C 21-steroids with parent and mutant strains of Curvularia lunata . Appl Biochem Biotech 46: 198–205. [PubMed] [Google Scholar]
- Kristan, K. , and Rižner, T.L. (2012) Steroid‐transforming enzymes in fungi. J Steroid Biochem Mol Biol 129: 79–91. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah, L. , Kraševec, N. , Trontelj, P. , and Komel, R. (2008) High diversity and complex evolution of fungal cytochrome P450 reductase: cytochrome P450 systems. Fungal Genet Biol 45: 446–458. [DOI] [PubMed] [Google Scholar]
- Lah, L. , Podobnik, B. , Novak, M. , Korošec, B. , Berne, S. , Vogelsang, M. , et al (2011) The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol Microbiol 81: 1374–1389. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Ma, B.C. , Hou, J.M. , and Zuo, Y.H. (2014) Isolation and characterization of the PKAr gene from a plant pathogen, Curvularia lunata . Indian J Microbiol 54: 310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. , and Schmittgen, T. (2001) Analysis of relative gene expression data using real‐ time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, W. , Du, L. , Wang, M. , Jia, X. , Wen, J. , Huang, Y. , et al (2007) Optimisation of hydrocortisone production by Curvularia lunata . Appl Biochem Biotechnol 142: 17–28. [DOI] [PubMed] [Google Scholar]
- Mahato, S. B. , and Garai, S. (1997) Review advances in microbial steroid biotransformation. Steroids 62: 332–345. [DOI] [PubMed] [Google Scholar]
- Manosroi, J. , Chisti, Y. , and Manosroi, A. (2006) Biotransformation of cortexolone to hydrocortisone by molds using a rapid color development assay. Prikl Biokhim Mikrobiol 42: 547–551. [PubMed] [Google Scholar]
- Nordberg, H. , Cantor, M. , Dusheyko, S. , Hua, S. , Poliakov, A. , Shabalov, I. , et al (2014) The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42: 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de Montellano, P.R. , and De Voss, J.J. (2002) Oxidizing species in the mechanism of cytochrome P450. Nat Prod Rep 19: 477–493. [DOI] [PubMed] [Google Scholar]
- Paraszkiewicz, K. , and Długon, J. (1998) Cortexolone 11 beta‐hydroxylation in protoplasts of Curvularia lunata . J Biotechnol 65: 217–224. [Google Scholar]
- Petric, S. , Hakki, T. , and Bernhardt, R. (2010) Discovery of a steroid 11 alpha ‐hydroxylase from Rhizopus oryzae and its biotechnological application. J Bacteriol 150: 428–437. [DOI] [PubMed] [Google Scholar]
- Rambaut, A. FigTree, a graphical viewer of phylogenetic trees. URL http//tree.bio.ed.ac.uk/software/figtree
- Sakaki, T. , Sawada, N. , Nonaka, Y. , Ohyama, Y. , and Inouye, K. (1999) Metabolic studies using recombinant Escherichia coli cells producing rat mitochondrial CYP24. Eur J Biochem 262: 43–48. [DOI] [PubMed] [Google Scholar]
- Santamaria, R.I. , Gil, J.A. , and Martín, J.F. (1985) High‐frequency transformation of Brevibacterium lactofermentum protoplasts by plasmid DNA. J Bacteriol 162: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer, L. , Anderko, S. , Hobler, A. , Hannemann, F. , Kagawa, N. , and Bernhardt, R. (2015) A recombinant CYP11B1 dependent Escherichia coli biocatalyst for selective cortisol production and optimization towards a preparative scale. Microb Cell Fact 14: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shet, M.S. , Fisher, C.W. , and Estabrook, R.W. (1997) The function of recombinant cytochrome P450s in intact Escherichia coli cells: the 17 alpha‐hydroxylation of progesterone and pregnenolone by P450c17. Arch Biochem Biophys 339: 218–225. [DOI] [PubMed] [Google Scholar]
- Suzuki, K. , Sanga, K.I. , Chikaoka, Y. and Itagaki, E. (1993) Purification and properties of cytochrome P‐450 (P‐450lun) catalyzing steroid 11β‐hydroxylation in Curvularia lunata . Biochim Biophys Acta (BBA)/Protein Struct Mol 1203, 215–223. [DOI] [PubMed] [Google Scholar]
- Szczebara, F.M. , Chandelier, C. , Villeret, C. , Masurel, A. , Bourot, S. , Duport, C. , et al (2003) Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat Biotechnol 21: 143–149. [DOI] [PubMed] [Google Scholar]
- Undisz, K. , Groh, H. , Stopsack, H. , and Horhold‐Schubert, C. (1992) Bioconversion of steroids by Cochliobolus lunatus–II. 11 beta‐hydroxylation of 17‐alpha, 21‐dihydroxypregna‐1,4‐diene‐3,20‐dione 17‐acetate in dependence of the inducer structure. J Steroid Biochem Mol Biol 43: 543–547. [DOI] [PubMed] [Google Scholar]
- Unthan, S. , Baumgart, M. , Radek, A. , Herbs, M. , Siebert, D. , Bruhl, N. , et al (2015) Chassis organism from Corynebacterium glutamicum – a top‐down approach to identify and delete irrelevant gene clusters. Biotechnol 10: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita, M. , Smith, K. , Rozman, D. , and Komel, R. (1994) Progesterone metabolism by the filamentous fungus Cochliobolus lunatus . J Steroid Biochem Mol Biol 49: 87–92. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Sui, P. , Hou, X. , Cao, T. , Jia, L. , Lu, F. , et al (2017) Cloning and identification of a novel steroid 11α‐hydroxylase gene from Absidia coerulea . J Steroid Biochem Mol Biol 171: 254–261. [DOI] [PubMed] [Google Scholar]
- Zakelj‐Mavric, M. , and Belic, I. (1987) Hydroxylation of steroids with 11 alpha‐hydroxylase of Rhizopus nigricans . J. Steroid Biochem. 28: 197–201. [DOI] [PubMed] [Google Scholar]
- Zuidweg, M.H. (1968) Hydroxylation of Reichstein's compound S with cell‐free preparations from Curvularia lunata . Biochim Biophys Acta 152: 144–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alignment of CYP103168 predict protein annotated in JGI database and the protein translate from the sequenced gDNA.
Fig. S2. Phylogenetic tree of CYP103168 homologous sequences. Neighbour‐joining tree shows the distances between the 100 nearest homologous sequences to CYP103168 contained in the GenBank database. H. sapiens homologous cytochrome (NP_001021384.1) was used as an outgroup (real distance to root = 7.51). Main bootstrap values (N = 100) are shown in their corresponding nodes. The two main sets of sequences have been called α and β.
Table S1. Oligonucleotides used in this study.
Table S2. CYPome of C. lunatus.


