Summary
In recent years, the marine environment has been the subject of increasing attention from biotechnological and pharmaceutical industries. A combination of unique physicochemical properties and spatial niche‐specific substrates, in wide‐ranging and extreme habitats, underscores the potential of the marine environment to deliver on functionally novel bioactivities. One such area of ongoing research is the discovery of compounds that interfere with the cell–cell signalling process called quorum sensing (QS). Described as the next generation of antimicrobials, these compounds can target virulence and persistence of clinically relevant pathogens, independent of any growth‐limiting effects. Marine sponges are a rich source of microbial diversity, with dynamic populations in a symbiotic relationship. In this study, we have harnessed the QS inhibition (QSI) potential of marine sponge microbiota and through culture‐based discovery have uncovered small molecule signal mimics that neutralize virulence phenotypes in clinical pathogens. This study describes for the first time a marine sponge Psychrobacter sp. isolate B98C22 that blocks QS signalling, while also reporting dual QS/QSI activity in the Pseudoalteromonas sp. J10 and Paracoccus JM45. Isolation of novel QSI activities has significant potential for future therapeutic development, of particular relevance in the light of the pending perfect storm of antibiotic resistance meeting antibiotic drug discovery decline.
Introduction
The spectre of a post‐antibiotic era is emerging as a very real and imminent threat as the discovery void in antibiotic development is met with an ongoing increase in antibiotic resistance among bacterial and fungal pathogens alike. The rise of antibiotic resistance has become one of the major global health issues of recent years (Fernández and Hancock, 2012; Tanwar et al., 2014), as the report in 2014 of the World Health Organization (WHO) describes (http://www.who.int/mediacentre/news/releases/2014/amr-report/en/). Therefore, there is an urgent need for new strategies to combat microbial diseases, which are more effective and less susceptible in generating of resistance as conventional antibiotics. One strategy is to target virulence systems in the pathogen, neutralizing key virulence factors and in some cases the basis for resistance and tolerance to antibiotics (Cegelski et al., 2008; Cooper and Shlaes, 2011; LaSarre and Federle, 2013). In this context, biofilm formation and strategies to counteract it have received much attention in recent years.
Biofilm formation is one of the major virulence factors associated with clinical human pathogens, and this mode of growth has been described to be directly involved with an increase in antibiotic resistance (Hoiby et al., 2011). As with many multicellular processes, biofilm formation is known to be controlled by a process called quorum sensing (QS). QS is a bacterial cell–cell communication signalling system that controls gene expression in response to changes in cell density. As part of this system, autoinducer molecules enable bacteria to coordinate and control behaviours in a synchronized way (Papenfort and Bassler, 2016). The signalling components of QS systems are different between Gram‐negative and Gram‐positive bacteria, primarily with respect to the autoinducer signalling molecules and their respective receptors. Although several distinct autoinducing systems have been described in Gram‐negative bacteria, by far, the most widely distributed are the N‐acyl‐homoserine lactones (AHLs) (Ng and Bassler, 2009; Schertzer et al., 2009). AHL–QS signalling is a key component of the biofilm mode of growth in many pathogens. It has not been shown to be required for essential cellular processes (Rasmussen and Givskov, 2006). While this was initially thought to support the claim that the selective pressure associated with conventional antibiotic use would not generally apply, recent studies have described the selection of QS mutants (Maeda et al., 2012; García‐Contreras et al., 2016). Furthermore, an adaptable QS circuitry has been described in variants from patients with Cystic Fibrosis, suggesting that circuit heterogeneity may apply for many target pathogens (Feltner et al., 2016). Notwithstanding this, molecules that are able to inhibit the QS system (quorum sensing inhibition molecules, QSIm), and by extension biofilm formation or other related virulence phenotypes, remain promising targets to fight opportunistic human pathogens (Rasmussen and Givskov, 2006; Atkinson and Williams, 2009; Njoroge and Sperandio, 2009; Kalia, 2013).
Targeting AHL‐based signalling systems has already been shown to be an effective anti‐biofilm strategy with compounds such as halogenated furanones, coumarin and enzymes such as acylases and lactonases, which have all been shown to have anti‐pathogenic activity (Dong et al., 2001, 2002; Hentzer et al., 2003; Park et al., 2005; O'Loughlin et al., 2013; Gutierrez‐Barranquero et al., 2015). AHL signalling is based on structure‐specific interactions between small molecular signals and their cognate receptor proteins. AHLs can vary in chain length from C4 to C18, with modifications on the homoserine‐lactone framework (Churchill and Chen, 2011) resulting in a diverse collection of potential signals. In addition, a new QS system using aryl‐homoserine lactones as autoinducer molecules has been reported in Rhodopseudomonas palustris and in Bradyrhizobium (Schaefer et al., 2008;. Ahlgren et al., 2011).
The discovery of AHL–QS systems in marine Gram‐negative bacteria (Nealson et al., 1970; Ruby, 1996; Hastings and Greenberg, 1999) has led to a concerted focus on this relatively unique ecological niche for cell signalling mimics. The marine environment is emerging as a rich and untapped source of novel bioactive molecules with invaluable biotechnological and pharmaceutical potential (Reen et al., 2015a,b). Specifically, marine sponges and their symbiotic microbiomes have been described to be one of the major sources of novel bioactive molecules (Hardoim and Costa, 2014; Blunt et al., 2016). Different studies support the potential for use of culturable bacteria from marine sponges as new platforms for the discovery of novel anti‐pathogenic compounds, including those with a QSI mode of action (Skindersoe et al., 2008; Dobretsov et al., 2011; Pejin B et al., 2014; Mai et al., 2015; Saurav et al., 2016). Therefore, unravelling the QSI potential of marine microorganisms, and subsequently, linking their bioactive potential to the inhibition of QS‐regulated virulence phenotypes of bacterial pathogens is a key research and translational biodiscovery goal.
In this study, we have profiled the potential QSI capabilities from a bacterial collection isolated from marine sponges sourced from multiple geographical locations. A combined high‐throughput biosensor‐based screening protocol was designed, encompassing both generalized disruption and subsequently structure‐specific interference with QS. QSI activities were found to be non‐enzymatic (QSI by enzymes: quorum quenching, QQ) and exhibited a significant degree of structural specificity for disruption of C4, 3OC8, 3OC10 and 3OC12 AHL signalling molecules. This activity was extended to anti‐biofilm activity against both the nosocomial pathogen P. aeruginosa PA14 and the biofouling agent Bacillus subtilis CH8a. In addition, disruption of other important QS virulence phenotypes in P. aeruginosa PA14 was demonstrated. In this work, we have described for the first time the marine sponge Psychrobacter sp. B98C22 with QSI activity, and interestingly, another two marine sponge isolates which showed dual QS/QSI activity (Pseudoalteromonas sp. J10 and Paracoccus sp. JM45). The results in this study further highlight the potential of marine sponge bacteria as a valuable source of diverse QSI compounds that could play a vital role in controlling the new era of emergence of multidrug‐resistant pathogens.
Results
Identification and phylogenetic analysis of QSI‐producing marine sponge bacteria
A screening and validation pipeline was designed to mine for QSI‐producing candidates from a collection of culturable bacteria isolated from a diverse array of marine sponge samples (Fig. S1). Three different biosensor reporter strains, S. marcescens SP15, C. violaceum DSM 30191 and A. tumefaciens NTL4 were used to detect QQ activity against short‐, medium‐ and long‐chain AHLs respectively. Analysis of a total of 440 bacterial isolates led to the identification of 18 isolates (4.1%) with the potential ability to inhibit the QS system of at least one biosensor reporter strain. After the initial screening, these 18 bacterial isolates were taxonomically identified by 16S rDNA sequencing, and a phylogenetic distribution of the QSI candidates was performed (Fig. 1). QSI isolates were identified as belonging primarily to the Gram‐negative Gammaproteobacteria class; five Pseudomonas sp. strains (B98C39, B98SK51b, B98SK53b, B98SK52 and B98SM8), five Pseudoalteromonas sp strains (J10, JC29, W3, W11 and W21) and one Psychrobacter sp. strain (B98C22). In addition, one strain belonging to the Alphaproteobacteria class (Paracoccus sp. JM45) was also identified. Furthermore, Gram‐positive QSI candidates belonging to the Phylum Firmicutes were also identified (five Bacillus sp. strains: AF46, AAF47, AF52, B9853 and CC32 and one Staphylococcus sp. strain: B98C566). A comparative analysis of QQ and QSI activities described in related bacteria to our study in previous reports could highlight the novelty of the activities uncovered in this work (Table 1).
Figure 1.
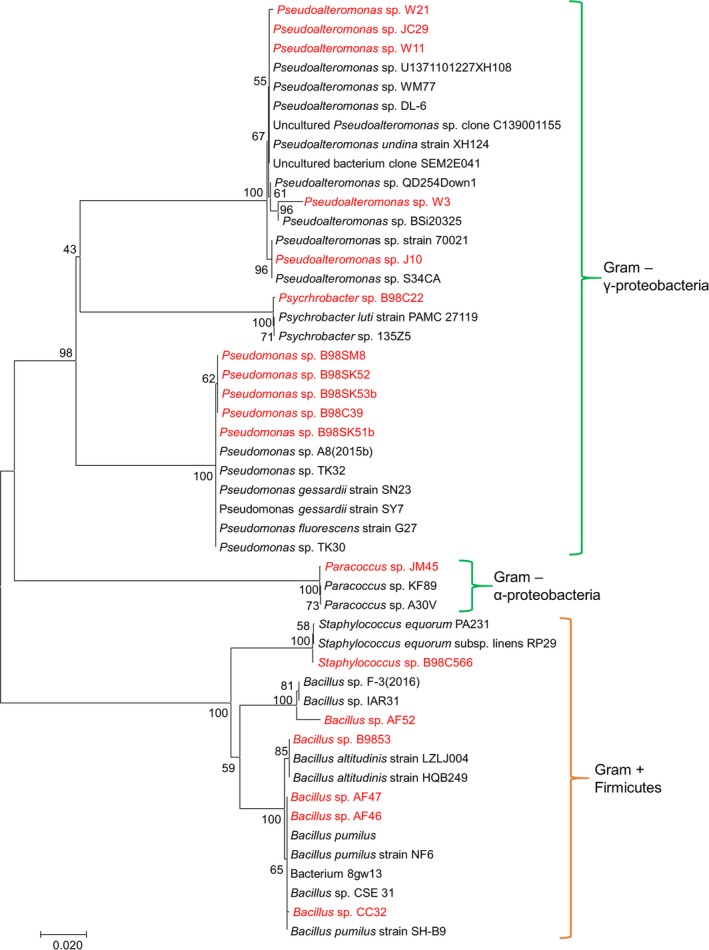
Phylogenetic distribution based on the 16S rRNA sequence of the QQ marine sponge bacteria. Isolates with QQ activity isolated from this study are highlighted in red. Gram‐negative bacteria from γ and α Proteobacteria classes are marked in green. Gram‐positive bacteria belonging to the Phylum Firmicutes, Bacilli Class, are marked in orange.
Table 1.
Comparative analysis of QQ/QSI activities from related bacteria to the novel activities from this study
| Bacterial species | Source | Proposed activity | Phenotypes/Bioassay used | References |
|---|---|---|---|---|
| Bacillus | Different non‐sponge marine origins | Lactonase | Biosensors | (Romero et al., 2011) |
| Bacillus sp. QSI‐1 | Fish gut | Lactonase |
C. violaceum
biosensor protease, haemolytic activity, biofilm |
(Chu et al., 2014) |
| B. cereus D28 | Marine sediment | Cyclic dipeptide | Biosensors, bioluminescence in Vibrio | (Teasdale et al., 2011) |
| B. cereus | Soil | Not analysed |
C. violaceum
biosensor, biofilm |
(Wahman et al., 2015) |
| Bacillus horikosshi | Coral | Extracts | Biosensors | (Thenmozhi et al., 2009) |
| Bacillus pumilus | Marine sediment | Acylase | Biosensors, P. aeruginosa and Serratia virulence | (Nithya et al., 2010) |
| Bacillus sonorensis | Soya sauce | Not analysed |
C. violaceum
biosensor |
(Yin et al., 2012a) |
| B. thuringiensis, B. cereus and B. mycoides | Soil | Lactonase | Biosensors | (Dong et al., 2002) |
| Bacillus Pseudalteromonas Pseudomonas | Brown algae | Not analysed | Serratia biosensor | (Kanagasabhapathy et al., 2009) |
| Bacillus Pseudomonas | Soil | Not analysed |
C. violaceum
biosensor |
(Chong et al., 2012) |
| Halobacillus salinus | Seagrass | Phenethylamide | Biosensors bioluminescence in Vibrio | (Teasdale et al., 2009) |
| P aeruginosa | Marine sponge | Cyclic dipeptide | Not directly related with QSI | (Jayatilake et al., 1996) |
| P. aeruginosa | Clinical sample | Acylase | Biosensors, AHL inhibition, Elastase, Pyocyanin | (Sio et al., 2006) |
| P aeruginosa | Clinical sample | QuiP acylase | Control its own QS system | (Huang et al., 2006) |
| P. aeruginosa | Clinical sample | PvdQ acylase | Control its own QS system | (Bokhove et al., 2010) |
| P. aeruginosa | Clinical sample | Lactonase SsoPox | Pyocyanin, protease, biofilm | (Guendouze et al., 2017) |
| Pseudomonas | Rhizosphere | Not analysed |
C. violaceum
biosensor with OC6, biocontrol activity |
(Alymanesh et al., 2016) |
| Staphylococus | Clinical sample | Small molecules ‘yayurea A, B’ | Biosensors, Pyocyanin. Biofilm | (Chu et al., 2013) |
| Staphylococcus | Marine sponge | Not analysed | Not tested | (Saurav et al., 2016) |
| Staphylococcus saprophyticus | Marine source | Cyclic dipeptide | Biosensors | (Li et al., 2013) |
| Staphylococcus saprophyticus | Sewage | Not analysed |
C. violaceum
biosensor with OC6 |
(Chan et al., 2015) |
| Paracoccus | Marine sponge | Three putative small molecules – structure unknown | Biosensors, Antimicrobial activity, Pyocyanin, Biofilm | (Saurav et al., 2016) |
| Pseudoalteromonas JG1 | Water to rear healthy turbot | Enzymes | Genomic data | (Yu et al., 2013) |
| Pseudoalteromonas | Marine eukaryotes | Not analysed |
C. violaceum
biosensor, biofilm |
(Busetti et al., 2015) |
| Pseudoalteromonas | Marine and estuarine waters | Not analysed | Serrattia and C. violaceum biosensors, bioluminescence in Vibrio | (Linthorne et al., 2015) |
| Pseudoalteromonas | Surface of different marine Eukarya | Enzymes | E. coli biosensor for AHL and AI‐2, biofilm | (Weiland‐Bräuer et al., 2016) |
| Alteromonas | Different non‐sponge marine origins | Acylase | Biosensors | (Romero et al., 2011) |
All QQ activities were subsequently validated on both Marine Agar (MA) and SYP supplemented with 1.5% (w/v) sea salt. The higher concentration of sea salt in MA affected pigment production in the case of S. marcescens SP15 and C. violaceum DSM 30191. As a result, the response of the biosensors was less intense, although the inhibition results were comparable in both media (Table 2). Bacillus sp. strains (AF46, AF47, AF52 and CC32) showed the most remarkable QSI activity, being able to block all three biosensors reporter strains, and more specifically, 3OC10 and 3OC12 AHLs when A. tumefaciens NTL4 was used. In contrast, Psychrobacter sp. B98C22 and Staphylococcus sp. B98C566 displayed the lowest promiscuity with regards to QSI activity, only showing activity against S. marcescens SP15. In general, QSI active strains did not exhibit antibacterial activity against the biosensor strains. However, all five Pseudomonas strains along with Pseudoalteromonas JC29 affected the growth of S. marcescens SP15, while three Pseudomonas strains (B98SK53b, B98SK52 and B98SM8) inhibited growth of C. violaceum DSM 30191 (Table 2). Although none of the other QSI isolates affected growth of the biosensor strains and do not appear to produce antibacterial activity under the conditions tested, several Bacillus strains did suppress the growth of two fish pathogens and a Staphylococcus aureus strain (Table 3). In addition, two of the QSI isolates (Pseudoalteromonas sp. J10 and Paracoccus sp. JM45) also possessed QS activity, being capable of activating the A. tumefaciens NTL4 biosensor in response to the production of long AHLs (Fig. S2). To our knowledge, this is the first description of dual‐acting QS and QSI isolates from the marine sponge environment.
Table 2.
Quorum quenching activity shown by marine sponge bacterial isolates
| QQ Isolate | Genus | Isolated from sponge | Biosensor reporter strains | |||
|---|---|---|---|---|---|---|
| SP15 | DSM 30191 | NTL4‐(3OC10)a | NTL4‐(3OC12)b | |||
| AF46 | Bacillus sp. | Genus Amphilectus | ++c | +d | + | ++ |
| AF47 | Bacillus sp. | Genus Amphilectus | ++ | + | + | ++ |
| AF52 | Bacillus sp. | Genus Amphilectus | + | −/+e | + | + |
| B9853 | Bacillus sp. | Class Hexactinellida | ++ | −/+ | ++ | −f |
| B98C39 | Pseudomonas sp. | Class Hexactinellida | IGa | + | −/+ | + |
| B98C22 | Psychrobacter sp. | Class Hexactinellida | + | − | − | − |
| B98C566 | Staphylococcus sp. | Class Hexactinellida | + | − | − | − |
| B98SK51b | Pseudomonas sp. | Class Hexactinellida | IG | + | −/+ | + |
| B98SK52 | Pseudomonas sp. | Class Hexactinellida | IG | IG | −/+ | −/+ |
| B98SK53b | Pseudomonas sp. | Class Hexactinellida | IG | IG | −/+ | −/+ |
| B98SM8 | Pseudomonas sp. | Class Hexactinellida | IG | IG | + | −/+ |
| CC32 | Bacillus sp. | Genus Cliona | ++ | −/+ | ++ | ++ |
| J10 | Pseudoalteromonas sp. | Genus Polymastia | + | + | + | + |
| JC29 | Pseudoalteromonas sp. | Genus Polymastia | IG | + | − | − |
| JM45 | Paracoccus sp. | Genus Polymastia | −/+ | −/+ | + | + |
| W3 | Pseudoalteromonas sp. | Genus Axinella | + | ++ | −/+ | −/+ |
| W11 | Pseudoalteromonas sp. | Genus Axinella | + | + | −/+ | −/+ |
| W21 | Pseudoalteromonas sp. | Genus Axinella | + | + | −/+ | −/+ |
a. 3OC10: N‐(3‐Oxodecanoyl)‐l‐homoserine lactone.
b. 3OC12: N‐(3‐Oxododecanoyl)‐l‐homoserine lactone.
c. ++: strong pigment inhibition, QQ activity. Inhibition halo > 20.0 mm.
d. +: pigment inhibition, QQ activity. Inhibition halo from ˃ 2.5 mm to ≤ 20.0 mm.
e. −/+: weak response of pigment inhibition. Inhibition halo from 1.0 mm to ≤ 2.5 mm.
f. −: no inhibition of pigment, no QQ activity.
IG: inhibition of growth.
Table 3.
Antimicrobial activity of marine Bacillus sp. strains
| Bacterial strains | Antimicrobial activity | ||
|---|---|---|---|
| Fish pathogens | Opportunistic human pathogen | ||
| V. anguillarum | E. tarda | S. aureus NCDO949 | |
| Bacillus | |||
| AF46 | +a | −c | + |
| AF47 | + | − | + |
| AF52 | + | − | − |
| B9853 | −/+b | − | + |
| CC32 | + | −/+ | + |
a. +: antimicrobial activity. Growth inhibition halo from ˃ 2.5 mm to ≤ 20.0 mm.
b. −/+: weak response of antimicrobial activity. Growth inhibition halo from 1.0 mm to ≤ 2.5 mm.
c. −: no antimicrobial activity.
Specific AHL inhibition by marine bacterial supernatants is not related to extracellular enzymatic activity
The role of AHLs in controlling adaptive behaviour in bacteria has been long established, with many virulence phenotypes displayed by bacterial pathogens under tight regulation by QS systems (Rutherford and Bassler, 2012). These systems are hierarchical and temporal in nature, with interconnecting downstream regulatory pathways the norm in many well‐studied bacterial pathogens. In order to decipher whether there was a structural specificity to the AHL inhibition activity of the different marine bacterial strains, the QS biosensor reporter assay was modified by adding specific exogenous AHLs. S. marcescens SP19 was used to detect structure‐specific inhibition against C4‐HSL, and C. violaceum CV026 was used to detect specific inhibition against C6‐HSL and 3OC8‐HSL. Six QSI strains (Bacillus sp. B9853, Paracoccus sp. JM45, Psychrobacter sp. B98C22, Staphylococcus sp. B98C566, Pseudomonas sp. B98SM8 and Pseudoalteromonas sp. J10) were able to inhibit activity towards a single AHL, namely 3OC8 (Table 4). Most of the marine strains (Bacillus sp.: AF46, AF47, AF52 and CC32, Pseudomonas: B98C39, B98SK51b, B98SK53b and B98Sk52 and Pseudoalteromonas sp. JC29) were able to disrupt the QS activity of two different AHLs, C4 and 3OC8. The remaining three Pseudoalteromonas sp. strains (W3, W11 and W21) displayed QSI activity against the three different AHLs tested (C4, C6 and 3OC8).
Table 4.
Inhibition of QS system by marine sponge bacterial supernatants
| Bacterial strains | Quorum quenching activity towards specific AHLs | ||
|---|---|---|---|
| S. marcescens SP19 | C. violaceum CV026 | ||
| C4a | C6b | 3OC8c | |
| Bacillus sp. | |||
| AF46 | +d | −e | + |
| AF47 | + | − | + |
| AF52 | −/+f | − | + |
| B9853 | − | + | + |
| CC32 | + | − | + |
| Paracoccus sp. | |||
| JM45 | − | − | + |
| Pseudoalteromonas sp | |||
| J10 | − | − | + |
| JC29 | + | − | + |
| W3 | + | + | + |
| W11 | + | −/+ | + |
| W21 | + | −/+ | + |
| Pseudomonas sp. | |||
| B98C39 | −/+ | − | + |
| B98SK51b | −/+ | − | + |
| B98SK53b | + | − | + |
| B98SK52 | + | − | + |
| B98SM8 | − | − | + |
| Psychrobacter sp. | |||
| B98C22 | − | − | + |
| Staphylococcus sp. | |||
| B98C566 | − | − | + |
a. C4: N‐Butyryl‐dl‐homoserine lactone.
b. C6: N‐Hexanoyl‐l‐homoserine lactone.
c. 3OC8: N‐(3‐Oxooctanoyl)‐l‐homoserine lactone.
d. +: AHL inhibition, inhibition of pigment production.
e. −: no AHL inhibition, no inhibition of pigment production.
f. −/+: weak response of AHL inhibition. Comparison of pigment production inhibition in respect to the control.
To determine if the QSI activity shown by the different marine bacterial supernatants was related with the action of extracellular QQ enzymes (mostly acylases and lactonases), a thermostability assay was performed using the biosensor C. violaceum CV026 and 3O8‐HSL, the only AHL whose activity was inhibited by all marine bacterial supernatants (Fig. S3). Heat treatment at 85°C for 1 h was not able to disrupt the QSI activity of the marine supernatants. It is well known that different Bacillus species possess a potent QQ activity due to the presence of extracellular enzymes (Lee et al., 2002). Therefore, an extra heat treatment of 95°C for 30 min was applied specifically to the marine Bacillus supernatants. As before, this extra heat treatment was not able to disarm the QSI activity of the marine Bacillus isolate supernatants. Therefore, a small molecule or other inhibitory factor would appear to underpin the QSI activity of the isolates presented in this study.
Biofilm disruption by marine bacterial supernatants
The finding that the QSI activity attributed to the marine sponge strains may be molecular rather than enzymatic, suggests that these compounds could have a potential application for anti‐biofilm development. To test this, the capacity of the marine bacterial strains to either inhibit biofilms or disarm preformed biofilms in the nosocomial pathogen P. aeruginosa PA14 and the marine biofouling agent B. subtilis CH8a was evaluated (Fig. 3). A wide range of QSI bacterial supernatants significantly impaired the formation of P. aeruginosa PA14 biofilms, reducing it by at least 50% (Bacillus AF52, the five Pseudomonas sp. strains, Psychrobacter sp. B98C22, Staphylococcus sp. B98C566, Pseudoalteromonas sp. J10 and Paracoccus sp. JM45) (Fig. 2A). The remaining four Bacillus sp. strains were not able to inhibit PA14 biofilm formation, while Pseudoalteromonas sp. W3, W11 and W21 increased biofilm formation in this pathogen. Importantly, no inhibition of PA14 growth was observed during biofilm formation in the presence of the different marine QSI supernatants (Fig. S4A). Regarding biofilm formation of Bacillus subtilis CH8a (Fig. 2B), a different pattern emerged. Bacillus sp. (all five strains), Pseudomonas sp. B98C39 and Pseudoalteromonas sp. JC29 W11 and W21) all suppressed biofilm formation, by up to 60% in the case of Pseudomonas sp. B98C39 and Pseudoalteromonas sp. JC29 and W21. Pseudoalteromonas sp. W3 strongly increased biofilm formation in CH8a. Planktonic CH8a growth was also affected in the presence of supernatants from the four marine Bacillus sp. (AF46, AF47, B9853 and CC32), perhaps explaining the apparent reduction in attached CH8a cells observed for these strains (Fig. S4B).
Figure 3.
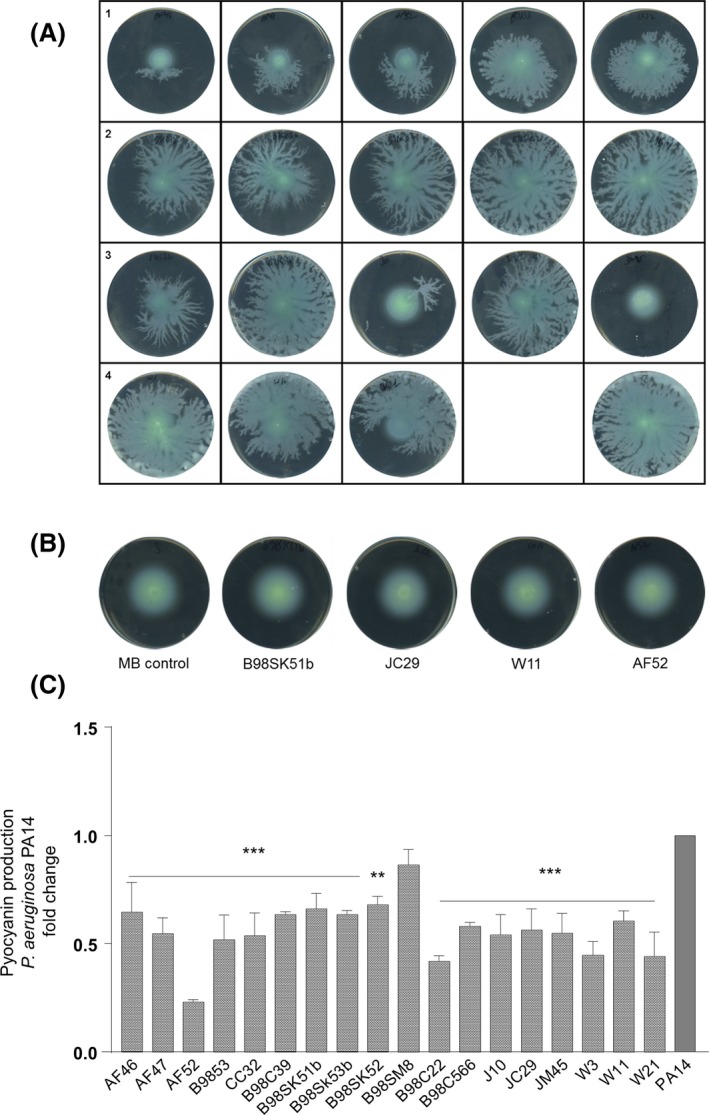
QQ marine bacterial supernatants impair different virulence phenotypes of P. aeruginosa PA14.
A. Impact of marine bacterial supernatants on PA14 swarming motility. Row 1, from left to right: Bacillus sp. AF46, AF47, AF52, B9853 and CC32. Row 2, from left to right: Pseudomomas sp. B98C39, B98SK51, B98SK53B, B98SK52 and B98SM8. Row 3, from left to right: Psychrobacter sp. B98C22, Staphylococcus sp. B98C566, Pseudoalteromonas sp J10 and JC29 and Paracoccus sp. JM45. Row 4, from left to right: Pseudoalteromonas sp. W3, W11 and W21. The untreated MB control is presented on the far right of this row.
B. Impact of marine bacterial supernatants on PA14 swimming motility.
C. Impact of marine bacterial supernatant on PA14 pyocyanin production.
Data presented are normalized to the PA14 control and are the mean (±SEM) of at three independent biological replicates. In each case, the individual replicates of the untreated control are normalized to the mean. Statistical analysis was performed using one‐way ANOVA with post hoc Bonferroni testing (**P ≤ 0.005, ***P ≤ 0.001).
Figure 2.
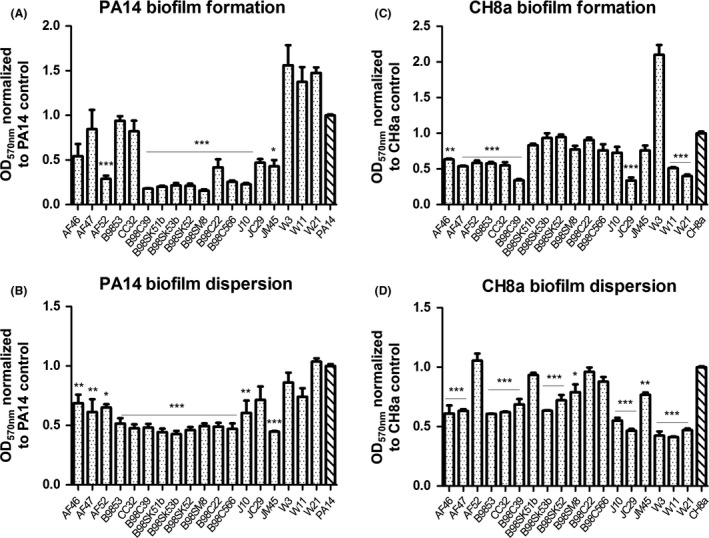
Biofilm formation inhibition and dispersion of P. aeruginosa PA14 and B. subtilis CH8a by marine bacterial supernatants.
A. Biofilm formation inhibition of PA14.
B. Biofilm formation inhibition of CH8a.
C. PA14 dispersion assay.
D. CH8a dispersion assay.
Data presented are the mean (±SEM) of at three independent biological replicates and are normalized to the PA14 or CH8a controls. In each case, the individual replicates of the untreated control are normalized to the mean. Statistical analysis was performed using one‐way ANOVA with post hoc Bonferroni testing (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.001).
With respect to the disruption of preformed biofilms, 13 of the 18 QSI marine strains (all Bacillus sp, all Pseudomonas sp., Psychrobacter sp. B98C22, Staphylococcus sp. B98C566 and Paracoccus sp. JM45) significantly reduced preformed biofilms of PA14 (Fig. 2C). Conversely, Pseudoalteromonas sp. JC29, W3, W11 and W21 did not measurably affect preformed PA14 biofilms. In relation to the ability of the marine QSI supernatants to disrupt B. subtilis CH8a‐preformed biofilms, 14 of the 18 marine QSI bacteria displayed a significant reduction (Fig. 2D). In this case, the most significant reductions in attached biomass were achieved with the addition of supernatants from Pseudoalteromonas sp. JC29, W3, W11 and W21. Bacillus sp. AF52, Pseudomonas sp. B98SK51b, Psychrobacter sp. B98C22 and Staphylococcus sp. B98C566 did not disrupt preformed biofilms of CH8a (Fig. 2D).
Pseudomonas aeruginosa PA14 virulence determinants are suppressed by marine bacterial supernatants
Pseudomonas aeruginosa is a leading nosocomial pathogen, and it represents a significant clinical challenge associated with morbidity and mortality in chronic disease. As antibiotic‐mediated control is currently under significant threat, new strategies to disarm virulence and persistence‐related behaviours in this pathogen are urgently required. P. aeruginosa has a well‐studied QS network that controls its virulence behaviour (Bjarnsholt et al., 2010; Jimenez et al., 2012), chiefly through two AHL systems, LasRI and RhlRI (Latifi et al., 1996; Papenfort and Bassler, 2016). The QSI and anti‐biofilm activity of the marine strains suggest they may interfere with other virulence behaviours in this pathogen, such as QS‐controlled phenazine production and swarming motility (Déziel et al., 2004; Caiazza et al., 2005). As with biofilm formation, most of the marine QSI supernatants were able to disrupt swarming motility in P. aeruginosa to some extent (Fig. 3A). For example, Bacillus sp. (AF46, AF47 and AF52), Pseudoalteromonas sp. J10 and Paracoccus sp. JM45 showed the highest reduction in swarming motility. However, no impact on swimming motility was observed for any of the marine bacterial supernatants tested in this study (Fig. 3B). The marine bacterial supernatants also displayed an inhibition towards the production of pyocyanin by P. aeruginosa PA14 (Fig. 3C). Four QSI candidates (Bacillus sp. AF52, Psychrobacter sp. B98C22 and Pseudoalteromonas W3 and W21) elicited a potent reduction in pyocyanin production (over 50%) with respect to the control.
Discussion
Our understanding of the importance of cell–cell communication among microbes is continuing to expand, as new systems and models are uncovered. While care with terminology must be observed, with distinction required between signals, cues and coercive factors, the paradigm of communication between cells is universal (Fuqua and Greenberg, 2002; Perbal, 2003). A recent review by Hmelo (2017) has summarized the phenomenon of quorum sensing in the marine environment, highlighting the importance of these networks in controlling community dynamics in this largely undiscovered natural ecosystem (Hmelo, 2017). Although QS systems in Gram‐negative bacteria were originally described over 40 years ago in a marine bacterium (Nealson et al., 1970), the intrigue of many research groups to decipher the role of AHLs molecules in marine biogeochemistry and ecology is quite recent (Hmelo, 2017). Marine sponges in particular have been described as an untapped reservoir of biodiversity and an increasingly important source of novel bioactives natural products (Blunt et al., 2016). Although initially attributed to the sponge itself, it has since emerged that sponge‐associated bacteria play a vital role in the production of these compounds (Thoms and Schupp, 2005; Mehbub et al., 2014; Wilson et al., 2014). Although QS signalling seems to be quite common in the marine environment (Hmelo, 2017), studies that describe the presence and possible role of AHL–QS system in marine sponge‐associated bacteria are limited (Taylor et al., 2004; Mohamed et al., 2008; Gardères et al., 2012; Zan et al., 2012). Equally, there is also a lack of data on the prevalence of marine sponge bacteria that are able to block the signalling capacity of quorum sensing system (Saurav et al., 2016). Our study brings new knowledge on the capacity of marine sponge bacteria to quench QS signalling, with the first description of a dual‐acting QS/QSI isolate from a marine sponge. Furthermore, we also describe QSI activity in a marine Psychrobacter isolate for the first time. While the discovery of novel QSI small molecules is of interest for the development of next‐generation antimicrobials to treat bacterial infections, the role of QS/QSI in shaping the microbe–sponge symbiotic relationship is also worthy of further investigation.
The bacterial isolates that were discovered to have QSI activity were identified as Bacillus, Staphylococcus, Pseudomonas, Pseudoalteromonas, Paracoccus and Psychrobacter. With the exception of Psychrobacter, QSI activities (either enzymatic or small molecule) have been previously described for these organisms. However, studies on isolates associated with the marine environment are limited (Romero et al., 2011; Yu et al., 2013; Saurav et al., 2016). Interestingly, as far as we know, this is the first description of a marine sponge Psychrobacter sp. strain that can interfere with QS in a model organism, thus broadening the potential for QS signalling among marine microbes. Although three genera accounted for 77% of the QSI activities observed, each isolate included in the study was at least phenotypically distinct (Table 2), a result that was confirmed when activity against specific AHLs was investigated (Table 4). While all marine supernatants inhibited 3OC8‐HSL, only a few of them were able to block C4 and C6‐HSL. We consider two possibilities that may underpin this specificity: (i) the presence of the OH group at the C3 position in the 3OC8‐HSL or (ii) that 3OC8‐HSL molecules are more abundant than other AHLs in the marine sponge environment, and thus select for a higher abundance of antagonist QSIm's. Furthermore, several Bacillus strains exhibited antibacterial activity against a panel of clinically relevant isolates suggesting that their QSI activity is only one mechanism used within the ordered symbiotic community of the marine sponge. Antimicrobial activity has been recently reported for other marine sponge Bacillus isolates (Matobole et al., 2017). In addition, two of the QSI isolates (Pseudoalteromonas sp. J10 and Paracoccus sp. JM45) also displayed QS activity, being capable of activating the A. tumefaciens NTL4 long‐chain biosensor. This dual activity has not been previously described in marine sponge isolates and adds another layer of complexity to the role of QS/QSI in shaping the population dynamics of this ecosystem.
Consistent with their ability to disrupt QS signalling in the biosensor strains, supernatants from the marine QSI strains were able to inhibit biofilm formation and preformed biofilms, (Fig. 2A and B) respectively, swarming motility (Fig. 3A) and pyocyanin production (Fig. 3C) at different levels (all data summarized in Fig. S5). Interestingly, swimming motility was unaffected in the presence of QSI supernatants, perhaps a reflection of the fact that it is not considered a multicellular behaviour (Kearns, 2010). There appears to be a genera‐specific signature to the QSI activity whereby Pseudoalteromonas sp. did not inhibit P. aeruginosa biofilm PA14 formation, but were quite active against B. subtilis CH8a. Conversely, Pseudomonas sp. inhibited PA14 biofilm formation, while it did not significantly reduce biofilm formation in CH8a. Future isolation and characterization of the active fraction underpinning the QSI activity will provide answers as to what governs this pattern of activity, with genome sequencing being required to unravel the molecular pathways involved in QSIm production.
Experimental procedures
Bacterial strains and culture conditions
The marine bacterial strains analysed in this study are summarized in Table 2. Marine agar or broth (MA or MB respectively) was routinely used to grow marine bacterial strains (Difco, Oxford, UK). SYP agar with 1.5% of sea salt and MA or MB were used in different experiments performed in this work to analyse the potential QSI capability of marine bacteria. All marine bacterial strains used in this study were grown routinely at 23°C. Different N‐Acyl homoserine lactones (AHLs) used in this work were purchased from Sigma‐Aldrich (Cambridge, UK) and were dissolved in DMSO (N‐butyryl‐dl‐homoserine lactone: C4‐HSL; N‐hexanoyl‐l‐homoserine lactone: C6‐HSL; N‐(3‐oxooctanoyl)‐l‐homoserine lactone: 3OC8‐HSL; N‐(3‐oxodecanoyl)‐l‐homoserine lactone: 3OC10‐HSL; and N‐(3‐oxododecanoyl)‐l‐homoserine lactone: 3OC12‐HSL).
Isolation and characterization of potential QSI marine isolates
A screening pipeline to mine QS antagonists from a collection of culturable bacteria isolated from marine sponge samples was designed (Fig. S1). A total of 440 marine bacterial isolates were previously selected from different marine sponge samples to present different colony morphologies and pigmentation on MA plates (MA) (Difco, Oxford, UK). These bacterial isolates were collected from different sponges belonging to different families such as Hexactinellida, Polymastia, Cliona, Haliclonas, Stelleta, Lissodendoryx, Poecillastra, Inflatella and Axinella. These sponge samples were collected using a remote‐operated vehicle on board the Celtic Explorer research vessel, 300 nautical miles off the west coast of Ireland as part of the marine biodiscovery cruise carried out in May 2010. These bacterial isolates were initially tested for their potential QSI activity against three different biosensor reporter strains. From this original screening, 18 bacterial isolates were selected as possible QSI candidates (Table 2).
Phylogenetic analysis
Genomic DNA from 18 bacterial isolates was extracted using UltraClean® Microbial DNA Isolation Kit (MO BIO, USA) following the manufacturer guidelines. PCR based on the amplification of 16S rRNA using the universal primers 27F (5′‐AGAGTTTGATCMTGGCTCAG‐3′) and 1492R (5′‐TACGGYTACCTTGTTACGACTT‐3′) was performed (da Silva et al., 2013, Lane, 1991). PCR amplicons were sequenced by MWG Eurofins, UK. The 16S rRNA sequences were assembled and edited with the software Vector NTI Suite 9, and then were compared with those available in NCBI database through BLASTn searches (Altschul et al., 1997). For the phylogenetic analysis, three of the closest sequences for each sequence obtained in this study were taken from Genbank, and subsequently, a multi‐alignment was carried out using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). A maximum‐likelihood phylogenetic tree was constructed by Mega 6.06 (Tamura et al., 2013) using the Kimura two‐parameter model and the option of complete deletion to eliminate positions containing gaps. Confidence levels of the branching points were determined using 100 bootstrap replicates.
Quorum sensing inhibition biosensor assay
The following biosensor reporter strains were used to identify the QSI ability of the different marine bacterial isolates: Serratia marcescens SP15 for the detection of QSI activity against short‐chain AHLs (Poulter et al., 2010), Chromobacterium violaceum DSM 30191 for the detection of QSI activity against medium‐chain AHLs (Morohoshi et al., 2008) and Agrobacterium tumefaciens NTL4 for the detection of QSI activity against long‐chain AHLs (A. tumefaciens NTL4 contains the plasmid pZLR4 carrying a traG::lacZ reporter fusion (Farrand et al., 1996; Yin et al., 2012a, b). Whole cells of marine bacterial strains were cultured on MA and SYP‐1.5 for 72 h at 23°C. Then, they were overlaid with LB soft agar (0.5% agar) inoculated with SP15 and DSM 30191 QSI biosensors strains at an OD 600 nm of 0.25. A. tumefaciens NTL4 was also inoculated at an OD600 nm of 0.25 but in MM minimal medium (K2HPO4: 10.5 g, KH2PO4: 4.5 g, MgSO4·7H2O: 0.2 g, calcium dehydrate: 0.01 g, (NH4)2SO4: 2 g, FeSO4: 0.005 g, MnCl2·4H2O, d‐Mannitol: 2 g and agarose: 5 g) supplemented with 50 μg ml−1 of 5‐bromo‐4chloro‐3‐indolyl‐β‐d‐galactopyranoside and 20 nM of 3OC10 and 3OC12. QSI activity was identified by a colour inhibition halo around the growth of each marine bacterial strain.
Generation of marine bacterial supernatants
One hundred millilitre of MB were inoculated from overnight MB fresh cultures of each marine bacterium at an OD600 nm of 0.02 and were incubated for 72 h at 23°C at 170 r.p.m.. Then, bacteria were removed by centrifugation and the supernatants were filter sterilized using a Sartorius AG vacuum filtration system (0.22 μm).
QSI activity of marine bacterial supernatants towards specific AHLs
To decipher the range of specific quorum quenching activity of each marine bacterial strain, an experimental approach was performed as described previously (Gutierrez‐Barranquero et al., 2015) with minor modifications. 15 ml of LB agar 1% mixed with 5 ml of supernatant from each marine bacterial strain were used to prepare the agar plates. Then, prior to pouring LB‐supernatant agar plates, the biosensors S. marcescens SP19 and C. violaceum CV026 were inoculated at an OD 600 nm of 0.25. Subsequently, wells were made in the LB‐supernatant agar plates, and 20 μl of a 50 μM concentration of different AHLs and 20 μl of DMSO as control were added. DMSO was the solvent where the different AHLs were previously dissolved. C4‐HSL was used for the plates inoculated with S. marcescens SP19, and C6‐HSL and 3OC8‐HSL for the plates inoculated with C. violaceum CV026. LB 1% (15 ml) mixed with 5 ml of marine broth and also inoculated with the different biosensors were used as control plates. Finally, plates were incubated for 24 h at 30°C. QSI activity was identified by pigment inhibition around the well where the different AHLs were inoculated, respect to the control plates.
Thermostability of marine bacterial supernatants
To stablish the thermostability of the putative QQ enzymes or QSIm in the different marine bacterial supernatants, supernatant from each one was heated to 85°C for 1 h. The supernatants of the five Bacillus strains included in this study were also subjected to an additional heat treatment of 95°C for 30 mins. After these heat treatments, the different supernatants were used to prepare LB‐supernatant agar plates inoculated with C. violaceum CV026 as described above. Wells were made in the LB‐supernatant agar plates, and 20 μl of a 50 μM concentration of 3OC8‐HSL was added. Supernatants not heat treated, and also, MB heat and not heat treated were used as control.
Biofilm formation of Pseudomonas aeruginosa PA14 and Bacillus subtilis CH8a
Biofilm formation experiments with minor modifications were carried out as previously described (O'Toole and Kolter, 1998). The opportunistic human pathogen P. aeruginosa PA14 and the biofouling agent Bacillus subtilis CH8 were used to analyse the anti‐biofilm activity of the marine bacterial supernatants. PA14 and CH8a from a fresh overnight culture (LB broth and MB respectively) were used to inoculate 500 μl of LB broth and MB, mixed with 500 μl of the different marine bacterial supernatant in 24‐well microtitre plates (Starstedt, UK) at an OD600 nm of 0.05. P. aeruginosa PA14 biofilms were incubated at 37°C for 24 h and Bacillus subtilis CH8a biofilms were incubated at 23°C for 72 h, both statically. Biofilm formation was measured by solubilization of crystal violet with 96% ethanol and quantified at 570 nm.
Dispersion assay of P. aeruginosa PA14 and Bacillus subtilis CH8a
The capability to inhibit preformed biofilm of P. aeruginosa PA14 and B. subtilis CH8a by the different marine bacterial supernatants was analysed. Fresh LB and MB overnight culture of P. aeruginosa PA14 and B. subtilis CH8a, respectively, were used to inoculate 150 μl of LB and MB at an OD600 nm of 0.05 in 96‐well microtitre plates (Sarstedt, Leicester, UK). Biofilms then were incubated at 37°C for 24 h for P. aeruginosa PA14 and 23°C for 72 h for B. subtilis CH8a, both statically. After this time of incubation, 150 μl of the different marine bacterial supernatants were added to the wells where the biofilms were developed. The preformed biofilm mixed with the marine bacterial supernatants were incubated for another 24 h at 37°C and for another 72 h at 23°C for P. aeruginosa PA14 and B. subtilis CH8a respectively. 150 μl of MB was used as positive control. Preformed biofilms inhibition was measured by solubilization of crystal violet with 96% ethanol and quantified at 570 nm.
PA14 swarming and swimming motility
Swarming and swimming motilities of P. aeruginosa PA14 were tested in the presence of the different marine bacterial supernatants. For swarming plates, 15 ml of Eiken‐swarming agar (0.8% Eiken nutrient broth, 0.6% Eiken agar and 0.5% glucose) were mixed with 5 ml of MB supernatant. For swimming plates, 15 ml LB agar 0.3% were mixed with 5 ml of MB supernatants The bacterial cells of P. aeruginosa PA14 were gently inoculated using a toothpick at the centre of the agar surface, and the plates were incubated at 37°C for 18 and 24 h for swarming and swimming respectively.
PA14 pyocyanin production
Overnight cultures of P. aeruginosa PA14 were inoculated into 2.5 ml of LB broth + 2.5 ml of the different marine bacterial supernatants at an OD600 nm of 0.05. PA14 cultures were then incubated at 37°C for 24 h at 180 r.p.m. The total volume of the culture was centrifuged at 3320 × g for 10 min. Chloroform (3 ml) was added to the supernatant fraction and vortexed. The bottom phase was then recovered following centrifugation at 3320 × g for 5 min, and 2 ml of 0.2 M HCl was added. The samples were then vortexed for 20 s, allowed to settle, and the absorbance of the pink layer upper phase was measured at 570 nm.
Quorum sensing screening assay
The following reporter strains were used to identify AHL production by the different marine bacterial strains: S. marcescens SP19 (Poulter et al., 2010) was used to detect the production of short‐chain AHLs; C. violaceum CV026 (McClean et al., 1997) was used to detect medium‐chain AHLs and A. tumefaciens NTL4 was used for the detection of long‐chain AHLs. Marine bacterial strains were cultured on MA and SYP‐1.5 for 72 h at 23°C. Then, they were overlaid with LB soft agar (0.5% agar) inoculated with the different biosensors strains at an OD 600 nm of 0.25, with the exception that for A. tumefaciens NTL4 a MM was used and supplemented with 50 μg ml−1 of 5‐bromo‐4chloro‐3‐indolyl‐β‐d‐galactopyranoside. Overlaid plates were incubated for 24 h at 30°C. Quorum sensing activity was identified by prodigiosin (red colour) due to the production of short‐chain AHLs, violacein (purple colour) by the production of medium‐chain AHLs and blue colour production by the breakdown of 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐galactopyranoside by the production of long‐chain AHLs.
Antimicrobial plate overlay assay
Whole cells of marine bacterial strains were cultured on MA plates for 72 h at 23°C. The different bacterial indicator strains were grown overnight in LB broth. Then, they were inoculated at an OD600 nm of 0.1 in LB soft agar (0.5%) and overlaid the marine bacterial strains. The fish pathogens V. anguillarum and E. tarda, an environmental hospital isolate E. coli MUH76317, food‐borne pathogen S. typhimurium C5369 and the opportunistic human pathogens P. aeruginosa PA14, B. cepacia NCTC10743 and S. aureus NCDO949 were used to test the potential antimicrobial activity of marine bacterial strains. Zones of inhibition were tested after 24 h of incubation at 37°C.
Statistical analysis
Data analysis was performed using GraphPad Prism 5.03 (San Diego, CA, USA). At least three independent biological replicates were performed for all experiments. Statistical analyses were performed using one‐way ANOVA with Bonferroni post hoc testing, or two‐tailed paired Student's t‐test. Differences were considered significant when the P value was ≤ 0.05 (*P ≤ 0.05, **P ≤ 0.005).
Nucleotide sequence accession numbers
The partial 16S rRNA sequences of the different marine sponge bacteria used in this study were deposited in GenBank (NCBI) under the accession numbers: MF289535‐MF289552.
Conflict of Interest
The authors declare there is no conflict of interest in the submission of this manuscript.
Supporting information
Fig. S1. Schematic representation of the screening pipeline protocol used to decipher the QQ potential of marine isolates.
Fig. S2. QS activation of the A. tumefaciens NTL4 biosensor by two marine bacteria, Pseudoalteromonas sp. J10 (up picture) and Paracoccus sp. JM45 (down picture).
Fig. S3. Thermostability of QQ marine bacterial supernatants. (A) Non‐heat treated plates. (B) Heat – treated plates.
Fig. S4. Biofilm (OD600 nm) of (A) P. aeruginosa PA14 and (B) B. subtilis CH8a.
Fig. S5. Suppression of primary virulence phenotypes regulated by QS in P. aeruginosa by marine sponge QQ isolates. Red colour: no inhibition of the virulence phenotype. Green light colour: inhibition of the virulence phenotype ≤ 50%. Green dark colour: inhibition of the virulence phenotype > 50%. Yellow colour: Promotes the increased of the virulence phenotype.
Acknowledgements
This research was supported in part by grants awarded by the European Commission (FP7‐PEOPLE‐2013‐ITN, 607786; FP7‐KBBE‐2012‐6, CP‐TP‐312184; FP7‐KBBE‐2012‐6, 311975; OCEAN 2011‐2, 287589; Marie Curie 256596; EU‐634486), Science Foundation Ireland (SSPC‐2, 12/RC/2275; 13/TIDA/B2625; 12/TIDA/B2411; 12/TIDA/B2405; 14/TIDA/2438, 15/TIDA/2977), the Department of Agriculture and Food (FIRM/RSF/CoFoRD; FIRM 08/RDC/629; FIRM 1/F009/MabS; FIRM 13/F/516), the Irish Research Council for Science, Engineering and Technology (PD/2011/2414; GOIPG/2014/647), the Health Research Board/Irish Thoracic Society (MRCG‐2014‐6), the Marine Institute (Beaufort award C2CRA 2007/082) and Teagasc (Walsh Fellowship 2013).
Microbial Biotechnology (2019) 12(5), 1049–1063
Funding Information
This research was supported in part by grants awarded by the European Commission (FP7‐PEOPLE‐2013‐ITN, 607786; FP7‐KBBE‐2012‐6, CP‐TP‐312184; FP7‐KBBE‐2012‐6, 311975; OCEAN 2011‐2, 287589; Marie Curie 256596; EU‐634486), Science Foundation Ireland (SSPC‐2, 12/RC/2275; 13/TIDA/B2625; 12/TIDA/B2411; 12/TIDA/B2405; 14/TIDA/2438, 15/TIDA/2977), the Department of Agriculture and Food (FIRM/RSF/CoFoRD; FIRM 08/RDC/629; FIRM 1/F009/MabS; FIRM 13/F/516), the Irish Research Council for Science, Engineering and Technology (PD/2011/2414; GOIPG/2014/647), the Health Research Board/Irish Thoracic Society (MRCG‐2014‐6), the Marine Institute (Beaufort award C2CRA 2007/082) and Teagasc (Walsh Fellowship 2013).
References
- Ahlgren, N.A. , Harwood, C.S. , Schaefer, A.L. , Giraud, E. , and Greenberg, P.E. (2011) Aryl‐homoserine lactone quorum sensing in stem‐nodulating photosynthetic bradyrhizobia . Proc Natl Acad Sci USA 108: 7183–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alymanesh, M.R. , Taheri, P. , and Tarighi, S. (2016) Pseudomonas as a frequent and important quorum quenching bacterium with biocontrol capability against many phytopathogens. Biocontrol Sci Technol 26: 1719–1735. [Google Scholar]
- Atkinson, S. , and Williams, P. (2009) Quorum sensing and social networking in the microbial world. J R Soc Interface 6: 959–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt, T. , Jensen, P.Ø. , Jakobsen, T.H. , Phipps, R. , Nielsen, A.K. , Rybtke, M.T. , et al (2010) Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 5: e10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt, J.W. , Copp, B.R. , Keyzers, R.A. , Munro, M.H.G. , and Prinsep, M.R. (2016) Marine natural products. Nat Prod Rep 33: 382–431. [DOI] [PubMed] [Google Scholar]
- Bokhove, M. , Jimenez, P.N. , Quax, W.J. , and Dijkstra, B.W. (2010) The quorum‐quenching N‐acyl homoserine lactone acylase PvdQ is an Ntn‐hydrolase with an unusual substrate‐binding pocket. Proc Natl Acad Sci USA 107: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busetti, A. , Shaw, G. , Megaw, J. , Gorman, S.P. , Maggs, C.A. , and Gilmore, B.F. (2015) Marine‐derived quorum‐sensing inhibitory activities enhance the antibacterial efficacy of Tobramycin against Pseudomonas aeruginosa . Mar Drugs 13: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza, N.C. , Shanks, R.M.Q. , and O'Toole, G.A. (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa . J Bacteriol 187: 7351–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski, L. , Marshall, G.R. , Eldridge, G.R. , and Hultgren, S.J. (2008) The biology and future prospects of antivirulence therapies. Nat Rev Micro 6: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K.‐G. , Sulaiman, J. , Yong, D.A. , Tee, K.K. , Yin, W.‐F. , and Priya, K. (2015) Draft genome perspective of Staphylococcus saprophyticus strain SU8, an N‐acyl homoserine lactone‐degrading bacterium. Genome Announc 3: e01097–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, T.M. , Koh, C.L. , Sam, C.K. , Choo, Y.M. , Yin, W.F. , and Chan, K.‐G. (2012) Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian Tropical montane forest. Sensors 12: 4846–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, W. , Zhou, S. , Zhu, W. and Zhuang, X. (2014) Quorum quenching bacteria Bacillus sp. QSI‐1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci Rep 4, 5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y.Y. , Nega, M. , Wölfle, M. , Plener, L. , Grond, S. , Jung, K. , and Götz, F. (2013) A new class of quorum quenching molecules from Staphylococcus species affects communication and growth of Gram‐negative bacteria. PLoS Pathog 9: e1003654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, M.E. , and Chen, L. (2011) Structural basis of acyl‐homoserine lactone‐dependent signaling. Chem Rev 111: 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, M.A. , and Shlaes, D. (2011) Fix the antibiotics pipeline. Nature 472: 32. [DOI] [PubMed] [Google Scholar]
- da Silva, M.A.C. , Cavalett, A. , Spinner, A. , Rosa, D.C. , Jasper, R.B. , Quecine, M.C. , et al. (2013) Phylogenetic identification of marine bacteria isolated from deep‐sea sediments of the eastern South Atlantic Ocean. SpringerPlus 2: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel, E. , Lépine, F. , Milot, S. , He, J. , Mindrinos, M.N. , Tompkins, R.G. , and Rahme, L.G. (2004) Analysis of Pseudomonas aeruginosa 4‐hydroxy‐2‐alkylquinolines (HAQs) reveals a role for 4‐hydroxy‐2‐heptylquinoline in cell‐to‐cell communication. Proc Natl Acad Sci USA 101: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov, S. , Teplitski, M. , Bayer, M. , Gunasekera, S. , Proksch, P. , and Paul, V.J. (2011) Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 27: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y.H. , Wang, L.H. , Xu, J.L. , Zhang, H.B. , Zhang, X.F. , and Zhang, L.H. (2001) Quenching quorum‐sensing‐dependent bacterial infection by an N‐acyl homoserine lactonase. Nature 411: 813–817. [DOI] [PubMed] [Google Scholar]
- Dong, Y.‐H. , Gusti, A.R. , Zhang, Q. , Xu, J.‐L. , and Zhang, L.‐H. (2002) Identification of quorum‐quenching N‐acyl homoserine lactonases from Bacillus species. Appl Environ Microbiol 68: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand, S.K. , Hwang, I. , and Cook, D.M. (1996) The tra region of the nopaline‐type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J Bacteriol 178: 4233–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltner, J.B. , Wolter, D.J. , Pope, C.E. , Groleau, M.C. , Smalley, N.E. , Greenberg, E.P. , et al (2016) LasR variant cystic fibrosis isolates reveal an adaptable quorum‐sensing hierarchy in Pseudomonas aeruginosa . mBio 7, e01513–e01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, L. , and Hancock, R.E.W. (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25: 661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua, C. , and Greenberg, E.P. (2002) Listening in on bacteria: acyl‐homoserine lactone signalling. Nat Rev Mol Cell Biol 3: 685–695. [DOI] [PubMed] [Google Scholar]
- García‐Contreras, R. , Maeda, T. , and Wood, T.K. (2016) Can resistance against quorum‐sensing interference be selected? ISME J 10: 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardères, J. , Taupin, L. , Saïdin, J.B. , Dufour, A. , and Le Pennec, G. (2012) N‐acyl homoserine lactone production by bacteria within the sponge Suberites domuncula (Olivi, 1792) (Porifera, Demospongiae). Mar Biol 159: 1685–1692. [Google Scholar]
- Guendouze, A. , Plener, L. , Bzdrenga, J. , Jacquet, P. , Rémy, B. , Elias, M. , et al (2017) Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front Microbiol 8: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez‐Barranquero, J.A. , Reen, F.J. , McCarthy, R.R. , and O'Gara, F. (2015) Deciphering the role of coumarin as a novel quorum sensing inhibitor suppressing virulence phenotypes in bacterial pathogens. Appl Microbiol Biotechnol 99: 3303–3316. [DOI] [PubMed] [Google Scholar]
- Hardoim, C. , and Costa, R. (2014) Microbial communities and bioactive compounds in marine sponges of the family Irciniidae—a review. Mar Drugs 12: 5089–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, J.W. , and Greenberg, E.P. (1999) Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol 181: 2667–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer, M. , Wu, H. , Andersen, J.B. , Riedel, K. , Rasmussen, T.B. , Bagge, N. , et al (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22: 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmelo, L.R. (2017) Quorum sensing in marine microbial environments. Ann Rev Mar Sci 9: 257–281. [DOI] [PubMed] [Google Scholar]
- Hoiby, N. , Ciofu, O. , Johansen, H.K. , Song, Z.J. , Moser, C. , Jensen, P.O. , et al (2011) The clinical impact of bacterial biofilms. Int J Oral Sci 3: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.J. , Petersen, A. , Whiteley, M. , and Leadbetter, J.R. (2006) Identification of QuiP, the product of gene PA1032, as the second acyl‐homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 72: 1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayatilake, G.S. , Thornton, M.P. , Leonard, A.C. , Grimwade, J.E. , and Baker, B.J. (1996) Metabolites from an Antarctic sponge associated bacterium, Pseudomonas aeruginosa . J Nat Prod 59: 293–296. [DOI] [PubMed] [Google Scholar]
- Jimenez, P.N. , Koch, G. , Thompson, J.A. , Xavier, K.B. , Cool, R.H. , and Quax, W.J. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa . Microbiol Mol Biol Rev 76: 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia, V.C. (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31: 224–245. [DOI] [PubMed] [Google Scholar]
- Kanagasabhapathy, M. , Yamazaki, G. , Ishida, A. , Sasaki, H. , and Nagata, S. (2009) Presence of quorum‐sensing inhibitor‐like compounds from bacteria isolated from the brown alga Colpomenia sinuosa . Lett Appl Microbiol 49: 573–579. [DOI] [PubMed] [Google Scholar]
- Kearns, D.B. (2010) A field guide to bacterial swarming motility. Nat Rev Microbiol 8: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D.J. (1991). 16S/23S rRNA sequencing In Nucleic acid techniques in bacterial systematics. Stackebrandt E. and Goodfellow M. (eds). New York: John Wiley and Sons, pp. 115–175. [Google Scholar]
- LaSarre, B. , and Federle, M.J. (2013) Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev 77: 73–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi, A. , Foglino, M. , Tanaka, K. , Williams, P. , and Lazdunski, A. (1996) A hierarchical quorum‐sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary‐phase sigma factor RpoS. Mol Microbiol 21: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Lee, S.J. , Park, S.‐Y. , Lee, J.‐J. , Yum, D.‐Y. , Koo, B.‐T. , and Lee, J.‐K. (2002) Genes encoding the N‐acyl homoserine lactone‐degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl Environ Microbiol 68: 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Huiru, Z. , Biting, D. , Yun, J. , Wei, J. , and Kunming, D. (2013) Study on the anti‐quorum sensing activity of a marine bacterium Staphylococcus saprophyticus 108. BioTechnol 7: 480–487. [Google Scholar]
- Linthorne, J.S. , Chang, B.J. , Flematti, G.R. , Ghisalberti, E.L. , and Sutton, D.C. (2015) A direct pre‐screen for marine bacteria producing compounds inhibiting quorum sensing reveals diverse planktonic bacteria that are bioactive. Mar Biotechnol 17: 33–42. [DOI] [PubMed] [Google Scholar]
- Maeda, T. , García‐Contreras, R. , Pu, M. , Sheng, L. , Garcia, L.R. , Tomás, M. , and Wood, T.K. (2012) Quorum quenching quandary: resistance to antivirulence compounds. ISME J 6: 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, T. , Tintillier, F. , Lucasson, A. , Moriou, C. , Bonno, E. , Petek, S. , et al (2015) Quorum sensing inhibitors from Leucetta chagosensis Dendy, 1863. Lett Appl Microbiol 61: 311–317. [DOI] [PubMed] [Google Scholar]
- Matobole, R.M. , van Zyl, L.J. , Parker‐Nance, S. , Davies‐Coleman, M.T. , and Trindade, M. (2017) Antibacterial activities of bacteria isolated from the marine sponges Isodictya compressa and Higginsia bidentifera collected from Algoa Bay, South Africa. Mar Drugs 15: 47. [Google Scholar]
- McClean, K.H. , Winson, M.K. , Fish, L. , Taylor, A. , Chhabra, S.R. , Camara, M. , et al (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N‐acylhomoserine lactones. Microbiology 143: 3703–3711. [DOI] [PubMed] [Google Scholar]
- Mehbub, M.F. , Lei, J. , Franco, C. , and Zhang, W. (2014) Marine sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar Drugs 12: 4539–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed, N.M. , Cicirelli, E.M. , Kan, J. , Chen, F. , Fuqua, C. , and Hill, R.T. (2008) Diversity and quorum‐sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10: 75–86. [DOI] [PubMed] [Google Scholar]
- Morohoshi, T. , Kato, M. , Fukamachi, K. , Kato, N. , and Ikeda, T. (2008) N‐acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol Lett 279: 124–130. [DOI] [PubMed] [Google Scholar]
- Nealson, K.H. , Platt, T. , and Hastings, J.W. (1970) Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.L. , and Bassler, B.L. (2009) Bacterial quorum‐sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithya, C. , Aravindraja, C. , and Pandian, S.K. (2010) Bacillus pumilus of Palk Bay origin inhibits quorum‐sensing‐mediated virulence factors in Gram‐negative bacteria. Res Microbiol 161: 293–304. [DOI] [PubMed] [Google Scholar]
- Njoroge, J. , and Sperandio, V. (2009) Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol Med 1: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loughlin, C.T. , Miller, L.C. , Siryaporn, A. , Drescher, K. , Semmelhack, M.F. , and Bassler, B.L. (2013) A quorum‐sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA 110: 17981–17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G.A. , and Kolter, R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461. [DOI] [PubMed] [Google Scholar]
- Papenfort, K. , and Bassler, B.L. (2016) Quorum sensing signal‐response systems in Gram‐negative bacteria. Nat Rev Microbiol 14: 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.‐Y. , Kang, H.‐O. , Jang, H.‐S. , Lee, J.‐K. , Koo, B.‐T. , and Yum, D.‐Y. (2005) Identification of extracellular N‐acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol 71: 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejin, B. , Talevska, A. , Ciric A, G.J. , Nikolic M, T.T. and M., S. (2014) Anti‐quorum sensing activity of selected sponge extracts: a case study of Pseudomonas aeruginosa . Nat Prod Res 28, 2330–2333. [DOI] [PubMed] [Google Scholar]
- Perbal, B. (2003) Communication is the key. Cell Commun Signal 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter, S. , Carlton, T.M. , Su, X. , Spring, D.R. , and Salmond, G.P.C. (2010) Engineering of new prodigiosin‐based biosensors of Serratia for facile detection of short‐chain N‐acyl homoserine lactone quorum‐sensing molecules. Environ Microbiol Rep 2: 322–328. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T.B. , and Givskov, M. (2006) Quorum sensing inhibitors: a bargain of effects. Microbiology 152: 895–904. [DOI] [PubMed] [Google Scholar]
- Reen, F. , Gutiérrez‐Barranquero, J. , Dobson, A. , Adams, C. and O′Gara, F. (2015a) Emerging concepts promising new horizons for marine biodiscovery and synthetic biology. Mar Drugs 13, 2924–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen, F.J. , Romano, S. , Dobson, A.D. , and O'Gara, F. (2015b) The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar Drugs 13: 4754–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, M. , Martin‐Cuadrado, A.‐B. , Roca‐Rivada, A. , Cabello, A.M. , and Otero, A. (2011) Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol Ecol 75: 205–217. [DOI] [PubMed] [Google Scholar]
- Ruby, E.G. (1996) Lessons from a cooperative, bacterial‐animal association: the Vibrio fischeri‐Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50: 591–624. [DOI] [PubMed] [Google Scholar]
- Rutherford, S.T. , and Bassler, B.L. (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2: a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurav, K. , Bar‐Shalom, R. , Haber, M. , Burgsdorf, I. , Oliviero, G. , Costantino, V. , et al (2016) In search of alternative antibiotic drugs: quorum‐quenching activity in sponges and their bacterial isolates. Front Microbiol 7: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, A.L. , Greenberg, E.P. , Oliver, C.M. , Oda, Y. , Huang, J.J. , Bittan‐Banin, G. , et al (2008) A new class of homoserine lactone quorum‐sensing signals. Nature 454: 595–599. [DOI] [PubMed] [Google Scholar]
- Schertzer, J.W. , Boulette, M.L. , and Whiteley, M. (2009) More than a signal: non‐signaling properties of quorum sensing molecules. Trends Microbiol 17: 189–195. [DOI] [PubMed] [Google Scholar]
- Sio, C.F. , Otten, L.G. , Cool, R.H. , Diggle, S.P. , Braun, P.G. , Bos, R. , et al (2006) Quorum quenching by an N‐acyl‐homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74: 1673–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skindersoe, M.E. , Alhede, M. , Phipps, R. , Yang, L. , Jensen, P.O. , Rasmussen, T.B. , et al (2008) Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa . Antimicrob Agents Chemother 52: 3648–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanwar, J. , Das, S. , Fatima, Z. , and Hameed, S. (2014) Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis 2014: 541340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M.W. , Schupp, P.J. , Baillie, H.J. , Charlton, T.S. , de Nys, R. , Kjelleberg, S. , and Steinberg, P.D. (2004) Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl Environ Microbiol 70: 4387–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, M.E. , Liu, J. , Wallace, J. , Akhlaghi, F. , and Rowley, D.C. (2009) Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing‐ controlled phenotypes in gram‐negative bacteria. Appl Environ Microb 75: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, M.E. , Donovan, K.A. , Forschner‐Dancause, S.R. , and Rowley, D.C. (2011) Gram‐positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar Biotechnol 13: 722–732. [DOI] [PubMed] [Google Scholar]
- Thenmozhi, R. , Nithyanand, P. , Rathna, J. , and Pandian, S.K. (2009) Antibiofilm activity of coral‐associated bacteria against different clinical M serotypes of Streptococcus pyogenes . FEMS Immunol Med Microbiol 57: 284–294. [DOI] [PubMed] [Google Scholar]
- Thoms, C. , and Schupp, P. (2005) Biotechnological potential of marine sponges and their associated bacteria as producers of new pharmaceuticals. JIBL 2: 257–264. [Google Scholar]
- Wahman, S. , Emara, M. , Shawky, R.M. , El‐Domany, R.A. , and Aboulwafa, M.M. (2015) Inhibition of quorum sensing‐mediated biofilm formation in Pseudomonas aeruginosa by a locally isolated Bacillus cereus . J Basic Microbiol 55: 1406–1416. [DOI] [PubMed] [Google Scholar]
- Weiland‐Bräuer, N. , Kisch, M.J. , Pinnow, N. , Liese, A. , and Schmitz, R.A. (2016) Highly effective inhibition of biofilm formation by the first metagenome‐derived AI‐2 quenching enzyme. Front Microbiol 7: 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M.C. , Mori, T. , Ruckert, C. , Uria, A.R. , Helf, M.J. , Takada, K. , et al (2014) An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506: 58–62. [DOI] [PubMed] [Google Scholar]
- Yin, W.‐F. , Tung, H.‐J. , Sam, C.‐K. , Koh, C.‐L. , and Chan, K.‐G. (2012a) Quorum quenching Bacillus sonorensis isolated from soya sauce fermentation brine. Sensors 12: 4065–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W.‐F. , Purmal, K. , Chin, S. , Chan, X.‐Y. , and Chan, K.‐G. (2012b) Long chain N‐acyl homoserine lactone production by Enterobacter sp. isolated from human tongue surfaces. Sensors 12: 14307–14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Tang, K. , Liu, J. , Shi, X. , Gulder, T.A. , and Zhang, X.‐H. (2013) Genome analysis of Pseudoalteromonas flavipulchra JG1 reveals various survival advantages in marine environment. BMC Genom 14: 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan, J. , Cicirelli, E.M. , Mohamed, N.M. , Sibhatu, H. , Kroll, S. , Choi, O. , et al (2012) A Complex LuxR‐LuxI type quorum sensing network in a Roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol Microbiol 85: 916–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic representation of the screening pipeline protocol used to decipher the QQ potential of marine isolates.
Fig. S2. QS activation of the A. tumefaciens NTL4 biosensor by two marine bacteria, Pseudoalteromonas sp. J10 (up picture) and Paracoccus sp. JM45 (down picture).
Fig. S3. Thermostability of QQ marine bacterial supernatants. (A) Non‐heat treated plates. (B) Heat – treated plates.
Fig. S4. Biofilm (OD600 nm) of (A) P. aeruginosa PA14 and (B) B. subtilis CH8a.
Fig. S5. Suppression of primary virulence phenotypes regulated by QS in P. aeruginosa by marine sponge QQ isolates. Red colour: no inhibition of the virulence phenotype. Green light colour: inhibition of the virulence phenotype ≤ 50%. Green dark colour: inhibition of the virulence phenotype > 50%. Yellow colour: Promotes the increased of the virulence phenotype.


