Abstract
Dinoflagellates are a general group of phytoplankton, ubiquitous in aquatic environments. Most dinoflagellates are non-obligate autotrophs, subjected to potential physical and chemical DNA-damaging agents, including UV irradiation, in the euphotic zone. Delay of cell cycles by irradiation, as part of DNA damage responses (DDRs), could potentially lead to growth inhibition, contributing to major errors in the estimation of primary productivity and interpretations of photo-inhibition. Their liquid crystalline chromosomes (LCCs) have large amount of abnormal bases, restricted placement of coding sequences at the chromosomes periphery, and tandem repeat-encoded genes. These chromosome characteristics, their large genome sizes, as well as the lack of architectural nucleosomes, likely contribute to possible differential responses to DNA damage agents. In this study, we sought potential dinoflagellate orthologues of eukaryotic DNA damage repair pathways, and the linking pathway with cell-cycle control in three dinoflagellate species. It appeared that major orthologues in photoreactivation, base excision repair, nucleotide excision repair, mismatch repair, double-strand break repair and homologous recombination repair are well represented in dinoflagellate genomes. Future studies should address possible differential DNA damage responses of dinoflagellates over other planktonic groups, especially in relation to possible shift of life-cycle transitions in responses to UV irradiation. This may have a potential role in the persistence of dinoflagellate red tides with the advent of climatic change.
Keywords: dinoflagellate, DNA damage, DNA repair
1. Introduction
Cellular DNA is continuously challenged with intracellular or exogenous agents that cause DNA damages. To preserve DNA integrity during cell division, cells have developed integrated signaling cascades linking DNA damage responses (DDRs) to cell-cycle transition, making judgement call as to damage repair per proliferation versus growth postponement, life-cycle transitions or cell death. DNA repair systems are generally conserved in nucleosomal eukaryotes.
Dinoflagellates are a major phytoplankton group in aquatic ecosystems, contributing significantly to ocean primary productivity and carbon cycling [1]. Members of the group are infamous for causing harmful algal blooms (red tides) [2], which may cause mortality or physiological impairment either due to toxin production, oxygen depletion or physical clotting of gills attributed to high biomass [3]. Proliferation of many dinoflagellates is favored with prolonged surface water stratification with the increase in seawater temperature, and global warming was predicted to expand the distribution of harmful algal blooms into higher latitudes [4]. An active cell death called paraptosis, different from apoptosis, was also observed in Amphidinium carterae under culture senescence and darkness [5], which implicated strong solar energy could differentially modulate dinoflagellate production relative to other eukaryotic groups. In addition to their ecological profoundness, dinoflagellates behold the only alternative chromosomal packaging system in their Liquid Crystalline Chromosomes (LCCs), composing of superhelical modules and cation-mediated anisotropic organization (reviewed in this issue [6]). LCCs are characterized by many unique features including no detectable architectural nucleosomes and substantial replacement of thymine with 5-hydroxymethyluracil (reviewed in this issue [6]), implicating they may have different susceptibility to DNA damage agents when compared with the typical nucleosomal chromosome structure. Likely attributed to susceptibility of LCCs’ anisotropic organization and their large chromosomes (many up to microns, and genome among the largest in eukaryotes), many dinoflagellates are susceptible to physical turbulences of the sea, with many species arresting in cell cycle in response to mechanical stresses [7]. The rising ocean temperature has been associated with loss of symbiotic dinoflagellates in corals and other invertebrates, which are co-exposed to increasing physical stresses in the subtidal-littoral zones, with increasing climatic extremes [8,9].
UV irradiation inhibited growth and motility of dinoflagellates Gyrodinium aureolum, Prorocentrum minimum and Heterocapsa triquetra [10,11], Symbiodinium californium and Symbiodinium microadriaticum [12]. UV irradiation caused chromosome breaks in Prorocentrum micans [13], despite not being characterized at the molecular level in dinoflagellates. There is a paucity of data as to the effect of DNA damage on cell-cycle delay, amid inhibition of proliferation. With this imperative, we conducted a global analysis of the DNA damage repair genes in dinoflagellates, from different transcriptomic databases, and compared to other eukaryotic orthologues. Most dinoflagellate species are capable of mixotrophy [14,15], implicating photo-inhibition (or sub-eutrophic zone) as a non-limiting factor to the group, which can be a common factor that modulates selective cell proliferation of surface groups, especially if strong irradiation were to cause cell deaths in other groups. Differential effects on different phytoplankton groups have been a recognized driver for future bio-oceanographic regimes in response to climatic changes [16,17]. The unique architectural organization of dinoflagellate liquid crystalline chromosomes will substantiate differences in their DDRs to increasing UV irradiation, acidity, and physical turbulences.
2. Materials and Methods
Orthologues (ORFs) of DDR proteins (the complete lists of these proteins are available from: https://www.mdanderson.org/documents/Labs/Wood-Laboratory/human-dna-repair-genes.html#Human%20D) from animal Homo sapiens and budding yeast Saccharomyces cerevisiae were retrieved from UniProt database and used as reference sequences. These protein sequences were queried against the transcriptome datasets of Crypthecodinium cohnii (unpublished datasets, the final extracted sequences are available in Table S1), Symbiodinum minutum [18] (published data from: https://marinegenomics.oist.jp/symb/viewer/info?project_id=21), and Lingulodinium polyedrum ([19], the final obtained sequences are available in Table S1) by tBLASTn algorithm with a cut-off E-value of 1e-5 using the TBtool software [20]. The three dinoflagellate species represent heterotrophic, symbiotic and autotrophic dinoflagellate species respectively. For reference polypeptides with no hits, we further included orthologues of fission yeast Schizosaccharomyces pombe and plant Arabidopsis thaliana as reference templates to query against the transcriptome datasets. The hit sequences were extracted out and further verified by running a BLASTX algorithm against NCBI non-redundant (nr) database. If the reference genes or its orthologues appeared on the top five reciprocal BLAST hits, we label herewith as an orthologue. Given the special features of LCCs and the vast evolutionary distances, further studies would be required to functionally characterize these orthologues.
Phylogenetic analysis based on neighbour-joining and maximum-likelihood algorithms were conducted using software MEGA 5.05 [21], and only nodes with bootstrap value over 0.5 (50%) were labelled. Cladding of phylogenetic groups with major expected sister groups, though not a proof, gives additional information on expected evolutionary rates; long branches, on the other hand, would be indicative of accelerated evolution.
3. Results and Discussion
3.1. DNA Damage Checkpoint Signaling Networks
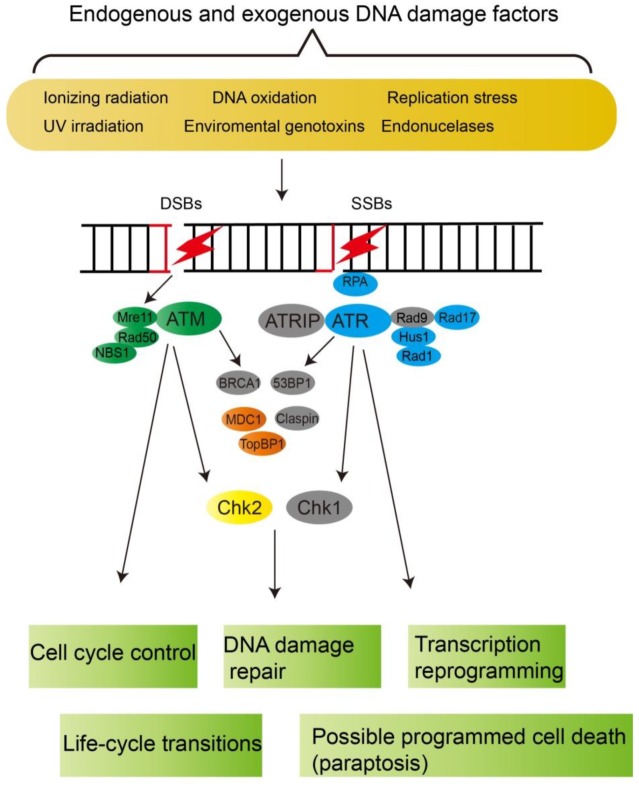
DNA damage checkpoint signaling is initiated by two conserved apical regulators Ataxia telangiectasia mutated (ATM) and ataxia telangiectasia mutated and Rad3-related (ATR), which are members of the phosphoinositide 3-kinase-related protein kinase (PIKKS) family, acting as major switches in DNA damage repair or apoptosis, senescence and even cell death [22,23,24,25].
In mammalian cells, ATM mainly responds to double-stranded DNA breaks (DSBs) generated by ionizing radiation [26]. The Mre11-Rad50-Nbs1 (MRN) complex recognizes the DSBs and stimulates the activation of ATM, which then triggers the rapid phosphorylation of the C-terminal tail (Ser 139) of the histone variant H2AX [27,28]. The phosphorylated histone variant γH2AX then interacts with Mdc1 through its C-terminal BRCT domain, which recruits more MRN complex and ATM, reinforcing γH2AX phosphorylation, which is taking as a common hallmark of DNA damage [29,30].
The ATR pathway is primarily triggered by replication protein A (RPA) coated single-stranded DNAs (ssDNAs) resulted from replication stress or UV exposure and other genotoxic agents [22], which recruits ATR-interacting protein (ATRIP) and ATR together to the lesion sites. The activation of ATR is mediated by ATR activators. TopBP1 is one of these ATR activators, which is also conserved in different organisms [31]. Its recruitment depends on the PCNA-like Rad9-Rad1-Hus1 (9-1-1) checkpoint clamp complex [32,33].
Following activation, ATM and ATR phosphorylates downstream proteins to amplify the signaling cascade for coordination of cell cycle, DNA repair and replication. A key amplification point is the two effector kinases, Chk2 and Chk1, two ATM/ATR substrates, which are cell-cycle control proteins: including phosphorylation of the cell-cycle phosphatase Cdc25, leading to cyclin-dependent kinase (CDK) inactivation and halting cell cycle [34,35,36,37]. Chk1 and Chk2 are conserved in metazoan and fungi, but both Chk1 and Chk2 orthologues are not present in plant kingdoms [38]. Chk1 and Chk2 have many overlapped substrates and non-overlapping substrates in different eukaryotes [39]. Although a previous study reported that Chk1 was found in Symibodinum and Lingulodinium [40], our reciprocal BLAST analysis showed that these putative genes were not true Chk1 orthologues. It seems that only Chk2 is present in dinoflagellates (Figure 1 and Table 1).
Figure 1.
Diagrammatic summary of the DNA damage response signaling network. The grey ellipses denote absence of putative dinoflagellate orthologues, whereas other colors indicate presence of putative dinoflagellate orthologues. For simplicity, nomenclatures differentiating genes, proteins and mutations are not enforced in this study.
Table 1.
Predicted checkpoint control genes in dinoflagellates.
| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Upstream factors | ||||
| ATR | Unigene24416_All (2e-022) | symbB.v1.2.040344.t1 (2e-067) | Lp_Unigene54547_All (9e-118) | apical checkpoint serine/threonine kinase |
| Unigene40907_All (1e-087) | symbB.v1.2.040393.t1 (1e-018) | |||
| ATRIP | * | * | * | ATR-interacting protein, necessary for checkpoint signaling upon DNA damage |
| ATM | Unigene1664_All (3e-091) | symbB.v1.2.029032.t1 (1e-087) | Lp_Unigene23700_All (6e-089) | apical checkpoint serine/threonine-protein kinase |
| Rad17 | Unigene30410_All (1e-015) | symbB.v1.2.000094.t1 (7e-008) | Lp_Unigene58859_All (4e-018) | involved in ATR-dependent checkpoint activation |
| Rad1 | CL2244.Contig1_All (2e-024) | - | Lp_Unigene59067_All (6e-006) | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| CL2244.Contig2_All (2e-029) | ||||
| Hus1 | Unigene57737_All (1e-008) | symbB.v1.2.041619.t1 (3e-014) | Lp_Unigene75437_All (2e-015) | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| Rad9 | * | * | * | subunits of the 9-1-1 complex which binds to DNA lesion after DNA damage |
| Mediators | ||||
| BRCA1 | * | * | * | E3 ubiquitin-protein ligase activity for formation of poly-ubiquitin chains |
| 53BP1 | * | * | * | p53-binding protein 1, binds damaged DNA |
| MDC1 | Unigene15380_All (9e-007) | * | Lp_CL4955.Contig1_All (5e-009) | mediator of DNA damage checkpoint, recruitment of repair proteins to DNA damage foci |
| Lp_CL4955.Contig2_All ((5e-009)) | ||||
| Claspin | * | * | * | upstream regulator of Chk1 |
| TopBP1 | * | * | Lp_Unigene64350_All (1e-006) | topoisomerase II beta interacting protein, ATR activator |
| Effectors | ||||
| Chk1 | * | * | * | serine/threonine-protein kinase |
| Chk2 | Unigene61604_All (3e-056) | symbB.v1.2.014603.t1 (4e-043) | Lp_CL13254.Contig1_All (2e-052) | serine/threonine-protein kinase |
| Unigene13287_All (2e-054) | symbB.v1.2.000403.t1 (3e-028) | Lp_Unigene47534_All (3e-041) | ||
| Unigene68813_All (2e-045) | Lp_CL1544.Contig1_All (6e-049) | |||
| Unigene52773_All (3e-048) | ||||
| Unigene5075_All (1e-041) | ||||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
Further down the signaling cascade (Figure 1 and Table 1), orthologues of some ATM accessory proteins MDC1, 53BP1, but not BRCA1, were found in dinoflagellate transcriptomes [26,41]. BRCA1 is only present in animals and plants [42]. Therefore, it is not unexpected to have no BRCA1 in dinoflagellates. Both orthologues of TopBP1 and Claspin, accessory proteins for ATR [24,25], are absent from our bioinformatics analysis.
Except for the ATRIP and Rad9, all other upstream factors including the central kinase ATM and ATR were found in C. cohnii, S. minutum and L. polyedrum (Figure 1 and Table 1). ATRIP, an obligate partner of ATR, and Rad9-Hus1-Rad1 complex, play an essential role for the recognition of RPA-ssDNA and subsequent activation of the ATR signaling respectively [24]. Therefore, the absence of ATRIP and Rad9 is surprising, which is probably due to sequence divergence. Phylogenetic analysis of the ATM and ATR of dinoflagellates suggested they formed a single clade respectively and clustered together with the apicomplexa (Figure S1A,B), consistent with their phylogenetic relationship under the super phylum alveolate [43]. Further investigations should address the bridging pathways between switches between vegetative growth, cell-cycle arrest and life-cycle transitions. These pathways would likely have group-specific genes specially adapted to dinoflagellate ecological niches.
3.2. DNA Repair Pathways
3.2.1. Direct Reversal of DNA Lesion
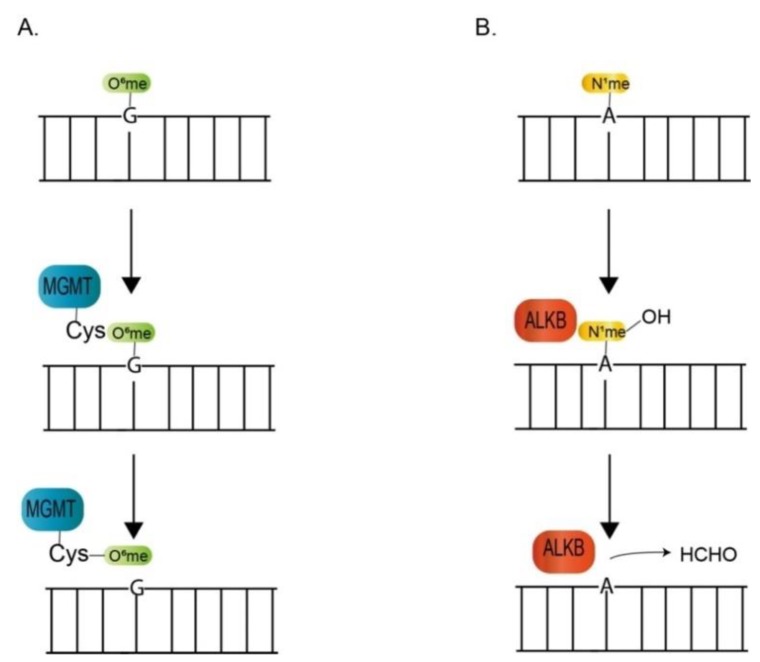
Alkylating agents—widely distributed reactive chemicals in intracellular and extracellular environments—react with DNA and produce various kinds of modifications on the DNA bases and backbone, leading to structure alterations and functional disruptions [44,45,46]. The alkylation attack on DNA mainly occurs at the ring nitrogen (N) and extracyclic oxygen (O) atoms of the DNA bases [46,47]. O6-methylguanine (O6-meG), a major deleterious base adduct produced from the reaction with the O6 position of guanine, will elicit a mispair with thymine during DNA duplication, causing the transition mutation of G:C to A:T. The O6-alkylguanine-DNA alkyltransferase, the Ada protein in E. coli and MGMT/AGT protein in mammalians, is responsible for direct repair of this type of lesion (Figure 2A). During repair process, alkyl group of O6-meG is transferred to Cys residues of MGMT protein, leading to MGMT protein’s inactivation and degradation [48]. N1-methyladenine (N1-meA) and N3-methylcytosine (N3-meC) are two other kinds of lesions occurred in the exposed DNA base of single-stranded DNA or replication fork. ALKB proteins, members of α-ketoglutarate/iron (II)-dependent dioxygenases, are involved in reversing these types of DNA lesions through oxidative dealkylation of the alkyl groups from N1-meA and N3-meC, leading to hydroxylmethylated products and the subsequent release of formaldehyde and the repaired base (Figure 2B) [47,49,50]. The orthologues of MGMT and ALKB protein are found in dinoflagellates transcriptomes (Table 2). The modified base N6-methyladenine (6mA), the most abundant modified base in eukaryotic RNAs [51,52], is also present naturally in DNA of dinoflagellates chromosomes, which was reported to accounting for 2–3% of the total nucleotides [53,54]. 6mA, a major type of modification in bacteria, is associated with restriction-modification system and discrimination between original and newly produced DNA [55]. 6mA is also conserved in eukaryotic genomes and linked with gene regulation events [56,57,58,59,60]. Both bacterial and human AlkB protein have activities for demethylating 6mA [60,61]. It remains to be further determined if dinoflagellate AlkB protein has activities towards 6mA.
Figure 2.
Diagrammatic summary of dinoflagellate orthologues in the direct reversal of DNA damage pathway. (A) Repair of O6-methylguanine by the MGMT protein. (B) Repair of N1-methyladenine and N3-methylcytosine by ALKB protein. Only N1-methyladenine is represented.
Table 2.
Predicted dinoflagellate orthologues in direct reversal of DNA damage and photoreactivation.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| direct reversal | ||||
| MGMT | * | * | Lp_Unigene68135_All (1e-009) | O6-methylguanine DNA methyltransferase |
| Lp_Unigene71190_All (5e-007) | ||||
| ALKB/APH | Unigene40709_All (5e-018) | symbB.v1.2.000566.t1 (1e-006) | Lp_Unigene17309_All (9e-015) | nucleic acid dioxygenase involved in alkylation damage |
| symbB.v1.2.012827.t1 (5e-017) | ||||
| photoreactivation | ||||
| 6-4 photolyase | CL2432.Contig2_All (2e-113) | symbB.v1.2.031430.t1 (3e-051) | Lp_Unigene48358_All (9e-128) | involved in repair of (6-4) pyrimidine–pyrimidine induced by UV irradiation |
| CL6843.Contig1_All (8e-119) | symbB.v1.2.033063.t1 (5e-114) | Lp_Unigene59149_All (2e-121) | ||
| CPD photolyase | Unigene1971_All (2e-151) | symbB.v1.2.000806.t1 (3e-022) | Lp_Unigene17168_All (3e-015) | involved in repair of cyclobutane-pyrimidine dimer (CPD) induced by UV irradiation |
| CL5100.Contig1_All (1e-80) | symbB.v1.2.012126.t1 (6e-027) | Lp_Unigene75562_All (9e-024) | ||
| symbB.v1.2.023060.t1 (5e-028) | Lp_Unigene21665_All (3e-013) | |||
| symbB.v1.2.037491.t1 (4e-038) | Lp_Unigene43660_All (3e-012) | |||
| symbB.v1.2.001247.t1 (7e-041) | Lp_Unigene4570_All (2e-069) | |||
| symbB.v1.2.030395.t1 (2e-043) | Lp_Unigene41185_All (3e-021) | |||
| symbB.v1.2.030396.t1 (3e-057) | Lp_Unigene47312_All (2e-074) | |||
| symbB.v1.2.030397.t1 (6e-022) | Lp_Unigene58611_All (3e-155) | |||
| symbB.v1.2.031429.t1 (4e-010) | Lp_Unigene63301_All (2e-020) | |||
| symbB.v1.2.011563.t1 (4e-035) | ||||
| cryptochrome DASH | Unigene5536_All (5e-47) | symbB.v1.2.014092.t1 (9e-017) | Lp_Unigene16311_All (3e-108) | removal of cyclobutane-pyrimidine dimers (CPD) in ssDNA |
| Unigene41133_All (4e-110) | symbB.v1.2.021362.t1 (8e-068) | Lp_Unigene17251_All (3e-068) | ||
| Unigene49976_All (5e-69) | symbB.v1.2.025343.t1 (1e-102) | Lp_Unigene20031_All (4e-074) | ||
| symbB.v1.2.028883.t1 (1e-043) | Lp_Unigene25347_All (9e-078) | |||
| symbB.v1.2.027391.t1 (7e-046) | Lp_Unigene63655_All (2e-075) | |||
| symbB.v1.2.021130.t1 (1e-084) | Lp_Unigene67177_All (9e-101) | |||
| symbB.v1.2.030740.t1 (1e-062) | Lp_CL14399.Contig1_All (2e-097) | |||
| symbB.v1.2.029072.t1 (1e-035) | Lp_CL14399.Contig2_All (3e-097) | |||
| symbB.v1.2.015001.t1 (3e-080) | Lp_Unigene7953_All (1e-092) | |||
| symbB.v1.2.006488.t1 (1e-084) | Lp_Unigene24084_All (4e-065) | |||
| symbB.v1.2.007315.t1 (2e-026) | Lp_Unigene39631_All (3e-071) | |||
| Lp_Unigene51086_All (1e-032) | ||||
| Lp_Unigene54913_All (6e-065) | ||||
| Lp_Unigene6860_All (8e-012) | ||||
| Lp_Unigene8874_All (6e-079) | ||||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
3.2.2. Photoreactivation
Photoreactivation, regarded as the most efficient and error-free pathway for reversal of UV-induced DNA damage, is present in bacterial, fungi, animals (except the placental mammals) and plants [62]. In plants, photoreactivation is the major and preferred pathway responsible for UV-induced lesions repair [63,64]. Photoreactivation depends on a single light-activated enzyme called photolyase to recognize and repair the photoproducts cyclobutane-pyrimidine dimers (CPDs) or 6-4 pyrimidine–pyrimidine photoproducts (6-4PPs) at lesion sites [65]. CPD photolyases and (6-4) photolyases, specific for the repair of CPDs and 6-4PPs respectively, belong to the cytochrome/photolyase family (CPF) and share similar biochemical activities on their substrates [66]. Those photolyases directly bind to damaged DNA substrates and rely on light-dependent reduction of their cofactor flavin adenine dinucleotide (FAD) to mediate lesion repair [67,68]. The activated FADH- then passes an electron to the lesions for the cleavage of the covalent bonds within pyrimidine dimers. In addition, Cry-DASH proteins, another group in CPF, are also suggested to be a group of single-stranded DNA photolyases [69,70,71]. They could bind to damaged sites of ssDNAs and have a preference for the repair of CPDs. Our bioinformatic analysis indicated putative orthologues are present in dinoflagellates (Table 2). Dimerization of 5-hydroxymethyluracil (5hmu), the major UV-induced adduct, is theoretically impossible; as 5hmu replaces substantial amount, but not all, genomic thymines, photoreactivation repair in LCCs is likely compartmentalized.
3.2.3. Three Excision Repair Pathways, Nucleotide Excision Repair (NER), Base Excision Repair (BER), and DNA Mismatch Repair (MMR), Confer Single-Strand DNA Damage Repair Through Excision-Coupled Resynthesis
BER and MMR pathways deal with non-bulky lesions in DNA, which are glycosylase-dependent and mismatch base-pair dependent respectively. NER pathway copes with the bulky DNA lesions, which is TFIIH protein complex-dependent. Althouth detailed mechanisms are different among these three excision repair pathways, they share some similar components of DNA resynthesis, including PCNA and DNA polymerase δ.
Base Excision Repair (BER)
BER rectifies a wide range of DNA damages that modify non-bulky bases such as DNA oxidation from reactive oxygen species (ROS) attack, hydrolysis, deamination and alkylation [72,73,74,75]. Impaired DNA bases are identified and removed by DNA glycosylases, generating an abasic (apurinic-apyrimidinic, AP) site in DNA. The dinoflagellates have both mono-functional and bi-functional glycosylases (Figure 3 and Table 3). The glycosylases targeting uracil and its derivatives including UNG [76], SMUG1 [77], MBD4 [78], TDG [77,78] and NTH1 [79], are of special interest. These genes were reported to have activities against 5hmu. The modified base 5hmu is a natural component of the DNA of dinoflagellates, which could replace 12–70% of thymine in genomes of dinoflagellates [53,54,80]. The existence of UNG, MBD4 and NTH1 implicates dinoflagellates must develop a mechanism to distinguish between the damage-induced and endogenous 5hmu, or a specific compartmentalization mechanism. The AP-site generated by mono-functional DNA glycosylases is targeted by AP-endonuclease (APE1), which then produces a single nucleotide nick, leading to the 3′OH and 5′deoxyribosephosphate(dRP) terminal in the DNA backbone. DNA polymerase Polβ is then engaged to remove the 5′-dRP group and produce 3′OH and 5′P ends. Alternatively, the bi-functional DNA glycosylases could cut the phosphodiester bond of the AP-site directly through its AP lyase activity and also create a single nucleotide nick. The nick is further converted into 3′OH and 5′P ends by extra enzymatic activities such as APE1 or polynucleotide kinase (PNKP). Later, the DNA polymerase Polβ and ligase LIG3/XRCC1 complex or ligase LIG1 is involved in gap-filling DNA synthesis and ligation sequentially. Additionally, the long-patch BER pathway is used to cope with 2–12 nucleotides lesions, in which DNA polymerases δ and ε (Polδ, Polε), FEN1, PCNA and DNA ligase I are involved [81,82]. For these steps, LIG3 and XRCC1 were not found in dinoflagellates (Figure 3 and Table 3). No genes equivalent to LIG3 were found in many eukaryotes including most plants and budding yeast [83]. Pol λ and LIG1 were functionally replaced by Polβ and LIG3 in plant Arabidopsis thaliana [84]. A similar mechanism could be adopted by dinoflagellates.
Figure 3.
Diagrammatic summary of dinoflagellate orthologues predicted in the base excision repair pathway. The ellipses filled with grey color mean the absence of putative dinoflagellate orthologues in the searched transcriptomes. The other colors in ellipses indicate the presence of putative dinoflagellate orthologues.
Table 3.
Predicted dinoflagellate orthologues in base excision repair.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| UNG | Unigene44697_All (2e-059) | symbB.v1.2.022126.t1 (9e-069) | Lp_Unigene44751_All (3e-078) | uracil-DNA glycosylases; removes uracil |
| Unigene70580_All (6e-071) | symbB.v1.2.022126.t2 (3e-070) | Lp_Unigene47003_All (4e-071) | ||
| symbB.v1.2.031173.t1 (6e-052) | ||||
| SMUG1 | * | * | * | single-strand selective mono-functional uracil-DNA glycosylase |
| MBD4 | Unigene51401_All (7e-008) | symbB.v1.2.021865.t1 (2e-014) | Lp_Unigene54655_All (3e-019) | DNA N-glycosylase involved in removal of thymine mismatch |
| TDG | * | * | * | thymine DNA glycosylase; removes thymine mismatch, uracil |
| OGG1 | Unigene85095_All (4e-019) | symbB.v1.2.040865.t1 (5e-011) | Lp_Unigene28278_All (1e-054) | 8-oxoG DNA glycosylase |
| Unigene82000_All (1e-013) | Lp_Unigene40876_All (4e-054) | |||
| MYH | Unigene67393_All (4e-036) | symbB.v1.2.014787.t1 (4e-067) | Lp_Unigene17300_All (4e-078) | adenine DNA glycosylase; removes A opposite G |
| NTH1 | Unigene62006_All (2e-034) | symbB.v1.2.007780.t1 (4e-050) | Lp_CL9519.Contig1_All (5e-006) | Bi-functional DNA N-glycosylase; removal of oxidized pyrimidines, formamidopyrimidines, 5-formyluracil |
| Unigene64896_All (2e-045) | symbB.v1.2.038247.t1 (2e-037) | Lp_Unigene32526_All (1e-061) | ||
| Lp_Unigene58890_All (6e-053) | ||||
| MPG/Mag1 | * | symbB.v1.2.024316.t1 (4e-007) | Lp_Unigene52978_All (4e-013) | DNA-3-methyladenine glycosylase |
| FPG | Unigene42128_All (6e-049) | symbB.v1.2.002485.t1 (2e-071) | Lp_Unigene4975_All (3e-064) | formamidopyrimidine-DNA glycosylase |
| CL7240.Contig3_All (3e-048) | Lp_Unigene48155_All (6e-008) | |||
| NEIL1 | * | * | * | DNA glycosylases; removal of oxidized pyrimidines, thymine glycol |
| NEIL2 | * | * | * | DNA glycosylases; removal of oxidized pyrimidines |
| NEIL3 | Unigene48672_All (1e-011) | symbB.v1.2.022143.t1 (5e-018) | Lp_Unigene13451_All (5e-014) | DNA glycosylases; removal of oxidized pyrimidines |
| APE1 | Unigene9067_All (1e-033) | symbB.v1.2.023749.t1 (4e-039) | Lp_CL8817.Contig2_All (3e-011) | apurinic/apyrimidinic (AP) endonuclease |
| Unigene17453_All (6e-006) | symbB.v1.2.024175.t1 (4e-012) | Lp_CL8837.Contig2_All (1e-036) | ||
| Unigene34000_All (2e-012) | symbB.v1.2.033468.t1 (1e-035) | Lp_Unigene2242_All (5e-025) | ||
| Unigene73707_All (2e-011) | symbB.v1.2.033468.t2 (1e-035) | Lp_Unigene33584_All (7e-035) | ||
| Unigene41862_All (3e-037) | ||||
| Unigene73755_All (1e-032) | ||||
| APE2 | CL7855.Contig3_All (1e-014) | symbB.v1.2.021745.t1 (1e-009) | Lp_Unigene55979_All (4e-008) | apurinic/apyrimidinic (AP) endonuclease |
| Lp_Unigene48320_All (7e-010) | ||||
| LIG1 | CL2462.Contig1_All (7e-151) | symbB.v1.2.029028.t1 (3e-011) | Lp_CL8189.Contig1_All (9e-022) | DNA ligase required for long-patch BER |
| Unigene56781_All (2e-051) | symbB.v1.2.029030.t1 (1e-055) | Lp_CL8189.Contig2_All (6e-043) | ||
| symbB.v1.2.007861.t1 (7e-021) | Lp_CL13983.Contig1_All (8e-060) | |||
| symbB.v1.2.007862.t3 (3e-094) | Lp_CL13983.Contig2_All (1e-059) | |||
| symbB.v1.2.007862.t4 (1e-011) | Lp_Unigene18447_All (6e-020) | |||
| symbB.v1.2.007862.t5 (5e-081) | Lp_Unigene44117_All (2e-154) | |||
| symbB.v1.2.007862.t6 (4e-081) | ||||
| XRCC1 | * | * | * | interacts with PARP, LIG3, and Polβ |
| LIG3 | * | * | * | DNA ligase required for short-patch BER |
| PKNP | CL5342.Contig1_All (5e-029) | Lp_Unigene21188_All (3e-025) | DNA 3′-phosphatase 5′-kinase required for restoration of 5′-phosphate and 3′-hydroxyl termini | |
| CL5342.Contig2_All (2e-029) | Lp_Unigene67913_All (4e-012) | |||
| CL5342.Contig3_All (3e-029) | Lp_Unigene67503_All (8e-027) | |||
| FEN1 | Unigene36893_All (6e-102) | symbB.v1.2.012846.t1 (2e-067) | Lp_Unigene74714_All (3e-108) | structure-specific nuclease |
| symbB.v1.2.005353.t1 (1e-047) | ||||
| symbB.v1.2.017794.t1 (9e-099) | ||||
| PCNA | CL2939.Contig1_All (1e-075) | symbB.v1.2.024689.t1 (1e-069) | Lp_CL16467.Contig1_All (3e-051) | loading platform of PolD1 and PolE1 |
| CL2939.Contig2_All (3e-075) | symbB.v1.2.005740.t1 (1e-072) | Lp_CL16467.Contig2_All (5e-072) | ||
| CL2939.Contig3_All (5e-076) | symbB.v1.2.003346.t1 (1e-036) | Lp_CL16467.Contig3_All (2e-039) | ||
| CL2939.Contig4_All (1e-075) | Lp_CL16467.Contig5_All (4e-051) | |||
| CL2939.Contig5_All (1e-075) | Lp_CL16467.Contig6_All (3e-056) | |||
| CL2939.Contig6_All (5e-076) | Lp_CL16467.Contig7_All (6e-057) | |||
| Lp_CL16467.Contig9_All (9e-058) | ||||
| Lp_Unigene31367_All (2e-075) | ||||
| Lp_Unigene31369_All (3e-072) | ||||
| Lp_CL9592.Contig1_All (1e-008) | ||||
| Lp_CL15065.Contig1_All (1e-011) | ||||
| Lp_CL15065.Contig2_All (1e-010) | ||||
| Lp_CL15065.Contig4_All (5e-010) | ||||
| Lp_Unigene7276_All (2e-033) | ||||
| Lp_Unigene31368_All (7e-029) | ||||
| Lp_Unigene31370_All (1e-027) | ||||
| Lp_Unigene31371_All (2e-022) | ||||
| PolB | Unigene65173_All (8e-055) | symbB.v1.2.005179.t1 (8e-051) | Lp_Unigene74664_All (5e-061) | DNA polymerase β/beta |
| Unigene70894_All (1e-009) | ||||
| POLE1 | Unigene29376_All (0.0) | symbB.v1.2.023008.t1 (4e-015) | Lp_CL7119.Contig1_All (0.0) | DNA polymerase ε/epsilon catalytic subunit |
| Unigene37482_All (3e-126) | symbB.v1.2.023011.t1 (3e-019) | Lp_CL7119.Contig2_All (0.0) | ||
| symbB.v1.2.017036.t1 (6e-006) | Lp_Unigene71337_All (5e-029) | |||
| symbB.v1.2.001887.t1 (7e-131) | ||||
| symbB.v1.2.001887.t2 (2e-130) | ||||
| symbB.v1.2.001887.t3 (3e-048) | ||||
| symbB.v1.2.001889.t1 (1e-042) | ||||
| POLD1 | Unigene4585_All (3e-050) | symbB.v1.2.029507.t1 (1e-045) | Lp_Unigene28445_All (5e-082) | DNA polymerase δ/delta catalytic subunit |
| Unigene44692_All (3e-029) | symbB.v1.2.037870.t1 (2e-089) | Lp_Unigene37178_All (3e-071) | ||
| Unigene50187_All (8e-055) | symbB.v1.2.036929.t1 (3e-144) | Lp_Unigene55814_All (1e-019) | ||
| Unigene61374_All (0.0) | symbB.v1.2.036930.t1 (3e-118) | Lp_Unigene70233_All (0.0) | ||
| Unigene77552_All (2e-148) | symbB.v1.2.033323.t1 (2e-014) | Lp_Unigene71779_All (0.0) | ||
| PARP1 | CL1934.Contig3_All (1e-015) | symbB.v1.2.024122.t1 (6e-011) | Lp_CL11756.Contig2_All (2e-006) | poly(ADP-Ribose) polymerase-1; necessary for recruitment of other DNA-repairing enzymes |
| Unigene13204_All (1e-027) | symbB.v1.2.018837.t1 (5e-048) | Lp_CL11756.Contig3_All (2e-007) | ||
| Unigene82709_All (1e-015) | Lp_Unigene29089_All (6e-048) | |||
| PARP2 | Unigene9060_All (7e-006) | symbB.v1.2.001263.t1 (4e-022) | Lp_CL1803.Contig2_All (5e-028) | poly(ADP-Ribose) polymerase-2; necessary for recruitment of other DNA-repairing enzymes |
| Unigene45362_All (7e-017) | Lp_Unigene35603_All (2e-045) | |||
| Lp_Unigene36239_All (5e-019) | ||||
| Lp_Unigene54635_All (2e-142) | ||||
| PARP3 | Unigene53685_All (3e-016) | symbB.v1.2.040205.t1 (2e-035) | Lp_Unigene5203_All (6e-032) | poly(ADP-Ribose) polymerase-3; necessary for recruitment of other DNA-repairing enzymes |
| symbB.v1.2.034465.t1 (1e-015) | ||||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
Nucleotide Excision Repair
Nucleotide Excision Repair (NER) is a multi-step process that recognizes and removes bulky structurally unrelated DNA lesions, such as CPDs and 6–4PPs induced by UV irradiation, endogenous oxidative DNA damage, and DNA adducts formed by environmental and chemical mutagens [85,86,87,88,89,90]. NER consists of two pathways: the global genome NER (GG-NER) and the transcription-coupled NER (TC-NER). GG-NER occurs in all regions of genomic DNA, while TC-NER is dedicated to repairing lesions that block the transcription of active genes. The two pathways adopt different strategies to initiate DNA damage recognition but share the same sets of proteins in the later stages of the repair process. In mammalian cells, with the help of UV-damaged DNA-binding complex composed of DDB1 and DDB2 subunits, XPC-Rad23B complex (with CETN2) is responsible for the recognition of the DNA lesions in GG-NER pathway [87,91]. Stalled RNA polymerase activates the TC-NER pathway and recruits CSA and CSB proteins to remove them at lesion sites [92,93]. TFIIH protein complex is involved in the next steps of the GG-NER and TC-NER pathways for DNA lesion verification, helix unwinding and incision. TFIIH comprises of 10 subunits and can be divided into a core complex consisting of XPB, XPD, p8, p34, p44, p52 and p62, and a cyclin-activated kinase (CAK) complex containing MAT1, CDK7 and cyclin H [94,95,96]. The helicase and ATPase activities of XPB and XPD are required for unwinding the DNA into a bubble-like structure at the lesions. Later, the pre-incision complex consisting of XPA, RPA, and XPG is formed at the damaged DNA sites, which help to recruit endonucleases. Incision action is thought to be initiated by the 5′ incision complex ERCC1-XPF, followed by 3′ incision of XPG, creating a single-strand DNA break. DNA polymerase δ, ε, and κ (Polδ, Polε and Pol κ), with the assistance of PCNA and RFC, are required for the subsequent DNA repair synthesis. DNA ligase LIG1 or LIG3/XRCC1 complex are involved in the final gap filling after DNA repair synthesis.
Except DDB2, XPA, the subunits of TFIIH complex including GTF2H1 and GTF2H5, and the RPA3 subunits of RPA complex, the other proteins involved in NER pathway were identified in dinoflagellates (Figure 4 and Table 4). XPA, an intrinsically unstructured protein, is a crucial component of the pre-incision complex for DNA damage recognition [97]. No putative orthologues were found in plants, although they have the ability to remove UV photoproducts in NER dependent pathway [98]. Only three subunits of TFIIH complex were found in L. polyedrum in an analysis of basal transcriptional factors [99]. The absence of putative XPA, RPA3 and GTF2H1 orthologues were also reported in Trypanosomatids [100].
Figure 4.
Diagrammatic summary of the dinoflagellate orthologues predicted in the nucleotide excision repair pathway. The ellipses filled with grey color mean the absence of putative dinoflagellate orthologues in the searched transcriptomes. The other colors in ellipses indicate the presence of putative dinoflagellate orthologues.
Table 4.
Predicted dinoflagellate orthologues in nucleotide excision repair.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| DDB1 | * | symbB.v1.2.027839.t1 (2e-058) | Lp_Unigene56329_All (2e-161) | component of UV-damaged DNA-binding protein complex |
| DDB2/XPE | * | * | * | component of UV-damaged DNA-binding protein complex |
| XPC/Rad4 | Unigene32549_All (8e-015) | symbB.v1.2.001527.t1 (2e-009) | Lp_Unigene33147_All (3e-017) | binds damaged DNA as XPC complex |
| Rad23 | Unigene21346_All (2e-006) | symbB.v1.2.023245.t1 (2e-022) | Lp_CL13062.Contig2_All (2e-007) | binds damaged DNA as XPC complex |
| Unigene68760_All (3e-019) | symbB.v1.2.021833.t1 (6e-017) | Lp_Unigene20125_All (6e-010) | ||
| symbB.v1.2.010422.t1 (6e-009) | Lp_Unigene72427_All (2e-012) | |||
| Lp_CL7915.Contig1_All (7e-009) | ||||
| Lp_CL7915.Contig2_All (8e-009) | ||||
| Lp_CL13062.Contig1_All (2e-007) | ||||
| Lp_Unigene36792_All (8e-006) | ||||
| Lp_Unigene50304_All (3e-006) | ||||
| CETN2 | Unigene29893_All (5e-049) | symbB.v1.2.018116.t1 (5e-043) | Lp_CL4243.Contig1_All (1e-043) | component of the XPC complex |
| Unigene38411_All (2e-047) | Lp_CL4243.Contig2_All (7e-043) | |||
| Lp_CL4243.Contig3_All (2e-043) | ||||
| ERCC8/CSA/Rad28 | Unigene20816_All (1e-009) | symbB.v1.2.020387.t1 (2e-010) | * | required for transcription-coupled nucleotide excision repair |
| symbB.v1.2.020387.t2 (2e-010) | ||||
| ERCC-6/Rad26/CSB | Unigene349_All (7e-166) | symbB.v1.2.032523.t1 (5e-084) | Lp_CL6060.Contig1_All (1e-038) | required for transcription-coupled excision repair |
| Unigene1702_All (8e-056) | symbB.v1.2.026497.t1 (7e-057) | Lp_CL6060.Contig2_All (2e-036) | ||
| CL1148.Contig1_All (1e-037) | symbB.v1.2.019527.t1 (1e-161) | Lp_CL12386.Contig2_All (8e-123) | ||
| Unigene61410_All (2e-041) | symbB.v1.2.019505.t1 (1e-033) | Lp_CL12386.Contig1_All (7e-123) | ||
| symbB.v1.2.019507.t1 (2e-024) | Lp_CL14187.Contig1_All (6e-157) | |||
| symbB.v1.2.026498.t1 (3e-021) | Lp_CL14187.Contig2_All (7e-157) | |||
| Lp_Unigene36232_All (2e-038) | ||||
| XPA | * | * | * | binds to DNA damage foci in pre-incision complex |
| RPA1 | Unigene13507_All (3e-041) | symbB.v1.2.000388.t1 (7e-036) | Lp_CL2032.Contig1_All (4e-047) | subunits of heterotrimeric replication protein A complex |
| Unigene40788_All (9e-018) | symbB.v1.2.016776.t1 (3e-023) | Lp_CL2032.Contig3_All (2e-048) | ||
| Unigene69752_All (7e-034) | symbB.v1.2.010569.t1 (9e-043) | Lp_CL2032.Contig4_All (2e-048) | ||
| Unigene72956_All (1e-043) | symbB.v1.2.015898.t1 (2e-051) | Lp_CL2227.Contig4_All (1e-054) | ||
| Lp_CL2227.Contig5_All (2e-057) | ||||
| Lp_CL2227.Contig6_All (2e-060) | ||||
| Lp_CL2227.Contig8_All (4e-058) | ||||
| Lp_CL2227.Contig9_All (1e-047) | ||||
| Lp_Unigene5237_All (3e-048) | ||||
| Lp_CL2032.Contig2_All (6e-047) | ||||
| Lp_CL2227.Contig1_All (3e-044) | ||||
| Lp_CL2227.Contig2_All (2e-042) | ||||
| Lp_CL2227.Contig3_All (2e-037) | ||||
| Lp_CL2227.Contig7_All (3e-045) | ||||
| Lp_CL14409.Contig1_All (2e-045) | ||||
| Lp_CL14409.Contig2_All (2e-046) | ||||
| Lp_CL14409.Contig3_All (2e-046) | ||||
| Lp_Unigene27814_All (2e-045) | ||||
| RPA2 | Unigene14481_All (5e-010) | symbB.v1.2.033139.t1 (3e-007) | Lp_CL6472.Contig2_All (1e-007) | subunits of heterotrimeric replication protein A complex |
| Lp_CL6472.Contig5_All (7e-008) | ||||
| Lp_CL6472.Contig6_All (6e-007) | ||||
| Lp_CL9761.Contig1_All (2e-008) | ||||
| Lp_CL9761.Contig2_All (1e-008) | ||||
| Lp_Unigene6245_All (8e-008) | ||||
| Lp_Unigene13916_All (9e-009) | ||||
| RPA3 | * | * | * | subunits of heterotrimeric replication protein A complex |
| RFC1 | CL4680.Contig1_All (1e-076) | symbB.v1.2.031425.t1 (2e-037) | Lp_CL3410.Contig1_All (2e-050) | required for strand displacement and DNA synthesis |
| CL4680.Contig2_All (2e-023) | symbB.v1.2.031427.t1 (9e-013) | Lp_CL3410.Contig3_All (1e-084) | ||
| CL4680.Contig3_All (2e-076) | symbB.v1.2.000210.t1 (1e-073) | Lp_CL3410.Contig5_All (2e-006) | ||
| Unigene45646_All (2e-052) | symbB.v1.2.025385.t1 (3e-054) | Lp_CL3410.Contig6_All (3e-016) | ||
| Unigene77812_All (3e-089) | Lp_Unigene33492_All (2e-008) | |||
| Lp_Unigene35510_All (6e-040) | ||||
| Lp_Unigene43360_All (3e-049) | ||||
| TFIIH complex | catalyzes DNA helix unwinding in pre-incision complex | |||
| GTF2H1 | * | * | * | core TFIIH subunit p62 |
| GTF2H2 | Unigene58152_All (5e-034) | * | Lp_Unigene48571_All (2e-051) | core TFIIH subunit p44 |
| GTF2H3 | * | symbB.v1.2.011909.t1 (2e-007) | Lp_Unigene75735_All (6e-014) | core TFIIH subunit p34 |
| GTF2H4 | Unigene45546_All (2e-030) | symbB.v1.2.036961.t2 (2e-016) | Lp_Unigene63874_All (5e-022) | core TFIIH subunit p52 |
| GTF2H5 | * | * | * | core TFIIH subunit p8 |
| ERCC3/XPB | Unigene17412_All (1e-177) | symbB.v1.2.000409.t1 (1e-150) | Lp_Unigene15679_All (7e-142) | core subunits of TFIIH complex, 3′ to 5′ DNA helicase |
| ERCC2/XPD/Rad3 | Unigene61441_All (0.0) | symbB.v1.2.043770.t1 (3e-017) | Lp_Unigene51894_All (0.0) | core subunits of TFIIH complex, 5′ to 3′ DNA helicase |
| symbB.v1.2.003111.t1 (5e-085) | ||||
| symbB.v1.2.003112.t1 (8e-033) | ||||
| symbB.v1.2.003109.t1 (2e-056) | ||||
| CDK7 | Unigene65253_All (6e-042) | * | Lp_CL3145.Contig1_All (3e-076) | subunit of kinase module of TFIIH complex |
| Unigene66528_All (1e-019) | Lp_CL3145.Contig2_All (4e-079) | |||
| Lp_CL3145.Contig3_All (3e-079) | ||||
| MNAT1 | Unigene73596_All (2e-014) | symbB.v1.2.023842.t1 (3e-018) | Lp_Unigene66408_All (2e-017) | subunit of kinase module of TFIIH complex |
| CCNH | * | * | Lp_Unigene28603_All (7e-010) | subunit of kinase module of TFIIH complex |
| ERCC1 | Unigene34115_All (1e-045) | symbB.v1.2.018882.t1 (1e-038) | Lp_Unigene75127_All (5e-053) | 5′ incision DNA-binding subunit of TFIIH complex |
| XPF/ERCC4 | Unigene21562_All (2e-035) | symbB.v1.2.008218.t1 (1e-056) | Lp_Unigene48147_All (2e-057) | 5′ incision catalytic subunit of TFIIH complex |
| Lp_Unigene63025_All (4e-049) | ||||
| XPG/ERCC5 | Unigene8568_All (2e-031) | symbB.v1.2.006008.t1 (4e-025) | Lp_Unigene10768_All (2e-023) | 3′ incision subunits of TFIIH complex |
| Unigene57291_All (2e-024) | symbB.v1.2.006011.t1 (2e-026) | Lp_Unigene47869_All (4e-033) | ||
| symbB.v1.2.006013.t1 (3e-008) | ||||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
Mismatch Repair
DNA mismatch repair is a DNA repair pathway specializing in repairing mismatched bases during DNA replication, process mismatch-containing heteroduplex during recombination, and correct DNA lesions resulting from either endogenous or exogenous sources [101,102,103,104]. In bacteria, MutS and MutL homodimers are responsible for the initial DNA mismatch recognition, followed by the endonuclease MutH’s nick on the newly synthesized strand. The nicked strand is further excised by the exonuclease and re-synthesized by DNA polymerase [105,106]. Similar eukaryotic mechanisms are also applied for the recognition and repair of DNA mismatches. MutS homologs-membered heterodimers MSH2-MSH6 (MutSα) or MSH2-MSH3 (MutSβ) are used to scan DNA mispair. MutSα has a preference of binding single base mismatches and 1 or 2 base insertions or deletions (indels), while MutSβ is capable of recognizing both base mispairs and small/large indels [101]. The MutL homologs also form heterodimers MLH1-PMS2 (MutLα, also known as MLH1-PMS1 complex in budding yeast cells), which are then recruited to form a ternary complex with MutS homologs to initiate the incision at lesion sites. PCNA protein is thought to activate the endonuclease activities of MutLα and help to distinguish the two DNA strands. Exonuclease 1 (Exo1) subsequently cleaved the nicked mispaired strands and the resulting gap is filled by DNA polymerase Polδ and DNA ligase LIG1. Except MSH3, which is also absent from the apicomplexan Plasmodium [107], all the other orthologues are present in dinoflagellates (Figure 5 and Table 5). Detailed mechanism without heterodimer MutSβ awaits further functional investigation.
Figure 5.

Diagrammatic summary of the dinoflagellate orthologues predicted in the DNA mismatch repair pathway. The rectangles and ellipses filled with grey color indicate the absence of putative dinoflagellate orthologues in the searched transcriptomes. The other colors in rectangles and ellipses indicate the presence of putative dinoflagellate orthologues.
Table 5.
Predicted dinoflagellate orthologues in DNA mismatch repair.
| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| MLH1 | Unigene48309_All (7e-103) | symbB.v1.2.021543.t1 (3e-111) | Lp_Unigene17816_All (2e-116) | MutL homolog, form heterodimer with PMS1, MLH2 and MLH3 |
| Unigene56264_All (6e-020) | ||||
| MLH3 | * | symbB.v1.2.038425.t1 (7e-014) | Lp_Unigene24248_All (7e-036) | MutL homolog, form heterodimer with MLH1 |
| PMS1 | CL7093.Contig1_All (1e-042) | symbB.v1.2.037608.t1 (1e-051) | Lp_CL13190.Contig3_All (4e-049) | MutL homolog, form heterodimer with MLH1, part of the DNA mismatch repair (MMR) complex |
| CL7093.Contig2_All (9e-044) | symbB.v1.2.037606.t1 (2e-014) | Lp_Unigene124_All (2e-049) | ||
| CL7093.Contig3_All (3e-041) | Lp_Unigene48112_All (9e-040) | |||
| MSH1 | Unigene56453_All (2e-010) | symbB.v1.2.040100.t1 (2e-012) | Lp_Unigene39523_All (1e-006) | MutS homolog, required for repair of mitochondrial DNA |
| Unigene41842_All (3e-007) | symbB.v1.2.011567.t1 (4e-019) | |||
| MSH2 | Unigene8950_All (3e-133) | symbB.v1.2.002929.t1 (1e-135) | Lp_Unigene75744_All (1e-138) | MutS homolog 2, form heterodimers with MSH3 or MSH6; component of the DNA mismatch repair (MMR) complex |
| symbB.v1.2.002929.t2 (2e-061) | ||||
| MSH3 | * | * | * | MutS homolog 3 |
| MSH6 | Unigene36848_All (2e-072) | symbB.v1.2.026874.t1 (1e-056) | Lp_CL3961.Contig1_All (6e-086) | MutS homolog 6 |
| Unigene62212_All (4e-011) | symbB.v1.2.038785.t1 (5e-012) | Lp_CL3961.Contig2_All (7e-007) | ||
| Unigene69195_All (3e-023) | symbB.v1.2.039566.t1 (1e-014) | Lp_CL3961.Contig3_All (5e-100) | ||
| Lp_CL3961.Contig4_All (2e-100) | ||||
| Lp_CL3961.Contig5_All (2e-082) | ||||
| Lp_CL3961.Contig6_All (7e-101) | ||||
| MSH4 | * | symbB.v1.2.013503.t1 (7e-060) | Lp_Unigene9260_All (8e-065) | MutS homolog 4, specific for meiosis |
| MSH5 | Unigene89588_All (4e-019) | symbB.v1.2.033801.t1 (7e-079) | Lp_Unigene70169_All (1e-039) | MutS homolog 5, specific for meiosis, formation of heterodimer with MSH4 |
| Lp_Unigene63036_All (2e-030) | ||||
| EXO1 | Unigene52745_All (3e-035) | symbB.v1.2.019143.t1 (1e-043) | Lp_CL3557.Contig1_All (9e-048) | 5′ exonuclease |
| CL691.Contig1_All (7e-037) | Lp_Unigene36569_All (2e-037) | |||
| Lp_Unigene71445_All (9e-056) | ||||
| Lp_Unigene77209_All (1e-007) | ||||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
3.2.4. DNA Double-Strand Breaks Repair
DNA double-strand breaks (DSBs) are one of the most cytotoxic chromosome lesions in which breaks occurs in both parallel strands of duplex DNA. DNA DSBs could be caused by many endogenous and exogenous insults including chromosome replication errors, unintended breakage by nuclear enzymes, reactive species during normal metabolism, ionizing radiation and certain chemical drugs [108]. It has been estimated that DSBs happened spontaneously at a frequency of about one per 108 base pair per cell cycle for many organisms [109]. Additionally, a single unrepaired DNA DSB could lead to cell unviability [110].
Two major distinct pathways are involved in the repair of DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ could occur in the whole cell cycle and particularly during the G1 phase. HR is mainly restricted to in the mid-S and G2 cell-cycle stages, when the nascent sister chromatids are available for recombination repair template.
Non-Homologous End Joining
Non-Homologous End Joining (NHEJ) refers to a type of DNA DSB repair that requires little or no sequence homology (≤4 nucleotides) to re-join two DSB ends [111,112]. Binding the DSB ends with the heterodimer Ku protein of Ku70 and Ku80 serves as a loading platform to promote recruitment of other NHEJ proteins [113,114]. Compatible DNA termini such as blunt-ends could be directly repaired with the binding of XRCC4-DNA ligase IV to those termini. For non-compatible ends, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is engaged in a complex with DNA nuclease Artemis to process these ends [115,116]. In the complex with DNA-PKcs, Artemis acquires endonuclease activity on the DNA hairpin, and both the 5' and 3' DNA overhangs. Other proteins including aprataxin, APLF and PNK could also execute its function in the modification of these non-compatible ends [117,118,119,120]. Pol X family DNA polymerase, Pol4 in budding yeast [121], PolL and PolM in mammalian cells [122,123], are involved in the nucleotide synthesis during the processing of the ends. In addition, end processing is a repetitive progress, of which modifications could be performed for multiple rounds, leading to a small sequence deletion or insertion [124]. Finally, the ligase IV complex including XRCC4-LIG4 and XLF is responsible for the ligation of the reconstituted compatible ends [125,126,127].
Key players involved in NHEJ, such as Ku70, Ku80, DNA-PK and LIG4 were identified, but orthologues of XLF, XRCC4 and Artemis were not found in dinoflagellates transcriptomes (Table 6). Fission yeast XRCC4 gene is highly divergent from human orthologues and has only been found in recent years [128], whereas XLF orthologues were not found in plants genomes [129]. The lack of full NHEJ pathway was reported in apicomplexan Plasmodium, with the absence of Ku70, Ku80, DNA-PK and LIG4 [130].
Table 6.
Predicted dinoflagellate orthologues in non-homologous end joining.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Ku70 | Unigene61676_All (3e-062) | symbB.v1.2.013334.t1 (2e-048) | Lp_Unigene63133_All (8e-069) | components of Ku heterodimer, which binds to damaged DNA ends |
| Ku80 | Unigene21225_All (8e-012) | symbB.v1.2.017317.t1 (2e-033) | Lp_Unigene60142_All (3e-041) | components of Ku heterodimer, which binds to damaged DNA ends |
| DNA-PK/PRKDC | Unigene41976_All (4e-076) | symbB.v1.2.025517.t1 (2e-071) | Lp_Unigene62602_All (1e-075) | catalytic subunit of DNA-dependent serine/threonine-protein kinase |
| Unigene56338_All (3e-006) | ||||
| Unigene72430_All (3e-006) | ||||
| LIG4 | Unigene30802_All (5e-018) | symbB.v1.2.033586.t1 (1e-055) | Lp_Unigene8556_All (7e-100) | DNA ligase which forms complex with XRCC4 and responsible for the NHEJ ligation |
| Unigene57597_All (3e-023) | symbB.v1.2.033587.t1 (2e-022) | |||
| Unigene73919_All (3e-029) | ||||
| XRCC4 | * | * | * | forms complex with LIG4 and responsible for the NHEJ ligation |
| XLF/Nej1 | * | * | * | end-joining factor; serve as bridge for XRCC4-LIG4 complex |
| Artemis | * | * | * | nuclease for DNA ends processing |
| APTX | Unigene85214_All (2e-008) | symbB.v1.2.019417.t1 (2e-018) | * | resolve of DNA single-strand interruptions |
| APLF | * | symbB.v1.2.012028.t1 (7e-006) | * | auxiliary factor for DNA end-joining |
| PolL | Unigene72723_All (3e-018) | symbB.v1.2.039928.t1 (8e-031) | Lp_Unigene32155_All (3e-036) | DNA polymerase λ/lambda, involved in NHEJ of nuclear DNA |
| symbB.v1.2.007119.t1 (2e-037) | Lp_Unigene47083_All (3e-040) | |||
| PolM | * | * | * | DNA polymerase μ/mu |
| Pol4 | * | * | * | DNA polymerase IV |
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
Homologous Recombination
Homologous Recombination (HR) refers to a high-fidelity DSB repair mechanism with the use of a homology sequence as the repair template. In HR, extensive 5′ to 3′ resection of one DNA strand is required at first to produce 3′-OH ended ssDNA tails after DSB formation. DNA termini resection is initiated by the combined action of Mre11-Rad50-NBS1 (MRN) complex and nuclease CtIP, creating a short stretch of ssDNA. Extensive resection is then carried out by additional nucleases and exonucleases including CtIP, DNA2, BLM and EXOI [131,132]. The length of the extensively resected ssDNA tail could range from hundreds to thousands of nucleotides [131]. The RPA protein then binds to the resulting resected ssDNAs to protect it from nucleolytic degradation and formation of secondary structures. In order to proceed to recombination, recombinase Rad51, the central player of HR, is needed to be loaded onto the RPA-coated ssDNA. Mediator proteins, such as Rad52 in yeast and BRCA2 in mammalian cell, are responsible for promoting the Rad51 nucleofilament formation through the displacement of RPA. Other proteins including Rad51 paralogs and Rad54 help to stabilize the Rad51 filaments [133]. Additionally, negative regulators, e.g., DNA helicase Srs2 in yeast, recQ5, BLM and FANCJ in mammalian cells, could suppress Rad51 function via the disassembly of Rad51-ssDNA filaments [134]. Following nucleofilament formation, the recognition of homology sequences and strand invasion ensues, generating a structure called displacement loops (D-loops), primed for DNA repair synthesis from the invading 3'-end ssDNA [135].
In canonical Double-Strand Break Repair (DSBR), the other ssDNA end pairs with the displaced template strand and forms a double Holiday junction (DHJ). The resolution of DHJs could lead to either cross-over or non-crossover recombination products [136]. Alternatively, DHJs formation is inhibited in the synthesis dependent strand annealing (SDSA) mode of HR, in which the D-loops are disrupted after the limited DNA synthesis from the invading 3′-end ssDNA. The displaced 3′-end ssDNA then recombine with the complementary strand of the second 3′-end ssDNA tail, followed by repair DNA synthesis, leading to the formation of non-crossover products. SDSA is the preferred recombination pathway during mitosis [134].
HR pathway, members of which are also important for meiosis, is uninvestigated at the mechanistic level in dinoflagellates. The essential components of HR including Rad51, MRN complex (MRE11-NBS1-RAD50) and RPA (except RPA3 subunit) could be found in their transcriptomes (Table 7). The mediator protein Rad52 was absent in Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, Plamodium [130] and Trypanosomatids [100], as in the case for dinoflagellates. A putative homolog BRCA2 was found in Symbiodinum, which could function as mediator for Rad51. Other putative orthologues such as RMI2, DNA2, SLX4 and EME1 were absent possibly due to the low sequence conservation in dinoflagellates. The recombinase RAD51 responsible for catalyzing the homology search and strand change is the essential protein in the HR pathway [137,138]. Comparative analysis with other eukaryote RAD51 orthologues showed that it contains the canonical Walker A and Walker B motif of the RECA/RAD51 superfamily (Figure 6). A phylogenetic tree constructed with selective eukaryotic orthologues exhibited dinoflagellate RAD51 orthologues forming an individual clade (Figure S2). The prokaryotic RecA orthologues, which are involved in maintaining the integrity of chloroplast and mitochondria genome [139,140], are also present in S. minutum and L. polyedrum transcriptomes.
Table 7.
Predicted dinoflagellate orthologues in homologous recombination.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| RAD51 | Unigene44686_All (1e-133) | symbB.v1.2.024814.t1 (4e-084) | Lp_CL13972.Contig1_All (8e-120) | recombinase, eukaryote RecA homologue |
| Lp_CL13972.Contig3_All (6e-123) | ||||
| Lp_CL13972.Contig4_All (6e-126) | ||||
| Lp_CL13972.Contig5_All (2e-124) | ||||
| Lp_Unigene24767_All (2e-110) | ||||
| DMC1 | Unigene64853_All (9e-113) | symbB.v1.2.000608.t1 (2e-109) | Lp_Unigene20110_All (1e-118) | RAD51 homologue specific for meiosis |
| symbB.v1.2.008353.t1 (3e-109) | ||||
| RecA | * | symbB.v1.2.015650.t1 (2e-063) | Lp_Unigene51570_All (4e-093) | bacterial recombinase |
| symbB.v1.2.028306.t1 (5e-016) | ||||
| RAD51B | Unigene1912_All (2e-016) | * | Lp_Unigene71304_All (1e-023) | RAD51 paralog |
| RAD51C | Unigene41837_All (3e-017) | * | Lp_Unigene66289_All (3e-034) | RAD51 paralog |
| RAD51D | * | * | * | RAD51 paralog |
| XRCC2 | * | symbB.v1.2.008271.t1 (2e-008) | * | DNA break and cross-link repair |
| XRCC3 | * | * | Lp_Unigene71572_All (2e-010) | DNA break and cross-link repair |
| RAD50 | CL6927.Contig1_All (2e-068) | symbB.v1.2.031416.t1 (3e-051) | Lp_Unigene51516_All (1e-065) | part of the MRN complex |
| CL6927.Contig2_All (6e-068) | symbB.v1.2.031416.t2 (3e-051) | |||
| Unigene31993_All (2e-021) | symbB.v1.2.025527.t1 (8e-044) | |||
| Unigene68915_All (7e-019) | ||||
| NBS1 | Unigene77295_All (5e-013) | symbB.v1.2.016565.t1 (3e-014) | Lp_CL4311.Contig2_All (9e-011) | part of the MRN complex |
| MRE11 | Unigene13700_All (8e-117) | symbB.v1.2.022929.t1 (1e-113) | Lp_Unigene63346_All (7e-111) | part of the MRN complex |
| symbB.v1.2.022929.t2 (2e-093) | ||||
| symbB.v1.2.022929.t3 (3e-010) | ||||
| RAD52 | * | * | * | accessory factor for recombination |
| RAD54L | Unigene77564_All (4e-149) | symbB.v1.2.021721.t2 (3e-127) | Lp_CL12947.Contig4_All (4e-148) | involved in recombination |
| symbB.v1.2.012979.t1 (9e-088) | Lp_Unigene46377_All (6e-147) | |||
| symbB.v1.2.021720.t1 (4e-077) | Lp_CL12947.Contig2_All (2e-148) | |||
| symbB.v1.2.021721.t1 (4e-066) | Lp_CL12947.Contig3_All (2e-148) | |||
| symbB.v1.2.021721.t3 (4e-066) | ||||
| RAD54B | * | symbB.v1.2.012978.t1 (6e-024) | Lp_Unigene31872_All (5e-096) | accessory factor for recombination |
| Lp_Unigene981_All (4e-037) | ||||
| BRCA2 | * | symbB.v1.2.003783.t1 (1e-008) | * | involved in recombination |
| DSS1/SHFM1 | * | * | * | BRCA2 accessary factor |
| CtIP | * | symbB.v1.2.018901.t1 (3e-006) | * | involved in DNA end resection |
| BLM/Sgs1 | * | symbB.v1.2.009075.t3 (2e-037) | Lp_Unigene24386_All (1e-018) | 3′-5′ DNA helicase |
| symbB.v1.2.009075.t4 (2e-037) | ||||
| symbB.v1.2.009075.t2 (2e-037) | ||||
| symbB.v1.2.009074.t1 (3e-041) | ||||
| symbB.v1.2.008709.t1 (7e-022) | ||||
| Top3α | Unigene32503_All (2e-163) | symbB.v1.2.026949.t1 (6e-160) | Lp_Unigene1654_All (3e-166) | DNA topoisomerase 3-alpha |
| symbB.v1.2.035328.t1 (2e-021) | Lp_Unigene44680_All (6e-022) | |||
| symbB.v1.2.037631.t1 (2e-006) | Lp_CL12602.Contig1_All (3e-011) | |||
| Lp_Unigene21536_All (4e-012) | ||||
| Lp_Unigene29037_All (1e-011) | ||||
| Lp_Unigene53944_All (3e-006) | ||||
| Lp_Unigene62514_All (3e-012) | ||||
| RMI1 | Unigene5006_All (6e-010) | symbB.v1.2.037665.t1 (1e-006) | Lp_Unigene13746_All (1e-010) | key component of the RMI complex |
| RMI2 | * | * | * | key component of the RMI complex |
| DNA2 | * | * | * | ATP-dependent helicase/nuclease |
| PARI/Srs2 | Unigene16314_All (1e-009) | * | * | inhibit inappropriate homologous recombination |
| MUS81 | Unigene1977_All (6e-016) | symbB.v1.2.018987.t1 (1e-017) | Lp_Unigene50884_All (1e-013) | subunit of structure-specific endonuclease |
| EME1/MMS4 | * | * | * | interaction with Mus81 |
| SLX1 | Unigene40330_All (4e-021) | symbB.v1.2.003012.t1 (1e-020) | Lp_Unigene20489_All (2e-020) | subunit of SLX1-SLX4 structure-specific nuclease |
| symbB.v1.2.009695.t1 (2e-012) | ||||
| SLX4 | * | * | * | subunit of SLX1-SLX4 structure-specific nuclease |
| GEN1/YEN1 | Unigene8182_All (2e-008) | * | Lp_Unigene2100_All (7e-019) | nuclease seperating Holliday junctions |
| Lp_Unigene17855_All (2e-012) | ||||
| SPO11 | Unigene18126_All (3e-012) | symbB.v1.2.038121.t1 (7e-025) | Lp_Unigene20651_All (2e-024) | meiosis specific endonuclease |
| Unigene24373_All (1e-012) | symbB.v1.2.012520.t1 (8e-031) | Lp_Unigene9316_All (2e-038) | ||
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
Figure 6.

Multi-alignment of dinoflagellate RAD51 proteins.
3.2.5. Translesion Synthesis
Translesion Synthesis (TLS)—a highly conserved pathway which repairs DNA damages associated with replication—directly mediates duplication of damaged DNA in the template strand that was blocked [141,142,143]. TLS is mediated by the specialized DNA polymerases that could tolerate DNA lesions. These specialized DNA polymerases, including DNA polymerase REV1, Rev3L, PolH, PolI and PolK, have active sites that accommodate impaired or distorted templates [143,144,145]. Execution of TLS involved the switching of canonical polymerases with TLS polymerases, which is thought to be conducted by the interaction with PCNA and Rev1 [146,147]. The low fidelity of TLS polymerase, orthologues of which are found in dinoflagellates transcriptomes (Table 8), makes the repair progress intrinsically error prone [148].
Table 8.
Dinoflagellate orthologues predicted in translesion synthesis.
| Genes/Alternate Name or Function Homolog | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| Rev3L/PolZ | Unigene7171_All (1e-017) | symbB.v1.2.025103.t1 (1e-067) | Lp_Unigene29950_All (8e-022) | DNA polymerase ζ/zeta catalytic subunit |
| Unigene32515_All (9e-131) | symbB.v1.2.025103.t2 (3e-067) | Lp_Unigene60556_All (1e-025) | ||
| Unigene56596_All (3e-083) | symbB.v1.2.025103.t3 (2e-067) | Lp_Unigene61611_All (1e-030) | ||
| symbB.v1.2.028081.t1 (7e-091) | Lp_Unigene76685_All (4e-026) | |||
| symbB.v1.2.028084.t1 (1e-024) | Lp_Unigene76716_All (1e-013) | |||
| Lp_Unigene81304_All (2e-008) | ||||
| REV1 | Unigene56396_All (3e-046) | symbB.v1.2.017539.t1 (2e-014) | Lp_Unigene31865_All (3e-008) | non-classical DNA polymerase, dCMP transferase |
| symbB.v1.2.017542.t1 (1e-017) | Lp_Unigene55084_All (5e-053) | |||
| Lp_Unigene62480_All (6e-044) | ||||
| PolH/Rad30 | Unigene678_All (9e-062) | symbB.v1.2.015189.t1 (3e-054) | Lp_Unigene8962_All (3e-049) | DNA polymerase η/eta involved in the DNA repair by translesion synthesis |
| Unigene54870_All (1e-008) | symbB.v1.2.015189.t2 (9e-051) | |||
| symbB.v1.2.017537.t1 (3e-027) | ||||
| PolI/Rad30B | Unigene46925_All (8e-036) | symbB.v1.2.027247.t1 (6e-058) | Lp_Unigene39489_All (1e-056) | error-prone DNA polymerase ι/iota involved in bypass of DNA lesions |
| PolK/DINB1 | Unigene49999_All (1e-044) | symbB.v1.2.024275.t1 (1e-016) | Lp_Unigene16086_All (8e-040) | error-prone DNA polymerase κ/kappa involved in bypass of DNA lesions |
#, E-value obtained from tBLASTn algorithm.
3.2.6. DNA Interstrand Crosslinks Repair
DNA interstrand cross-link (ICL), forming covalent bond between two opposite strands of DNA, can be generated from several sources including bi-functional alkylating agents (such as nitrogen mustard), by-products of lipid peroxidation, abasic sites, and natural psoralens [149]. ICLs prevent complimentary DNA strands separation and thus will impose damages at DNA replication and transcription, making it one of the most toxic DNA damages. In eukaryotes, ICL repair occurs through different mechanisms for non-dividing (G1 phase) and dividing cells (S or G2/M phase) [150,151,152]. However, both mechanisms share similar steps, which include nuclease-mediated detachment from one DNA strand, coupled with TLS polymerase-dependent synthesis across the ICL-containing DNA region, rendering a complete DNA template to finish the repair.
Fanconi anemia is a rare genetic disease associated with the mutation of one of the 19 known FANC genes [153]. In cooperation with NER, TLS and HR pathway, the FANC proteins play important roles in signaling and repair of the replication-dependent ICLs [152,154,155]. ICLs recognition is mediated through binding of FANCM to the damaged sites, which function as a landing platform for the recruitment of heptameric FANC core complex (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG and FANCL). The FANC core complex further interacts with many other proteins including other FANC proteins and repair factors to repair the ICLs. It should be mentioned that the full Fanconi anemia pathway genes could to be only found in mammals but not in other organisms. In the yeast Saccharomyces cerevisiae and the plant Arabidopsis thaliana, a partial Fanconi pathway associated with FANCM was used to repair the ICLs [156,157]. Surprisingly, none of the FANC core complexs, FANCM, and FANCM accessory factors MHF1 and MHF2, were identified in dinoflagellates transcriptomes (Table 9), although we are not certain if their levels at vegetative life cycles may be too rare for mRNA isolation.
Table 9.
Predicted dinoflagellate orthologues predicted in interstrand crosslinks repair.
| Genes | Gene ID (E-Value #) | Activity/Remarks | ||
|---|---|---|---|---|
| C. cohnii | S. minutum | L. polyedrum | ||
| FANCA | * | * | * | core complex member required for interstrand DNA cross-link repair |
| FANCB | * | * | * | ibid |
| FANCC | * | * | * | ibid |
| FANCE | * | * | * | ibid |
| FANCF | * | * | * | ibid |
| FANCG | * | * | * | ibid |
| FANCL | * | * | * | ibid |
| FANCM | * | * | * | lesion recognition and recruitment of the core complex |
| MHF1 | * | * | * | involved in the damage-dependent DNA binding of FANCM |
| MHF2 | * | * | * | involved in the damage-dependent DNA binding of FANCM |
| SNM1 | Unigene68129_All (9e-006) | symbB.v1.2.005478.t1 (5e-046) | Lp_Unigene56381_All (2e-063) | required for interstrand DNA cross-link repair |
| Unigene48769_All (6e-023) | ||||
| SNM1B | * | symbB.v1.2.023872.t2 (1e-024) | Lp_Unigene44216_All (4e-036) | related to SNM1 |
*, no orthologues could be found; #, E-value obtained from tBLASTn algorithm.
The Snm1 family of widely conserved proteins, encoding metallo-b-lactamases (MBL) with nuclease activities, is believed to function in the end procession of the detached ICLs [158,159]. Snm1 deletion in budding yeast showed a hypersensitivity to ICL reagents psoralen and nitrogen mustard [159]. Mammalian cells have three SNM1 orthologues, SNM1A, SNM1B and SNM1C [154], although only SNM1A is thought to be the functional homolog. Both SNM1 and SNM1B were identified in dinoflagellates transcriptomes (Table 9).
4. Conclusions and Perspectives
Eukaryotic DNA damage responses (DDR) evolved from—and build on—prokaryotic counterparts, with modifications for nucleosomal accessibility; whether there is a reversal of evolution in dinoflagellates would not only be of interest in evolutionary biology, and potentially bears a biotechnology element for up-scale synthetic biology. We explored the presence of putative orthologues of DDRs genes in three dinoflagellate transcriptomes. We found most of orthologues of DNA damage checkpoint signaling networks, DNA repair pathways including direct reversal of DNA lesion, photoreactivation, BER, NER, mismatch repair, and NHEJ, homologous recombination repair and translesion synthesis, with one exception of Fanconi pathway required for repair of interstrand crosslinks. We speculate that this may be attributed to different forms of DNA damage in anisotropically-aligned supercoiled domains.
Our current knowledge of dinoflagellate DDR is still in its infancy, with a paucity of investigations addressing the specific effect on cell-cycle control, which will directly impact dinoflagellate productivity, and could potentially overlap with the effect of photo-inhibition. The present study represents a starting point for further research in the molecular biology of dinoflagellate DNA damage responses. How DDR coordinates with organelle genome damage processing, in which nuclear genomes seem to have disproportionate representation (especially plastid minicircle) will need to be addressed. In nucleosomal eukaryotes, core histones have major roles in DDRs [160,161]. Their substantially lower expression levels, as well as the lack of nucleosomal nuclease resistance pattern, will have major implications to the molecular mechanism of DNA repair.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/7/7/191/s1. Table S1. Sequences of extracted putative DDR orthologues from C. cohnii and L. polyedrum.
Author Contributions
J.T.Y.W. conceived the study. C.L. performed the analyses. C.L. and J.T.Y.W. wrote and edited the manuscript.
Funding
The present work was supported in part by GRF16101415 and IEG17SC05, respectively from Hong Kong Research Grant Council and HKUST, to J.T.Y.W.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Matthiessen J., Schreck M. Dinoflagellates. In: Harff J., Meschede M., Petersen S., Thiede J., editors. Encyclopedia of Marine Geosciences. Springer; Dordrecht, The Netherlands: 2014. pp. 1–7. [Google Scholar]
- 2.Saldarriaga J.F., Taylor F.J.R. Dinoflagellata. In: Archibald J.M., Simpson A.G.B., Slamovits C.H., Margulis L., Melkonian M., Chapman D.J., Corliss J.O., editors. Handbook of the Protists. Springer International Publishing; Cham, Switzerland: 2017. pp. 1–54. [Google Scholar]
- 3.Anderson D.M., Cembella A.D., Hallegraeff G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 2011;4:143–176. doi: 10.1146/annurev-marine-120308-081121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells M.L., Trainer V.L., Smayda T.J., Karlson B.S., Trick C.G., Kudela R.M., Ishikawa A., Bernard S., Wulff A., Anderson D.M., et al. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae. 2015;49:68–93. doi: 10.1016/j.hal.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franklin D.J., Berges J.A. Mortality in cultures of the dinoflagellate Amphidinium carterae during culture senescence and darkness. Proc. R. Soc. Lond. B Biol. Sci. 2004;271:2099–2107. doi: 10.1098/rspb.2004.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong J.T.Y. Architectural organization of dinoflagellate liquid crystalline chromosomes. Microorganisms. 2019;7:27. doi: 10.3390/microorganisms7020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeung P.K., Wong J.T. Inhibition of cell proliferation by mechanical agitation involves transient cell cycle arrest at G1 phase in dinoflagellates. Protoplasma. 2003;220:173–178. doi: 10.1007/s00709-002-0039-2. [DOI] [PubMed] [Google Scholar]
- 8.Lesser M.P. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs. 1997;16:187–192. doi: 10.1007/s003380050073. [DOI] [Google Scholar]
- 9.Hoegh-Guldberg O., Mumby P.J., Hooten A.J., Steneck R.S., Greenfield P., Gomez E., Harvell C.D., Sale P.F., Edwards A.J., Caldeira K., et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- 10.Ekelund N.G.A. Effects of UV-B radiation on growth and motility of four phytoplankton species. Physiol. Plant. 1990;78:590–594. doi: 10.1111/j.1399-3054.1990.tb05246.x. [DOI] [Google Scholar]
- 11.Ekelund N.G.A. The effects of UV-B radiation on dinoflagellates. J. Plant Physiol. 1991;138:274–278. doi: 10.1016/S0176-1617(11)80287-7. [DOI] [Google Scholar]
- 12.Banaszak A.T., Trench R.K. Effects of ultraviolet (UV) radiation on marine microalgal-invertebrate symbioses. I. Response of the algal symbionts in culture an in hospite. J. Exp. Mar. Biol. Ecol. 1995;194:213–232. doi: 10.1016/0022-0981(95)00072-0. [DOI] [Google Scholar]
- 13.Dodge J. Effects of ultra-violet light on the survival and nuclear division of a Dinoflagellate. Protoplasma. 1965;59:485–493. doi: 10.1007/BF01252452. [DOI] [Google Scholar]
- 14.Lessard E.J. The trophic role of heterotrophic dinoflagellates in diverse marine environments. Mar. Microb. Food Webs. 1991;5:49–58. [Google Scholar]
- 15.Schnepf E., Elbrächter M. Nutritional strategies in dinoflagellates: A review with emphasis on cell biological aspects. Eur. J. Protistol. 1992;28:3–24. doi: 10.1016/S0932-4739(11)80315-9. [DOI] [PubMed] [Google Scholar]
- 16.Hallegraeff G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. J. Phycol. 2010;46:220–235. doi: 10.1111/j.1529-8817.2010.00815.x. [DOI] [Google Scholar]
- 17.Gao K., Zhang Y., Häder D.-P. Individual and interactive effects of ocean acidification, global warming, and UV radiation on phytoplankton. J. Appl. Phycol. 2018;30:743–759. doi: 10.1007/s10811-017-1329-6. [DOI] [Google Scholar]
- 18.Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., Takeuchi T., Hisata K., Tanaka M., Fujiwara M., et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 19.Chan W.S., Kwok A.C.M., Wong J.T.Y. Knockdown of dinoflagellate cellulose synthase CesA1 resulted in malformed intracellular cellulosic thecal plates and severely impeded cyst-to-swarmer transition. Front. Microbiol. 2019;10:546. doi: 10.3389/fmicb.2019.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C., Chen H., He Y., Xia R. TBtools, a Toolkit for Biologists integrating various biological data handling with a user-friendly interface. BioRXiv. 2018:289660. [Google Scholar]
- 21.Tamura K., Peterson D., Stecher G., Peterson N., Kumar S., Nei M. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saldivar J.C., Cortez D., Cimprich K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017;18:622. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackford A.N., Jackson S.P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Cimprich K.A., Cortez D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maréchal A., Zou L. DNA damage sensing by the ATM and ATR kinases. CSH Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiloh Y., Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 27.So S., Davis A.J., Chen D.J. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 2009;187:977–990. doi: 10.1083/jcb.200906064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 29.Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., Van Deursen J., Nussenzweig A., Paull T.T., et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Savic V., Yin B., Maas N.L., Bredemeyer A.L., Carpenter A.C., Helmink B.A., Yang-Iott K.S., Sleckman B.P., Bassing C.H. Formation of dynamic γ-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 32.Lee J., Kumagai A., Dunphy W.G. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 33.Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K.-i., Karnitz L.M. The Rad9–Hus1–Rad1 (9–1–1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tapia-Alveal C., Calonge T.M., O’Connell M.J. Regulation of Chk1. Cell. Div. 2009;4:8. doi: 10.1186/1747-1028-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen R.D., Cimprich K.A. The ATR pathway: Fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Besteiro M.A.G., Gottifredi V. The fork and the kinase: A DNA replication tale from a CHK1 perspective. Mutat. Res. 2015;763:168–180. doi: 10.1016/j.mrrev.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zannini L., Delia D., Buscemi G. CHK2 kinase in the DNA damage response and beyond. J. Mol. Cell Biol. 2014;6:442–457. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Z.B., Cools T., de Veylder L. Mechanisms used by plants to cope with DNA damage. In: Merchant S.S., editor. Annual Review of Plant Biology. Volume 67. Annual Reviews; Palo Alto, CA, USA: 2016. pp. 439–462. [DOI] [PubMed] [Google Scholar]
- 39.Stracker T.H., Usui T., Petrini J.H.J. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair. 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse D., Daoust P., Benribague S. A transcriptome-based perspective of cell cycle regulation in dinoflagellates. Protist. 2016;167:610–621. doi: 10.1016/j.protis.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Stewart G.S., Wang B., Bignell C.R., Taylor A.M.R., Elledge S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 42.Pfeffer C.M., Ho B.N., Singh A.T.K. The evolution, functions and applications of the breast cancer genes BRCA1 and BRCA2. Cancer Genom. Proteom. 2017;14:293–298. doi: 10.21873/cgp.20040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachvaroff T.R., Gornik S.G., Concepcion G.T., Waller R.F., Mendez G.S., Lippmeier J.C., Delwiche C.F. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogenet. Evol. 2014;70:314–322. doi: 10.1016/j.ympev.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee N., Walker G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017;58:235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyatt M.D., Pittman D.L. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu D., Calvo J.A., Samson L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishina Y., Duguid E.M., He C. Direct reversal of DNA alkylation damage. Chem. Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedgwick B., Bates P.A., Paik J., Jacobs S.C., Lindahl T. Repair of alkylated DNA: Recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Falnes P.Ø., Rognes T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003;154:531–538. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 50.Yang C.-G., Garcia K., He C. Damage detection and base flipping in direct DNA alkylation repair. ChemBioChem. 2009;10:417–423. doi: 10.1002/cbic.200800580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vågbø C.B., Shi Y., Wang W.L., Song S.H., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rae P. Hydroxymethyluracil in eukaryote DNA: A natural feature of the pyrrophyta (dinoflagellates) Science. 1976;194:1062–1064. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- 54.Rae P.M.M., Steele R.E. Modified bases in the DNAs of unicellular eukaryotes: An examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978;10:37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 55.Heyn H., Esteller M. An adenine code for DNA: A second life for N6-methyladenine. Cell. 2015;161:710–713. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 56.Fu Y., Luo G.Z., Chen K., Deng X., Yu M., Han D., Hao Z., Liu J., Lu X., Doré L., et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizábal-Corrales D., Hsu C.H., Aravind L., He C., Shi Y. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J., et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Liang Z., Shen L., Cui X., Bao S., Geng Y., Yu G., Liang F., Xie S., Lu T., Gu X., et al. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell. 2018;45:406–416. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 60.Xiao C.L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F., et al. N6-methyladenine DNA modification in the human genome. Mol. Cell. 2018;71:306–318. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 61.Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch M., Lucas-Lledó J.I. Evolution of mutation rates: Phylogenomic analysis of the photolyase/cryptochrome family. Mol. Biol. Evol. 2009;26:1143–1153. doi: 10.1093/molbev/msp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dany A.-L., Douki T., Triantaphylides C., Cadet J. Repair of the main UV-induced thymine dimeric lesions within Arabidopsis thaliana DNA: Evidence for the major involvement of photoreactivation pathways. J. Photochem. Photobiol. B. 2001;65:127–135. doi: 10.1016/S1011-1344(01)00254-8. [DOI] [PubMed] [Google Scholar]
- 64.Britt A.B. Repair of DNA damage induced by solar UV. Photosynth. Res. 2004;81:105–112. doi: 10.1023/B:PRES.0000035035.12340.58. [DOI] [Google Scholar]
- 65.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003;103:2203–2238. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 66.Fortunato A.E., Annunziata R., Jaubert M., Bouly J.-P., Falciatore A. Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 2015;172:42–54. doi: 10.1016/j.jplph.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 67.Brettel K., Byrdin M. Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 2010;20:693–701. doi: 10.1016/j.sbi.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z., Wang L., Zhong D. Dynamics and mechanisms of DNA repair by photolyase. Phys. Chem. Chem. Phys. 2015;17:11933–11949. doi: 10.1039/C4CP05286B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selby C.P., Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J., Du X., Pan W., Wang X., Wu W. Photoactivation of the cryptochrome/photolyase superfamily. J. Photochem. Photobiol. C. 2015;22:84–102. doi: 10.1016/j.jphotochemrev.2014.12.001. [DOI] [Google Scholar]
- 71.Zhang M., Wang L., Zhong D. Photolyase: Dynamics and mechanisms of repair of sun-induced DNA damage. Photochem. Photobiol. 2017;93:78–92. doi: 10.1111/php.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sung J.-S., Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 73.Whitaker A.M., Schaich M.A., Smith M.R., Flynn T.S., Freudenthal B.D. Base excision repair of oxidative DNA damage: From mechanism to disease. Front. Biosci. 2017;22:1493–1522. doi: 10.2741/4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.David S.S., O’Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baute J., Depicker A. Base excision repair and its role in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2008;43:239–276. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- 76.Papaluca A., Wagner J.R., Saragovi H.U., Ramotar D. UNG-1 and APN-1 are the major enzymes to efficiently repair 5-hydroxymethyluracil DNA lesions in C. elegans. Sci. Rep. 2018;8:6860. doi: 10.1038/s41598-018-25124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olinski R., Starczak M., Gackowski D. Enigmatic 5-hydroxymethyluracil: Oxidatively modified base, epigenetic mark or both? Mutat. Res. 2016;767:59–66. doi: 10.1016/j.mrrev.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Sjolund A.B., Senejani A.G., Sweasy J.B. MBD4 and TDG: Multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yonekura S.I., Nakamura N., Doi T., Sugiyama H., Yamamoto K., Yonei S., Zhang Q.M. Recombinant Schizosaccharomyces pombe Nth1 protein exhibits DNA glycosylase activities for 8-oxo-7,8-dihydroguanine and thymine residues oxidized in the methyl group. J. Radiat. Res. 2007;48:417–424. doi: 10.1269/jrr.07042. [DOI] [PubMed] [Google Scholar]
- 80.Rae P.M.M. 5-Hydroxymethyluracil in the DNA of a Dinoflagellate. Proc. Natl. Acad. Sci. USA. 1973;70:1141–1145. doi: 10.1073/pnas.70.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klungland A., Lindahl T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robertson A.B., Klungland A., Rognes T., Leiros I. Base excision repair: The long and short of it. Cell. Mol. Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simsek D., Jasin M. DNA ligase III: A spotty presence in eukaryotes, but an essential function where tested. Cell Cycle. 2011;10:3636–3644. doi: 10.4161/cc.10.21.18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Córdoba-Cañero D., Roldán-Arjona T., Ariza R.R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011;68:693–702. doi: 10.1111/j.1365-313X.2011.04720.x. [DOI] [PubMed] [Google Scholar]
- 85.Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. doi: 10.1016/S0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 86.Wood R.D., Araujo S.J., Ariza R.R., Batty D.P., Biggerstaff M., Evans E., Gaillard P.H., Gunz D., Köberle B., Kuraoka I., et al. DNA damage recognition and nucleotide excision repair in mammalian cells. CSH Symp. Quant. Biol. 2000;65:173–182. doi: 10.1101/sqb.2000.65.173. [DOI] [PubMed] [Google Scholar]
- 87.Schärer O.D. Nucleotide excision repair in eukaryotes. CSH Perspect. Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kamileri I., Karakasilioti I., Garinis G.A. Nucleotide excision repair: New tricks with old bricks. Trends Genet. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Mullenders L.H.F. Solar UV damage to cellular DNA: From mechanisms to biological effects. Photochem. Photobiol. Sci. 2018;17:1842–1852. doi: 10.1039/C8PP00182K. [DOI] [PubMed] [Google Scholar]
- 90.Tatum D., Li S. Nucleotide excision repair in S. cerevisiae. In: Storici F., editor. DNA Repair-On the Pathways to Fixing DNA Damage and Errors. IntechOpen; Rijeka, Croatia: 2011. pp. 97–122. [Google Scholar]
- 91.Spivak G. Nucleotide excision repair in humans. DNA Repair. 2015;36:13–18. doi: 10.1016/j.dnarep.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li S. Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26. DNA Repair. 2015;36:43–48. doi: 10.1016/j.dnarep.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 93.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 94.Compe E., Egly J.-M. TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012;13:343. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 95.Rimel J.K., Taatjes D.J. The essential and multifunctional TFIIH complex. Protein Sci. 2018;27:1018–1037. doi: 10.1002/pro.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sugasawa K. Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair. 2016;44:110–117. doi: 10.1016/j.dnarep.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 97.Iakoucheva L.M., Kimzey A.L., Masselon C.D., Bruce J.E., Garner E.C., Brown C.J., Dunker A.K., Smith R.D., Ackerman E.J. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001;10:560–571. doi: 10.1110/ps.29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Canturk F., Karaman M., Selby C.P., Kemp M.G., Kulaksiz-Erkmen G., Hu J., Li W., Lindsey-Boltz L.A., Sancar A. Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. USA. 2016;113:4706–4710. doi: 10.1073/pnas.1604097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roy S., Morse D. Transcription and maturation of mRNA in Dinoflagellates. Microorganisms. 2013;1:71–99. doi: 10.3390/microorganisms1010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Genois M.M., Paquet E.R., Laffitte M.C.N., Maity R., Rodrigue A., Ouellette M., Masson J.Y. DNA repair pathways in Trypanosomatids: From DNA repair to drug resistance. Microbiol. Mol. Biol. Rev. 2014;78:40–73. doi: 10.1128/MMBR.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reyes G.X., Schmidt T.T., Kolodner R.D., Hombauer H. New insights into the mechanism of DNA mismatch repair. Chromosoma. 2015;124:443–462. doi: 10.1007/s00412-015-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spies M., Fishel R. Mismatch repair during homologous and homeologous recombination. CSH Perspect. Biol. 2015;7:a022657. doi: 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kunz C., Saito Y., Schär P. Mismatched repair: Variations on a theme. Cell. Mol. Life Sci. 2009;66:1021–1038. doi: 10.1007/s00018-009-8739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 105.Li G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007;18:85. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 106.Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 107.Tarique M., Ahmad M., Chauhan M., Tuteja R. Genome wide in silico analysis of the mismatch repair components of Plasmodium falciparum and their comparison with human host. Front. Microbiol. 2017;8:130. doi: 10.3389/fmicb.2017.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mehta A., Haber J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. CSH Perspect. Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bennett C.B., Lewis A.L., Baldwin K.K., Resnick M.A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiruvella K.K., Liang Z., Wilson T.E. Repair of double-strand breaks by end joining. CSH Perspect. Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Emerson C.H., Bertuch A.A. Consider the workhorse: Nonhomologous end-joining in budding yeast. Biochem. Cell Biol. 2016;94:396–406. doi: 10.1139/bcb-2016-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin S., Weaver D.T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 115.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 116.Goodarzi A.A., Yu Y., Riballo E., Douglas P., Walker S.A., Ye R., Härer C., Marchetti C., Morrice N., Jeggo P.A., et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li S., Kanno S.I., Watanabe R., Ogiwara H., Kohno T., Watanabe G., Yasui A., Lieber M.R. Polynucleotide Kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3’ exonuclease and can participate in DNA end joining in a biochemical system. J. Biol. Chem. 2011;286:36368–36377. doi: 10.1074/jbc.M111.287797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grundy G.J., Rulten S.L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., Oliver A., Caldecott K.W. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 120.Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 121.Wilson T.E., Lieber M.R. Efficient processing of DNA ends during yeast nonhomologous end joining: Evidence for a DNA polymerase β (pol4)-dependent pathway. J. Biol. Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 122.Bertocci B., de Smet A., Weill J.-C., Reynaud C.-A. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination In Vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 123.Moon A.F., Garcia-Diaz M., Batra V.K., Beard W.A., Bebenek K., Kunkel T.A., Wilson S.H., Pedersen L.C. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair. 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pannunzio N.R., Watanabe G., Lieber M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018;293:10512–10523. doi: 10.1074/jbc.TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T.E., Mann M., Lieber M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 126.Gu J., Lu H., Tsai A.G., Schwarz K., Lieber M.R. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Modesti M., Hesse J.E., Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li J., Yu Y., Suo F., Sun L.-L., Zhao D., Du L.-L. Genome-wide screens for sensitivity to ionizing radiation identify the fission yeast nonhomologous end joining factor Xrc4. G3-Genes Genom. Genet. 2014;4:1297–1306. doi: 10.1534/g3.114.011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Manova V., Gruszka D. DNA damage and repair in plants-from models to crops. Front. Plant Sci. 2015;6:885. doi: 10.3389/fpls.2015.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee A.H., Symington L.S., Fidock D.A. DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol. Mol. Biol. Rev. 2014;78:469–486. doi: 10.1128/MMBR.00059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Symington L.S. End resection at double-strand breaks: Mechanism and regulation. CSH Perspect. Biol. 2014;6:a016436. doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodgers K., McVey M. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016;231:15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Spirek M., Altmannova V., Zhao X., Krejci L. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wright W.D., Shah S.S., Heyer W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018;293:10524–10535. doi: 10.1074/jbc.TM118.000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Renkawitz J., Lademann C.A., Jentsch S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 2014;15:369. doi: 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- 136.Kaniecki K., de Tullio L., Greene E.C. A change of view: Homologous recombination at single-molecule resolution. Nat. Rev. Genet. 2017;19:191. doi: 10.1038/nrg.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sung P., Krejci L., van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 138.Kawabata M., Kawabata T., Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Med. Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- 139.Miller-Messmer M., Kühn K., Bichara M., le Ret M., Imbault P., Gualberto J.M. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 2012;159:211–226. doi: 10.1104/pp.112.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rowan B.A., Oldenburg D.J., Bendich A.J. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J. Exp. Bot. 2010;61:2575–2588. doi: 10.1093/jxb/erq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 142.Waters L.S., Minesinger B.K., Wiltrout M.E., D’Souza S., Woodruff R.V., Walker G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sale J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. CSH Perspect. Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao L., Washington M.T. Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes. 2017;8:24. doi: 10.3390/genes8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marians K.J. Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 2018;87:217–238. doi: 10.1146/annurev-biochem-062917-011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Powers K.T., Washington M.T. Eukaryotic translesion synthesis: Choosing the right tool for the job. DNA Repair. 2018;71:127–134. doi: 10.1016/j.dnarep.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chang D.J., Cimprich K.A. DNA damage tolerance: When it's OK to make mistakes. Nat. Chem. Biol. 2009;5:82. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McCulloch S.D., Kunkel T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shen X., Li L. Mutagenic repair of DNA interstrand crosslinks. Environ. Mol. Mutagen. 2010;51:493–499. doi: 10.1002/em.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hashimoto S., Anai H., Hanada K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016;38:9. doi: 10.1186/s41021-016-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muniandy P.A., Liu J., Majumdar A., Liu S.-t., Seidman M.M. DNA interstrand crosslink repair in mammalian cells: Step by step. Crit. Rev. Biochem. Mol. Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ceccaldi R., Sarangi P., D’Andrea A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016;17:337. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 154.Clauson C., Schärer O.D., Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. CSH Perspect. Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dangel N.J., Knoll A., Puchta H. MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014;78:822–833. doi: 10.1111/tpj.12507. [DOI] [PubMed] [Google Scholar]
- 157.Daee D.L., Ferrari E., Longerich S., Zheng X.F., Xue X., Branzei D., Sung P., Myung K. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J. Biol. Chem. 2012;287:26563–26575. doi: 10.1074/jbc.M112.369918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cattell E., Sengerová B., McHugh P.J. The SNM1/Pso2 family of ICL repair nucleases: From yeast to man. Environ. Mol. Mutagen. 2010;51:635–645. doi: 10.1002/em.20556. [DOI] [PubMed] [Google Scholar]
- 159.Munari F.M., Guecheva T.N., Bonatto D., Henriques J.A.P. New features on Pso2 protein family in DNA interstrand cross-link repair and in the maintenance of genomic integrity in Saccharomyces cerevisiae. Fungal Genet. Biol. 2013;60:122–132. doi: 10.1016/j.fgb.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 160.van Attikum H., Gasser S.M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 161.Vidanes G.M., Bonilla C.Y., Toczyski D.P. Complicated tails: Histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.