Abstract
Background: Microorganisms are widespread in all environments, including in and on animal bodies. The gut microbiome has an essential influence on fish health, and is affected by several persistent and harmful organic and inorganic contaminants. Considering the shifts in gut microbiota composition observed in those studies, we hypothesized that certain microbial groups in the gut can serve as indicators of pollution. To test this hypothesis, we explored the possibility of identifying key microbial players that indicate environmental contamination. Methods: Published 16S rRNA gene amplicon sequencing data generated from the gut microbiota of Atlantic cod caught in geographically different Norwegian waters were used for bacterial diversity comparison. Results: Different microbiomes were identified between the northern Norway and southern Norway samples. Several bacterial genera previously identified as polycyclic aromatic hydrocarbon degraders were present only in the samples collected in the southern Norway area, suggesting fish contamination with oil-related compounds. Conclusions: The results contribute to the identification of bacterial taxa present in the Atlantic cod gut that indicate fish exposure to contaminants in the marine environment.
Keywords: gut, prokaryote, animal, marine, microbiome
1. Introduction
Approximately 108 microbial cells populate the fish digestive tract, influencing the development, metabolism, immune system, ecology, and evolution of the host [1,2,3,4]. Gut microbial communities have a pivotal role on how fish respond to environmental stressors, and specific bacteria (holding specific genomic repertoire) might be used as a proxy to evaluate the fish health condition and the environmental contamination status. Most studies have examined microbial community changes in commercially relevant fish species with the aim of better understanding how gut microbiota can be modulated to produce healthier fish in aquaculture settings [5,6,7]. However, in the past years, interest in the effect of pollutants on the gut microbiota and their interplay with the host in responding to these has grown as well. Few studies have addressed such questions employing high-throughput amplicon sequencing, nor have many so far shown shifts in community composition upon the laboratory exposure of various fish species to contaminants, including pentachlorophenol, polybrominated diphenyl ethers, titanium dioxide nanoparticles, bisphenol A, and crude oil [8,9,10,11].
Offshore oil production on the Norwegian continental shelf and the shipping of crude oil pose continuous risks of exposing the marine ecosystem to oil contamination. Norway is the largest producer of oil and gas among all the 16 OSPAR (Oslo-Paris commission for protecting and conserving the North-East Atlantic and its resources) contracting parties [12], and has increased the total oil gross production in the last few years (15% between 2012–2016) [13], with a possibility of expansion after recent new discoveries of hydrocarbon reserves and wells (particularly in the Arctic region) [14]. Short-term and long-term impacts over habitats and populations are likely to increase with the continued production of petroleum-derived compounds. Therefore, it is of particular interest to monitor the presence of polycyclic aromatic hydrocarbons (PAHs), which are common constituents of crude oil complex mixtures originating from natural sources and anthropogenic activities [15,16]. PAHs have been circulating through biogeochemical cycles for millions of years by the occurrence of natural seeps [17]. Nevertheless, anthropogenic activities have considerably increased PAH levels in the ecosystems [18], and several of them feature on the European Union and the United States lists of priority pollutants to monitor in aquatic and terrestrial ecosystems [19,20], due their carcinogenic, mutagenic, and toxic effects in biota [21,22,23,24,25]. In fish, crude oil–PAHs and residual fuel oils exposure promote several organs disorders, such as toxic hepatic lesions [26] and heart failure [27,28]. The cardiotoxicity effect leads to the direct disruption of fish cardiac function, cardiac edema, arrhythmia, and impaired swimming performance [28,29,30,31,32]. Furthermore, controlled experiments have shown that oil exposure-induced immunosuppression increases susceptibility to bacterial infection and high mortality rates [33].
Atlantic cod (Gadus morhua) is commonly used as a bioindicator species for monitoring the presence and effects of contaminants in the marine environment [34]. It has been selected in our study due to its commercial value and its use in ecotoxicological studies and monitoring surveys [35,36,37,38,39,40]. Different biomarkers have been successfully used to trace PAH compounds in fish organs, greatly improving the knowledge of PAHs effects and persistence. While the mechanisms of PAH detoxification through liver enzyme activities in fish are well established and used as biomarkers of pollution [41], less is known about the fate and effect of PAH metabolites entering the intestinal tract of the fish via the bile content.
High-throughput sequencing technologies have contributed to increasing our knowledge regarding the identities of microbial key players growing in the presence of petroleum-derived PAH mixtures [42,43,44,45,46,47], as well as identifying which metabolic activities of resident microbes may exacerbate or mitigate the toxic effects of these metabolites. Atlantic cod gut microbiota was shown to be affected by crude oil exposure at 0.05 mg·L−1 and 0.1 mg·L−1 in a laboratory study [11]. The results of this high-throughput sequencing analysis suggested that a set of bacterial taxa that are present in the gut of Atlantic cod can be used as novel indicators of PAH contamination. The microbiome sequence analysis was the selected approach in our work to trace contamination. The sequence analysis strategy is faster and highly sensitive in comparison with traditional methodologies, bringing more valuable detailed information besides confirming former results obtained using, e.g., the traditional set of fish health biological markers [48,49].
Here, we explore the potential of specific microbial taxa that are present in the Atlantic cod gut microbiome as oil contamination markers using publicly available data. Our goals were: (i) to perform an overall comparison of the microbial community composition of gut samples collected in a presumably pristine northern area of Norway and in a more anthropogenic impacted one located in southern Norway; and (ii) to analyze the relative abundance of known marine hydrocarbon degraders bacteria in the different gut samples.
2. Materials and Methods
2.1. Samples
Publicly available sequencing data were obtained from wild cod caught in northern Norway (Lofoten and Sørøya locations) and in southern Norway (Kvitsøy, Rogaland) (Table 1). Cod from the southern Norway area were maintained in laboratory conditions and only fish exposed to no oil and low concentration of oil (0.01 ppm) were used in this study, as these were highly similar according to a preliminary analysis [11], and the exposure levels were the closest to real environmental contamination conditions. A total of seven samples were obtained from Riiser et al. [50] (northern Norway: four Lofoten, and three Sørøya) and six were obtained from Bagi et al. [11] (southern Norway: three concentration samples with no oil, and three with low oil). Both datasets were based on high-throughput Illumina sequencing of the 16S rRNA V4 region, and were generated based on fish intestinal content and mucosa. A limited amount of Atlantic cod gut microbiome samples using the same sequencing technology and the same 16S rRNA region is publicly available at present. The use of 16S rRNA sequence data allows the analysis of the prokaryote components, which are directly influenced by environmental changes [51]. Here, we focused on the microbial PAH-degraders (Bacteria domain) found in the gastrointestinal organ.
Table 1.
Summary of the main features of the study areas.
| Area | Latitude | Longitude | Samples | |
|---|---|---|---|---|
| Northern Norway | Lofoten | 68°10′19″ N | 13°45′00″ E | 4 |
| Sørøya | 70°29′33″ N | 22°09′51″ E | 3 | |
| Southern Norway | Kvitsøy | 59°03′52″ N | 05°24′19″ E | 6 |
2.2. Quality Filtering of Sequence Data and Analysis
Raw sequences data were downloaded from the European Nucleotide Archive (PRJEB22384 for [50] and PRJEB21667 for [11]) database and processed under the same methodology to minimize bias. Sequencing reads of 16S rRNA gene were checked for quality with FastQC v. 0.11.2 [52] before being filtered and processed through the DADA2 package v. 1.6 [53], implemented in R software environment v. 3.6.0 [54], to generate amplicon sequence variants (ASVs) corresponding to high-quality sequences at single-nucleotide resolution. ASV table is generated after several steps: quality check, trimming, filtering (Supplementary Figure S1a,b), dereplicating sequences, learning error rates (Figure S2a,b), removing sequences potentially containing errors (denoising), merging paired-end reads as contigs, screening contigs for mismatches to reduce errors, sequence variant abundance identification, and removing chimeric sequences (using the “bimera” method). Each ASV was taxonomically classified to the closest reference sequence of the Silva bacterial database (release 132) [55,56]. The assigned results were classified at domain to genus level.
Alpha diversity was applied to analyze taxa diversity in a sample through the number of observed ASVs, Chao1 richness, Shannon diversity index, Simpson diversity index, inverse Simpson, and ACE. Relative taxa abundances were represented in stacked barplots. To analyze how closely related the samples were to each other, beta diversity analyses were determined based on the Bray–Curtis dissimilarities metric and weighted UniFrac phylogenetic distances metric. Both distances were visualized using the ordination method principal coordinate analysis (PCoA) into two-dimensional plots. Also, Ward’s hierarchical clustering method based on Euclidean distance was applied to interpret the distance matrix using average linkages. Differences in the samples’ beta diversity were measured using the permutational multivariate analysis of variance (PERMANOVA) test with the adonis function from the vegan package v. 2.5-5 [57] in R. The Microbiome Analyst [58] tool was used to perform the diversity and compositional analysis, as well as comparative analysis based on the ASVs table from the 16S rRNA sequencing data used in the present study. The ASVs abundance table was filtered, and a normalization step using the minimum library size and data transformation based on the relative log expression (RLE) method was used to account for the compositional differences. To identify differences in taxa abundances of fish gut samples from two different locations, we used differential expression analysis for sequence count data v. 2 (DESeq2) [59].
The ASVs abundance table corresponds to the traditional operational taxonomic units (OTUs) table. Although the ASV-based method has been shown to generate more accurate results for analysis of 16S rRNA [60,61], we also inferred the taxonomical composition of the samples using OTUs. 16S rRNA gene sequences were filtered and processed using the quantitative insights into microbial ecology (QIIME) v. 1.8 [62] that uses the USEARCH algorithm v. 9.2 [63] to find the closest taxa in the reference database. To process raw Illumina data, paired-end reads were merged with a minimum overlap of 10 bp and a 5% error rate in the overlapping bases. Singletons were removed. Joined reads were trimmed by quality, removing low complexity sequences with a Phred score less than 25, as well as length and ambiguous bases. Then, samples were then screened for chimeric sequences using the default parameters in UCHIME algorithm v. 2011-11-02 [64], and the detected chimerae were removed. Merged sequences that passed quality filtering thresholds were subsequently clustered into OTUs, based on 97% identity using the UPARSE command [65] in USEARCH. Taxonomic information was provided using the Silva bacterial database (release v.119), and the assigned results were classified at the domain to genus level.
3. Results
The resulting high-quality non-chimeric-reads table retained 392 ASVs, which were used for downstream analysis. Overall, the observed error rates from the learning error rates step tracked well with the estimated error rates (Figure S2a,b), indicating a suitable quality control based on Phred quality scores (Figure S1a,b).
3.1. Overview of the bacterial community profiles
Richness estimates (Chao1, ACE, Fisher, and number of ASVs) and alpha diversity were calculated as Shannon and Simpson’s indices, showed fluctuations across fish gut from northern Norway and southern Norway (Figure S3). In addition, fish gut from northern Norway had a lower microbial community richness compared to the fish gut from southern Norway (Figure S3). The same was observed when analyzing OUT-based results (data not shown).
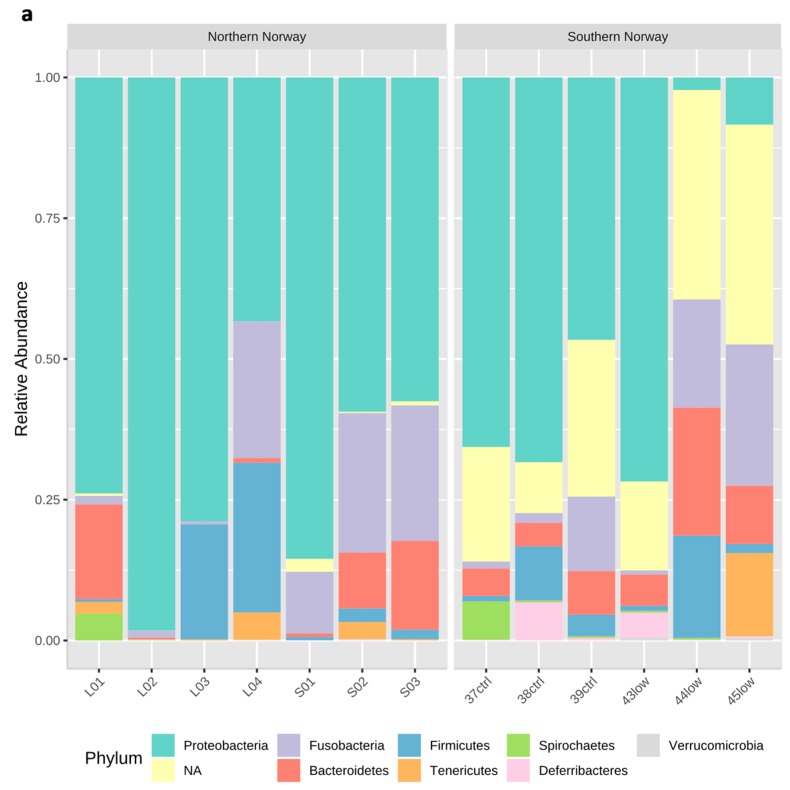
Firmicutes, Proteobacteria, Bacteroidetes, and Fusobacteria were the four most abundant phyla in all the samples (Figure 1a). There were multiple specific differences in the proportions of the relative abundance of various taxa from phylum to genus level when comparing the northern and southern samples (Figure 1a–c and Figure S4). Whereas for the area designed as pristine, i.e., northern Norway, Fusobacteria and Proteobacteria were the most abundant, followed by Firmicutes and Bacteroidetes. In the fish from southern Norway, we observed the dominance of Firmicutes, followed by Proteobacteria, Bacteroidetes, and Fusobacteria. Vibrionales was the most abundant order identified (Figure 1b). The Photobacterium genus was highly frequent in the fish gut from northern Norway (Figure 1c) and notably, non-annotated ASVs could be detected specially in the fish gut from southern Norway (Figure 1c).
Figure 1.
Microbial community composition in Atlantic cod gut samples from geographically different areas. Relative abundance of the sequences is expressed as percentage (%) and are presented at the phylum level (a), order level (b), and genus level (c).
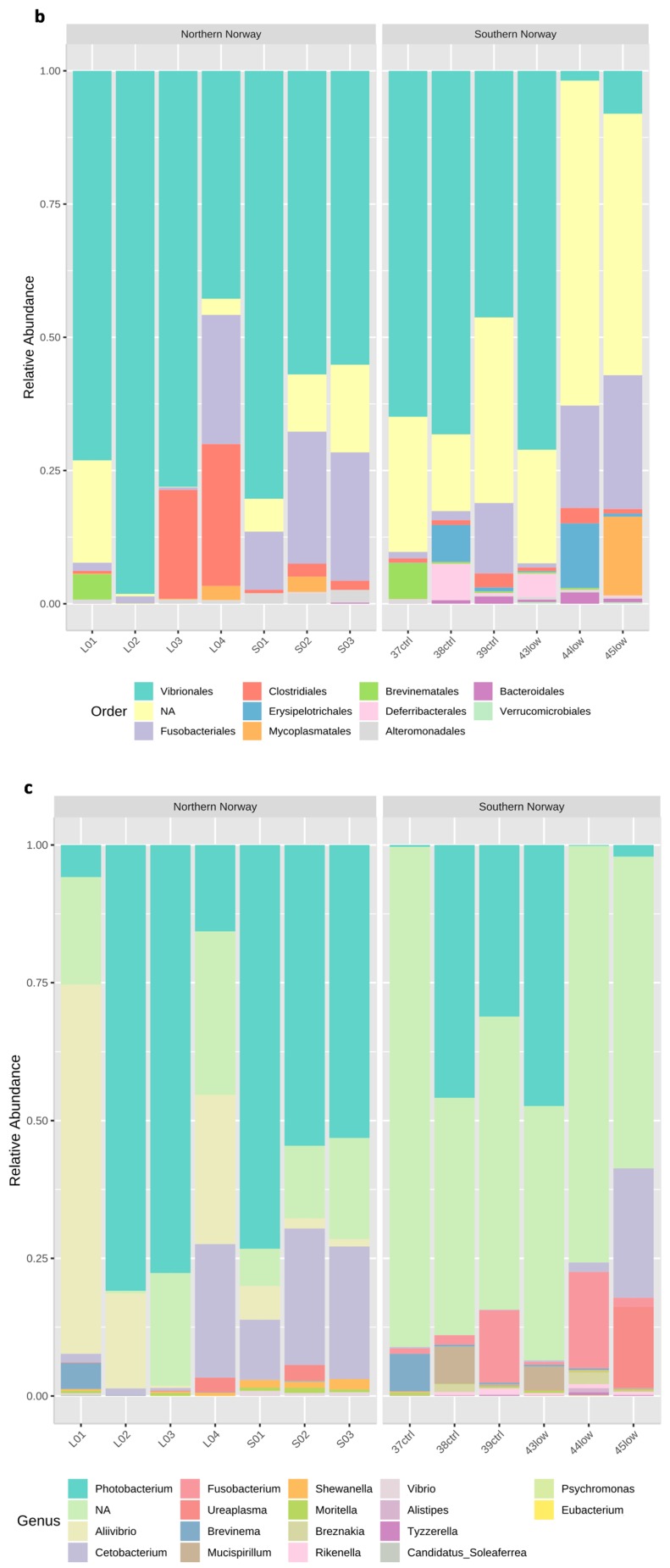
A hierarchical clustering of samples based on a beta diversity analysis of ASV abundances showed cluster relationships between samples, indicating differences in bacterial composition between the northern Norway and southern Norway samples (Figure 2a). This hypothesis is supported by the ordination analysis using PCoA on Bray–Curtis (Figure 2b) and PCoA on ordinated weighted UniFrac distances of individual samples across locations (Figure 2c), where the PERMANOVA results displayed that location accounts for a substantial variation in beta diversity (R2 = 0.467; p < 0.002). The two first axes of the Bray–Curtis distance metric-based PCoA plot explained 63.8% of the taxonomic variation among the microbiomes, while determining by a phylogenetic weighted UniFrac distance metric-based PCoA plot explained 69.4% of the taxonomic variation. The samples from northern Norway clustered together, and samples from southern Norway tended to form a separate group (Figure 2a–c). Location has been observed to influence gut microbial beta diversity. The ordinated beta diversity analysis based on the ASV abundance supports the hypothesis that gut microbiomes from northern Norway fish differ from the gut microbiomes from southern Norway fish (Figure 2a–c).
Figure 2.
Microbial community composition in Atlantic cod gut samples from northern and southern Norway. (a) Hierarchical clustering of amplicon sequence variant (ASVs) abundances generated through Bray–Curtis distance metric and Ward’s linkage method. (b) Principal coordinate analysis (PCoA) ordination of all the samples based on the beta diversity analysis of ASV abundances generated using the Bray–Curtis distance metric. (c) Nonmetric multidimensional scaling (NMDS) beta diversity ordination of individual samples calculated with weighted UniFrac distances. Oil exposure treatment codes: ctrl = control (no oil), low = low concentration of oil [11]; L = Lofoten; S = Sørøya.
3.2. Differences in Most Abundant Gut Microbiota
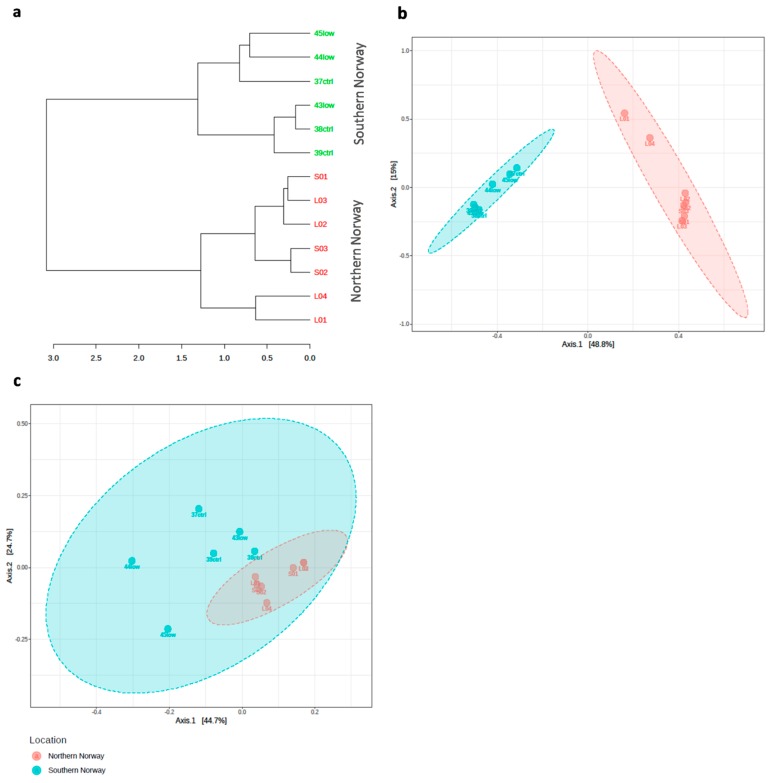
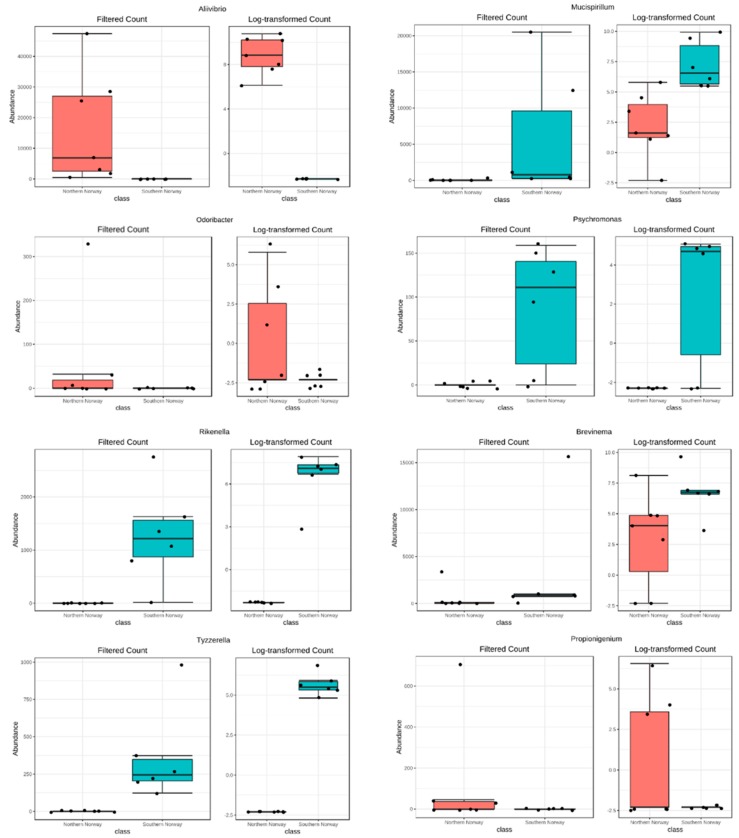
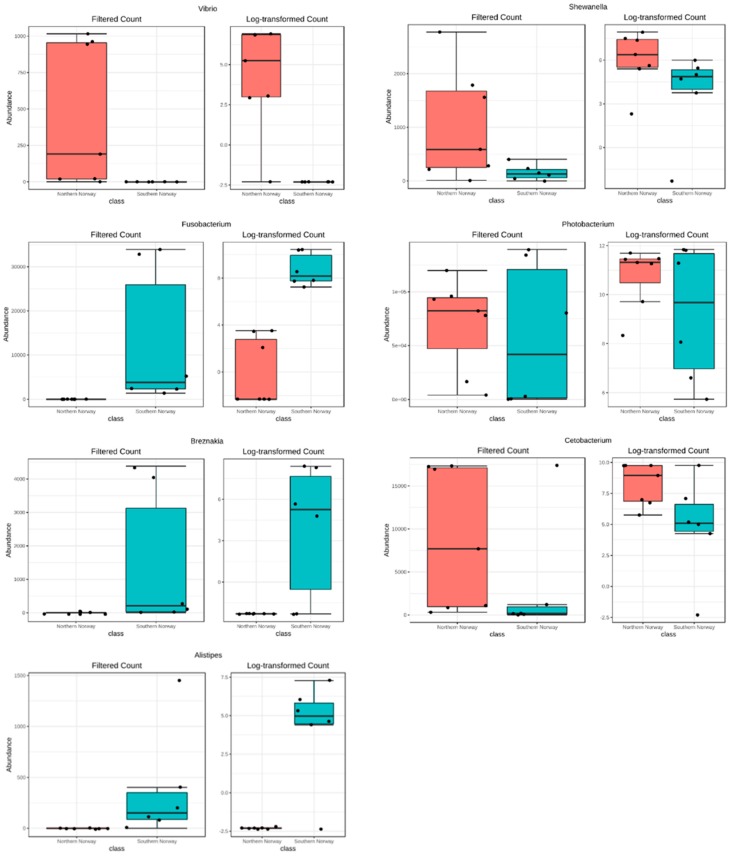
Wald tests of log2 fold differential abundances of ASVs revealed significant fluctuations in the abundances of several genera (Figure 3). A total of 16 genera with significant features showed differential abundance, and among them, 14 were ranked among the 22 most abundant genera (Figure 1c). Most of the genera that were ranked as significant, based on a p value of 0.05, through differential analysis of DESeq2 were located in southern Norway (Figure 3), such as Rikenella, Tyzzerella, Fusabacterium, Breznakia, Alistipes, Mucispirilum, and Psychromonas.
Figure 3.
Differential abundance of bacteria in Atlantic cod gut samples from northern and southern Norway. Wald tests of log2 fold differential abundances of amplicon sequence variants (ASVs). Sixteen genera revealed significant features, based on p values, with differential abundance.
3.3. Contribution of PAH Degraders Bacteria
A total of 65 bacterial genera, distributed in 24 orders and four phyla (Table S1) that had been previously identified as PAH degraders [12] were found in all the gut microbiota samples. The presence of PAH degraders bacteria was presented here as OTUs results. Previous work [11] reported eight significantly different OTUs in their relative abundance in the gut microbial community of fish exposed to 0.05 mg·L−1 and 0.1 mg·L−1 of dispersed crude oil. Their presence and abundance were analyzed in the gut microbiomes. Members of Deferribacteraceae responded, increasing their relative abundance with rising oil exposure levels. No Deferribacteraceae sequences were detected in the gut of fish from the northern Norway areas. Analyzing the potential biomarker taxa from [11], Alistipes (p = 0.04), Anaerotruncus (p = 0.01), Parabacteroides (p = 0.03), Porphyromonas (p = 0.02), and Ruminococcus (p = 0.01) genera were significantly different in the relative abundance between southern and northern Norway fish gut microbiomes, according to the analysis performed in this study. In addition, Ornithobacterium (p = 0.05), Pedobacter (p = 0.02), Psychromonas (p = 0.02), Lactobacillus (p = 0.02), and Salinivibrio (p = 0.02) were also differentially abundant between those geographic areas. Members of known marine hydrocarbon-degrading groups, namely the Oceanospirillales and Thiotrichales orders, were also present through the Alcanivorax and Cycloclasticus genera [66,67]. Sequences annotated as Novosphingobium, Sphingobium, and Sphingomonas, potentially representing alphaproteobacterial PAH degraders, were also present only in the samples from fish collected in southern Norway [68]. Although these genera were not identified as significant through the ASVs method, with the exception of Alistipes (Figure 3), the presence of these bacteria should be considered, suggesting them as an important signal for potential oil-degradation contributors.
4. Discussion
Pollutants present in the environment (e.g., persistent organic pollutants such as PAHs) are becoming strong and pivotal factors contributing to shape the individual’s gastrointestinal microbiotype [69]. Therefore, the monitoring of the microbial communities in the gut of marine species as sentinel organisms has the value of indicating the habitat health condition. Bacteria with the potential to degrade hydrocarbons, including PAHs, express genes for oxygenases or peroxidases, as found in Pseudomonas (alkB, alkane monooxygenase; and ndoB, naphthalene monooxygenase), Rhodococcus (alkB1 and alkB2), Mycobacterium (nidA, pyrene dioxygenase), Cycloclasticus (rhd, hydroxylating dioxygenases), and Stenotrophomonas (sdr, dehydrogenase reductase). These genes take part in intricate pathways for the degradation of PAHs and other hydrocarbons, potentially doing so in the fish gut as well.
Although some genera showed significant differences in the relative abundance between the southern and northern Norway samples, results analyzed for false discovery rate by Benjamini–Hochberg tests were all above the corrected p value. Nevertheless, the presence and the likely role held by specific bacteria in the fish gut of the two geographically different populations verified in the present study should not be neglected. That is why we were motivated to interpret the results beyond the traditional p value. Although we know that geographic location, salinity, seasonality and developmental stage are factors that could influence differences in the gut microbiome of fish, as well as dietary input in the aquaculture conditions [70,71,72,73], the presence of well-known bacteria that degrade PAHs compounds reflects the potential presence of oil in surrounding waters. Bacterial genera such as Novosphingobium, Sphingobium, and Sphingomonas present only in the southern Norway fish gut samples might suggest exposure to oil-related compounds, since these Alphaproteobacteria members hold genes that are involved in the degradation of PAHs [74,75]. In addition, OTUs of representative other hydrocarbon-degrading bacteria were present only in the southern Norway fish gut microbiomes, such as Oleispira antarctica (Oceanospirillales) and genera Marinobacter (Alteromonadaceae), Rhodococcus (Actinomicetales), Microbacterium (Actinomicetales), Alcanivorax (Oceanospirillales), and Cycloclasticus (Thiotrichales). For the last two genera, few reads (1–3) were found in some samples from northern Norway. Alkane-degrading bacteria, such as Alcanivorax spp., are biological signals of the alkane distribution. A recent study [76] showed that strains belonging to Alcanivorax spp., Marinobacter spp., and Planomicrobium sp. are able to grow on certain nonhydrocarbons, as well as organic compounds such as peptone, glutamic acid, pyruvic acid, sucrose, mannose and others, raising the question of whether there are obligate hydrocarbonoclastic bacteria or not, as previously thought [77,78,79,80]. Although it is important to mention that these bacteria grow on crude oil, n-octadecane and phenanthrene, as sole carbon substrates, and they have an important role in the effective removal of hydrocarbons that have spilled in the environment. Indeed, the alkane-degrading Alcanivorax spp. were frequently the first to increase in response to oil spill [43,44,81], whereas Cycloclasticus spp. are capable of degrading more complex hydrocarbons (including both aromatic and aliphatic hydrocarbon components), usually increasing their population later [46,77,81,82]. Cycloclasticus and Alcanivorax members, as other PAH degraders, have mechanisms to overcome the low bioavailability of oil droplets. For example, an increased surface hydrophobicity is generated through a biosurfactant produced by Alcanivorax borkumensis that facilitates direct contact between bacteria and the contaminant, such as the very hydrophobic n-alkane substrate [82]. Such features allow n-alkane oxygenase systems, which are located in the membrane, to act directly with their substrates [83]. Species belonging to these genera might act together to degrade PAHs, growing and operating in succession after the fish ingestion of oil droplets through changes in the hydrocarbon supply [84,85,86,87,88].
Acetoclastic iron-reducers that are members of Deferribacteraceae showed significantly different relative abundance in the gut microbiomes of fish exposed to 0.05 mg·L−1 and 0.1 mg·L−1 of dispersed crude oil compared to samples from fish exposed to 0.01 mg·L−1 and no oil [11], and they were absent in all the gut microbiomes from fish collected in northern Norway. The absence of Deferribacteraceae member sequences in the pristine northern Norway microbiomes reinforce the role of these taxa [79,80], and are potential biomarkers of oil pollution. Overall, these genera may be considered key players to guide analyses of fish contamination. Although no statistical significance was found for several taxa, the presence of specific genera only in the southern Norway fish gut microbiomes might suggest a specific role linked to the specific anthropogenic contamination. Shipping, coastal industry activities, and even small-scale oil leakage on the bustling Stavanger fjord (southern Norway) might have an accentuated effect, especially considering the nature of the fjord stagnant waters. Exclusive OTUs could reveal interesting functional effects, including negative displacements, in the host. Considering the contemporary effort on the establishment of an appropriate method to analyze microbiome-sized data, accommodating unique characteristics is under discussion. These include sparsity, sequencing depth variation, and the nonconformity of read counts to theoretical distributions [89]—besides normalization effects—as well as the use and interpretation of traditional p-value thresholds [89,90,91].
Shotgun metagenomics and metatranscriptomics surveys are seen as the next step. This approach will help us assess a broader taxon identification, including Fungi and Archaea members potentially involved in PAHs degradation. Furthermore, the functional component might be revealed, allowing the detection of specific metabolic genes involved in different functional preferences for the degradation pathways of PAHs.
5. Conclusions
The gut microbiome represents a key determinant to a variety of host metabolic and immune functions, including the reaction to oil pollutant exposure. In this study, we have compared the relative abundance of bacterial communities in the gut of Atlantic cod populations from two geographical areas of the Norwegian Continental Shelf. In addition, we verified the presence of PAH-degrading bacteria in the gut microbiome of fish collected in southern Norway. The presence of hydrocarbonoclastic taxa in the southern Norway fish gut microbiome even in the control samples can be explained by the higher anthropogenic impact in this area, in comparison to the northern Norway area. It reinforces that internal microbiota signatures are associated with PAH contaminants, and certain bacterial taxa can be used as indicators to monitor environmental and fish health. Although the 16S rRNA-based approach provides the composition of microbiomes to the genus level, an increasing number of studies are focusing on functional aspects of the whole community using metagenomics and metatranscriptomics sequencing. Through such approaches, dominant metabolic processes can be explored, and the contribution of the whole prokaryotic and eukaryotic communities can be accessed. Using metagenomics and metatranscriptomics approaches to Atlantic cod gut studies, in the context of monitoring polluted environments, will be the subject of further studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/7/7/209/s1. Table S1: Bacterial genera able to grow using hydrocarbons as a sole source of carbon and energy.
Author Contributions
Conceptualization, D.M.P., A.B. and J.M.W.; methodology, J.M.W.; software, J.M.W.; validation, D.M.P., A.B. and J.M.W.; formal analysis, J.M.W.; investigation, D.M.P., A.B. and J.M.W.; resources, D.M.P.; data curation, D.M.P., A.B. and J.M.W.; writing—original draft preparation, J.M.W.; writing—review and editing, D.M.P., A.B. and J.M.W.; visualization, J.M.W.; supervision, D.M.P.; project administration, D.M.P.; funding acquisition, D.M.P.
Funding
The Nor-Bra2020: Gearing the future generations toward a sustainable use of the environment project funded by DIKU (Norwegian Agency for International Cooperation and Quality Enhancement in Higher Education), project number UTF-2016-long-term/10088.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Rawls J.F., Samuel B.S., Gordon J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero J., Navarrete P. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch) Microb. Ecol. 2006;51:422–430. doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- 3.Fraune S., Bosch T.C. Why bacteria matter in animal development and evolution. Bioessays. 2010;32:571–580. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 4.Kohl K.D., Carey H.V. A place for host-microbe symbiosis in the comparative physiologist’s toolbox. J. Exp. Biol. 2016;219:3496–3504. doi: 10.1242/jeb.136325. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Porchas M., Vargas-Albores F. Microbial metagenomics in aquaculture: A potential tool for a deeper insight into the activity. Rev. Aquac. 2017;9:42–56. doi: 10.1111/raq.12102. [DOI] [Google Scholar]
- 6.Ringø E., Sperstad S., Myklebust R., Refstie S., Krogdahl Å. Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.): The effect of fish meal, standard soybean meal and a bioprocessed soybean meal. Aquaculture. 2006;261:829–841. doi: 10.1016/j.aquaculture.2006.06.030. [DOI] [Google Scholar]
- 7.Li T.T., Long M., Li H., Gatesoupe F.-J., Zhang X., Zhang Q., Feng D., Li A. Multi-omics analysis reveals a correlation between the host phylogeny, gut microbiota and metabolite profiles in cyprinid fishes. Front. Microbiol. 2017;8:454. doi: 10.3389/fmicb.2017.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Guo Y., Hu C., Lam P.K.S., Lam J.C.W., Zhou B. Dysbiosis of gut microbiota by chronic coexposure to titanium dioxide nanoparticles and bisphenol A: Implications for host health in zebrafish. Environ. Pollut. 2018;234:307–317. doi: 10.1016/j.envpol.2017.11.074. [DOI] [PubMed] [Google Scholar]
- 9.Chen L., Hu C., Lok-Shun Lai N., Zhang W., Hua J., Lam P.K.S., Lam J.C.W., Zhou B. Acute exposure to PBDEs at an environmentally realistic concentration causes abrupt changes in the gut microbiota and host health of zebrafish. Environ. Pollut. 2018;240:17–26. doi: 10.1016/j.envpol.2018.04.062. [DOI] [PubMed] [Google Scholar]
- 10.Kan H., Zhao F., Zhang X.-X., Ren H., Gao S. Correlations of Gut Microbial Community Shift with Hepatic Damage and Growth Inhibition of Carassius auratus Induced by Pentachlorophenol Exposure. Environ. Sci. Technol. 2015;49:11894–11902. doi: 10.1021/acs.est.5b02990. [DOI] [PubMed] [Google Scholar]
- 11.Bagi A., Riiser E.S., Molland H.S., Star B., Haverkamp T.H.A., Sydnes M.O., Pampanin D.M. Gastrointestinal microbial community changes in Atlantic cod (Gadus morhua) exposed to crude oil. BMC Microbiol. 2018;18:25. doi: 10.1186/s12866-018-1171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.OSPAR Commission Protecting and conserving the North-East Atlantic and its resources. [(accessed on 11 July 2019)]; Available online: https://www.ospar.org/
- 13.OSPAR Commission Assessment of the discharges, spills and emissions from offshore installations on the Norwegian Continental Shelf in 2012–2016. [(accessed on 11 July 2019)]; Available online: https://www.ospar.org/documents?d=38985.
- 14.NPD, Norwegian Petroleum Directorate Factpages. [(accessed on 11 July 2019)]; Available online: www.npd.no/en/
- 15.Prince R. The Microbiology of Marine Oil Spill Bioremediation. In: Ollivier B., Magot M., editors. Petroleum Microbiology. ASM Press; Washington, DC, USA: 2005. pp. 317–335. [Google Scholar]
- 16.Pampanin D.M., Sydnes M.O. Polycyclic aromatic hydrocarbons a constituent of petroleum: Presence and influence in the aquatic environment. In: Kutcherov V., Kolesnikov A., editors. Hydrocarbon. InTech; Rijeka, Croatia: 2013. pp. 83–118. [Google Scholar]
- 17.Henner P., Schiavon M., Morel J.L., Lichtfouse E. Polycyclic aromatic hydrocarbon (PAH) occurrence and remediation methods. Analusis. 1997;25:M56–M59. [Google Scholar]
- 18.National Research Council (US) Committee on Oil in the Sea . Oil in the Sea III: Inputs. Fates and Effects. National Academies Press; Washington, DC, USA: 2003. [PubMed] [Google Scholar]
- 19.US EPA Toxic Release Inventory Public Data Release. [(accessed on 11 July 2019)]; Available online: https://www.epa.gov/triinter/tridata/index.htm.
- 20.Yan J., Wang L., Fu P.P., Yu H. Photomutagenicity of 16 polycyclic aromatic hydrocarbons from the US EPA priority pollutant list. Mutat. Res. 2004;557:99–108. doi: 10.1016/j.mrgentox.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aas E., Baussant T., Balk L., Liewenborg B., Andersen O. PAH metabolites in bile, cytochrome P4501A and DNA adducts as environmental risk parameters for chronic oil exposure: A laboratory experiment with Atlantic cod. Aquat. Toxicol. 2000;51:241–258. doi: 10.1016/S0166-445X(00)00108-9. [DOI] [PubMed] [Google Scholar]
- 22.Aas E., Beyer J., Jonsson G., Reichert W., Andersen O. Evidence of uptake, biotransformation and DNA binding of polyaromatic hydrocarbons in Atlantic cod and corkwing wrasse caught in the vicinity of an aluminium works. Mar. Environ. Res. 2001;52:213–229. doi: 10.1016/S0141-1136(00)00269-5. [DOI] [PubMed] [Google Scholar]
- 23.Samanta S.K., Singh O.V., Jain R.K. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. Trends Biotechnol. 2002;20:243–248. doi: 10.1016/S0167-7799(02)01943-1. [DOI] [PubMed] [Google Scholar]
- 24.Hylland K. Polycyclic aromatic hydrocarbon (PAH) ecotoxicology in marine ecosystems. J. Toxicol. Environ. Health. 2006;69:109–123. doi: 10.1080/15287390500259327. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-Pinos M.-C., Vila-Costa M., Arrieta J.M., Morales L., González-Gaya B., Piña B., Dachs J. Dysregulation of photosynthetic genes in oceanic Prochlorococcus populations exposed to organic pollutants. Sci. Rep. 2017;7:8029. doi: 10.1038/s41598-017-08425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers M.S., Johnson L.L., Olson O.P., Stehr C.M., Horness B.H., Collier T.K., McCain B.B. Toxicopathic hepatic lesions as biomarkers of chemical contaminant exposure and effects in marine bottomfish species from the Northeast and Pacific Coasts, USA Mar. Pollut. Bull. 1998;37:92–113. doi: 10.1016/S0025-326X(98)00135-0. [DOI] [Google Scholar]
- 27.Incardona J.P., Collier T.K., Scholz N.L. Oil spills and fish health: Exposing the heart of the matter. J. Expo Sci. Environ. Epidemiol. 2011;21:3–4. doi: 10.1038/jes.2010.51. [DOI] [PubMed] [Google Scholar]
- 28.Incardona J.P., Collier T.K., Scholz N.L. Defects in cardiac function precede morphological abnormalities in fish embryos exposed to polycyclic aromatic hydrocarbons. Toxicol. Appl. Pharmacol. 2004;196:191–205. doi: 10.1016/j.taap.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Incardona J.P., Carls M.G., Teraoka H., Sloan C.A., Collier T.K., Scholz N.L. Aryl hydrocarbon receptor-independent toxicity of weathered crude oil during fish development. Environ. Health Perspect. 2005;113:1755–1762. doi: 10.1289/ehp.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatlen K., Sloan C.A., Burrows D.G., Collier T.K., Scholz N.L., Incardona J.P. Natural sunlight and residual fuel oils are an acutely lethal combination for fish embryos. Aquat. Toxicol. 2010;99:56–64. doi: 10.1016/j.aquatox.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Jung J.-H., Hicken C.R., Boyd D., Anulacion B.F., Carls M.G., Shim W.J., Incardona J.P. Geologically distinct crude oils cause a common cardiotoxicity syndrome in developing zebrafish. Chemosphere. 2013;91:1146–1155. doi: 10.1016/j.chemosphere.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Cherr G.N., Fairbairn E., Whitehead A. Impacts of Petroleum-Derived Pollutants on Fish Development. Annu. Rev. Anim. Biosci. 2017;5:185–203. doi: 10.1146/annurev-animal-022516-022928. [DOI] [PubMed] [Google Scholar]
- 33.Bayha K.M., Ortell N., Ryan C.N., Griffitt K.J., Krasnec M., Sena J., Ramaraj T., Takeshita R., Mayer G.D., Schilkey F., et al. Crude oil impairs immune function and increases susceptibility to pathogenic bacteria in southern flounder. PLoS ONE. 2017;12:e0176559. doi: 10.1371/journal.pone.0176559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.OSPAR JAMP, Joint Assessment and Monitoring Program. Guidelines for Monitoring Contaminants in Biota. [(accessed on 12 July 2019)]; Available online: https://www.ospar.org/documents?d=32414.
- 35.Sundt R.C., Ruus A., Jonsson H., Skarphéðinsdóttir H., Meier S., Grung M., Beyer J., Pampanin D.M. Biomarker responses in Atlantic cod (Gadus morhua) exposed to produced water from a North Sea oil field: Laboratory and field assessments. Mar. Pollut. Bull. 2012;64:144–152. doi: 10.1016/j.marpolbul.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Nahrgang J., Brooks S.J., Evenset A., Camus L., Jonsson M., Smith T.J., Lukina J., Frantzen M., Giarratano E., Renaud P.E. Seasonal variation in biomarkers in blue mussel (Mytilus edulis), Icelandic scallop (Chlamys islandica) and Atlantic cod (Gadus morhua): implications for environmental monitoring in the Barents Sea. Aquat. Toxicol. 2013;127:21–35. doi: 10.1016/j.aquatox.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Holth T.F., Eidsvoll D.P., Farmen E., Sanders M.B., Martínez-Gómez C., Budzinski H., Burgeot T., Guilhermino L., Hylland K. Effects of water accommodated fractions of crude oils and diesel on a suite of biomarkers in Atlantic cod (Gadus morhua) Aquat. Toxicol. 2014;154:240–252. doi: 10.1016/j.aquatox.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Karlsen O.A., Sheehan D., Gpksøyr A. Alteration in the Atlantic cod (Gadus morhua) hepatic thiol-proteome after methylmercury exposure. J. Toxicol. Environ. Health. 2014;77:650–662. doi: 10.1080/15287394.2014.887427. [DOI] [PubMed] [Google Scholar]
- 39.Pampanin D.M., Larssen E., Øysæd K.B., Sundt R.C., Sydnes M.O. Study of the bile proteome of Atlantic cod (Gadus morhua): Multi biological markers of exposure to polycyclic aromatic hydrocarbons. Mar. Environ. Res. 2014;101:161–168. doi: 10.1016/j.marenvres.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Pampanin D.M., Goff J.L., Skogland K., Marcucci C.R., Øysæd K.B., Lorentzen M., Jørgensen K.B., Sydnes M.O. Biological effects of polycyclic aromatic hydrocarbons (PAH) and their first metabolic products in in vivo exposed Atlantic cod (Gadus morhua) J. Toxicol. Environ. Health. 2016;79:633–646. doi: 10.1080/15287394.2016.1171993. [DOI] [PubMed] [Google Scholar]
- 41.Oost R.V.D., Beyer J., Vermeulen N.P.E. Fish bioaccumulation and biomarkers in environmental risk assessment: A review. Environ. Toxicol. Pharmacol. 2003;13:57–149. doi: 10.1016/S1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- 42.Geiselbrecht A.D., Hedlund B.P., Tichi M.A., Staley J.T. Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of puget sound Cycloclasticus strains. Appl. Environ. Microbiol. 1998;64:4703–4710. doi: 10.1128/aem.64.12.4703-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harayama S., Kasai Y., Hara A. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 2004;15:205–214. doi: 10.1016/j.copbio.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Kostka J.E., Prakash O., Overholt W.A., Green S.J., Freyer G., Canion A., Delgardio J., Norton N., Hazen T.C., Hettel M. Hydrocarbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potts L.D., Perez-Calderon L.J., Gontikaki E., Keith L., Gubry-Rangin C., Anderson J.A., Witte U. Effect of spatial origin and hydrocarbon composition on bacterial consortia community structure and hydrocarbon biodegradation rates. FEMS Microbiol. Ecol. 2018;94:fiy127. doi: 10.1093/femsec/fiy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez T., Morris G., Ellis D., Bowler B., Jones M., Salek K., Mulloy B., Teske A. Hydrocarbon-degradation and MOS-formation capabilities of the dominant bacteria enriched in sea surface oil slicks during the Deepwater Horizon oil spill. Marine Pollution Bulletin. 2018;135:205–215. doi: 10.1016/j.marpolbul.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 47.Ribicic D., Netzer R., Hazen T.C., Techtmann S.M., Drabløs F., Brakstad O.G. Microbial community and metagenome dynamics during biodegradation of dispersed oil reveals potential key-players in cold Norwegian seawater. Mar. Pollut. Bull. 2018;129:370–378. doi: 10.1016/j.marpolbul.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 48.Kroon F., Streten C., Harries S. A protocol for identifying suitable biomarkers to assess fish health: A systematic review. PLoS ONE. 2017;12:e0174762. doi: 10.1371/journal.pone.0174762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanni S., Björkblom C., Jonsson H., Godal B.F., Liewenborg B., Lyng E., Pampanin D.M. Biomarker quantification in fish exposed to crude oil as input to species sensitivity distributions and threshold values for environmental monitoring. Mar. Environ. Res. 2017;125:10–24. doi: 10.1016/j.marenvres.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Riiser E.S., Haverkamp T.H.A., Borgan Ø., Jakobsen K.S., Jentoft S., Star B. A Single Vibrionales 16S rRNA Oligotype Dominates the Intestinal Microbiome in Two Geographically Separated Atlantic cod Populations. Front. Microbiol. 2018;9:1561. doi: 10.3389/fmicb.2018.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ager D., Evans S., Li H., Lilley A.K., van der Gast C.J. Anthropogenic disturbance affects the structure of bacterial communities. Environ Microbiol. 2010;12:670–678. doi: 10.1111/j.1462-2920.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 52.Andrews S. FastQC: A Quality Control Tool for High throughput Sequence Data. [(accessed on 11 July 2019)];2010 Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 53.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R Development Core Team R: A Language and Environment for Statistical Computing. [(accessed on 11 July 2019)]; Available online: ftp://ftp.uvigo.es/CRAN/web/packages/dplR/vignettes/intro-dplR.pdf.
- 55.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’hara R.B., Simpson G.L., Solymos P., et al. Vegan: Community Ecology Package. R package Version 2.5–5. [(accessed on 11 July 2019)];2019 Available online: https://cran.r-project.org/package=vegan.
- 58.Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Callahan B.J., McMurdie P.J., Holmes S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knight R., Vrbanac A., Taylor B.C., Aksenov A., Callewaert C., Debelius J., Gonzalez A., Kosciolek T., McCall L., McDonald D., et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 62.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman. F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 64.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 66.Terrisse F., Cravo-Laureau C., Noël C., Cagnon C., Dumbrell A.J., McGenity T.J., Duran R. Variation of Oxygenation Conditions on a Hydrocarbonoclastic Microbial Community Reveals Alcanivorax and Cycloclasticus Ecotypes. Front. Microbiol. 2017;8:1549. doi: 10.3389/fmicb.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong C., Bai X., Sheng H., Jiao L., Zhou H., Shao Z. Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences. 2015;12:2163–2177. doi: 10.5194/bg-12-2163-2015. [DOI] [Google Scholar]
- 68.Ghosal D., Ghosh S., Dutta T.K., Ahn Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front Microbiol. 2016;7:1369. doi: 10.3389/fmicb.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Claus S.P., Guillou H., Ellero-Simatos S. The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes. 2016;2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nayak S.K. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010;41:1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x. [DOI] [Google Scholar]
- 71.Sullam K.E., Essinger S.D., Lozupone C.A., O’Connor M.P., Rosen G.L., Knight R., Kilham S.S., Russell J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kormas K.A., Meziti A., Mente E., Frentzos A. Dietary differences are reflected on the gut prokaryotic community structure of wild and commercially reared sea bream (Sparus aurata) Microbiol. Open. 2014;3:718–728. doi: 10.1002/mbo3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zarkasi K.Z., Abell G.C.J., Taylor R.S., Neuman C., Hatje E., Tamplin M.L., Katouli M., Bowman J.P. Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J. Appl. Microbiol. 2014;117:18–27. doi: 10.1111/jam.12514. [DOI] [PubMed] [Google Scholar]
- 74.Rehmann K., Hertkon N., Kettrup A.A. Fluoranthene metabolism in Mycobacterium sp. strain KR20: Identify of pathway intermediates during degradation and growth. Microbiology. 2001;147:2783–2794. doi: 10.1099/00221287-147-10-2783. [DOI] [PubMed] [Google Scholar]
- 75.Mahjoubi M., Jaouani A., Guesmi A., Ben-Amor S., Jouini A., Cherif H., Najjari A., Boudabous A., Koubaa N., Cherif A. Hydrocarbonoclastic bacteria isolated from petroleum contaminated sites in Tunisia: Isolation, identification and characterization of the biotechnological potential. New Biotechnol. 2013;30:723–733. doi: 10.1016/j.nbt.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Radwan S.S., Khanafer M.M., Al-Awadhi H.A. Ability of the So-Called Obligate Hydrocarbonoclastic bacteria to utilize nonhydrocarbon substrates thus enhancing their activities despite their misleading name. BMC Microbiol. 2019;19:41. doi: 10.1186/s12866-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yakimov M.M., Golyshin P.N., Moore E.R., Abraham W.R., Lümsdorf H., Timmis K.N. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 1998;48:339–348. doi: 10.1099/00207713-48-2-339. [DOI] [PubMed] [Google Scholar]
- 78.Yakimov M.M., Timmis K.N., Golyshin P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotech. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 79.Gittel A., Kofoed M.V., Sørensen K.B., Ingvorsen K., Schramm A. Succession of Deferribacteres and Epsilonproteobacteria through a nitrate-treated high temperature oil production facility. Syst. Appl Microbiol. 2012;35:165–174. doi: 10.1016/j.syapm.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 80.Vigneron A., Alsop E.B., Lomans B.P., Kyrpides N.C., Head I.M., Tsesmetzis N. Succession in the petroleum reservoir microbiome through an oil field production lifecycle. ISME J. 2017;1:2141–2154. doi: 10.1038/ismej.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naether D.J., Slawtschew S., Stasik S., Engel M., Olzog M., Wick L.Y., Timmis K.N., Heipieper H.J. Adaptation of the Hydrocarbonoclastic Bacterium Alcanivorax borkumensis SK2 to Alkanes and Toxic Organic Compounds: A Physiological and Transcriptomic Approach. Appl. Environ. Microbiol. 2013;79:4282–4293. doi: 10.1128/AEM.00694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Golyshin P.N., Dos Santos V., Kaiser O., Ferrer M., Sabirova Y.S., Lunsdorf H., Chernikova T.N., Golyshina O.V., Yakimov M.M., Puhler A., et al. Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon-degrading bacterium that plays a global role in oil removal from marine systems. J. Biotechnol. 2003;106:215–220. doi: 10.1016/j.jbiotec.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 83.Wang W., Shao Z. Diversity of flavin-binding monooxygenase genes (almA) in marine bacteria capable of degradation long-chain alkanes. FEMS Microbiol Ecol. 2012;80:523–533. doi: 10.1111/j.1574-6941.2012.01322.x. [DOI] [PubMed] [Google Scholar]
- 84.Yakimov M.M., Giuliano L., Gentile G., Crisafi E., Chernikova T.N., Abraham W.-R., Lünsdorf H., Timmis K.N., Golyshin P.N. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003;53:779–785. doi: 10.1099/ijs.0.02366-0. [DOI] [PubMed] [Google Scholar]
- 85.Deppe U., Richnow H.-H., Michaelis W., Antranikian G. Degradation of crude oil by an arctic microbial consortium. Extremophiles. 2005;9:461–470. doi: 10.1007/s00792-005-0463-2. [DOI] [PubMed] [Google Scholar]
- 86.Head I.M., Jones D.M., Röling W.F. 2006: Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 87.Hazen T.C., Dubinsky E.A., DeSantis T.Z., Andersen G.L., Piceno Y.M., Singh N., Jansson J.K., Probst A., Borglin S.E., Fortney J.L. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 88.Dubinsky E.A., Conrad M.E., Chakraborty R., Bill M., Borglin S.E., Hollibaugh J.T., Mason O.U., Piceno Y.M., Reid F.C., Stringfellow W.T., et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the Deepwater Horizon oil spill in the Gulf of Mexico. Environ. Sci. Technol. 2013;47:10860–10867. doi: 10.1021/es401676y. [DOI] [PubMed] [Google Scholar]
- 89.Thorsen J., Brejnrod A., Mortensen M., Rasmussen M.A., Stokholm J., Al-Soud W.A., Sørensen S., Bisgaard H., Waage J. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome. 2016;4:62. doi: 10.1186/s40168-016-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baker M. Statisticians issue warning on P values. Nature. 2016;531:151. doi: 10.1038/nature.2016.19503. [DOI] [PubMed] [Google Scholar]
- 91.Wasserstein R.L., Schirm A.L., Lazar N.A. Moving to a World Beyond “p < 0.05”. Am. Stat. 2019;73:1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.